Abstract
In this study, culture supernatnats of Bacillus subtilis T‐1 growing on brewery effluents and molasses was used for silver nanoparticles (Ag‐NPs) synthesis. The biosurfactant production of B. subtilis T‐1 was confirmed by the detection of genes in the genome and by the identification of the product in the supernatants. The genes for synthesis of surfactin (sfp, srfAA) and iturin (ituC) were noted by PCR reactions. Also, in examined culture supernatants the presence of C13, C14 and C15 surfactin homologues with the sodiated molecules [M + Na]+ at m /z 1030, 1044 and 1058 was confirmed using LC/MS/MS analysis. The formation of NPs in the culture supernatants was confirmed by UV–vis spectroscopy. The dynamic light scattering measurements and transmission electron microscopy images showed the nanometric sizes of the biosynthesised Ag‐NPs which ranged from several nm to several tens of nm depending on the used culture supernatant. Biological properties of Ag‐NPs were evaluated by binding of Ag‐NPs with DNA isolated from the Escherichia coli ATCC 25922 and B. subtilis ATCC 6633. Biogenic Ag‐NPs were actively bound to DNA in increased concentration which could be the one important mode of antibacterial action of the Ag‐NPs.
Inspec keywords: silver, nanoparticles, nanofabrication, materials preparation, microorganisms, antibacterial activity, industrial waste, agrochemicals, surfactants, breweries, genomics, genetics, chromatography, mass spectroscopic chemical analysis, ultraviolet spectroscopy, visible spectroscopy, spectrochemical analysis, light scattering, transmission electron microscopy, DNA, bonds (chemical), biochemistry, molecular biophysics, nanobiotechnology, biological techniques, particle size, enzymes
Other keywords: silver nanoparticle synthesis, Bacillus subtilis T‐1 growth, agro‐industrial waste, biosurfactant production, brewery effluent, molasses, Ag‐NP synthesis, B. subtilis T‐1, gene detection, genome, supernatant product identification, surfactin synthesis, sfp, srfAA, iturin synthesis, ituC, PCR reaction, C13 surfactin homologue, C14 surfactin homologue, C15 surfactin homologue, sodiated molecules, LC‐MS‐MS analysis, UV‐vis spectroscopy, dynamic light scattering measurement, transmission electron microscopy image, Ag‐NP nanometric size range, Ag‐NP biosynthesis, used culture supernatant dependence, biological properties, DNA isolation, Escherichia coli ATCC 25922, B. subtilis ATCC 6633, biogenic Ag‐NP‐DNA binding, Ag‐NP antibacterial action, Ag
1 Introduction
During the last decade, biosynthesis of metal nanoparticles (MeNPs) has emerged and is being developed as an alternative environmentally benign procedure. Biological methods of NP synthesis belong to new green generation processes, which are eco‐friendly being often designed as ‘green‐synthesis’ or ‘green chemistry’ procedures. As such they provide a credible alternative to chemical and physical methods. It has been reported that NPs can be synthesised by biological sources such as plant extracts, fungi, algae, cyanobacteria, bacteria, yeasts and actinomycetes [1, 2, 3, 4, 5]. Biosynthesis of NPs is carried out by microorganisms which grab target ions from solutions and then accumulate the reduced metal in its elemental form through enzymatic activities (as bio‐reducing agent) generated by microbial cell metabolic activities. The process is then categorised as intracellular or extracellular synthesis according to where NPs formation took place [6, 7].
Recognising the importance of developing environmentally friendly, biological methods of NPs synthesis, scientists have recently started looking into research related to synthesis of metallic NPs with the use of biosurfactants as capping agents [8, 9, 10, 11, 12, 13]. It was observed that biosurfactants produced by microorganisms can play a very important role in aggregation and stabilisation processes. Thus, the use of biosurfactant has emerged as a green alternative for enhancing both NP synthesis and stabilisation. The mechanism of surfactant adsorption depends on the type of surfactant (ionic, non‐ionic, polymeric, etc.) and the thickness of the adsorbed layer [14, 15, 16, 17]. Different commercial or bacteria produced biosurfactants have been examined as stabiliser and modifier in the synthesis of metallic NPs [18, 19, 20, 21, 22, 23]. So far, no comparative study has been published concerning the influence of biosurfactants’ nature and composition on the properties and ability to control the formation of MeNPs in biosynthesis.
Different strains of Bacillus are capable of producing a broad spectrum of biosurfactants, mainly lipopeptides from surfactin, iturin and fengicin family. Despite their great chemical diversity, lipopeptides belong to one family which is further subdivided to isoforms and homologues differing in the composition of the peptide part and the length of the fatty acid chain of the lipid tail, respectively. The present work aims at investigating silver nanoparticles (Ag‐NPs) synthesised in culture supernatants of Bacillus subtillis T‐1 growing on agro‐industrial wastes (brewery effluents and molasses) and producing lipopeptide biosurfactant. The production and properties of the produced Ag‐NPs have been verified by UV–vis and transmission electron microscopy (TEM) techniques. In addition, the biological properties of Ag‐NPs, e.g. interaction of synthetised Ag‐NPs with DNAs were evaluated.
2 Materials and methods
2.1 Bacterial strain identification and characterisation
The Bacillus strain (T‐1) used in this study was isolated from sludge of a 100‐year‐old oil refinery in Czechowice‐Dziedzice (Poland) and was identified based on the 16S rRNA gene sequence analysis using universal primers 27F and 1492R as described by Berry et al. [24]. The biosurfactant production of the isolate growing on the agro‐industrial wastes was well characterised by Płaza et al. [25, 26, 27].
