Abstract
Biosynthesis of novel therapeutic nano‐scale materials for biomedical and pharmaceutical applications has been enormously developed, since last decade. Herein, the authors report an ecological way of synthesising the platinum nanoparticles (PtNPs) using Streptomyces sp. for the first time. The produced PtNPs exhibited the face centred cubic system. The fourier transform infrared spectrum revealed the existence of amino acids in proteins which serves as an essential reductant for the formation of PtNPs. The spherical morphology of the PtNPs with an average size of 20–50 nm was observed from topographical images of atomic force microscopy and field emission scanning electron microscopy. The X‐ray fluorescence spectrum confirms the presence of PtNPs with higher purity. The PtNPs size was further confirmed with transmission electron microscopy analysis and the particles were found to exist in the same size regime. Additionally, PtNPs showed the characteristic surface plasmon resonance peak at 262 nm. Dynamic light scattering studies report that 97.2% of particles were <100 nm, with an average particle diameter of about 45 nm. Furthermore, 3‐(4, 5‐dimethyl‐2‐thiazolyl)‐2, 5‐diphenyl‐tetrazolium assay based in vitro cytotoxicity analysis was conducted for the PtNPs, which showed the inhibitory concentration (IC50) at 31.2 µg/ml against Michigan Cancer Foundation‐7 breast cancer cells.
Inspec keywords: biomedical materials, materials preparation, nanoparticles, nanomedicine, nanofabrication, cellular biophysics, microorganisms, cancer, platinum, Fourier transform infrared spectra, proteins, atomic force microscopy, scanning electron microscopy, fluorescence, transmission electron microscopy, surface plasmon resonance, light scattering
Other keywords: cytotoxic potentials; biologically fabricated platinum nanoparticles; Streptomyces sp; MCF‐7 breast cancer cells; biosynthesis; therapeutic nanoscale materials; biomedical applications; pharmaceutical applications; Fourier transform infrared spectrum; amino acids; spherical morphology; topographical images; atomic force microscopy; field emission scanning electron microscopy; X‐ray fluorescence spectrum; transmission electron microscopy analysis; surface plasmon resonance; dynamic light scattering; 3‐(4, 5‐dimethyl‐2‐thiazolyl)‐2, 5‐diphenyl‐tetrazolium assay; cytotoxicity analysis; Pt
1 Introduction
In recent years, eco‐friendlier nanomaterials development plays several key role in most of the fields including biomedical, electronics, drug delivery and energy applications [1, 2, 3, 4]. Noble metal nanoparticles (NPs) especially platinum NPs (PtNPs) have explored well in the domain of pharmaceutical applications due to their multifunctional properties such as antibactericidal [5] and anticancer activities [6]. Since the PtNPs show extraordinary behaviour in various application ends, their synthetic strategies and their characterisation analyses became much essential. Various synthetic routes for obtaining the PtNPs include colloidal systems, reduction, bacterial cellulose, microemulsion, sol–gel, sonochemical method and electrode position synthesis [7]. Among the aforementioned synthetic mechanisms, bacterial cellulose, i.e. biological processing of PtNPs is an ecological, harmless and simpler technique. Amino acids which are essential constituent of proteins present in the microorganisms will act as the reducing agent for obtaining the PtNPs [8]. In recent days, syntheses of NPs are mediated via microorganisms [9, 10]. Streptomyces sp. is renowned as the initiator of bioactive metabolites [11]. From the best of our knowledge, no reports are available for the biosynthesis of PtNPs using Streptomyces sp. So, herein we report the biosynthesis of PtNPs using Streptomyces sp. In this paper, we have isolated the Streptomyces sp. which is selected as the bio‐reductant for the synthesis of PtNPs by the aforesaid substantial prominences. The NPs synthesised by the members of actinomycetes present good polydispersity and stability and possess enhanced biocidal activities against various pathogens due to proteins involved in the