Abstract
Biological routes of synthesising metal nanoparticles (NPs) using microbes have been gaining much attention due to their low toxicity and eco‐friendly nature. Pseudomonas aeruginosa JP2 isolated from metal contaminated soil was evaluated towards extracellular synthesis of silver NPs (AgNPs). Cell‐free extract (24 h) of the bacterial isolate was reacted with AgNO3 for 24 h in order to fabricate AgNPs. Preliminary observations were recorded in terms of colour change of the reaction mixture from yellow to greyish black. UV‐visible spectroscopy of the reaction mixture has shown a progressive increase in optical densities that correspond to peaks near 430 nm, depicting reduction of ionic silver (Ag+) to atomic silver (Ag0) thereby synthesising NPs. X‐ray diffraction spectra exhibited the 2θ values to be 38.4577° confirming the crystalline and spherical nature of NPs [9.6 − 26.7 (Ave. = 17.2 nm)]. Transmission electron microscopy finally confirmed the size of the particles varying from 5 to 60 nm. Moreover, rhamnolipids and proteins were identified as stabilising molecules for the AgNPs through Fourier transform‐infrared spectroscopy. Characterisation of bacterial crude and purified protein fractions confirmed the involvement of nitrate reductase (molecular weight 66 kDa and specific activity = 3.8 U/mg) in the Synthesis of AgNPs.
Inspec keywords: microorganisms, silver, nanoparticles, enzymes, molecular biophysics, ultraviolet spectra, visible spectra, X‐ray diffraction, transmission electron microscopy, Fourier transform infrared spectra, catalysis, biochemistry, nanobiotechnology
Other keywords: catalytic protein, stabilising agents, Pseudomonas aeruginosa, metal nanoparticles, UV–visible spectroscopy, optical densities, ionic silver, atomic silver, X‐ray diffraction spectra, transmission electron microscopy, nitrate reductase, rhamnolipids, Fourier transform‐infrared spectroscopy, Ag
1 Introduction
Nanotechnology, an emerging field in science and technology, has led to the synthesis of nanoscale materials with enormous implications in electronics, photonics, nanomedicine, biolabelling, biomedical engineering and environment [1]. Various physical and chemical strategies have been used to synthesise well‐defined nanomaterials of noble metals [2]. However, these conventional methods despite their commercial importance are facing challenges in terms of high cost and hazardous nature. Hence there is growing demand for the use of environmentally benign procedures for the synthesis of nanoparticles (NPs) [3]. Recently, there has been an upsurge of biological techniques where metallic ions are reduced to NPs [4] by different plants [5] and microorganisms [6]. In this context, new enzymatic approaches have been viewed as playing an important role for both intracellular [7] as well as extracellular syntheses [7] of NPs in batch and continuous cultures of bacteria and fungi [8].
Generally, efficacies associated with NPs in different applications and procedures have been linked with their sizes and stable monomeric nature on a large scale [9]. The higher stability of biogenic AgNPs can be attributed to the capping agents [10]. The nature of the capping molecule depends on the organisms used for enzyme source. Various capping molecules such as proteins and rhamnolipids have been identified in bacteria‐ and fungus‐mediated synthesis of NPs. In addition, elucidation of molecular mechanisms involved in the biological synthesis of metal NPs with controlled size, shape and monodispersity are some of the challenges to deal with. The pathways involved in the biosynthesis of AgNPs are also of prime importance for the development and upscaling of production methodologies for other elements. [11]. Participation of cellular proteins in the synthesis of AgNPs has been investigated earlier [12]. Extracellularly secreted nitrate reductase from different microbes has been identified as the reducing agent that converts silver ions to AgNPs [8]. Moreover, similar reducing activities have been reported intracellularly in different studies [13].
Biological reduction processes of metals are not only important in natural metal transformation processes, but they are also equally important in understanding the bioleaching processes associated with metal ores [14] and in the bioremediation of metal‐contaminated sites [15]. In this study, efforts were directed towards exploring the basic mechanism involved in the synthesis of AgNPs. In this perspective, the participation of nitrate reductase and metal capping or stabilising proteins was typically perused.
