Abstract
Azo dyes are widely used in industries and their release in the environment contributes to the pollution of effluents. The authors aim to develop a new eco‐friendly water treatment method for the degradation of azo dyes based on in situ magnetic separation and immobilisation of bacterial cells. The immobilisation was achieved using superparamagnetic Fe3 O4 nanoparticles and offers the possibility of reusing bacteria by magnetic separation for several degradation cycles. The iron– oxide nanoparticles were synthesised by reverse co‐precipitation. The Gram‐positive bacteria Bacillus subtilis were immobilised using iron–oxide nanoparticles by adsorption and then separated with an external magnetic field. Transmission electron microscopy observation showed that the particles' diameter was ∼20 nm with a narrow size distribution. Moreover, the iron–oxide nanoparticles were adsorbed onto the surface in order to coat the cells. B. subtilis has proved its ability to decolorise and degrade several azo dyes at different values of pH, with the highest decolorisation rate for Congo red. Furthermore, immobilised cells have a degradation activity similar to that of free cells. The system provided a degradation rate up to 80% and could be reused for seven batch cycles.
Inspec keywords: biotechnology, microorganisms, pH, adsorption, iron compounds, superparamagnetism, transmission electron microscopy, ultraviolet spectra, chemical technology, wastewater treatment, effluents, dyes, magnetic separation, iron, magnetic particles, decontamination
Other keywords: degradation rate, immobilisation, magnetic nanoparticles, azo dyes, eco‐friendly water treatment method, degradation cycles, Gram‐positive bacteria, iron–oxide nanoparticles, external magnetic field, immobilised cells, degradation activity, magnetic separation, effluents, bacterial cells, Fe3 O4
1 Introduction
Azo dyes represent ∼70% of the weight of all the dyes used in the world, making them the largest group of synthetic dyes produced, as well as the ones that are most released in the environment [1, 2, 3]. They are widely used in the textile, leather, paper, food, cosmetics and pharmaceutical industries because of their stability, ease and cost‐effectiveness of their synthesis as well as the variety of colours available compared with natural dyes [4]. Synthetic azo dye stuffs released by the industrial sector pose a threat to environmental safety due to their toxicity [5, 6]. The textile industry is one of the most polluting industries in terms of generated liquid effluents, containing, among other things, azo dyes and their metabolites [7]. When released into the aquatic ecosystem, they lead to a reduction in the penetration of sunlight and thus to the destabilisation of the photosynthesis process. The presence of azo dyes in effluents has also an adverse impact on total organic carbon (TOC), biological oxygen demand (BOD) and chemical oxygen demand (COD) [8, 9, 10]. In addition, most of these dyes and their metabolites are carcinogenic, mutagenic and toxic to humans [5, 11, 12].
Physico‐chemical methods such as electrolysis, ozonation, reverse osmosis and coagulation–flocculation are used to degrade dyes, but these methods are complex, expensive and generate wastes, which cause secondary pollution problems [13, 14].
Other methods are therefore needed such as ones involving enzymes for the biotransformation of azo dyes and their metabolites [11]. Two types of bacterial enzymes are involved in these processes: oxidising enzymes that cleave asymmetrically the azo dye molecule, and reducing enzymes that are responsible for a symmetric cleavage of the azo band using azoreductase. In the case of reducing enzymes, the modification leads to the transfer of four electrons giving rise to colourless solution of aromatic amines [13]. After fragmentation of aromatic amines, bacteria use the fragments as carbon, nitrogen and oxygen sources for their nutrition and growth [14].
However, the use of such biological methods requires the addition of large amounts of enzyme‐producing bacteria within the environment to be treated, which are ultimately discharged in the effluents. To avoid the bacterial contamination of effluents and be able to use the enzyme‐producing bacteria for several degradation cycles, we have investigated the immobilising and recovering potential of bacteria fixed onto magnetic iron–oxide nanoparticles. This method is called ‘magnetic recovery’, and it allows the separation of immobilised bacteria by the effect of an external magnetic field produced by a permanent magnet [15, 16].
Iron–oxide nanoparticles with a strong magnetic moment have an attractive and novel potential since they are detectable, remotely manipulated and responsive by a magnetic field. In addition, iron–oxide nanoparticles exhibit chemical inertness as well as an oxidation resistance, which explain their non‐toxicity with regard to the bacteria [17, 18]. In addition to this, the nanoparticles possess hydroxyl functions on their surface, allowing their adsorption on the surface of bacteria [19, 20].