capping of PtNPs. A well‐established non‐toxic and eco‐friendly potent methodology for the synthesis of NPs has mounted to a level of supreme importance. Hence, the use of Streptomyces sp. for the synthesis of PtNPs with potential anticancer activity seems to be very promising approach for the development of newer nano‐antimicrobials by extracellular method. Nanotechnology offers the way to point treatments directly and selectively at cancerous cells. Routine chemotherapy utilises drugs that are known to kill cancer cells successfully. Be that as it may, these cytotoxic medications slaughter healthy cells notwithstanding tumour cells, prompting antagonistic symptoms, for example, neuropathy, hair loss and compromised immune function. PtNPs can be utilised as medication transporters for chemotherapeutics to convey medication specifically to the tumour while sparing healthy tissue. Not too far off are PtNPs that will effectively target medications to cancerous cells, the particles that they express on their cell surface. Molecules that predicament specific cell receptors can be appended to a PtNP to effectively target cells expressing the receptor. The utilisation of nanotechnology in cancer treatment offers some energising conceivable outcomes including the likelihood of decimating cancer tumours with minimal harm to healthy tissues and additionally the identification and elimination of cancer cells before they frame tumours [12]. In general, NPs have the nature of anticancer effect [13, 14] especially PtNPs have the nature of cytotoxic effects against the cancer cells [15, 16, 17]. Furthermore, the obtained PtNPs was characterised for its structural, morphological and anticancer activities.
2 Materials and methods
2.1 Materials
The potassium hexachloroplatinate (IV) (K2 PtCl6) was purchased from Fisher Inorganics & Aromatics Ltd., India. The microbiological nutrient media was procured from Hi‐Media, India. Double distilled water was used in all experiments. The strain Streptomyces sp. was isolated from the Kodiyakarai marine segment of Tamil Nadu, India. All chemicals used were of analytical grade.
2.2 Biosynthesis of PtNPs
The isolated Streptomyces sp. were centrifuged well at 10,000 rpm and then filtered through 0.2 µm polyethersulphone filters. The resulted cell free supernatant (CFS) was used for graded bioassay against the Michigan Cancer Foundation‐7 (MCF‐7) cell line. Various concentrations of CFS of Streptomyces sp. were added to the aqueous potassium hexachloroplatinate (IV) K2 PtCl6 (1 mM in 30 ml water) and incubated at 30°C for 24 h.
2.3 Characterisation of PtNPs
Ultraviolet–visible spectrometric measurements were performed (Shimadzu‐UV‐2450, Japan) in 200–800 nm range. The bioactive molecules responsible for the reduction of Pt ions were gauged with Fourier transform infrared (FTIR) spectroscopy in 400–4000 cm−1 range (Perkin‐Elmer, UK, Paragon‐500). The crystalline phases of PtNPs were identified with X‐ray diffraction studies (PANalytical Make) in 20–90° (2θ), 40 kV and 30 mA. Surface topography was determined by non‐compact mode atomic force microscopy (NCAFM‐PARK XE70 model). The PtNPs size distribution and zeta potential studies were interpreted using dynamic light scattering (DLS) spectroscopy (Malvern Zeta sizer, V7.10, MAL1087926). The surface morphology was studied by using field emission scanning electron microscopy (FESEM) (Carl Zeiss microscopy Ltd., UK & SIGMA) under the magnification of 469.13KX, 10 kV in 20 and 100 nm. The transmission electron microscopic (TEM) images were recorded on an Field Emission Gun‐TEM (Phillips CM 200 field emission gun TEM). The Streptomyces sp. stabilised PtNPs were prepared for TEM measurement by pouring a drop of the NPs on carbon coated copper grid followed by drying in vacuum.
2.4 Cell line and culture
Breast cancer cell line MCF‐7 was obtained from National Centre for Cell Science, Pune, India. The cells were cultured in minimal essential medium supplemented with 10% foetal bovine serum, penicillin (100 μg/ml) and streptomycin (100 μg/ml) and maintained in a humidified (atmosphere of 50 μg/ml) CO2 incubator 37°C.