2 Experimental
In this study, environmental isolate from the metal‐contaminated soil was used as a source of enzyme, which was microbiologically and biochemically characterised as Pseudomonas aeruginosa (P. aeruginosa) JP2. The specific isolate produced fluorescence at Cetrimide agar (Sigma‐Aldrich). For the synthesis of AgNPs, bacterial culture was aerobically cultivated in the slightly modified MGYP media containing malt extract 3 g/l, glucose 10 g/l, yeast extract 3 g/l and peptone 5 g/l for a 100 ml growth medium at 37°C on a rotary shaker (150 rpm) for 24 h and harvested by centrifugation (centrifuge Model H‐251, Kokusan Co., Ltd., Tokyo, Japan) at 8000 rpm at 4°C for 20 min to obtain the cell‐free extract.
2.1 Biosynthesis of silver NPs (AgNPs)
To synthesise AgNPs, 50 ml of cell‐free extract and 50 ml of an aqueous solution of silver nitrate (AgNO3) (10 mM) were mixed in an Erlenmeyer flask (250 ml). The flask containing the reaction mixture along with positive control (cell‐free extract) and negative controls (AgNO3) were incubated in a rotary shaker (150 rpm) under dark conditions at 37°C for 24 h. The preliminary detection of the synthesised AgNPs was done by perceiving the colour change from pale yellow to greyish black. Besides, another investigation was processed in which bacterial extract was heated (4 h at 98°C) to inactivate the possible biological reducing agent.
2.2 Characterisation of NPs
Visual observation indicated the colour change, from yellow to greyish black. The biological reduction of silver ions in the solution and preliminary characterisation was detected by measuring the ultraviolet–visible (UV–vis) spectra of the sampling aliquots subsequently removed (at equal intervals of an hour) from the reaction mixture using UV–vis spectrophotometer (Agilent 8453 UV–Vis, Agilent Technologies, Santa Clara, CA, USA) from 250 to 750 nm. The reaction mixture was centrifuged (centrifuge Model H‐251, Kokusan Co., Ltd., Tokyo, Japan) at 10,000 rpm for 8 min and AgNPs were precipitated. AgNPs were washed with chilled ethanol and let them air dry. Crystalline nature and the mean particle size were confirmed by X‐ray diffraction (XRD) analysis. XRD spectrum was obtained when dry powder‐coated films of dried sample containing NPs on silica were subjected to XRD analysis (X'pert PRO XRD, PANalytical BV, Almelo, The Netherlands) operating in a transmission mode, at 30 kV, 20 mA with CuKα radiation.
The morphology of the synthesised NPs was determined by transmission electron microscopy (TEM). The AgNPs film was created on carbon‐coated copper grids that were examined by TEM (JEM‐1010, JEOL Ltd., Tokyo, Japan) at an accelerating voltage of 80 kV. Fourier transform‐infrared spectroscopy (FTIR) analysis was done to identify the nature of capping molecules on the basis of functional groups vibration and their interaction with AgNPs. For the sample preparation, the reaction mixture was concentrated by ultracentrifugation (centrifuge Model H‐251, Kokusan Co., Ltd., Tokyo, Japan) at 12,000 rpm for 20 min. After concentrating, the pellet was washed three times with pure ethanol and double distilled water followed by air drying in a petri dish. Then powder of AgNPs was pressed into a clear disc, which was mounted and examined directly by FTIR (Perkin‐Elmer, Shelton, CT).
2.3 Enzyme (nitrate reductase) purification and characterisation
All the steps of enzyme purification and characterisation were performed at 4°C.
2.4 Ammonium sulphate precipitation
Cell‐free extract was precipitated with 60% ammonium sulphate saturation and the precipitates were concentrated by centrifugation (centrifuge Model H‐251, Kokusan Co., Ltd., Tokyo, Japan) at 6000 rpm for 20 min. This crude protein extract was suspended in 50 mM phosphate buffer (pH 7.4) and later on nitrate reductase activity and total protein concentrations were determined.