Identification of the isolate was performed by GEN III MicroPlate™ test panel of the Biolog system. The GEN III MicroPlates™ enable testing of Gram‐negative and Gram‐positive bacteria in the same test panel. The test panel contains 71 carbon sources and 23 chemical sensitivity assays. GEN III dissects and analyses the ability of the cell to metabolise all major classes of compounds, in addition to determining other important physiological properties such as pH, salt and lactic acid tolerance, reducing power and chemical sensitivity. The commercially available OmniLog ID system, fully automated, was used according to the manufacturer's specifications. All the reagents applied were from Biolog, Inc. (Hayward, CA, USA). The bacterial suspension was prepared by removing bacterial colonies from the plate surface with a sterile cotton swab and agitating it in 5 ml of 0.85% saline. The bacterial suspension was adjusted in IF‐0a to achieve a 90–98% transmittance (T90) using a Biolog turbidimeter. The 150 µl of the suspension was dispensed into each well of a Biolog GEN III microplate. The plate was incubated at 30°C in an Omnilog Reader/Incubater (Biolog). Readings were recorded for 48 h, and data were analysed with Omnilog software (Biolog), which generated the plate information for tetrazolium colour formation. An average height threshold of 100 omnilog units (OUs) was chosen as a positive reaction.
2.2 Detection of genes encoding enzymes involved in biosurfactants synthesis
Genomic DNA was isolated from B. subtilis T‐1 by the standard protocol of Roche PCR Master Kit Ref. 11 636 103 001. The primers of genes used in the PCR amplifications are presented in Table 1. PCR amplifications were carried out in 25 μl reaction mixtures containing: 12.5 μl PCR Master Mix, 1 μl primer first, 1 μl primer second, 4 μl bacterial DNA (isolated by kit Roche). The PCR amplifications were performed using a Mastercycle® nexus GSX1 (Eppendorf) with the following cycle conditions: initial activation at 95°C for 4 min; 35 cycles of 94°C for 1 min, followed by annealing for 30 s at different temperatures (Table 1) depending on the primers used, extension step of 1 min for 70°C and final extension step of 5 min at 70°C. The experiment included control reaction mixture without added DNA. A total of 8 μl of each amplification reaction were analysed by electrophoresis using a 1% agarose gel followed by ethidium bromide staining and ultraviolet visualisation.
Table 1.
PCR primers for biosurfactant biosynthesis genes used in the work
| Biosurfactant | Gene | Sequence | PCR product size, bp | Annealing temperature, °C |
|---|---|---|---|---|
| surfactin | Sfp |
F‐5′ ATGAAGATTTACGGAATTTA 3′ R‐5′ TTATAAAAGCTCTTCGTACG 3′ |
675 | 48–52 |
| srfAA |
F‐5′ TCGGGACAGGAAGACATCAT 3′ R‐5′ CCACTCAAACGGATAATCCTGA 3′ |
201 | 58–60 | |
| fengycin | fenB |
F‐5′ CCTGGAGAAAGAATATACCGTACCY 3′ R‐5′ GCTGGTTCAGTT KGATCACAT 3′ |
670 | 58–65 |
| fenD |
F‐5′ GGCCCGTTCTCTAAATCCAT 3′ R‐5′ GTCATGCTGACGAGAGCAAA 3′ |
269 | 58 | |
| iturin | ituD |
F‐5′ TTGAAYGTCAGYGCSCCTTT 3′ R‐5′ TGCGMAAATAATGGSGTCGT 3′ |
482 | 57–58 |
| ituC |
F‐5′GGCTGCTGCAGATGCTTTAT 3′ R‐5′ TCGCAGATAATCGCAGTGAG 3′ |
423 | 58 |
2.3 Lipopeptides isolation and liquid chromatography–mass spectrometry (LC–MS) analysis
Cell‐free supernatant obtained by centrifugation (10.000 × g, for 10 min, at 4°C) of B. subtilis T‐1 culture (10 ml) was acidified with 6 N HCl to pH 2.0 and stored overnight at 4°C. The produced precipitate (collected by centrifugation: 10.000 × g, for 20 min, at 4°C) was suspended in 10 ml of distilled water and adjusted to pH 7.0 with 1 N NaOH. The sample was extracted with 10 ml of the mixture containing: ethyl acetate:methanol (4:1, vol/vol). Anhydrous sodium sulphate was added to the polled organic phase, and, after filtration, the solvent was evaporated. The experiments were carried out in duplicate. For surfactin measurement three independent samples were prepared in each experiment. Arithmetic averages and standard deviation were calculated.
Measurement of surfactin isomers was performed using an Agilent 1200 HPLC (Santa Clara CA, USA) system and a 3200 Q Trap mass spectrometer (AB Sciex, Framingham, MA, USA) with an ESI source. For reversed‐phase chromatographic analysis, the extract was dissolved in 2 ml of methanol and then in methanol: water solution (6:4, v/v). Five µl of sample were injected onto an Allure® PFP Propyl column (50 mm × 2.1 mm, 5 μm particle size; Restek, Bellefonte, PA, USA). The mobile phase consisted of water (A) and methanol (B), 2 mM ammonium formate and 0.2% formic acid was also used in all solvents as an additive. The solvent gradient was initiated at 60% B, after 1 min increased to 100% B over 1 min, and maintained at 100% B for four additional minutes before returning to the initial solvent composition over 2 min. The run time was 6 min. The column temperature was maintained at 37°C and the flow rate was 600 ml min−1. The instrumental settings were as follows: spray voltage 5500 V, curtain gas (CUR) 25, nebuliser gas (GS1) 50, turbo gas (GS2) 60 and ion source temperature of 600°C. Data analysis was performed with the Analyst™ v1.5.1 software (AB Sciex, Framingham, MA, USA). The monitored multiple reaction monitoring (MRM) pairs are given in Table S1.