2.4.1 In vitro assay for anticancer activity
The cytotoxic activity of PtNPs against MCF‐7 cell line was determined by using 3‐(4, 5‐dimethyl‐2‐thiazolyl)‐2, 5‐diphenyl‐tetrazolium (MTT) bromide assay. In brief, MCF‐7 cells (1 × 105 /well) were plated in 24‐well plates and incubated overnight at 37°C with 5% CO2, Then, PtNPs at increasing concentration (0–100 μg/7 ml were added to the cells and incubated at 37°C with 5% CO2. Following the treatment period, 100 μl/well (5 gm/ml) of 0.5% MTT was added to each well and incubated for 4 h. About 1 ml of dimethyl sulphoxide (DMSO) were added in all wells after the removal of the MTT with culture medium. The absorbance of purple formazon crystals were measured at 570 nm in an UV‐spectrophotometer using DMSO as the blank. The concentration of PtNPs showing 50% inhibition of visibility (IC50) was calculated as previously described [18]. Cell control and sample control are included in each assay to compare full cell viability and cell death assessments.
3 Results and discussion
3.1 Biosynthesis of PtNPs
The hypothetical mechanism of extracellular production of PtNPs in Streptomyces sp. is that the chloride reductase enzyme is involved in nitrogen cycle and responsible for reduction of chloride to chlorine. It is known that nicotinamide adenine dinucleotide dependent chloride reductase enzyme is an important factor in biogenic synthesis of NPs [8]. The possible mechanism, which may involve the reduction of Pt ions, is probably the electron shuttle enzymatic metal reduction process
| (1) |
Chloride reductase like chemical reaction might be responsible for the bioreduction of Pt ions to metallic Pt and the subsequent formation of PtNPs. The formation mechanism (1) represents the presence of Pt ions and it was confirmed through X‐ray fluorescence (XRF) analysis being discussed in Section 3.6.
3.2 Optical properties of PtNPs
The absorption spectrum of the colloidal NPs prepared via biosynthetic route is an essential to determine their distinguished plasmonic properties. Various concentrations (250, 500, 1000, 1500 and 2000 µl) of CFS were added to each 30 ml of stock solution and incubated at 30°C for 72 h. The optimised CFS volume was identified to be 1500 µl from the spectra which exhibited hyperchromic shift. The peaks obtained after 1500 µl started to aggregate which showed lower in the intensity levels. The spectra obtained for the PtNPs (Fig. 1 a) have showed a symmetric peak at 262 nm, which is the characteristic surface plasmon resonance (SPR) of the PtNPs [19]. This SPR peak could be mainly observed from the spectrum of the PtNPs colloid, which indicate the existence of PtNPs produced by utilising the CFS. Inset Fig. 1 a shows the absorbance peak at 237 nm indicates the presence of CFS which is confirmed that the CFS was acted as a reducing agent for the synthesis of PtNPs.
Fig. 1.
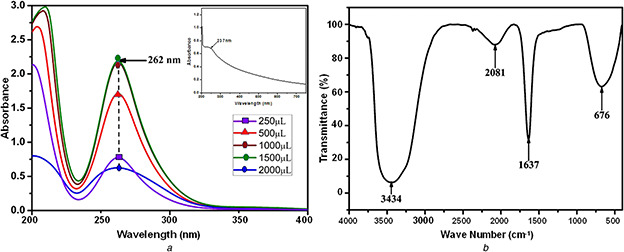
Spectrum obtained for the PtNPs
(a) UV absorbance spectrum of biologically synthesised PtNPs, inset: UV absorbance spectrum of Streptomyces sp. CFS, (b) FTIR spectroscopy spectrum of biologically synthesised PtNPs
3.3 FTIR spectroscopy analysis
The obtained FTIR spectrum of the NPs indicated the existence of amino acids (Fig. 1 b). Intensive vibrations were received at the band positions of 676, 1637, 2081 and 3434 cm−1. The vibration bands received at 1637 and 3434 cm−1 could be attributed to –C=C– and free N–H vibrations, respectively, which could also be assigned to the heterocyclic compounds such as proteins as assigned in earlier reports [20]. The –C=C– stretch was dispensed alkene function group and N–H stretch was allotted primary amine group which may act as a reducing agent for the synthesis of PtNPs. The band for 676 cm−1 is assigned to alkyl halides (C–X) functional groups chloride and 2081 cm−1 is assigned to amines functional group (R–N=C=S).