2.5 Gel filtration chromatography
For further purification, 2 ml of the partially purified proteins was subjected to size exclusion chromatography using a column packed with sephadex G‐100. The protein sample was eluted by phosphate buffer (pH 7.4) that was continuously added and fractions of 3 ml (each) were obtained with an average speed of 15 min/fraction. Gel filtration chromatography is a standard technique [16] to purify and separate biomolecules on the basis of their sizes. Initially larger molecules are excluded, then smaller molecules. Each fraction is supposed to contain specific type and size of the proteins (enzyme). Nitrate reductase assay and Bradford assay were performed to determine the enzyme activity and the total protein content, respectively. Evaluation of the reducing agent that might be responsible for reduction as well as synthesis of AgNPs was done by treating the purified fraction of enzyme with equal volumes of 10 mM AgNO3 solution. After incubation at 37°C for 24 h the reduction was measured at 420 nm.
2.6 Sodium dodecyl sulphate (SDS)‐polyacrylamide gel electrophoresis
The purified enzyme (nitrate reductase) was analysed by SDS‐PAGE for the determination of molecular weight. Proteins were visualised by staining with Coomassie brilliant blue R‐250; their molecular weights were determined by comparison with standard protein marker (Bio‐Rad, USA) (Laemmli 1970).
3 Results and discussion
When the extracellular filtrate of Pseudomonas aerogenosa JP2 was reacted with AgNO3 solution (10 mM) at 37°C for 24 h AgNPs were synthesised. Preliminary observation showed a visible colour change of the reaction mixture from yellow to greyish black (Fig. 1 a) indicating the formation of AgNPs, due to the reduction of silver metal ions into AgNPs via the biomolecules present in the extract. This observation highlighted the involvement of some reducing agent present in the cell filtrate [17]. While the negative control (Fig. 1 d) and positive control (Fig. 1 c) along with the heat‐killed (Fig. 1 b) reaction mixture did not show any colour change; the specific colour change was probably due to the excitation of metal electrons by surface plasmon resonance (SPR) indicating the formulation of AgNPs [18].
Fig. 1.
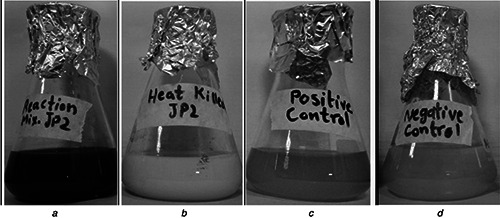
Preliminary observation showed a visible colour change of reaction mixture from yellow to greyish black
a Reaction mixture containing cell filtrate of P. aerogenosa JP2 and AgNO3 solution (10 mM) showed the visible colour change from yellow to greyish black
b Heat‐killed cell filtrate and AgNO3 solution showed no colour change
c Positive control (only cell filtrate), no change in colour
d Negative control (only AgNO3 solution), no colour change
3.1 UV–vis spectroscopy
UV–vis absorption spectroscopy has been an established method to characterise the formation of AgNPs [19]. UV–vis spectrum of the reaction mixture exhibited a characteristic SPR band in the region near 430 nm (Fig. 2), depicting the synthesis of AgNPs [20]. The comparative analysis of the peaks regarding the AgNO3 solution and the reaction mixture noticeably indicated the reduction of Ag+ to Ag0. The SPR bands for AgNPs in the range of 410–450 nm have been well documented [21]. The sharpness and area of the band depict the size, shape and amount of AgNPs [22]. The intensity of the SPR bands kept increasing till the eighth hour suggesting the accomplishment of the reaction which was considerably efficient in terms of time than reported before [23].
Fig. 2.