The quantitative analysis was performed using surfactin standard (Sigma‐Aldrich, Germany). The linear regressions for three surfactin homologues determined in the standard were found to be >0.998 over the range of 0.05–7.5 µg/ml. Surfactin concentration was measured as the sum of three homologues.
2.4 Growth conditions of B. subtilis T‐1
The bacterial suspensions, obtained from a nutrient agar slant incubated for 24 h at 30°C, in the liquid Standard Methods medium of the following composition: peptone – 8 g/l, yeast extract – 2.5 g/l, glucose – 1 g/l was adjusted to OD600 nm 0.65 (ca. 107 –108 CFU/ml). Then, 2 ml of the bacterial suspensions were inoculated in 300 ml Erlenmeyer flasks containing 100 ml of the sterilised brewery effluents no. 4 and no. 6, which were collected from two breweries, and molasses (2%, v/v) as nutrients sources. The composition of the wastes is presented in [26, 27]. Luria Broth (LB) medium was used as control. The cultures were grown aerobically at 30°C for 96 h with constant shaking (110 rpm) (Innova 42 Incubator, New Brunswick Scientific Co., Inc., USA).
2.5 Preparation of Ag‐NPs
100 ml freshly grown bacterial culture were centrifuged at 10,000 rpm (Eppendorf) for 10 min. After centrifuged the samples were filtered through 0.2 µm strainer to sterile flasks. In the synthesis protocol, a silver nitrate solution (Sigma‐Aldrich) was added to 50 ml of the B. subtilis strain T‐1 culture supernatant to final concentration of 1 mM, and kept for stirring at 200 rpm at room temperature for 48 h. Controls (without the silver ions, only supernatant) were also evaluated during the experiment. The bioreduction of silver ions was monitored at regular intervals (2, 24 and 48 h) by sampling 1 ml of the reaction mixture. UV–vis spectra of these sample aliquots were recorded as a function of time of the reaction from 300 to 600 nm on a UV–vis spectrophotometer (Lange DR5000 with a resolution of 0.72 nm) for the formation of peaks in the visible region of electromagnetic spectrum to confirm the synthesis of Ag‐NPs.
2.6 Characterisation of Ag‐NPs
The formation of the synthesised Ag‐NPs was confirmed by UV–vis absorption spectroscopy with the Specord S 600 spectro‐photometer (Analytik Jena, Germany) observing surface plasmon resonance absorption peak of Ag‐NPs. Sizes of the obtained NPs were estimated by dynamic light scattering technique (DLS) on a Brookhaven BI‐200 goniometer with vertically polarised incident light of wavelength λ = 632.8 nm, supplied by a He–Ne laser operating at 35 mW, and a Brookhaven BI‐9000 AT digital autocorrelator. The autocorrelation functions were analysed using the constrained regularised algorithm CONTIN. The measurements were made at a 90° angle. The dispersity of NP sizes was given as , where is the average value of relaxation rates Γ and μ 2 is its second moment. These values were obtained from the cumulant analysis. Before DLS analysis, the NP solutions were filtered through membrane filters with nominal pore sizes of 0.8 µm (Minisart, Sartorius Stedim Biotech) to remove remains of bacteria and other impurities.
Zeta potential measurements were performed on a Zetasizer Nano ZS 90 (Malvern Instruments) in disposable folded capillary cells in triplicate.
The morphology and size distribution of the biosynthesised Ag‐NPs were determined by TEM using the JEM‐2010 (Jeol Ltd., Japan) operated at 200 kV. Distribution diagrams which are presented together with the TEM images were obtained via analysis of these images using the Gwyddion program (Sourceforge.net, http://www.gwyddion.net/ Free and Open Source software, covered by GNU General Public License) which serves as two‐dimensional (2D) data processing and analysis software. The dispersity of NP sizes was calculated by the same method as is referred in the case of DLS measurements.
2.7 Interaction of prepared Ag‐NPs with isolated DNA
Chromosomal DNAs from reference strains Escherichia coli ATCC 25922 and B. subtilis ATCC 6633 were isolated by the standard protocol of Roche High Pure PCR Template Preparation kit Ref. 11 796 828 001. In order to study the interaction between DNA and synthesised Ag‐NPs, 2.5 µl of chromosomal bacterial DNAs were mixed with 7 µl of Ag‐NPs synthesised from each supernatants, and incubated for 30 min in 37°C. For the optimisation interaction, different volumes of Ag‐NPs (2, 4, 6 and 10 µl) were mixed with 2.5 µl of DNA. The reaction mixtures were loaded onto 1% agarose gel. Electrophoresis was carried under 85 V for 90 min. DNA bands were observed and photographed under benchtop UV transilluminator and PhotoDoc‐it™ gel image analysis system.
