Abstract
Nanoparticles (NPs) are gaining special interest due to their recent applications as antimicrobial agents to defeat the massive threat of resistant pathogens. This study focused on the utilisation of Streptomyces isolate S12 purified from waste discharge soil in the biological synthesis of silver (Ag), selenium (Se), and zinc oxide (ZnO) NPs. The isolate S12 was related to Streptomyces enissocaesilis according to 16S rRNA sequence analysis, morphological characteristics, and biochemical reactions. The cell‐free supernatant has been used for the synthesis of Ag, Se, and ZnO NPs. The synthesised NPs were characterised using ultraviolet–visible spectroscopy, dynamic light scattering (DLS), transmission electron microscopy, and Fourier transform infrared spectroscopy. The biogenic NPs were evaluated for antimicrobial effects against different Gram‐positive and Gram‐negative resistant isolates using the broth microdilution method. They showed antibacterial effect against standard and resistant isolates; Bacillus cereus, Staphylococcus aureus ATCC 29213, S. aureus S1.1, methicillin resistant S. aureus (MRSA 303, 402 and 807), Escherichia coli ATCC 12435, E. coli E7, Klebsiella pneumoniae ATCC 51503, K. pneumoniae K5, K112, Pseudomonas aeruginosa PAO1, and P. aeruginosa P8. This study showed the green synthesis of various NPs using Streptomyces isolate S12 which demonstrated diverse activities against multi‐drug resistant isolates.
Inspec keywords: nanoparticles, nanofabrication, silver, selenium, zinc compounds, II‐VI semiconductors, wide band gap semiconductors, nanomedicine, biomedical materials, microorganisms, cellular biophysics, antibacterial activity, RNA, molecular biophysics, molecular configurations, ultraviolet spectra, visible spectra, biochemistry, light scattering, transmission electron microscopy, Fourier transform infrared spectra
Other keywords: Gram‐positive resistant isolates, Gram‐negative resistant isolates, broth microdilution method, Bacillus cereus, Staphylococcus aureus ATCC 29213, S. aureus S1.1, methicillin resistant S. aureus MRSA 303, methicillin resistant S. aureus MRSA 402, methicillin resistant S. aureus MRSA 807, Escherichia coli ATCC 12435, E. coli E7, Klebsiella pneumoniae ATCC 51503, K. pneumoniae K5, Pseudomonas aeruginosa PAO1, P. aeruginosa P8, green synthesis, Streptomyces isolate S12, multidrug resistant isolates, Ag, Se, ZnO, biogenic NPs, Fourier transform infrared spectroscopy, transmission electron microscopy, dynamic light scattering, ultraviolet‐visible spectroscopy, cell‐free supernatant, biochemical reactions, morphological characteristics, 16S rRNA sequence analysis, biological synthesis, waste discharge soil, Streptomyces isolate S12 purification, antimicrobial agents, waste isolate Streptomyces enissocaesilis, resistant pathogens, antimicrobial activities, nanoparticles
1 Introduction
Antibiotic resistance is an omnipresent clinical challenge; the incidence of infection with multidrug‐resistant isolates has been elevated due to the rareness of new curative agents to control life‐threatening pathogens. This prompted the development of alternative strategies to control the dilemma of resistance and therapy failure [1].
Nanotechnology is a branch of science with various applications in several fields. Nanoparticles (NPs) are distinct from conventional materials, as they exhibit unique features such as ultra‐small particle size (1–100 nm), large surface area, and distinct morphological characters [2]. The revolution of nanotechnology has led to the synthesis of nano‐sized particles with potential applications in medicine, electronics, textiles, and therapeutics [3]. Nano‐constructed particles exhibit significant antimicrobial properties. Large surface area and rapid penetration of the NPs through the bacterial cells exert strong effects on bacterial targets and allow more efficient interaction [4]. Numerous types of NPs, either metals, semiconductors or metal oxides, have been approved for antimicrobial activities including; silver (Ag), selenium (Se), Ag oxide (Ag2 O), zinc oxide (ZnO), copper oxide (CuO), and iron oxide (Fe3 O4) [1].
Biological and chemical processes can be applied to the synthesis of those NPs, but using chemical methods encounters toxic and dangerous by‐products. Hence, there is a necessity to develop eco‐friendly and reproducible approaches to the production of NPs. Consequently, the use of biological methods for NP synthesis is more advantageous to avoid the accumulation of toxic chemicals. Microbial reduction of metal ions into nanoform involves the use of metabolic and enzymatic products of microorganisms like NADH‐reductase or nitrate reductase [5]. Furthermore, certain bacteria and fungi can synthesise nanometals through non‐enzymatic reaction with cell wall functional groups that induce reduction under appropriate environmental conditions such as pH and temperature [6, 7]. Plant extracts and phytochemicals have also been utilised for the biobased construction of the NPs [8]. Ag NPs are the prime nanometals with wide applications in medical and veterinary fields [9]. Numerous studies have discussed the green synthesis of Ag NPs from various sources; fungi, bacteria, plants and metabolites [10, 11]. Other NPs such as Se and ZnO are getting more attention for defending microbial colonisation and bacterial resistance, hence, there is an increasing demand to improve their synthesis processes [12, 13].
Actinomycetes and Streptomycetes are terrestrial sources of antimicrobial agents and secondary metabolites, however, they are less explored for their ability to green synthesis of NPs especially Se and ZnO. This study aims to biosynthesise nanomaterials such as Ag, Se, and ZnO using Streptomyces isolate obtained from soil, followed by characterisation of these NPs with a UV Spectrophotometer, transmission electron microscopy (TEM), particle size analysis, and Fourier transform infrared spectroscopy (FTIR). The antimicrobial activities of the produced NPs were evaluated against multidrug resistant pathogens.