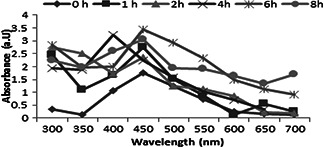
Absorption spectra of reaction mixture depicting the synthesis of AgNPs
3.2 XRD analysis
XRD analysis confirmed the crystalline nature of the AgNPs (Fig. 3). XRD analysis of the purified dried powder form of reaction mixture (Fig. 3) exhibited characteristic Bragg peaks at 2θ values of 38.2°, 44.3°, 64.8° and 77.4° corresponding to the 111, 200, 220 and 311 planes of the face‐centred cubic (fcc) AgNPs, respectively [24]. Debye–Scherrer equation was used to calculate the mean diameter range 17.23 nm (10–27 nm) of particles (Fig. 4). Similar observations have been made when extracellular AgNPs (31.7 nm) were synthesised by Pseudomonas putida [25]. A small insignificant peak was also observed that might be ascribed to other organic substances present in the reaction mixture.
Fig. 3.

XRD spectrum of AgNPs synthesised by P. aerogenosa JP2
Fig. 4.
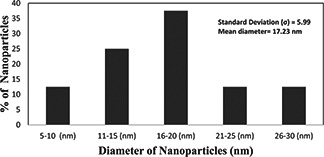
Histogram showing the particle distribution of NPs
Debye–Scherrer equation
3.3 Transmission electron microscopy
TEM analysis of the purified NPs showed spherical‐shaped AgNPs with an estimated size ranging from 5 to 60 nm (Fig. 5) This size range was much closely related to AgNPs synthesised by some other bacteria such as Lactobacillus bulgaricus (40–50 nm) [26], Pseudomonas aerogenosa 8–24 [27], 20–50 [23] and E scherichia coli 40–50 nm [28]. Moreover, AgNPs were separately clustered and close to monodispersed nature (Figs. 5 a and b). Previously, this sort of organization has been linked to certain capping peptides and proteins in the cell filtrate that were involved in the stabilisation of NPs and preventing them from aggregation [27].
Fig. 5.
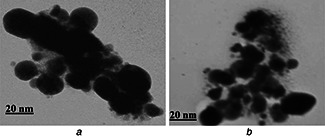
TEM of AgNPs synthesised by P. aerogenosa JP2
a Higher magnification (200 K)
b Lower magnification (150 K)
3.4 FTIR spectroscopy
The nature of the capping molecule was identified by FTIR analysis. FTIR spectra of the AgNPs showed strong IR bands at 782, 815, 1632, 1520, 3264, 2950 cm−1 and a few others (Fig. 6). Peaks at 782 and 815 cm−1 were assigned to C–H group and aromatic groups of proteins. Peaks at 1520 and 1632 cm−1 corresponded to the bending vibration of amide I and amide II of the protein [29], while their stretching vibrations were indicated by peaks at 3264 cm−1 [30]. The wave number 1022 cm−1 indicated the stretching vibrations of C–O–C bond in rhamnose sugar [31] Aliphatic bonds CH3, CH2 were identified in the peak range 2930–2875 cm−1 [32]. The presence of carboxylic acid functional group in the molecule was confirmed by the bending of the hydroxyl (O–H) by 1447 and 1383 cm−1, which also provided evidence for the presence of rhamnolipids on the surface of AgNPs [32]. The functional groups (C–H) have been identified in the characterisation of purified rhamnolipids [33]. Another peak at 1049 cm−1 attributed to the C–O–H bending vibrations due to certain capping proteins (Fig. 6), which might be responsible for the stability of AgNPs. FTIR spectrum clearly indicated the presence of proteins and rhamnolipids on the surface of biologically synthesised AgNPs. Similarly, involvement of proteins and rhamnolipids as a stabilising agent of AgNPs has been investigated earlier [27]. Capping molecules might have bounded to the surface of NPs through cysteine residues or similar amino acids, which prevent NPs from forming aggregations [33]. These results were also in agreement with those obtained with TEM.
Fig. 6.