3 Results and discussion
3.1 Identification of B. subtilis T‐1 and genes involved in biosurfactants synthesis
On the basis of the previous results [26] the 16S rRNA gene sequences showed that the isolate T‐1 was identified as Bacillus sp. The 16S rRNA gene sequencing could not clearly assign isolates T‐1 to any species of Bacillus as the analysed isolate showed >99% similarity to two distinct species B. subtilis and Bacillus licheniformis. The morphological and biochemical characteristics of the strain were presented by Płaza et al. [26]. Analysis of the 16S rDNA sequences is a powerful tool for the classification of microorganisms; however, the presence of highly conserved sequences in this gene sometimes disables the distinction among closely related Bacillus species and subspecies [28]. Another method which was used to clarify an ambiguous 16S rDNA identification of the isolate was the Biolog system. The metabolic profile of 94 biochemical tests as measured by the BIOLOG™ system, showed identification matches for B. subtilis. T‐1 strain was assigned to the species B. subtilis with SIM value of 0.749, based on its utilisation pattern of 23 substrates.
Bacillus species produce lipopeptides which were classified into three main families: surfactins, iturins and fengycins (or plipastatins). The potential ability of the isolate to synthesise surfactin, iturin and fengycin was noted. This ability was confirmed by polymerase chain reaction of selected genes: sfp, srfAA, fenB, fenD, ituD and ituC. When the primers were used, single DNA fragments of the expected size were amplified in B. subtilis T‐1 (Fig. 1). The strain contains the genes responsible for the synthesis of surfactin (sfp, srfAA) and iturin (ituC). Genes responsible for fengycin and iturin – ituD were not detected in the DNA of the isolate. Non‐ribosomal peptide synthesis is responsible for the biosynthesis of lipopeptides from Bacillus spp. and it involves multienzymatic proteins called non‐ribosomal peptide synthetases. Three genes, i.e. srfA, sfp and comS are required for the biosynthesis of the cyclic lipopeptide surfactin. In the genomic DNA of B. subtilis T‐1, two genes, i.e. sfp and srfAA were detected. Gene designated comS is located within the coding region of the domain of srfA and thus co‐expressed with the srfA operon. This gene is required for competence development in B. subtilis but not directly involved in the biosynthesis of surfactin [29].
Fig. 1.
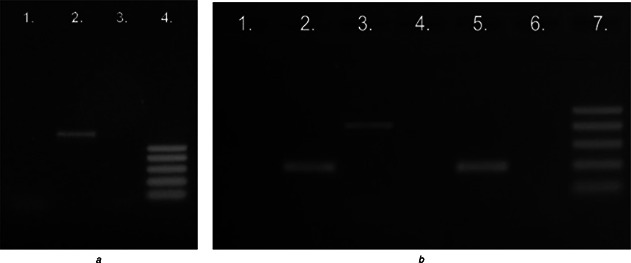
Agarose gel electrophoresis of PCR products for the genes of surfactin, iturin and fengycin
a PCR products of amplification reactions with the following primers fenB (line 1), sfp (line 2) and ituD (line 3). DNA ladder 100–500 bp (line 4)
b PCR products of amplification reactions with the following with primers fenD (line 1), srfAA (line 2), ituC (line 3) and DNA from the reference strain B. subtilis with primers fend (line 4), srfAA (line 5) and ituC (line 6). DNA ladder 100–500 bp (line 7)
3.2 Characterisation of biosurfactant produced by B. subtilis T‐1
Lipopeptides produced by the T‐1 strain were characterised using enhanced mass spectrum (EMS) analysis in the negative full scan mode (Figure S1A). Three main signals at m /z 1006, 1020 and 1034 corresponded to the deprotonated molecules [M −H]−; therefore the molecular weights of the molecules were 1007, 1021 and 1035, respectively. The obtained results are in agreement with LC–MS analysis of surfactin standard (Figure S1B). Three main signals were determined in positive full scan mode at m /z 1030, 1044 and 1058, corresponding to the sodiated molecules [M + Na]+ (data not shown). The sodiated molecules at m /z 1030, 1044 and 1058 were used as precursor ions for further EMS analyses and mass spectrum determination (Figure S2A). These data were in accordance with mass spectra of Sigma–Aldrich surfactin standard (data not shown) and mass spectra obtained by Pecci et al. [30]. The peak at m /z 481.3 observed in the spectrum could be assigned as an internal fragment ion [Val4–Asp5–Leu6–Leu7(OH) + H + Na]+. The peaks at m /z 707.5 and 594.3 could be assigned as internal peptide fragment ions [Leu2–Leu3–Val4–Asp5–Leu6–Leu7(OH) + H + Na]+ and [Leu3–Val4–Asp5–Leu6–Leu7(OH) + H + Na]+, respectively. The presence of these fragments in MS/MS spectra of surfactin homologues resulted from the cleavage of peptide bond after double hydrogen transfer [31].
The relative abundance of each homologue within the surfactin family in T‐1 extract fraction was inferred from [M + Na]+ extracted MRM pair by summing the peak areas of three homologues and then expressing the peak area of each homologue as a percentage of this sum. The relative content (Table S2) determined by this method showed a similar abundance of the surfactin homologues produced by T‐1 strain grown on the LB medium and brewery effluents in comparison to the composition of the commercial Sigma–Aldrich standard. Interestingly, surfactin extracted from the molasses medium presented the highest abundance of C15 surfactin, pointed as the most biologically active surfactin homologue [32]. The potential great impact of the medium components on the content and types of Bacillus lipopeptide homologues as well as isoforms have been also reported by others [30, 32, 33, 34].