2 Materials and methods
2.1 Isolation and characterisation of Streptomyces
Soil samples were collected from waste discharge areas and processed for isolation and purification of Streptomyces. The samples were dried for 10 min at 50°C. Each dried sample (0.5 g) was suspended in normal saline 0.9%. Serial dilutions 1:10 of the soil suspension were prepared in saline up to 10−5. One loop of the diluted suspension was cultured onto ISP‐2 media (malt extract: 10 g/l; yeast extract: 4 g/l; glucose: 4 g/l, pH 7) and the plates were propagated at 30°C for 14 days. The Streptomyces colonies were picked and purified on new ISP‐2 plates. Pure colonies were subsequently transferred to fresh medium and a lawn culture of Streptomyces was grown at 30°C for 7 days and kept at 4°C until needed [14]. One isolate S12 was selected for the synthesis of nanoscale Ag, Se, and ZnO.
2.2 Molecular characterisation of Streptomyces isolate S12
The characterisation of Streptomyces isolate S12 was carried out using the 16S rRNA molecular method [15]. Genomic DNA of the isolate S12 was extracted according to Nikodinovic et al. [16]. The 16S rRNA was amplified and sequenced using primers forward: 5´‐AGAGTTTGATCMTGGCTCAG‐3´ and reverse: 5´‐ACGAGCTGACGACARCCATG‐3´) (ABI 3730xl sequencer, Applied Biosystems). Nucleotide sequences were resolved using the basic local alignment search tool on the National Center for Biotechnology Information (NCBI) and the sequences of related taxa were retrieved. The sequences alignments were conducted and the phylogenetic tree was generated using the neighbour‐joining method [17] and Molecular Evolutionary Genetics Analysis (MEGA) Version 5.0 [18]. Data analyses were performed on a bootstrapped set with 1000 replicates.
2.3 Morphological and biochemical characterisation of Streptomyces isolate S12
Morphological and cultural features of S12 were examined; also, colours of the spores, aerial mycelium, substrate mycelium, diffusible pigment, and melanoid pigment were detected. Furthermore, utilisation of carbon sources and biochemical characteristics was evaluated according to Bergey's Manual of Determinative Bacteriology [19]. The spore chain morphology of S12 was evaluated by scanning electron microscopy (JEOL JSM 6510 lv).
2.4 Synthesis of Ag, Se and ZnO NPs
The biosynthesis of NPs was performed by inoculating the spores of Streptomyces S12 in 50 ml ISP‐2 media in 250 ml Erlenmeyer flasks and the flasks were incubated at 30°C with shaking at 150 rpm for 72 h. The cell‐free supernatant was obtained by centrifugation at 8.000 × g for 10 min and the harvested mycelial mass was 2.7 mg/ml (wet weight). Then, aqueous solutions of AgNO3, SeO2, and ZnSO4 were added to the cell‐free suspension until a final concentration of 5 mM. A control flask (without AgNO3, SeO2, and ZnSO4, only culture supernatant) was also run along with the other flasks [20]. Also, ISP‐2 medium mixed with 5 mM of AgNO3, SeO2 or ZnSO4 was included. Samples of the reaction solution were collected at 24, 48 and 72 h to be used for characterisation of the formed NPs. The synthesised NPs were collected by centrifugation of the bacterial‐free supernatant treated with AgNO3, SeO2 or ZnSO4 at a high speed 18.000 rpm for 30 min and washed twice with distilled water [20]. The precipitate was dried and the weight of the precipitated NPs was quantified as µg/ml.
2.5 Characterisation of NPs
2.5.1 Colour change and ultraviolet–visible (UV–Vis) spectroscopy
The biological formation of NPs was initially detected through a colour change of the cell free supernatants. The absorption spectra of the formed NPs were measured at a wavelength in the range of 200–600 nm using a UV‐Vis spectrophotometer 1601 pc at room temperature and a resolution of 1 nm [21]. The blank solution (media treated only with AgNO3, SeO2, and ZnSO4) were utilised for each nano‐preparation.
2.5.2 Detection of particle size analysis and zeta potential
The size of the formed NPs was detected by the dynamic light scattering (DLS) technique of laser light using a particle size analyser (Microtrac, nanotrac wave II Q) [21]. Each sample was diluted 1:10 in distilled water and sonicated for 5 min to avoid aggregation of the NPs. The zeta potentials of the synthesised NPs were detected from their electrophoretic mobilities using Microtrac, nanotrac wave II Q.
2.5.3 TEM imaging
The shape and dispersion of the formed NPs were morphologically perceived using 200 K TEM (JEOL JEM‐2100, UK) [22]. One drop of each bacterial supernatant treated with Ag+, Se4+, and Zn2+ ions was applied on a carbon coated copper grid, excess solution was absorbed on a filter paper and the grid was left for complete dryness. The NPs adsorbed on the grid were examined using TEM with magnification power X 1.000–800.000 (JEOL JEM‐2100).
2.5.4 Assessment of attenuated total reflectance (ATR)‐FTIR spectroscopy
FTIR spectroscopy mode was estimated using a Thermo‐Nicolet 6700 FTIR spectrophotometer provided with ATR and with full integration with the OMNIC software. The FTIR spectrum was obtained in the range from 400 to 4000 cm−1 at room temperature and a resolution of 4 cm−1 [23]. The peaks were plotted as the wavelength (cm−1) on the Y ‐axis and % transmittance on the X ‐axis.
2.6 Antimicrobial activities of the synthesised NPs
The antimicrobial activity of the synthesised NPs was assessed against various pathogens. The minimal bactericidal/inhibitory concentrations (MBCs/MICs) of Ag, Se and ZnO NPs were evaluated against different organisms such as: Bacillus cereus, Staphylococcus aureus ATCC 29213, S. aureus S1.1, methicillin resistant S. aureus (MRSA 303, MRSA 402 and MRSA 807), Escherichia coli ATCC 12435, E. coli E7, Klebsiella pneumoniae ATCC 51503, K. pneumoniae K5, K112, Pseudomonas aeruginosa PAO1, and P. aeruginosa P8 by the broth microdilution method according to the guidelines of the clinical laboratory standard institute [24]. The prepared NPs were diluted 1:1 in a 96‐well microtitre plate, then, each well was inoculated by the diluted cultures of the assessed organism. Both positive and negative controls were also run on each plate. The plates were incubated at 37°C for 24 h, and the minimum inhibitory concentrations were calculated as the lowest concentration of the NPs showing no visible microbial growth.