The existence of amino acids in proteins along with the PtNPs also confirms biosynthesis assisted formation of PtNPs (Fig. 2) and proteins as capping agent [21].
Fig. 2.
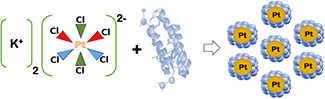
Formation mechanism of biologically synthesised PtNPs
3.4 X‐ray diffraction pattern analysis
The biosynthesised PtNPs were characterised with X‐ray diffractometer to study the crystalline information (Fig. 3). The obtained pattern belongs to face centred cubic (FCC) system space group of Fm3m of Pt in accordance to the standard Joint Committee on Powder Diffraction Standards (JCPDS) card number 04‐0802. The peaks observed at the 2θ positions of 39.7°, 45.5°, 67.6°, 81.6° and 86.2° are indexed to the crystalline planes (111), (200), (220), (222) and (311), respectively. The average crystalline size of the PtNPs were estimated using the Debye–Scherrer's equation
| (2) |
where D is the average crystalline size, λ denotes X‐ray wavelength, β is the width of half maximum intensity and θ is the Bragg's angle in degrees.
Fig. 3.
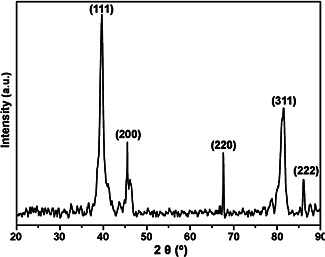
X‐ray diffraction pattern of crystalline PtNPs
Thus calculated crystalline size is 24.1 nm, which showed the formation of smaller crystallites as an added advantage of the utilised synthetic route. The unit cell parameters of PtNPs is under FCC crystal system and the α = β = γ = 90°. The atomic radius is in 0.139 nm with four atoms per unit cell. The lattice constant is calculated based on the formula
| (3) |
where R is the atomic radius with the value of 0.139 nm. Then the lattice constant is 3.9288 Å. The atomic facing factor is in 0.74. The PtNPs having the density of 21.47 g/cm3 and the volume of 60.38 μpm3. All the above will well match with the JCPDS card number 04‐0802.
3.5 AFM of PtNPs
The PtNPs dispersed over the glass substrate was scanned with an AFM in its non‐contact mode for their morphological information. The attained two‐dimensional (2D) and 3D surface topographical images were showed in Fig. 4. The images depict the agglomeration free spherical shaped NPs with size ranges from 20 to 50 nm.
Fig. 4.
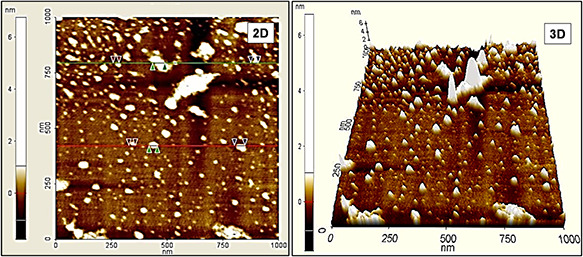
Topographical images of biologically synthesised PtNPs
The formation of smaller size particles as well as the crystalline size evidenced in X‐ray diffraction pattern designates the effective contribution of the amino acids against the higher‐order growth during the materials formation process and also the avoidance of agglomeration by serving as capping agent [8].