The quantitative analysis of surfactin homologues was carried out using MRM transition. For each surfactin homologue a MRM pair was prepared (Figure S3). Thus, the quantitative assay was specific, accurate and sensitive. Then, surfactin concentration was measured as the sum of three homologues. Establishing the total concentration of the surfactin in 96‐h‐old cultures of B. subtilis T‐1 carried out in the LB medium, brewery effluents (no. 4 and no. 6) and molasse medium was rather similar and reached 1.3 ± 0.12, 1.2 ± 0.24, 1.3 ± 0.14 and 1.9 ± 0.39, respectively.
3.3 Preparation and characterisation of Ag‐NPs
The formation of Ag‐NPs from silver nitrate by the supernatants of B. subtilis T‐1 cultured on brewery effluents (no. 4 and no. 6), molasses and LB medium was investigated.
The appearance of yellowish‐brown colour in the reaction vessels suggested formation of Ag‐NPs [13]. Production of Ag‐NPs was monitored by UV–vis spectroscopy. The spectra were recorded as a function of reaction time of the culture supernatants with silver nitrate. The NPs exhibited an absorption peak around 450–500 nm after 24 h of the incubation (Fig. 2). The appearance of the peak related to formation of Ag‐NPs was well‐documented for various MeNPs with sizes ranging from 2 to 100 nm [7].
Fig. 2.
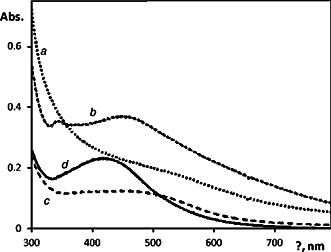
UV–vis spectra of the biosynthesised Ag‐NPs in aqueous dispersion prepared using B. subtilis cultivated on
a LB medium
b Molasses
c Sterilised brewery effluents no. 4
d Sterilised brewery effluents no. 6
Spectra were recorded after dilution of synthesised Ag‐NPs dispersions to concentration of silver 12 mg/l
The sizes of the obtained NPs were determined using DLS. The measurements were performed for solutions of silver nitrate incubated in the supernatants for 48 h. The values of hydrodynamic diameter D h 90° of Ag‐NPs from at least three measurements were averaged. When DLS distributions of dispersions were expressed as number weighted (Figures S4A, C, E, G), mainly NPs with the sizes of several nm were present. Only in supernatants of LB medium Ag‐NPs with sizes about 50 nm were observed. In the case of intensity‐weighed distribution (Figures S4B, D, F, H) the second populations of NPs with sizes ranging from 50 to 350 nm appeared. Taking into account both distributions it was clear that the amount of Ag‐NPs belonging to the second population was negligible. The average sizes are summarised in Table S3. The distribution of the sizes as indicated by values is rather broad.
Formation of Ag‐NPs from silver nitrate was reported in the supernatants of different bacteria Klebsiella pneumonia [35], E. coli [35, 36], Enterobacter cloacae [35], Bacillus licheniformis [37] cultured at the standard commercial medium. In all cases, spherical NPs with average size about 50 nm were obtained. We have detected Ag‐NPs of similar size in supernatants of the standard LB medium. In the case of brewery effluent supernatants, Ag‐NPs were much smaller (4–5 nm) than reported in [35, 36, 37]. TEM images of the studied Ag‐NPs supernatant solutions showed the presence of NPs of a broad range of sizes (Fig. 3).
Fig. 3.

TEM images of the biosynthesised Ag‐NPs in the case of B. subtilis cultivated on
a LB medium
b Molasses
c Sterilised brewery effluents no. 4
d Sterilised brewery effluents no. 6
Insets in TEM images represent the size distribution diagrams of the prepared NPs
Average diameters of the prepared Ag‐NPs obtained from the TEM observations with drained NPs were in the range of 1–100 nm, with average sizes of 13–19 nm (Table 2). The large NPs visible in TEM images seemed to be formed as aggregates of those with smaller sizes. The sizes of Ag‐NPs obtained by TEM were comparable for all supernatants, only in the case of brewery effluents no. 6 a broader distribution of sizes could be noticed. The differences between DLS and TEM could result from the calculation method. Distribution diagrams for TEM pictures were obtained from the pictures of a selected place of the samples when the results in DLS measurements were the sum of particles in the whole sample.
Table 2.
Average diameters and dispersities of the Ag‐NPs synthesised by B. subtilis T‐1 grown on brewery effluents and molasses obtained from TEM images
| Sample Ag‐NPs | Average diameter, nm | Variability, nm | Polydispersity |
|---|---|---|---|
| LB medium | 19.0 | 9.8 | 0.267 |
| brewery effluent no. 4 | 13.1 | 5.4 | 0.169 |
| brewery effluent no. 6 | 13.5 | 9.1 | 0.449 |
| molasses | 13.7 | 6.5 | 0.226 |
The values of zeta potential (Table S3) were substantially higher for Ag‐NPs prepared using supernatants obtained from B. subtilis T‐1 cultured on brewery effluents (around −30–40 mV). Simultaneously, the UV–vis spectra of these dispersions showed sharp absorption maxima of the surface plasmon confirming smaller tendency of these Ag‐NPs to form aggregates in dispersion. All measurements indicate formation of smaller particles in the supernatants of brewery effluents.