FTIR spectrum of AgNPs produced by P. aeruginosa JP2
3.5 Characterisation of nitrate reductase and its involvement in the synthesis of AgNPs
To understand the underlying mechanism of the synthesis of NPs evaluation of the reducing agent was done by crude and partially purified fraction of the nitrate reductase from the cell‐free extract of P. aeruginosa (Fig. 7 a). The crude extract was initially screened out by nitrate reductase assay. Subsequently, total protein was precipitated out and fractionised by gel‐filtration chromatography. Specific activity was determined for all fractions (Fig. 7 a) and fraction with the highest specific activity (3.8 U/mg) was subjected to SDS‐PAGE analysis. SDS‐PAGE results depicted the molecular weight (66 kDa) of purified nitrate reductase (Fig. 8), which might belong to periplasmic nitrate reductases. Molecular weight of nitrate reductase proved to be much closer to the previously reported one (70 kDa) [34]. A positive correlation (r = 0.715) was observed, when purified protein fractions was treated with AgNO3 solution to reconfirm the participation of nitrate reductase in the synthesis of AgNPs (Figs. 7 b and c). The involvement of NADH‐dependent reductase in Fusarium oxysporum has been reported [13]. It has also been suggested that the reduction of metal ions took place perhaps due to the conjugation between the electron shuttles and an NADPH‐dependent reductase [17]. Respiratory nitrate reductases (Nar) [35] and periplasmic nitrate reductases (Nap) [36] have been linked with the synthesis of AgNPs.
Fig. 7.
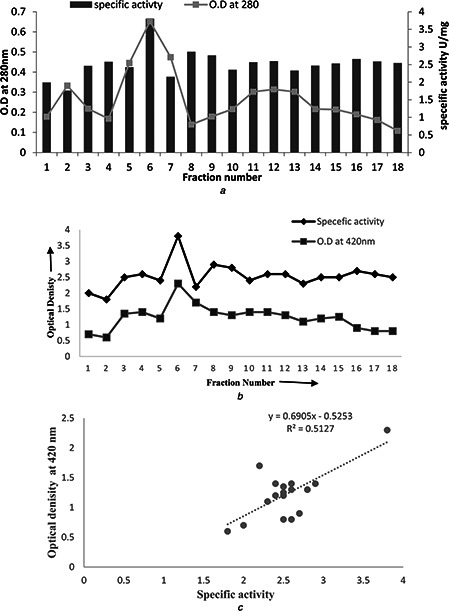
Characterisation of nitrate reductase and its involvement in synthesis of AgNPs
a Fractionation of proteins and associated specific nitrate reductase activity from total extracellular protein extract of JP2 strain using gel‐filtration chromatography
b Comparison of specific activity of purified fractions and their enzyme activities for AgNO3 solution
c Positive correlation (r = 0.715) indicating the involvement of nitrate reductases in the reduction of AgNO3 salt and synthesising AgNPs
Fig. 8.
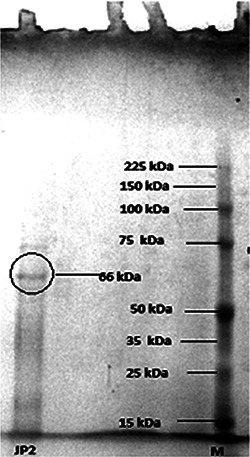
SDS‐PAGE of purified protein (catalytic) from crude extract of P. aeruginosa JP2 involved in the synthesis of AgNPs
Heat treated crude extract (reductase) of the bacteria did not reduce (Fig. 1 B). That was another indication of the involvement of a catalytic agent in the transformation of Ag to NPs. The additional evidence to the mechanistic theory was provided by the correlation results (Fig. 7 c) of the AgNO3 reduction with purified fractions of protein from crude extract of bacteria. Consequently, it could be inferred that the purified nitrate reductase might be the principle agent that reduced the metal ions and formulated the NPs, which also acted as an electron shuttle and took the electrons from nitrate molecule and transferred to the metal ion that converted it into an NP. The results were in accordance with Ahmed et al. (2003), where he described the enzyme mediated synthesis of extracellular AgNPs.
4 Conclusion
In this study, a simple, cost‐effective and eco‐friendly method for extracellular synthesis of AgNPs has been reported. The synthesised NPs were much stable and monodispersed, consequently, having good potential for biomedical applications. Involvement of nitrate reductase as a reducing agent in the synthesis of AgNPs was confirmed. Moreover, participation of proteins and rhamnolipid in the formation and stabilisation of NPs has been indicated. This understanding could further help in exploring the detailed mechanism of synthesis of NPs.