In this work, the choice was made on Bacillus subtilis, a Gram‐positive bacterium that is able to secrete enzymes like azoreductase at a temperature of 37°C [21, 22]. We have synthesised magnetic iron–oxide nanoparticles by a reverse co‐precipitation method and characterised their structural properties using X‐ray diffraction (XRD) and Fourier‐transform infrared spectroscopy (FTIR). The morphology and the distribution of magnetite Fe3 O4 nanoparticles were visualised by transmission electron microscopy (TEM) and scanning electronic microscopy (SEM). After the synthesis of the nanoparticles, bacteria were immobilised by adsorption. Finally, we realised the decolorisation of several dyes using B. subtilis at different values of pH, and we studied the degradation of Congo red (CR) by immobilised bacteria for several decolorisation cycles (see Fig. 1).
Fig. 1.
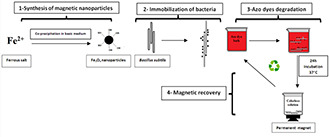
Azo dyes degradation process using B. subtilis immobilised by iron–oxide nanoparticles
2 Materials and methods
2.1 Chemicals
The following chemicals were used without further treatment and/or purification: iron chloride (FeCl2 ·6H2 O) (Sigma‐Aldrich); ammonium hydroxide (NH4 OH; 25 %w) (Sigma‐Aldrich); Direct orange 34 (DO 34, Mondial Chemistry); CR (Sigma‐Aldrich); Orange II sodium salt (Orange II) (Sigma‐Aldrich) and Procion® red MX‐5B (Red MX‐5B) (Sigma‐Aldrich).
2.2 Experimental
2.2.1 Synthesis of iron–oxide nanoparticles by reverse co‐precipitation
In this technique, we used a single precursor (Fe2+ ions). The solution of the ferrous ions was directly added to the alkaline solution, unlike the direct co‐precipitation method in which the base is added to the solutions containing a mixture of both iron ions (Fe2+, Fe3+) under an inert atmosphere [23, 24]. To this end, we chose to repeat the protocol described in [25] with some modifications: a volume of 50 mL of the deionised water was added to 50 mL of ammonium hydroxide. The pH of the solution is ∼13. The iron ion precursor (Fe2+) at a concentration of 0.05 M was prepared separately by dissolving 0.89 g of FeCl2 ·4H2 O in 90 mL of deionised water. The solution was then magnetically stirred for 15 min and sonicated for 10 min to ensure a complete dissolution of the ferrous salt. The solution of the Fe2+ ions was poured at once to the base solution and left under magnetic stirring for 1 h. Later, the black solution was washed several times with deionised water in order to remove the excess of ammonia, and then decanted using a permanent magnet and finally dried in an oven.
2.2.2 Bacterial strain and cultivation
B. subtilis ATCC6633 was grown in Lysogeny Broth (LB) culture medium containing 10 g of tryptone, 5 g of yeast extract and 10 g of NaCl solubilised in 950 mL of deionised water. The bacteria were allowed to grow in this medium overnight on a rotary shaker at 150 rpm in an incubator at 37°C. The growth rate of the cells was monitored by determining the optical density at 600 nm (OD600) using a UV/VIS spectrophotometer.
2.2.3 Immobilisation of bacteria and in‐situ magnetic separation
Bacterial cells from an overnight preculture were centrifuged for 10 min at 7000g. Bacteria were suspended in saline and different amounts of Fe3 O4 nanoparticles (4 g/L; 10 g/L and 40 g/L) were added to the suspensions. The mixtures were stirred at 150 rpm in an incubator at 37°C for 30 min and then washed with physiological water and separated with a permanent magnet to remove unbound bacteria and the remainder of the medium culture.
2.2.4 Plating and enumeration of immobilised bacteria
The number of viable bacteria (colony forming unit, CFU) was determined by plating serial dilutions of the bacterial suspensions onto LB agar plates and expressed as the number of bacteria/mL (CFU/mL). The number of bacteria in the inoculum was taken as a control sample.