3.4 Interaction of synthestised Ag‐NPs with isolated DNAs
Interaction of Ag‐NPs with DNA isolated from the reference strains E. coli and B. subtilis was evaluated using agarose gel electrophoresis. When optimising the DNA with AG‐NPs binding pattern, faint bands were observed in the lane in which 10 µl of Ag‐NPs solution inoculated (Figs. 4 A and B). A visual observation of the agarose gel electrophoresis images confirmed the binding of Ag‐NPs with the isolated DNA fragments of both bacteria. It was found that the intensity of the bands decreased and the mobility of the bands changed with the increase of the Ag‐NPs concentration. A slower mobility of the DNA bands with the Ag‐NPs was observed compared with the pure DNA. Similar results were observed in the reactions of both extracted DNAs and Ag‐NPs produced in the culture supernatants from brewery effluents and molasses. Biogenic silver NPs were actively bound to DNA in an increased concentration.
Fig. 4.

Agarose gel‐electrophoresis image presenting the different concentration (2, 4, 6 and 10 µl) of Ag‐NPs synthesised by B. subtilis T‐1 with DNA isolated from the reference strains
a E. coli
b B. subilis
Bands of DNA with Ag‐NPs synthesised in the culture supernatant from the brewery effluent no. 4 (lines 1–4), from the brewery effluent no. 6 (lines 6–9) and from the molasses (lines 11–14). 1A – control samples – E. coli DNA without the Ag‐NPs (lines 5, 10, 15); 1B – control samples – B. subtilis DNA without the Ag‐NPs (lines 5, 10, 15)
A similar study on the interaction of gold NPs synthetised by photosynthetic algal species, Coelastrella sp. (eukaryotic) and Phormidium sp. (prokaryotic) with DNA extracted from herring sperm was described by MubarakAli et al. [4]. They suggest that the DNA is wrapped (bent) around the NPs, and via binding to cationic MeNPs DNA is protected by enzymatic and physical degradation. Mishra et al. [38] described fungus‐mediated synthesis of gold NPs and their conjugation with genomic DNA isolated from E. coli and Staphylococcus aureus. The authors observed slight aggregation of gold NPs conjugated with isolated DNA. This fact was also supported from the UV–vis spectrum which showed slight red shift with an increase in the peak intensity. According to the authors, genomic DNA formed a complete single/multiple layer on the gold NPs surface. It could stabilise the particles against aggregation owing to negatively charged phosphate backbone. However, a detailed mechanism still remains unclear and needs extensive investigation.
4 Conclusions
The culture supernatants of B. subtilis T‐1 growing on brewery effluents and molasses was used for Ag‐NPs synthesis. The lipopeptide biosurfactant was produced by B. subtilis T‐1. The interaction of DNA with biological synthesised Ag‐NPs was detected by agarose gel. However, further studies on the investigation of the role of biosurfactant(s) in Ag‐NPs biosynthesis and DNA‐binding affinity with biologically synthesised metallic NPs are currently pursued.
5 Acknowledgments
This paper was prepared in connection with the work done under the project no. 2013/09/B/NZ9/01759 (decision no. 2013/09/B/NZ9/01759) sponsored by the National Science Center (Poland), the projects of the bilateral cooperation between Poland and Czech Republic (project numbers 9005/2014‐2015 and 7AMB14 PL025), and project no. LO1305 of the Ministry of Education, Youth and Sports of the Czech Republic.
6 References
- 1. Thakkar K.N. Mhatre S.S. Parikh R.Y.: ‘Biological synthesis of metallic nanoparticles’, Nanomed.: Nanotechnol. Biol. Med., 2010, 6, (2), pp. 257 –262 (doi: 10.1016/j.nano.2009.07.002) [DOI] [PubMed] [Google Scholar]
- 2. Narayanan K.B. Sakthive N.: ‘Biological synthesis of metal nanoparticles by microbes’, Add. Colloid Interface Sci., 2010, 156, (1), pp. 1 –13 (doi: 10.1016/j.cis.2010.02.001) [DOI] [PubMed] [Google Scholar]
- 3. Quester K. Avalos‐Borja M. Castro‐Longoria E.: ‘Biosynthesis and microscopic study of metallic nanoparticles’, Micron, 2013, 54–55, (1), pp. 1 –27 (doi: 10.1016/j.micron.2013.07.003) [DOI] [PubMed] [Google Scholar]
- 4. MubarakiAli D. Arunkumar J. Nag K.H. et al.: ‘Gold nanoparticles from Pro and eukaryotic photosynthetic microorganisms – Comparative studies on synthesis and its application on biolabelling’, Colloids Surf. B: Biointerfaces, 2013, 103, (1), pp. 166 –173 (doi: 10.1016/j.colsurfb.2012.10.014) [DOI] [PubMed] [Google Scholar]