Table 1.
Overall protein purification scheme along with fold purification and percentage yield and nitrate reductase activity of P. aerogenosa JP2
| Purification step | Enzyme activity, U/ml | Total protein, mg/ml | Specific activity, U/mg | Purification fold | Yield, % |
|---|---|---|---|---|---|
| crude extract | 0.239 | 0.337 | 0.709 | 1 | 100 |
| ammonium sulphate precipitation | 0.658 | 0.331 | 1.987 | 2.802 | 94 |
| purified enzyme | 1.002 | 0.2635 | 3.802 | 5.362 | 78 |
5 References
- 1. Yantasee W. Warner C.L. Sangvanich T. et al.: ‘Removal of heavy metals from aqueous systems with thiol functionalized superparamagnetic nanoparticles’, Environ. Sci. Technol., 2007, 41, (14), pp. 5114 –5119 (doi: 10.1021/es0705238) [DOI] [PubMed] [Google Scholar]
- 2. Darroudi M. Ahmad M.B. Abdullah A.H. et al.: ‘Green synthesis and characterization of gelatin‐based and sugar‐reduced silver nanoparticles’, Int. J. Nanomedicine, 2011, 6, (1), pp. 569 –574 (doi: 10.2147/IJN.S16867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kulkarni N. Muddapur U.: ‘Biosynthesis of metal nanoparticles: a review’, J. Nanotechnol., 2014, 2014, pp. 1 –8 (doi: 10.1155/2014/510246) [DOI] [Google Scholar]
- 4. Jafar Ali S.Z. Ali N.: ‘Green synthesis of metal nanoparticles by microorganisms; a current prospective’, J. Nanoanal., 2015, 2, (1), p. 7 [Google Scholar]
- 5. Panda K.K. Achary V.M.M. Krishnaveni R. et al.: ‘In vitro biosynthesis and genotoxicity bioassay of silver nanoparticles using plants’, Toxicol. In Vitro, 2011, 25, (5), pp. 1097 –1105 (doi: 10.1016/j.tiv.2011.03.008) [DOI] [PubMed] [Google Scholar]
- 6. Sadowski Z. Maliszewska I. Grochowalska B. et al.: ‘Synthesis of silver nanoparticles using microorganisms’, Mater. Sci., 2008, 26, (2), pp. 419 –424 [Google Scholar]
- 7. Ahmad A. Mukherjee P. Senapati S. et al.: ‘Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum ’ , Colloids Surf. B, Biointerfaces, 2003, 28, (4), pp. 313 –318 (doi: 10.1016/S0927-7765(02)00174-1) [DOI] [Google Scholar]
- 8. Jain N. Bhargava A. Majumdar S. et al.: ‘Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavus Njp08: a mechanism perspective’, Nanoscale, 2011, 3, (2), pp. 635 –641 (doi: 10.1039/C0NR00656D) [DOI] [PubMed] [Google Scholar]
- 9. Narayanan K.B. Sakthivel N.: ‘Biological synthesis of metal nanoparticles by microbes’, Adv. Colloid Interface Sci., 2010, 156, (1), pp. 1 –13 (doi: 10.1016/j.cis.2010.02.001) [DOI] [PubMed] [Google Scholar]
- 10. Vigneshwaran N. Ashtaputre N. Varadarajan P. et al.: ‘Biological synthesis of silver nanoparticles using the fungus Aspergillus flavus ’, Mater. Lett., 2007, 61, (6), pp. 1413 –1418 (doi: 10.1016/j.matlet.2006.07.042) [DOI] [Google Scholar]
- 11. Iravani S.: ‘Green synthesis of metal nanoparticles using plants’, Green Chem., 2011, 13, (10), pp. 2638 –2650 (doi: 10.1039/c1gc15386b) [DOI] [Google Scholar]