2.2.5 Azo dye decolorisation assays
The immobilised bacteria were introduced into LB supplemented with different solutions of azo dyes (DO 34, CR, Orange II and Red MX‐5B) of a concentration of 100 ppm at pH 5, 7 and 9, then incubated at 37°C for 24 h under agitation at 150 rpm and under aerobic conditions. The bacteria were harvested by centrifugation at 7000g for 10 min. The supernatants were filtered with 0.45 μm sterile syringe filters to assure the separation of non‐immobilised bacteria, which could interfere with the decolorisation rate. The filtrate was then used to determine the azo dye reduction by measuring the residual absorption at the appropriate wavelength of each azo dye according to (1):
| (1) |
2.2.6 Extracellular enzymatic measurement
Determination of the enzymatic activity was done as described in [26] using B. subtilis ATCC6633 grown at 37°C overnight. The samples (10 mL) were centrifuged at 7000g for 10 min. The supernatant was filtered through a 0.45 μm sterile syringe (Millipore) filter and the resulting filtrate, i.e. the extracellular fraction, was mixed with 10 mL of azo dye solution. The mixture was then incubated for 24 h at 37°C, and the azo dye decolorisation rate was determined as previously described.
2.2.7 Extraction and analysis of CR dye biotransformation metabolites
The extraction of CR dye (100 ppm) metabolites and their separation was done after a treatment using Bacillus subtilis ATCC6633 immobilised by Fe3 O4 nanoparticles. For this, the treated dye samples were collected and suspended solids were filtered through 0.45 µm pore size cellulose acetate filters and removed by centrifugation (10,000g for 15 min, 4 ± 0.2°C). The resulted clear supernatant was extracted with a half volume of dichloromethane; the organic phase was collected and injected into GC‐MS.
2.2.8 Analytical methods
The crystalline phases of iron–oxide nanoparticles were identified using a Panalytical X'pert diffractometer with Cu(Kα) radiation (λ = 1.5406 Å). The characteristic vibration bands of magnetic nanoparticles were recorded using FTIR spectroscopy on a Thermo Nicolet 5700 in an attenuated total reflectance configuration. The morphology and microstructure of the magnetic nanoparticles as well as the immobilised bacteria were analysed by using a Hitachi H‐7650 transmission electron microscope. The surface areas of the bacteria immobilised by iron–oxide nanoparticles were observed with a Zeiss SUPRA® VP 5 field‐effect scanning electron microscope (FE‐SEM). UV‐vis spectroscopy was used to determine the decolorisation rate of azo dyes by means of a UV‐vis spectrophotometer Thermo‐Scientific Evolution 300. The separation and analysis of CR dye metabolites was performed by using a Shimadzu GC‐MS – QP 2010SE.
3 Results and discussion
3.1 Characteristics of iron–oxide nanoparticles
The XRD patterns confirmed the formation of cubic magnetite (Fe3 O4) (Fig. 2). We noted the presence of all peaks corresponding to this phase. The magnetite peaks were very broad which is due to the small crystalline size of nanoparticles as confirmed by TEM results. The morphology of Fe3 O4 nanoparticles was characterised using TEM. As shown in Fig. 3, the iron–oxide nanoparticles displayed a large size distribution and more regular morphology with a range of size between 15 and 20 nm. The size of the nanoparticles calculated from the XRD peaks (Fig. 2) using the Scherrer formula is of a mean value of 16 nm, which is in agreement with the TEM images. FTIR analysis was performed to characterise Fe3 O4 magnetite nanoparticles. As depicted in Fig. 4, the hydroxyl bands appearing between 3400–3300 and 1620–1640 cm−1 were related to the –OH vibration and free H2 O, respectively.
Fig. 2.
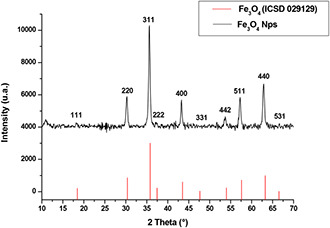
XRD patterns of magnetic nanoparticles obtained by reverse co‐precipitation
Fig. 3.

TEM images of iron–oxide nanoparticles synthesised by reverse co‐precipitation
Fig. 4.