2.7 Statistical analysis
All the experiments were performed in triplicate, mean of the readings was calculated using Excel spread sheet. The paired t test was used for comparison of MICs of the obtained NPs (Ag, Se, and ZnO) to the MIC of the control antibiotic cefotaxime and the data were considered significant when P < 0.05.
3 Results and discussion
3.1 Collection and purification of Streptomyces
Twelve Streptomyces isolates were purified from soil samples gathered from areas near waste discharges. Pure colonies with leathery, chalky, hard features, and various colours were characterised and transferred to pure an ISP‐2 medium [25]. The microscopic examination of the isolates indicated a branched network of hyphae with dispersed spores. The pure isolates were propagated on the ISP‐2 medium supplemented with AgNO3, SeO2, and ZnSO4 in concentrations 1–10 mM. The growth of isolate S12 has not been affected by Ag+, Se4+, and Zn2+ ions. Hence, isolate S12 was selected for the synthesis of the NPs. Isolation of the Streptomyces isolate S12 from waste soil has enhanced its ability to survive in the presence of heavy metals and various elements [26, 27].
3.2 Molecular identification of isolate S12
The partial sequence of 16S rRNA gene of the S12 isolate was submitted to GenBank (NCBI) under accession number KY285186 and the sequence analysis stipulated that S12 belongs to genus Streptomyces. The neighbour‐joining method [17] was adopted for generating dendrogram (phylogenetic tree) as shown in Fig. 1. The sequence analysis showed that isolate S12 formed a distinct branch with other Streptomyces; Streptomyces rochei strain NRRL B‐1559 (NR_116078.1), Streptomyces enissocaesilis strain NRRL B‐16365 (NR_115668.1), Streptomyces plicatus strain NBRC 13071(NR_112357.1) and Streptomyces vinaceusdrappus strain NRRL 2363 (NR_043383.1) with 99% similarity.
Fig. 1.
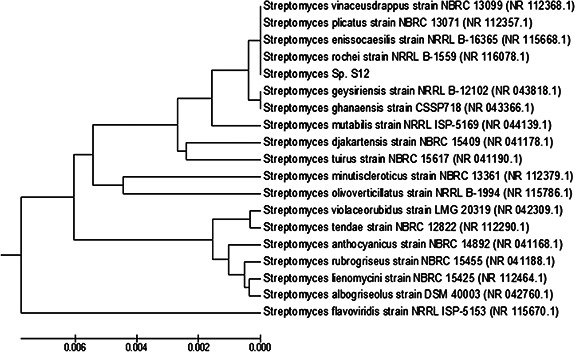
Phylogenetic tree showing the relationship between Streptomyces isolate S12 and representative species of genus Streptomyces based on 16S rRNA gene sequences. The sequences were aligned and the phylogenetic tree was generated using the neighbour‐joining method in MEGA Version 5.0
3.3 Morphological and biochemical characteristics of Streptomyces isolate S12
Isolate S12 revealed microscopic and macroscopic features like Streptomyces species. The morphological, cultural, biochemical, colour, and carbon utilisation of Streptomyces isolate S12 were summarised in Table 1. Isolate S12 was positive for citrate utilisation, starch hydrolysis, and casein hydrolysis but negative for H2S and indole production. Also, it can utilise glucose, sucrose, maltose, xylose, and fructose as carbon sources but cannot utilise mannitol and rhamnose. The isolate is a filamentous, Gram‐positive aerobic bacterium that has rugose, wrinkled colonies with an aerial mycelium which appeared as a powder colony with no melanoid or diffusible pigment. Scanning electron microscopy examination indicated that S12 has spiral spore chains with a smooth spore surface (Fig. 2).
Table 1.
Morphological and biochemical properties of Streptomyces isolate S12
| Characteristics | Results |
|---|---|
| spore chain morphology | spiral |
| colony colour on ISP2 | white‐grey |
| spore surface | smooth |
| production of diffusible pigment | − |
| production of melanoid pigment | − |
| starch hydrolysis | + |
| casein hydrolysis | + |
| citrate utilisation | + |
| H2 S production | − |
| indole | − |
| utilisation of carbon sources | |
| glucose | + |
| sucrose | + |
| xylose | + |
| fructose | + |
| mannitol | − |
| rhamnose | − |
| maltose | + |
‘+’ positive and ‘−’ negative.
Fig. 2.
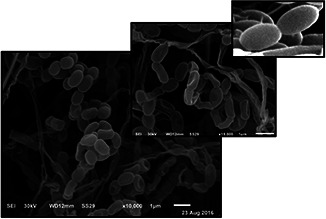
Scanning electron micrograph of spores of Streptomyces isolate S12
From the above‐mentioned results, it could be recoded that the S12 isolate belongs to Streptomyces and it may be a candidate of S. enissocaesilis species as indicated previously by Sirisha et al. [28]. In addition, S12 exhibited enzymatic activities such as starch hydrolysis and protein degradation accompanied by amylases and proteases enzymes that assist in the microbial reduction into the nanoform.
3.4 Characterisation of the formed NPs
3.4.1 Detection of the colour change
Ag, Se, and ZnO NPs were biologically synthesised using the cell‐free filtrate of Streptomyces S12. Initially, the colour of the supernatant converted into brown, orange and deep yellow as a primary indication for the reduction of the ions and the formation of Ag NPs, Se NPs, and ZnO NPs, respectively (Fig. 3). Furthermore, no colour change was detected in the control flasks. The intensity of each colour increased during the incubation period with marked deep coloration after 72 h incubation, especially with Ag and ZnO NPs. This colour change is due to the excitation of the surface of plasmon vibrations of the deposited NPs and considered as an indication for biogeneration of the NPs. The organic moiety of the soluble extract is assumed to act as both stabilising agent and reducing agent. The synthesis of NPs using a culture supernatant provides a great advantage since it assists in the enzymatic reduction of the supplied elements to the nanoforms and facilitates the collection of the formed NPs.