- 12. Barwal I. Ranjan P. Kateriya S. et al.: ‘Cellular oxido‐reductive proteins of Chlamydomonas reinhardtii control the biosynthesis of silver nanoparticles’, J. Nanobiotechnol., 2011, 9, p. 56 (doi: 10.1186/1477-3155-9-56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar S.A. Abyaneh M.K. Gosavi S. et al.: ‘Nitrate reductase‐mediated synthesis of silver nanoparticles from AgNO3 ’, Biotechnol. Lett., 2007, 29, (3), pp. 439 –445 (doi: 10.1007/s10529-006-9256-7) [DOI] [PubMed] [Google Scholar]
- 14. Pugazhenthiran N. Anandan S. Kathiravan G. et al.: ‘Microbial synthesis of silver nanoparticles by Bacillus Sp’, J. Nanoparticle Res., 2009, 11, (7), pp. 1811 –1815 (doi: 10.1007/s11051-009-9621-2) [DOI] [Google Scholar]
- 15. Durán N. Marcato P.D. Alves O.L. et al.: ‘Ecosystem protection by effluent bioremediation: silver nanoparticles impregnation in a textile fabrics process’, J. Nanoparticle Res., 2010, 12, (1), pp. 285 –292 (doi: 10.1007/s11051-009-9606-1) [DOI] [Google Scholar]
- 16. Sachiko S.‐C. Ishimoto M.: ‘Studies on nitrate reductase of clostridium perfringens I. Purification, some properties, and effect of tungstate on its formation’, J. Biochem., 1977, 82, (6), pp. 1663 –1671 [DOI] [PubMed] [Google Scholar]
- 17. Ahmad A. Senapati S. Khan M.I. et al.: ‘Intracellular synthesis of gold nanoparticles by a novel alkalotolerant Actinomycete, Rhodococcus species’, Nanotechnology, 2003, 14, (7), p. 824 (doi: 10.1088/0957-4484/14/7/323) [DOI] [Google Scholar]
- 18. Vinod V. Saravanan P. Sreedhar B. et al.: ‘A facile synthesis and characterization of Ag, Au and Pt nanoparticles using a natural hydrocolloid gum kondagogu (Cochlospermum gossypium)’, Colloids Surf. B, Biointerfaces, 2011, 83, (2), pp. 291 –298 (doi: 10.1016/j.colsurfb.2010.11.035) [DOI] [PubMed] [Google Scholar]
- 19. Gangula A. Podila R. Karanam L. et al.: ‘Catalytic reduction of 4‐nitrophenol using biogenic gold and silver nanoparticles derived from Breynia rhamnoides ’, Langmuir, 2011, 27, (24), pp. 15268 –15274 (doi: 10.1021/la2034559) [DOI] [PubMed] [Google Scholar]
- 20. Sarkar R. Kumbhakar P. Mitra A.: ‘Green synthesis of silver nanoparticles and its optical properties’, Dig. J. Nanomater. Biostruct., 2010, 5, (2), pp. 491 –496 [Google Scholar]
- 21. Kim K.‐J. Sung W.S. Suh B.K. et al.: ‘Antifungal activity and mode of action of silver nano‐particles on candida albicans’, Biometals, 2009, 22, (2), pp. 235 –242 (doi: 10.1007/s10534-008-9159-2) [DOI] [PubMed] [Google Scholar]
- 22. Pal S. Tak Y.K. Song J.M.: ‘Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram‐negative bacterium Escherichia coli ’, Appl. Environ. Microbiol., 2007, 73, (6), pp. 1712 –1720 (doi: 10.1128/AEM.02218-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oza G. Pandey S. Shah R. et al.: ‘Extracellular fabrication of silver nanoparticles using Pseudomonas aeruginosa and its antimicrobial assay’, Pelagia Res. Lib. Adv. Appl. Sci. Res., 2012, 3, pp. 1776 –1783 [Google Scholar]
- 24. Kannan P. John S.A.: ‘Synthesis of mercaptothiadiazole‐functionalized gold nanoparticles and their self‐assembly on Au substrates’, Nanotechnology, 2008, 19, (8), p. 085602 (doi: 10.1088/0957-4484/19/8/085602) [DOI] [PubMed] [Google Scholar]