- 5. Mittal A.K. Chisti Y. Banerjee U.C.: ‘Synthesis of metallic nanoparticles using plant extracts’, Biotechnol. Adv., 2013, 31, (3), pp. 346 –356 (doi: 10.1016/j.biotechadv.2013.01.003) [DOI] [PubMed] [Google Scholar]
- 6. Mandal D. Bolander M.E. Mukhopadhyay D. et al.: ‘The use of microorganisms for the formation of metal nanoparticles and their application’, Appl. Microbiol. Biotechnol., 2006, 69, (4), pp. 485 –492 (doi: 10.1007/s00253-005-0179-3) [DOI] [PubMed] [Google Scholar]
- 7. Zhang X. Yan S. Tyagi R.D. et al.: ‘Synthesis of nanoparticles by microorganisms and their application in enhancing microbiological reaction rates’, Chemosphere, 2011, 82, (3), pp. 489 –494 (doi: 10.1016/j.chemosphere.2010.10.023) [DOI] [PubMed] [Google Scholar]
- 8. Xie Y. Ye R. Liu H.: ‘Synthesis of silver nanoparticles in reverse micelles stabilized by natural biosurfactant’, Colloids Surf. A: Physicochem. Eng. Aspects, 2006, 2, (1), pp. 175 –178 (doi: 10.1016/j.colsurfa.2005.12.056) [DOI] [Google Scholar]
- 9. Reddy A.S. Chen C.Y. Chen C.C. et al.: ‘Synthesis of gold nanoparticles via an environmentally benign route using a biosurfactant’, J. Nanosci. Nanotechnol., 2009, 9, (14), pp. 6693 –6699 (doi: 10.1166/jnn.2009.1347) [DOI] [PubMed] [Google Scholar]
- 10. Reddy A.S. Chen C.Y. Baker S.C. et al.: ‘Synthesis of silver nanoparticles using surfactin: a biosurfactant as stabilizing agent’, Mater. Lett., 2009, 63, (10), pp. 1227 –1230 (doi: 10.1016/j.matlet.2009.02.028) [DOI] [Google Scholar]
- 11. Kiran G.S. Sabu A. Selvin J.: ‘Synthesis of silver nanoparticles by glicolipid biosurfactant produced from marine Brevibacterium casei MSA 19’, J. Biotechnol., 2010, 148, (2), pp. 221 –225 (doi: 10.1016/j.jbiotec.2010.06.012) [DOI] [PubMed] [Google Scholar]
- 12. Singh B.R. Dwivedi S. Al‐Khedhairy A.A. et al.: ‘Synthesis of stable cadmium sulfide nanoparticles using surfactin produced by Bacillus amyloliquifaciens strain KSU‐109’, Colloids Surf. B: Biointerfaces, 2011, 85, (1), pp. 207 –213 (doi: 10.1016/j.colsurfb.2011.02.030) [DOI] [PubMed] [Google Scholar]
- 13. Rangarajan V. Majumder S. Sen R.: ‘Biosurfactant‐mediated nanoparticles synthesis. A green and sustainable approach’, in Mulligan C.N. Sharma S.K. Mudhoo A. (Eds.): ‘Biosurfactants. research trends and applications’ (CRC Press Taylor & Francis Group, 2014), pp. 217 –229 [Google Scholar]
- 14. Kasture M. Singh S. Patel P. et al.: ‘Multiutility sophorolipids as nanoparticle capping agents: synthesis of stable and water dispersible Co nanoparticles’, Langmuir, 2007, 23, (15), pp. 11409 –11412 (doi: 10.1021/la702931j) [DOI] [PubMed] [Google Scholar]
- 15. Kasture M.B. Patel P. Prabhune A.A. et al.: ‘Synthesis of silver nanoparticles by sophorolipids: effects of temperature and sophorolipid structure on size of particles’, J. Chem. Sci., 2008, 120, (4), pp. 515 –520 (doi: 10.1007/s12039-008-0080-6) [DOI] [Google Scholar]
- 16. Worakitsiri P. Pornsunthorntawee O. Thanpitcha T. et al.: ‘Synthesis of polyaniline nanofibers and nanotubes via rhamnolipid biosurfactant templating’, Synth. Methods, 2011, 161, (3), pp. 298 –306 (doi: 10.1016/j.synthmet.2010.11.039) [DOI] [Google Scholar]
- 17. Saikia J.P. Bharali P. Konwar B.K.: ‘Possible protection of silver nanoparticles against salt by using rhamnolipid’, Colloids Surf. B: Biointerfaces, 2013, 104, (4), pp. 330 –332 (doi: 10.1016/j.colsurfb.2012.10.069) [DOI] [PubMed] [Google Scholar]
- 18. Palanisamy P.: ‘Biosurfactant mediated synthesis of NiO nanorods’, Mater. Lett., 2008, 62, (8), pp. 743 –746 (doi: 10.1016/j.matlet.2007.06.053) [DOI] [Google Scholar]
- 19. Palanisamy P. Raichur A.M.: ‘Synthesis of spherical NiO nanoparticles through a novel biosurfactant mediated emulsion technique’, Mater. Sci. Eng., 2009, 29, (1), pp. 199 –204 (doi: 10.1016/j.msec.2008.06.008) [DOI] [Google Scholar]
- 20. Ganesh K.C. Mamidyala S.K. Das B. et al.: ‘Synthesis of biosurfactant‐based silver nanoparticles with purified rhamnolipids isolated from Pseudomonas aeruginosa BS‐161R’, J. Microbiol. Biotechnol., 2010, 20, (11), pp. 1061 –1068 [DOI] [PubMed] [Google Scholar]
- 21. Narayanan J. Ramji R. Sahu H. et al.: ‘Synthesis, stabilization and characterization of rhamnolipid‐capped ZnS nanoparticles in aqueous medium’, IET Nanotechnol., 2010, 4, (1), pp. 29 –34 (doi: 10.1049/iet-nbt.2009.0010) [DOI] [PubMed] [Google Scholar]
- 22. Kumar G.G. Mamidyala S.K.: ‘Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa ‘, Colloids Surf. B: Biointerfaces, 2011, 84, (3), pp. 462 –466 (doi: 10.1016/j.colsurfb.2011.01.042) [DOI] [PubMed] [Google Scholar]