3.6 Morphological analysis
The surface morphology (Fig. 5 a) of the PtNPs was studied by using FESEM. The NPs synthesised from Streptomyces sp. were spherical in shape because of the presence of different groups of phytochemicals which help in reducing and stabilising the NPs during the synthesis procedure. The size of PtNPs were in 20–50 nm which lacking monodispersity. The higher concentration of bioactive compound in the colloidal solution may assist the formation of NPs [22]. The particle size distribution analysis (Fig. 5 b) confirms that most of the particles present between 20 and 50 nm. The XRF spectrum of PtNPs (Fig. 5 c) indicates the strong signal in the Pt region and serves as strong evidence for the formation of PtNPs with higher purity.
Fig. 5.
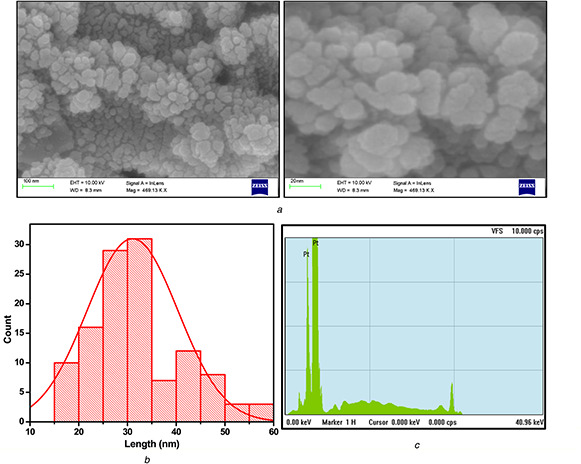
Morphological analysis of PtNPs
(a) Surface morphology analysis using FESEM, (b) Size distribution of PtNPs, (c) Energy dispersive X‐ray spectrum of PtNPs
The morphology and the size of the PtNPs were further examined by the TEM analysis (Fig. 6 a). It was evident that the biologically synthesised PtNPs using Streptomyces sp. possess somewhat uniform spherical shape and are well distributed within the size range of 20–50 nm. The size ranges obtained from the TEM results also concurred with the other analytical and topographical results.
Fig. 6.
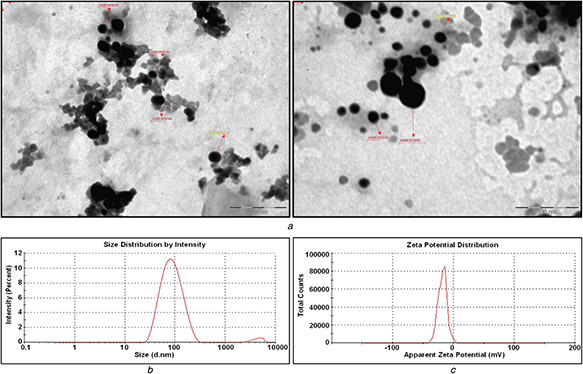
Morphology and the size analysis of the PtNPs
(a) Transmission electron microscopic analysis for biologically synthesised PtNPs, (b) Particle size distribution graphs measured using DLS for PtNPs, (c) Surface zeta potential graph showing negative zeta potential value for PtNPs synthesised by CFS of Streptomyces sp.
3.7 DLS analysis
The size distribution plot reports 97.2% of the particles were below 100 nm, with an average particle diameter of 44.98 nm. The biosynthesised NPs were indicative of monodispersive in nature by the polydispersity index value of 0.232 obtained for the colloidal solution (Fig. 6 b). The zeta potential (Fig. 6 c) for the PtNPs is in −18.4 mV, which specifies that the biologically synthesised PtNPs were abstemiously stable. The zeta potential of the biosynthesised PtNPs indicates the relatively neutral nature of the NPs. The neutral behaviour of the biosynthesised PtNPs suggest the presence of amino acids and proteins on its surface. Relatively, the average particle size measured in DLS is slightly in higher order than the average particle size calculated in AFM, which measures the particle size directly, whereas DLS measures the hydrodynamic diameter.