- 25. Prabhawathi V. Sivakumar P.M. Doble M.: ‘Green synthesis of protein stabilized silver nanoparticles using Pseudomonas fluorescens, a marine bacterium, and its biomedical applications when coated on polycaprolactam’, Ind. Eng. Chem. Res., 2012, 51, (14), pp. 5230 –5239 (doi: 10.1021/ie2029392) [DOI] [Google Scholar]
- 26. Sarvamangala D. Kondala K. Murthy U. et al.: ‘Biogenic synthesis of Agnp's using pomelo fruit–characterization and antimicrobial activity against gram + Ve and Gram–Ve bacteria’
- 27. Kumar C.G. Mamidyala S.K.: ‘Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa ’, Colloids Surf. B, Biointerfaces, 2011, 84, (2), pp. 462 –466 (doi: 10.1016/j.colsurfb.2011.01.042) [DOI] [PubMed] [Google Scholar]
- 28. Gurunathan S. Kalishwaralal K. Vaidyanathan R. et al.: ‘Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli ’, Colloids Surf. B, Biointerfaces, 2009, 74, (1), pp. 328 –335 (doi: 10.1016/j.colsurfb.2009.07.048) [DOI] [PubMed] [Google Scholar]
- 29. Vimala K. Sundarraj S. Paulpandi M. et al.: ‘Green synthesized doxorubicin loaded zinc oxide nanoparticles regulates the bax and Bcl‐2 expression in breast and colon carcinoma’, Process Biochem., 2014, 49, (1), pp. 160 –172 (doi: 10.1016/j.procbio.2013.10.007) [DOI] [Google Scholar]
- 30. Li S. Shen Y. Xie A. et al.: ‘Green synthesis of silver nanoparticles using Capsicum annuum L. Extract’, Green Chem., 2007, 9, (8), pp. 852 –858 (doi: 10.1039/b615357g) [DOI] [Google Scholar]
- 31. Arab F. Mulligan C.N.: ‘BIOSURFACTANTS research trends and applications’, in ‘Rhamnolipids characteristics, production, applications, and analysis’ 2014, p. 49
- 32. Heyd M. Kohnert A. Tan T.‐H. et al.: ‘Development and trends of biosurfactant analysis and purification using rhamnolipids as an example’, Anal. Bioanal. Chem., 2008, 391, (5), pp. 1579 –1590 (doi: 10.1007/s00216-007-1828-4) [DOI] [PubMed] [Google Scholar]
- 33. Kumar C.G. Mamidyala S.K. Das B. et al.: ‘Synthesis of biosurfactant‐based silver nanoparticles with purified rhamnolipids isolated from pseudomonas aeruginosa Bs‐161r’, J. Microbiol. Biotechnol., 2010, 20, pp. 1061 –1068 (doi: 10.4014/jmb.1001.01018) [DOI] [PubMed] [Google Scholar]
- 34. Kathiresan K. Manivannan S. Nabeel M. et al.: ‘Studies on silver nanoparticles synthesized by a marine fungus, Penicillium fellutanum isolated from coastal mangrove sediment’, Colloids Surf. B, Biointerfaces, 2009, 71, (1), pp. 133 –137 (doi: 10.1016/j.colsurfb.2009.01.016) [DOI] [PubMed] [Google Scholar]
- 35. Deepak V. Kalishwaralal K. Pandian S.R.K. et al.: ‘An insight into the bacterial biogenesis of silver nanoparticles, industrial production and scale‐up’, in M. Rai N. Duran (Eds.): ‘Metal nanoparticles in microbiology’ (Springer, 2011) [Google Scholar]
- 36. Stewart V. Lu Y. Darwin A.J.: ‘Periplasmic nitrate reductase (Napabc eEnzyme) supports anaerobic respiration by Escherichia coli K‐12’, J. Bacteriol., 2002, 184, (5), pp. 1314 –1323 (doi: 10.1128/JB.184.5.1314-1323.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]