- 23. Baccile N. Noiville R. Stievano L. et al.: ‘Sophorolipids‐functionalized iron oxide nanoparticles’, Phys. Chem., 2013, 15, (14), pp. 1606 –1620 [DOI] [PubMed] [Google Scholar]
- 24. Berry C.J. Story S. Altman D.J. et al.: ‘Biological treatment of petroleum and radiological contaminated soil’, in Clayton C. Lindner A. (Eds.): ‘Innovative approaches for the remediation of subsurface‐contaminated hazardous waste sites: bridging flask and field scales’ (Oxford University Press, 2006), pp. 87 –104 [Google Scholar]
- 25. Płaza G. Zjawiony I. Banat I.: ‘Use of different methods for detection of thermophilic biosurfactant‐producing bacteria from hydrocarbon‐contaminated and bioremediated soil’, J. Petrol. Sci. Eng., 2006, 50, (1), pp. 71 –77 (doi: 10.1016/j.petrol.2005.10.005) [DOI] [Google Scholar]
- 26. Płaza G. Gawior K. Jangid K. et al.: ‘Characterization of surface active properties of Bacillus strains growing in brewery effluent’, in Pawłowski L. Dudzińska M.R. Pawłowski A. (Eds.): ‘Environmental engineering III’ (Taylor and Francis Group, London, 2010), pp. 221 –226 [Google Scholar]
- 27. Płaza G.A. Pacwa‐Płociniczak M. Piotrowska‐Seget Z. et al.: ‘Agroindustrial wastes as unconventional substrates for growing of Bacillus strains and production of biosurfactants’, Environ. Prot. Eng., 2011, 37, (1), pp. 65 –71 [Google Scholar]
- 28. Wu X.Y. Walker M.J. Hornitzky M. et al.: ‘Development of a group‐specific PCR combined with ARDRA for the identification of Bacillus species of environmental significance’, J. Microbiol. Methods, 2006, 64, (1), pp. 107 –119 (doi: 10.1016/j.mimet.2005.04.021) [DOI] [PubMed] [Google Scholar]
- 29. Jacques P.: ‘Surfactin and other lipopeptides from Bacillus spp’, in Soberon‐Chavez G. (Ed.): ‘Biosurfactants. From genes to application’ (Springer‐Verlag, Berlin, Heidelberg, 2011), pp. 57 –91 [Google Scholar]
- 30. Pecci Y. Rivardo F. Martinotti M.G. et al.: ‘LC/ESI‐MS/MS characterisation of lipopeptide biosurfactants produced by the Bacillus licheniformis V9T14 strain’, J. Mass Spectrom., 2010, 45, (6), pp. 772 –778 (doi: 10.1002/jms.1767) [DOI] [PubMed] [Google Scholar]
- 31. Pathak K.V. Keharia H.: ‘Identification of surfactins and iturins produced by potent fungal antagonist, Bacillus subtilis K1 isolated from aerial roots of banyan (Ficus benghalensis) tree using mass spectrometry’, Biotechnology, 2014, 4, (2), pp. 283 –295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bacon C.W. Hinton D.M. Mitchell T.R. et al.: ‘Characterization of endophytic strains of Bacillus mojavensis and their production of surfactin isomers’, Biol. Control, 2012, 62, (1), pp. 1 –9 (doi: 10.1016/j.biocontrol.2012.03.006) [DOI] [Google Scholar]
- 33. Snook M.E. Mitchell T. Hinton D.M. et al.: ‘Isolation and characterization of leu7‐surfactin from the endophytic bacterium Bacillus mojavensis RRC 101, a biocontrol agent for Fusarium verticillioides ‘, J. Agric. Food Chem., 2009, 27, (14), pp. 4287 –4292 (doi: 10.1021/jf900164h) [DOI] [PubMed] [Google Scholar]
- 34. Li Y.M. Haddad N.I.A. Yang S.Z. et al.: ‘Variants of lipopeptides produced by Bacillus licheniformis HSN221 in different medium components evaluated by a rapid method ESI‐MS’, Int. J. Pept. Res. Ther., 2008, 14, (2), pp. 229 –235 (doi: 10.1007/s10989-008-9137-0) [DOI] [Google Scholar]
- 35. Shahverdi A.R. Minaeian S. Shahverdi H.R. et al.: ‘Rapid synthesis of silver nanoparticles using culture supernatants of Enterobacteria: a novel biological approach’, Process Biochem., 2007, 42, (8), pp. 919 –923 (doi: 10.1016/j.procbio.2007.02.005) [DOI] [Google Scholar]
- 36. Gurunathan S. Kalishwaralal K. Vaidyanathan R. et al.: ‘Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli ‘, Colloid Surf. B, 2009, 74, (2), pp. 328 –335 (doi: 10.1016/j.colsurfb.2009.07.048) [DOI] [PubMed] [Google Scholar]
- 37. Kalishwaralal K. Deepak V. Ramkumarpandian S. et al.: ‘Extracellular biosynthesis of silver nanoparticles by culture supernatant of Bacillus licheniformis ‘, Mater. Lett., 2008, 62, (16), pp. 4411 –4413 (doi: 10.1016/j.matlet.2008.06.051) [DOI] [Google Scholar]
- 38. Mishra A. Tripathy S.K. Yun S.I.: ‘Fungus mediated synthesis of gold nanoparticles and their conjugation with genomic DNA isolated from Escherichia coli and Staphylococcus aureus ‘, Process Biochem., 2012, 47, (6), pp. 701 –711 (doi: 10.1016/j.procbio.2012.01.017) [DOI] [Google Scholar]