Fig. 3.
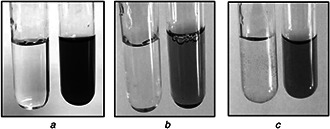
Visual detection of the biogenic synthesis of NPs using growth supernatant of Streptomyces isolate S12
(a) Ag NPs with brown colour, (b) Se NPs with orange colour, (c) ZnO NPs with yellow colour
3.4.2 UV–Vis spectra analysis
NPs strongly interact with specific light wavelengths and the unique optical properties of these materials are the basis for the area of plasmonic. The bioreduction of Ag+, Se4+ and Zn2+ ions in the aqueous filtrate was monitored by using UV–Vis spectroscopy [29]. We detected an increase in the formation of Ag and ZnO NPs over the time; however, the bio‐reduced Se NPs were constant over the time (Fig. 4).
Fig. 4.
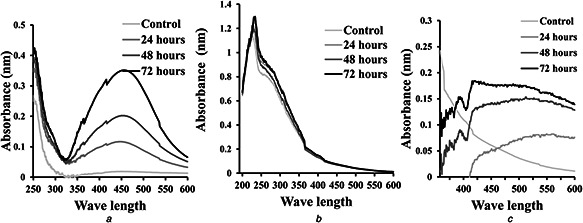
UV–Vis spectra of the synthesised NPs
(a) Ag NPs, (b) Se NPs, (c) ZnO NPs
Ag NPs produced the surface plasmon resonance band around 450 nm (Fig. 4 a). Likewise, Ag NPs synthesised using other Streptomyces sp. have maximum peaks at 450 nm [21, 22]. The biological reduction of Ag ions into metal nanoform could be processed through the metabolic products and the secreted enzymes by a microorganism in the culture supernatant. The nano Se showed a sharp absorption peak at 254 nm (Fig. 4 b). Similarly, the synthesised Se NPs using K. pneumoniae and Pseudomonas alcaliphila have UV absorption at 254 nm [30, 31].
The formation of ZnO NPs was increasing over time reaching a maximum peak after 72 h and an absorption peak at 392 nm (Fig. 4 c). This was closely related to results obtained by Balraj et al. [32] who revealed an absorption peak at 364 nm of ZnO NPs synthesised from Streptomyces sp. Other studies reported that the absorption peak of the biogenic ZnO NPs is at 310 nm [33].
Generally, NPs possess optical properties that are sensitive to size, shape, concentration, agglomeration state, and refractive index near the NP surface. Therefore, the shift in the absorption spectra observed in different studies could be related to the variation in shape and size of the NPs, in addition, the dielectric constant of the formed particles and the surrounding medium could participate in this variability [34].
3.4.3 Analysis of particle size and zeta potential
Fig. 5 a shows the size distribution measured by the laser light scattering method for Ag, Se, and ZnO NPs. For Ag NPs, they synthesised in small size and their size gradually increased until reaching the maximum value at 277 nm (Fig. 5 a). The size of Ag NPs produced by the S12 was in the size range of Ag NPs obtained by E. coli [35], and Bacillus CS 11 [36]. However, the particles size of Ag NPs produced from another Streptomyces sp. is very small (5–50 nm) [22, 29].
Fig. 5.
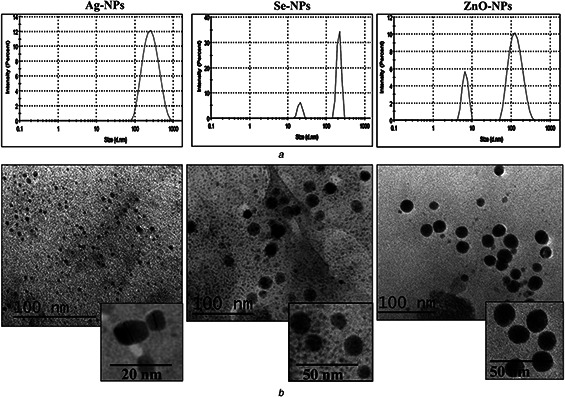
Size and shape of the synthesised NPs
(a) Particle size distribution for Ag, Se and ZnO NPs, (b) TEM of Ag, Se and ZnO NPs
The particle size range of Se NPs was 20–211 nm as determined by DLS (Fig. 5 a). In the same instance, the size distribution of Se NPs obtained by Streptomyces sp. ES2‐5 was between 28 and 123 nm [27]. For ZnO NPs, two peaks were clearly observed at 6.7 and 135 nm, and nano‐ZnO with particle size of 135 nm had the highest frequency. In the study of Balraj et al. [32], the particle size of the biogenic ZnO NPs obtained using the cell‐free supernatant of marine Streptomyces sp. was 20–50 nm. Broadening of size distribution peak observed with Se NPs and ZnO NPs (Fig. 5 a) could be due to the aggregation of NPs. Also, increase in the size of the particles with growth time could be the cause of the expansion of the UV–Vis spectrum of the microbial synthesised Se NPs [37].
The stability of nano‐systems was evaluated by the detection of the zeta potential, it represents the electrokinetic potential surrounding the particles. In general, the magnitude of the zeta potential indicates the potential stability, i.e. the degree of electrostatic repulsion between adjacent particles in a colloidal dispersion.
Herein, the zeta potentials for Ag, Se, and ZnO NPs were −162, −220 and +200 mV, respectively. These high values indicate the potential stability of the synthesised NPs. The negative zeta value may be due to the adsorption of OH− ions on the surface of the NPs which maintain particles in a small size. Likewise, the zeta potential value of Ag NPs biosynthesised by marine Streptomyces parvulus SSNP11 is −81.0 mV with monodispersity and high stability [38]. Also, Se NPs synthesised by pulsed laser ablation showed enhanced stability with a zeta value of −45.6 mV [13].