3.8 Cytotoxic potential of PtNPs against breast cancer cells in vitro
Fig. 7 illustrates the step‐by‐step mechanism for the cytoxicity activity against MCF‐7 cell line using PtNPs.
Fig. 7.
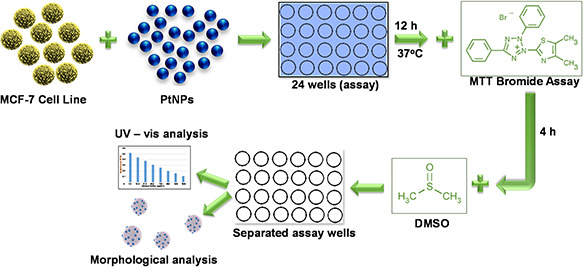
Schematic mechanism to illustrate the anticancer activity of PtNPs
The present study confirms the promising anticancer activity of PtNPs on human breast cancer cell line (MCF‐7) in vitro. Phase contrast photomicrographs of MCF‐7 cell line (Fig. 8 a) obtained at various concentrations showed abnormal cellular morphologies including detachment of cells from the surface, rounding and shrinkage with a significant reduction in the cell viability. The in vitro anticancer activity of biosynthesised PtNPs were studied using MTT assay at different concentrations and the inhibitory effect (IC50) in MCF‐7 cancer cell lines were determined by plotting the chart (Fig. 8 b) of different concentrations versus percentage (%) of cell viability and percentage (%) cell death. The analysis for concentration versus absorbance (Fig. 8 c) confirmed that the absorbance of the cell line decreased while increasing the concentration because of the higher toxicity against the cancer cells.
Fig. 8.
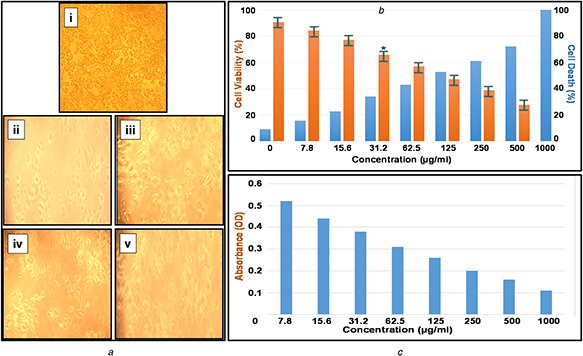
Phase contrast photomicrographs and graphical representation of MCF‐7 cell line
(a) Morphological changes in MCF‐7 cells treated with PtNPs. The changes in the cell morphology due to toxicity of PtNPs were determined by phase contrast microscopical analysis: (i) normal breast cancer cell line, (ii) Toxicity‐1000 µg/ml, (iii) Toxicity‐125 µg/ml, (iv) Toxicity‐62.5 µg/ml and (v) Toxicity‐31.2 µg/ml, (b) Assessment of PtNPs induced cell death in MCF‐7 breast cancer. The cells were treated with a range of concentration of PtNPs and incubated for 24 h, the per cent of cell viability and cell death was determined by MTT assay, (c) Absorbance analysis for MCF‐7 cell line with various concentrations of biological synthesised PtNPs
The IC50 analysis confirmed the biosynthesised PtNPs produced a significant cytotoxic effect. The in vitro analysis showed that there was 52.77% cell viability in MCF‐7 human cell lines, after 24 h of incubation. This reduction in percentage cell viability of MCF‐7 cell lines specifies that the IC50 value is 31.2 µg/ml. As concentration increases, there was a decrease in percentage cell viability demonstrating a direct dose‐dependent relationship and increase in cell death.