3.4.4 TEM imaging
The TEM images show that Ag and Se NPs were spherical in shape (Fig. 5 b). Also, the formed NPs were well‐dispersed indicating enhanced stability of the prepared NPs as the presence of extracellular proteins in the microbial supernatants could assist the stabilisation of NPs. Ag NPs are characterised by spherical shapes as previously formed by Streptomyces hygroscopicus and S. parvulus SSNP11 [29, 38]. Se nanospheres are also synthesised from Duganella sp. and Pseudomonas putida KT2440 [39]. Concerning ZnO NPs the particles were quasi‐spherical resembling ZnO NPs produced from Candida albicans [40]. The TEM micrograph revealed that the size of NPs was in the range of 5–20 nm. In DLS, the scattered intensity is proportional in size to the power 6 consequently, larger particles have high scattering, making the ‘average’ size look larger than the light scattered by the smallest particles [41, 42].
3.4.5 Analysis of FTIR spectroscopy
FTIR was performed in order to determine the biomolecules responsible for reducing, capping, and stabilisation of Ag, Se, and ZnO NPs synthesised by Streptomyces S12.
For Ag, Se, and ZnO NPs, the FTIR spectrum shown in Fig. 6 elucidates intense bands at 3423.59, 3442.04, and 3422.38 cm−1, respectively, which were corresponding to the stretch in the OH band (3200–3600) suggesting the presence of hydroxyl groups as a reducing agent. Peaks such as 2918.35 and 2850.09 cm−1 for Ag NPs, 2918.20 and 2850.13 cm−1 for Se NPs and 2918.87 and 2850.47 cm−1 for ZnO NPs assigned for (C–H stretch) alkane stretching vibration. The level of C=O ranges was observed at 1724.09 cm−1 for Ag NPs. The peaks at 1630.16, 1638.59 and 1635.95 cm−1 for Ag, Se, and Zn NPs, respectively, were characteristic for the amide band leads primarily to bending vibrations of the (NH) C=O group arising due to the carbonyl stretch in proteins [43].
Fig. 6.
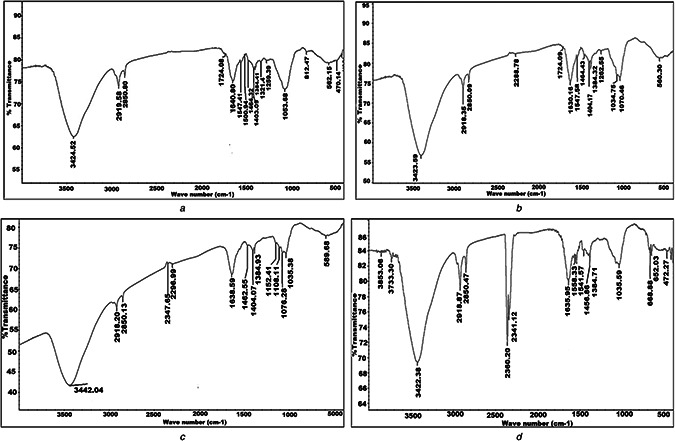
FTIR of
(a) Streptomyces isolate S12 cell‐free supernatant, (b) Synthesised Ag NPs, (c) Se NPs, (d) ZnO NPs
The peaks at 1034.75, 1053.68 and 1035.59 cm−1 for Ag, Se, and ZnO NPs, respectively, were corresponding to the C–N stretching vibration of the amine. For ZnO NPs, two peaks were observed at 2360.20 and 2341.12 cm−1 which may be assigned for alkyne —C≡C or cyano —C≡N groups. The small shifting in the position of different peaks in the NPs from the control may be due to the progression of the reduction reaction with capping and stabilisation of the NPs. The above‐mentioned results of FTIR spectra of the NPs indicated the presence of protein in the used samples which act as a capping protein for the bioreduced Ag, Se and ZnO NPs, hence responsible for the stability of the biosynthesised oxide NPs and well dispersed NPs in the reaction mixture as indicated by the TEM and zeta potential [44].
3.5 Antimicrobial activity of the formed NPs
Initial antibacterial screening indicated the antimicrobial potential of the nano‐synthesised particles. The MICs for the biosynthesised NPs were assessed and compared with that obtained for cefotaxime. In the present research, Ag, Se and ZnO NPs exhibited antimicrobial properties against various pathogens as determined by the broth microdilution method. The biologically synthesised NPs using cell‐free extract of S12 were more effective against Gram‐positive and Gram‐negative isolates than the third‐generation cefotaxime (P > 0.05). Particularly, Ag NPs had enhanced antibacterial activity against all tested strains with significant antibacterial activity against Gram‐positive isolates such as S. aureus (8.4–16.9 µg/ml) and B. cereus (133.7 µg/ml) and against Gram‐negative pathogens; E. coli (16.9 µg/ml), K. pneumoniae (33.75–67.5 µg/ml) and P. aeruginosa (8.4–16.9 μg/µl) (Table 2). The synthesised NPs inhibited MRSA isolates with very low MICs. All tested isolates exhibited tolerance to cefotaxime with MBC/MIC ratio >32; however, none of the tested isolates showed tolerance to the tested NPs as MBC/MIC ratio ≤8 which indicate the superior bactericidal effects of the synthesised NPs. NPs could overcome the existing antibiotic resistance, these effects could be attributed to the bactericidal activity of the NPs, which are associated with their nano‐size and physicochemical characteristics. The nano‐size of the prepared particles facilitates the penetration of the particles through cell membranes this is associated with damage to the membrane permeability [45, 46]. Another mechanism could be involved, the nano‐formed particles bind with DNA with the distribution of nucleic acid synthesis [47] and termination of enzymatic transcription.
Table 2.