4 Conclusion
PtNPs were synthesised biologically using Streptomyces sp. The absorption spectrum of the PtNPs showed its characteristic SPR peak at 262 nm authenticating the nano‐scaled Pt particles. The FTIR spectrum revealed that the action of amino acids as both reducing and capping agent for PtNPs. The biosynthesised PtNPs exhibited the FCC crystal system and their average crystal size was 24.1 nm. The PtNPs were spherical shaped with 20–50 nm size, which is evidenced from the AFM and FESEM topographical images. The elemental confirmation for PtNPs was authenticated with the XRF spectrum. Furthermore, the size and shape confirmations were envisaged by TEM, DLS and zeta potential studies which found to be in coexistence with the previous analytic investigations. Anticancer activity of synthesised PtNPs against MCF‐7 breast cancer cells was evaluated and the inhibitory concentration (IC50) was found to be 31.2 µg/ml. It has comparatively [23, 24, 25] lesser IC50 value and lower cell viability, which has revealed that the biosynthesised PtNPs possess strong cytotoxicity against human breast cancer cell lines (MCF‐7) and proved to be an effective drug for breast cancer therapy.
5 References
- 1. Riggio C. Raffa V. Cuschieri A.: ‘Synthesis, characterisation and dispersion of zinc oxide nanorods for biomedical applications’, IET Micro Nano Lett., 2010, 5, (5), pp. 355 –360 [Google Scholar]
- 2. Reddy M.V. Tse L.Y. Bruce W.K.Z. et al.: ‘Low temperature molten salt preparation of nano‐SnO2 as anode for lithium‐ion batteries’, Mater. Lett., 2015, 138, pp. 231 –234 [Google Scholar]
- 3. Luximon A.B. Jhurry D.: ‘New avenues for improving pancreatic ductal adenocarcinoma (PDAC) treatment: selective stroma depletion combined with nano drug delivery’, Cancer Lett., 2015, 369, pp. 266 –273 [DOI] [PubMed] [Google Scholar]
- 4. Li Y. Yang J. Song J.: ‘Nano‐energy system coupling model and failure characterization of lithium ion battery electrode in electric energy vehicles’, Renew. Sustain. Energy Rev., 2016, 54, pp. 1250 –1261 [Google Scholar]
- 5. Kumari A. Guliani A. Singla R. et al.: ‘Silver nanoparticles synthesised using plant extracts show strong antibacterial activity’, IET Nanobiotechnol., 2015, 9, (3), pp. 142 –152 [DOI] [PubMed] [Google Scholar]
- 6. Govindaraju K. Krishnamoorthy K. Alsagaby S.A. et al.: ‘Green synthesis of silver nanoparticles for selective toxicity towards cancer cells’, IET Nanobiotechnol., 2015, 9, (6), pp. 325 –330 [DOI] [PubMed] [Google Scholar]
- 7. Castr L. Blázquez M.L. González F.: ‘Biosynthesis of silver and platinum nanoparticles using orange peel extract: characterisation and applications’, IET Nanobiotechnol., 2015, 9, (5), pp. 252 –258 [DOI] [PubMed] [Google Scholar]
- 8. Sivalingam P. Antony J.J. Siva D. et al.: ‘Mangrove Streptomyces sp. BDUKAS10 as nanofactory for of bactericidal silver nanoparticles fabrication’, Colloids Surf. B, 2012, 98, pp. 12 –17 [DOI] [PubMed] [Google Scholar]
- 9. Nejad M.S. Khatami M. Bonjar G.H.S.: ‘Extracellular synthesis gold nanotriangles using biomass of Streptomyces microflavus’, IET Nanobiotechnol., 2016, 10, (1), pp. 33 –38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Busi S. Rajkumari J. Ranjan B. et al.: ‘Green rapid biogenic synthesis of bioactive silver nanoparticles (AgNPs) using Pseudomonas aeruginosa ’, IET Nanobiotechnol., 2014, 8, (4), pp. 267 –274 [DOI] [PubMed] [Google Scholar]