MBCs/MICs of the biogenic NPs against both Gram‐negative and Gram‐positive isolates performed in triplicates (n = 3)
| Organism | Organism type | MBCs/MICs of NPs, µg/ml | Cefotaxime, µg/ml | ||
|---|---|---|---|---|---|
| Se | Ag | ZnO | |||
| S. aureus ATCC 29213 | Gram positive | 1580/395 | 8.4/8.4 | 400/200 | 15.1 |
| S. aureus S1.1 | 196/49 | 16.9/16.9 | 400/200 | 156.25 | |
| MRSA 33 | 58.8/14.7 | 1.8/1.8 | 25/12.5 | 2500 | |
| MRSA 42 | 240/60 | 4.9/4.9 | 6.5/3.25 | 2500 | |
| MRSA 87 | 240/60 | 9.9/9.9 | 100/50 | 2500 | |
| B. cereus | Gram positive | 98/49 | 534.8/133.7 | 100/100 | 5000 |
| E. coli ATCC 12435 | Gram negative | 197.5/197.5 | 135.2/16.9 | 200/200 | 625 |
| E. coli (E7) | 197.5/197.5 | 135.2/16.9 | 200/200 | 156.25 | |
| K. pneumoniae ATCC 51503 | Gram negative | 99/99 | 33.75/33.75 | 50/50 | 625 |
| K. pneumoniae (K5) | 99/99 | 67.5/67.5 | 50/50 | 625 | |
| K. pneumoniae (K112) | 99/99 | 33.75/33.75 | 50/50 | 625 | |
| P. aeruginosa PAO1 | Gram negative | 395/395 | 33.8/8.4 | 400/200 | 156.25 |
| P. aeruginosa (P8) | 395/395 | 67.6/16.9 | 400/200 | 625 | |
MRSA; methicillin resistant S. aureus.
Also, Ag NPs bind the thiol groups (−SH) followed by the inactivation of proteins and inhibition of bacterial growth [48]. Additionally, the antibacterial action of Ag NPs could be related to gradual oxidation of Ag NPs and release of Ag+ ions that exhibit biocidal effects [49]. Previous studies indicated the antibacterial action of biologically formulated Ag NPs against Gram‐negative and Gram‐positive bacteria [9, 22]. Ag NPs eliminated microbial infection caused by K. pneumonia e [38], E. coli [29] and pathogenic P. aeruginosa isolates [50].
In the same instance, Se NPs displayed effective antibacterial against Gram‐negative pathogens; E. coli, K. pneumoniae and P. aeruginosa with MIC values of 197.5, 99, and 395 µg/ml, respectively. The MIC values of Se NPs against MRSA were 14.7–60 µg/ml and the MIC value against B. cereus was 49 µg/ml (Table 2).
ZnO NPs were effective against standard K. pneumoniae ATCC 51503 and pathogenic isolates K5 and K112 with very low MIC value of 50 µg/ml, these nano‐oxides were also effective against E. coli E7 and P. aeruginosa P8 with a MIC value of 200 µg/ml. Also, ZnO NPs were effective against S. aureus ATCC 29213 with a MIC value of 200 µg/ml and against MRSA clinical isolates with MIC values from 3.25 to 50 µg/ml. The biogenic ZnO NPs were highly effective against multi‐drug resistant Gram‐negative isolates compared with a previous study [51].
The mechanisms of antibacterial involved by Se NPs and ZnO NPs include the formation of reactive oxygen species with DNA damage and protein dysfunction [52]. Furthermore, they could disrupt the cellular integrity with the extrusion of the cytoplasmic contents and inhibition of bacterial growth [12].
4 Conclusion
In conclusion, this work demonstrates the synthesis of Ag, Se, ZnO NPs from Streptomyces isolate S12 purified from a waste region. The percentage yields of Ag, Se, and ZnO NPs were 85, 98 and 50%, respectively. The formed NPs were characterised by UV‐Vis spectroscopy, particle size, zeta potential, and TEM with small size and highly stable characteristics. The synthesised NPs showed a broad spectrum antimicrobial activity. They were capable of inhibiting both Gram‐positive and Gram‐negative pathogens.
5 Acknowledgments
Thanks and appreciation for the Clinical Microbiology Laboratory for providing clinical isolates used for the antibacterial study.
6 References
- 1. Rudramurthy G.R. Swamy M.K. Sinniah U.R. et al.: ‘Nanoparticles: alternatives against drug‐resistant pathogenic microbes’, Molecules, 2016, 21, p. 836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sahoo S.K. Parveen S. Panda J.J.: ‘The present and future of nanotechnology in human health care’, Nanomedicine, 2007, 3, (1), pp. 20 –31 [DOI] [PubMed] [Google Scholar]
- 3. Doria G. Conde J. Veigas B. et al.: ‘Noble metal nanoparticles for biosensing applications’, Sensors, 2012, 12, pp. 1657 –1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hajipour J.M. Fromm K.M. Ashkarran A.A. et al.: ‘Antibacterial properties of nanoparticles’, Trend Biotechnol., 2012, 30, pp. 499 –511 [DOI] [PubMed] [Google Scholar]
- 5. Mandal D. Bolander M.E. Mukhopadhyay D. et al.: ‘The use of microorganisms for formation of metal nanoparticles and their application’, Appl. Environ. Microbiol., 2006, 69, pp. 485 –492 [DOI] [PubMed] [Google Scholar]
- 6. Fu J.K. Liu Y.Y. Gu P.Y. et al.: ‘Spectroscopic characterization on the biosorption and bioreduction of Ag(I) by Lactobacillus sp. A09’, Acta Phys. Chim. Sin., 2000, 16, pp. 779 –782 [Google Scholar]
- 7. Lin Z.Y. Fu J.K. Wu J.M. et al.: ‘Preliminary study on the mechanism of non‐enzymatic bioreduction of precious metal ions’, Acta Phys. Chim. Sin., 2001, 17, pp. 477 –480 [Google Scholar]
- 8. Kharissova O.V. Dias H.V. Kharisov B.I. et al.: ‘The greener synthesis of nanoparticles’, Trends Biotechnol., 2013, 31, (4), pp. 240 –248 [DOI] [PubMed] [Google Scholar]
- 9. Rai M. Yadav A. Gade A.: ‘Silver nanoparticles as a new generation of antimicrobials’, Biotechnol. Adv., 2009, 27, pp. 76 –83 [DOI] [PubMed] [Google Scholar]
- 10. Sadowski Z. Maliszewska H.I. Grochowalska B. et al.: ‘Synthesis of silver nanoparticles using microorganisms’, Mater. Sci., 2008, 26, (2), pp. 419 –424 [Google Scholar]
- 11. Manivasagan P. Venkatesan J. Sivakumar K. et al.: ‘Actinobacteria mediated synthesis of nanoparticles and their biological properties: a review’, Crit. Rev. Microbiol., 2016, 42, (2), pp. 209 –221 [DOI] [PubMed] [Google Scholar]
- 12. Hsueh Y.H. Ke W.J. Hsieh C.T. et al.: ‘Zno nanoparticles affect Bacillus subtilis cell growth and biofilm formation’, PLoS One, 2015, 10, (6), p. e0128457, doi: 10.1371/journal.pone.0128457. eCollection 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guisbiers G. Wang Q. Khachatryan E. et al.: ‘Inhibition of E. coli and S. aureus with selenium nanoparticles synthesized by pulsed laser ablation in deionized water’, Int. J. Nanomed., 2016, 11, pp. 3731 –3736, doi: 10.2147/IJN.S106289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeffrey L.S.H.: ‘Isolation, characterization and identification of actinomycetes from agriculture soils at Semongok, Sarawak’, Afr. J. Biotechnol., 2008, 7, pp. 3697 –3702 [Google Scholar]
- 15. Maidak B.L. Cole J.R. Lilburn T.G. et al.: ‘The RDP (ribosomal database project) continues’, Nucleic Acids Res., 2000, 1, pp. 173 –174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nikodinovic J. Barrow K.D. Chuck J.A.: ‘High yield preparation of genomic DNA from Streptomyces’, Biotechniques, 2003, 35, pp. 932 –934 [DOI] [PubMed] [Google Scholar]
- 17. Saitou N. Nei M.: ‘The neighbor‐joining method: a new method for reconstructing phylogenetic trees’, Mol. Biol. Evol., 1987, 4, pp. 406 –425 [DOI] [PubMed] [Google Scholar]
- 18. Tamura K. Peterson D. Peterson N. et al.: ‘MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods’, Mol. Biol. Evol., 2011, 28, (10), pp. 2731 –2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holt J.G. Krieg N.R. Sneath P.H.A. et al.: ‘Bergey's manual of determinative bacteriology’ (Williams and Wilkins, Baltimore, 1994, 9th edn.) [Google Scholar]
- 20. Zonaro E. Lampis S. Turner R.J. et al.: ‘Biogenic selenium and tellurium nanoparticles synthesized by environmental microbial isolates efficaciously inhibit bacterial planktonic cultures and biofilm’, Front Microbiol., 2015, 6, pp. 1 –11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saminathan K.: ‘Biosynthesis of silver nanoparticles using soil Actinomycetes Streptomyces sp.’, Int. J. Curr. Microbiol. App. Sci., 2015, 4, (3), pp. 1073 –1083 [Google Scholar]
- 22. Dattu S. Vandana R. Fatima L. et al.: ‘Biologically reduced silver nanoparticles from Streptomyces sp. VDP‐5 and its antibacterial efficacy’, Int. J. Pharm. Pharm. Sci. Res., 2014, 4, (2), pp. 31 –36 [Google Scholar]
- 23. Umoren S.A. Obot I.B. Gasem Z.M.: ‘Green synthesis and characterization of silver nanoparticles using red apple (Malus domestica) fruit extract at room temperature’, J. Mater. Environ. Sci., 2014, 5, (3), pp. 907 –914 [Google Scholar]
- 24. Clinical and Laboratory Standards Institute : ‘Performance standards for antimicrobial susceptibility testing: twenty‐fifth informational supplement, M100‐S25’, vol. 33 (CLSI, Wayne, PA, USA, 2015) [Google Scholar]
- 25. Williams S.T. Cross T.: ‘Isolation, purification, cultivation and preservation of actinomycetes’, Methods Microbiol., 1971, 4, pp. 295 –334 [Google Scholar]
- 26. Bajaj M. Schmidt S. Winter J.: ‘Formation of Se (0) nanoparticles by Duganella sp. and Agrobacterium sp. Isolated from Se‐laden soil of North‐East Punjab, India’, Microb. Cell Fact., 2012, 11, p. 64, doi: 10.1186/1475‐2859‐11‐64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan Y. Yao R. Wang R. et al.: ‘Reduction of selenite to Se(0) nanoparticles by filamentous bacterium Streptomyces sp. ES2‐5 isolated from a selenium mining soil’, Microb. Cell Fact., 2016, 15, p. 157, doi: 10.1186/s12934‐016‐0554‐z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sirisha B. Haritha R. Jagan Mohan Y.S.Y.V. et al.: ‘Molecular characterization of marine Streptomyces enissocaesilis capable of L‐asparaginase production’, Bacteriol. J., 2013, 4, (1), pp. 1 –11 [Google Scholar]