- 11. Ndlovu T.M. Ward A.C. Glassey J. et al.: ‘Bioprocess intensification of antibiotic production by Streptomyces coelicolor A3(2) in micro‐porous culture’, Mater. Sci. Eng. C, 2015, 49, pp. 799 –806 [DOI] [PubMed] [Google Scholar]
- 12. Porcel E. Liehn S. Remita H. et al.: ‘Platinum nanoparticles: a promising material for future cancer therapy?’, Nanotechnology, 2010, 21, pp. 1 –7 [DOI] [PubMed] [Google Scholar]
- 13. Palaniappan P. Sathiskumar G. Sankar R.: ‘Fabrication of nano‐silver particles using Cymodocea serrulata and its cytotoxicity effect against human lung cancer A549 cells line’, Spectrochim. Acta A, 2014, doi: 10.1016/j.saa.2014.10.072 [DOI] [PubMed] [Google Scholar]
- 14. Ramar M. Manikandan B. Marimuthu P.N. et al.: ‘Synthesis of silver nanoparticles using Solanum trilobatum fruits extract and its antibacterial, cytotoxic activity against human breast cancer cell line MCF 7’, Spectrochim. Acta A, 2015, 140, pp. 223 –228 [DOI] [PubMed] [Google Scholar]
- 15. Ghosh S. Nitnavare R. Dewle A. et al.: ‘Novel platinum–palladium bimetallic nanoparticles synthesized by Dioscorea bulbifera: anticancer and antioxidant activities’, Int. J. Nanomed., 2015, 10, pp. 7477 –7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mohammadi H. Abedi A. Akbarzadeh A. et al.: ‘Evaluation of synthesized platinum nanoparticles on the MCF‐7 and HepG‐2 cancer cell lines’, Int. Nano Lett., 2013, 3, p. 28 [Google Scholar]
- 17. Butler J.S. Sadler P.J.: ‘Targeted delivery of platinum‐based anticancer complexes’, Curr. Opin. Chem. Biol., 2013, 17, pp. 175 –188 [DOI] [PubMed] [Google Scholar]
- 18. Mosmann T.: ‘Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays’, J. Immunol. Methods, 1983, 65, pp. 55 –63 [DOI] [PubMed] [Google Scholar]
- 19. Saion E.G.E.: ‘Influence of dose on particle size and optical properties of colloidal platinum nanoparticles’, Int. J. Mol. Sci., 2012, 13, pp. 1472 –1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gole A. Dash C. Ramakrishnan V. et al.: ‘Pepsin gold colloid conjugates: preparation, characterization and enzymatic activity’, Langmuir, 2001, 17, pp. 1674 –1679 [Google Scholar]
- 21. Mandal S. Phadtate S. Sastry M.: ‘Interfacing biology with nanoparticles’, Curr. Appl. Phys., 2005, 5, pp. 118 –127 [Google Scholar]
- 22. Sankar R. Karthik A. Prabhu A. et al.: ‘ Origanum vulgare mediated biosynthesis of silver nanoparticles for its antibacterial and anticancer activity’, Colloids Surf. B, 2013, 108, pp. 80 –84 [DOI] [PubMed] [Google Scholar]
- 23. Borse V. Kaler A. Banerjee U.C.: ‘Microbial synthesis of platinum nanoparticles and evaluation of their anticancer activity’, Int. J. Emerging Trends Electr. Electron., 2015, 11, pp. 26 –31 [Google Scholar]
- 24. Watanabe A. Kajita M. Kim J. et al.: ‘ In vitro free radical scavenging activity of platinum nanoparticles’, Nanotechnology, 2009, 20, (45), p. 455105 [DOI] [PubMed] [Google Scholar]
- 25. Mironava T. Simon M. Rafailovich M.H. et al.: ‘Platinum folate nanoparticles toxicity: cancer vs. normal cells’, Toxicol. In Vitro, 2013, 27, (2), pp. 882 –889 [DOI] [PubMed] [Google Scholar]