- 29. Sadhasivam S. Shanmugam P. Yun K.: ‘Biosynthesis of silver nanoparticles by Streptomyces hygroscopicus and antimicrobial activity against medically important pathogenic microorganisms’, Colloids Surf. B, Biointerfaces, 2010, 81, pp. 358 –362 [DOI] [PubMed] [Google Scholar]
- 30. Fesharaki P.J. Nazari P. Shakibaie M. et al.: ‘Biosynthesis of selenium nanoparticles using Klebsiella pneumoniae and their recovery by a simple sterilization process’, Braz. J. Microbiol., 2010, 41, pp. 461 –466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang W. Chen Z. Liu H. et al.: ‘Biosynthesis and structural characteristics of selenium nanoparticles by Pseudomonas alcaliphila ’, Colloids Surf. B, Biointerfaces, 2011, 88, pp. 196 –201 [DOI] [PubMed] [Google Scholar]
- 32. Balraj B. Senthilkumar N. Siva C. et al.: ‘Synthesis and characterization of zinc oxide nanoparticles using marine Streptomyces sp. with its investigations on anticancer and antibacterial activity’, Res. Chem. Intermed., 2017, 43, pp. 2367 –2376 [Google Scholar]
- 33. Ashajyothi C. Manjunath N.R. Chandrakanth R.K.: ‘Antibacterial activity of biogenic zinc oxide nanoparticles synthesized from Enterococcus faecalis ’, Int. J. Chem. Tech. Res., 2014, 6, (5), pp. 3131 –3136 [Google Scholar]
- 34. Gonzalo J. Serna R. Sol J. et al.: ‘Morphological and interaction effects on the surface plasmon resonance of metal nanoparticles’, J. Phys. Condens. Matter., 2003, 15, (42), pp. 3001 –3002 [Google Scholar]
- 35. Gurunathan S. Kalishwaralal K. Vaidyanathan R. et al.: ‘Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli ’, Colloids Surf B, 2009, 74, pp. 328 –335 [DOI] [PubMed] [Google Scholar]
- 36. Das V.L. Thomas R. Varghese R.T. et al.: ‘Extracellular synthesis of silver nanoparticles by the Bacillus strain CS 11 isolated from industrialized area’, 3 Biotech, 2014, 4, (2), pp. 121 –126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saha S. Bera K. Jana P.C.: ‘Growth time dependence of size of nanoparticle of ZnS’, Int. J. Soft Comput. Eng., 2011, 5, pp. 23 –26 [Google Scholar]
- 38. Prakasham R.S. Kumar B.S. Kumar Y.S. et al.: ‘Production and characterization of protein encapsulated silver nanoparticles by marine isolate Streptomyces parvulus SSNP11’, Indian J. Microbiol., 2014, 54, (3), pp. 329 –336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Avendaño R. Chaves N. Fuentes P. et al.: ‘Production of selenium nanoparticles in Pseudomonas putida KT2440’, Sci. Rep., 2016, 6, p. 37155, 10.1038/srep37155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uzzaman S. Mashrai A. Khanam H. et al.: ‘Biological synthesis of ZnO nanoparticles using C. albicans and studying their catalytic performance in the synthesis of steroidal pyrazolines’, Arab. J. Chem., 2013, 10.1016/j.arabjc.2013.05.004 [DOI] [Google Scholar]
- 41. Linkov P Artemyev M. Efimov A.E. et al.: ‘Comparative advantages and limitations of the basic metrology methods applied to the characterization of nanomaterials’, Nanoscale, 2013, 7, pp. 8781 –8798 [DOI] [PubMed] [Google Scholar]
- 42. Matos R.A. Courrol L.C.: ‘Saliva and light as templates for the green synthesis of silver nanoparticles’, Colloids Surf. A, Physicochem. Eng. Aspects, 2014, 441, pp. 539 –543 [Google Scholar]
- 43. Mandal S. Phadtate S. Sastry M.: ‘Interfacing biology with nanoparticles’, Curr. Appl. Phys., 2005, 5, pp. 118 –127 [Google Scholar]
- 44. Wang L. Hu C. Shao L.: ‘The antimicrobial activity of nanoparticles: present situation and prospects for the future’, Int. J. Nanomed., 2017, 12, pp. 1227 –1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morones J.R. Elechiguerra J.L. Camacho A. et al.: ‘The bactericidal effect of silver nanoparticles’, Nanotechnology, 2005, 16, pp. 2346 –2353 [DOI] [PubMed] [Google Scholar]
- 46. Feng Q.L. Wu J. Chen G.Q. et al.: ‘Mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus ’, J. Biomed. Mater. Res., 2000, 52, pp. 662 –668 [DOI] [PubMed] [Google Scholar]
- 47. Klueh U. Wagner V. Kelly S. et al.: ‘Efficacy of silver‐coated fabric to prevent bacterial colonization and subsequent device‐based biofilm formation’, J. Biomed. Mater. Res., 2000, 53, pp. 621 –631 [DOI] [PubMed] [Google Scholar]
- 48. Tran Q.H. Nguyen Y.Q. Le A.T.: ‘Silver nanoparticles: synthesis, properties, toxicology, applications and perspectives’, Adv. Nat. Sci. Nanosci. Nanotechnol., 2013, 4, 033001, pp. 1 –20 [Google Scholar]
- 49. Sukanya M. Saju K. Praseetha P. et al.: ‘Therapeutic potential of biologically reduced silver nanoparticles from actinomycete cultures’, J. Nanosci., 2013, 2013, pp. 1 –8 [Google Scholar]
- 50. Sultan A. Khan H.M. Malik A. et al.: ‘Antibacterial activity of ZnO nanoparticles against ESBL and Amp‐C producing gram negative isolates from superficial wound infections’, Int. J. Curr. Microbiol. App. Sci., 2015, e‐1, pp. 38 –47 [Google Scholar]
- 51. Lemire J.A. Harrison J.J. Turner R.J.: ‘Antimicrobial activity of metals: mechanisms, molecular targets and applications’, Nat. Rev. Microbiol., 2013, 11, pp. 371 –384 [DOI] [PubMed] [Google Scholar]
- 52. Chudobova D. Cihalova K. Dostalova S. et al.: ‘Comparison of the effects of silver phosphate and selenium nanoparticles on Staphylococcus aureus growth reveals potential for selenium particles to prevent infection’, FEMS Microbiol. Lett., 2014, 351, (2), pp. 195 –201 [DOI] [PubMed] [Google Scholar]