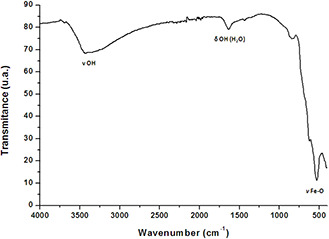
Infrared spectrum of magnetic nanoparticles obtained by reverse co‐precipitation
The ν Fe–O bands that characterise magnetite appeared between 700 and 450 cm−1 and centred at 622 cm−1. FTIR analysis indicated the presence of iron–oxide bands and hydroxyl groups on the surface of nanoparticles.
3.2 Immobilisation of B. subtilis ATCC6633 using iron–oxide nanoparticles
The immobilisation of the bacteria by the iron–oxide nanoparticles was achieved. TEM and SEM observations confirmed that B. subtilis cells were covered by iron–oxide nanoparticles (Figs. 5 and 6). TEM and SEM observations showed that the size of nanoparticles deposited on the surface of the bacteria was between 15 and 20 nm in diameter, while the size of the B. subtilis cell was 1000 nm in width and 1740 nm in length. The small size of the Fe3 O4 nanoparticles was beneficial for an efficient deposition on the large surface of B. subtilis. Indeed, the density of the nanoparticles on the surface of the bacteria increased with their concentration. At the highest concentration tested (40 g/L), we obtained a dense and uniform coverage of up to 50% of the outside surface area of bacteria as evidenced in Fig. 6.
Fig. 5.
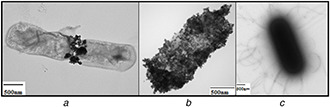
TEM Images of iron–oxide nanoparticles coating B. subtilis
(a) 4 g/L, (b) 40 g/L, (c) Free cell
Fig. 6.

SEM images of immobilised bacteria incubated with different concentrations of magnetic iron–oxide nanoparticles
(a) Free cells, (b) 4 g/L, (c) 10 g/L, (d) 40 g/L
The large specific surface area and the high surface energy of the Fe3 O4 nanoparticles (i.e. the nano‐size effect) facilitated a strong adsorption on the surface of microbial cells [27, 28, 29]. To assess the effect of the concentration of nanoparticles on the immobilisation efficiency, we quantified the number of viable bacterial cells present in the immobilisation medium (grafted cells) and in the supernatant (free cells) (Fig. 7).
Fig. 7.
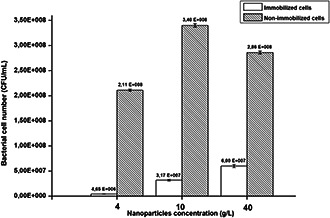
Enumeration results of immobilised bacteria by iron–oxide nanoparticles
Data points indicate the mean of three independent replicates; ±standard errors of mean (SEM) is indicated by error bars
The enumeration results showed that when increasing the concentration of nanoparticles in the solution, the number of immobilised cells increased. Moreover, the total enumeration of cells including non‐immobilised and immobilised cells corresponds nearly to that of the control sample (inoculums: 4.17 × 108 CFU/mL) and proved the non‐toxicity of nanoparticles towards B. subtilis. These results were in agreement with TEM and SEM observations showing that the immobilisation efficiency increases with the concentration of nanoparticles involved in the reaction medium.
3.3 Azo dye degradation
3.3.1 Bacterial degradation of azo dyes using free B. subtilis ATCC6633
To investigate the azo dye decolorisation activity of B. subtilis ATCC6633 at different values of pH (5; 7 and 9), we proceeded to a decolorisation test on four azo dyes: DO 34; CR; Orange II and Red MX‐5B with a concentration of 100 mg/L (Fig. 8 a). After 24 h of incubation under aerobic conditions, the bacterial decolorisation ability was visually noticed by the clear degradation of coloration intensity from most samples (Fig. 8 b). CR and DO 34 solutions showed highest decolorisation rates as only a partial decolorisation of the Orange II solution was observed and a very low decolorisation for the Red MX‐5B. Therefore, further investigations were carried out to determine azo dye‐degrading activity by measuring the decrease in absorbance of each dye and by calculating the decolorisation rate using (1) (Fig. 8 c). B. subtilis achieved a decolorisation rate of up to 60% in all pH ranges for DO 34; CR and Orange II. However, Red MX‐5B showed a decolorisation rate lower than 25%. These results confirmed the visual observations. In the literature, the majority of the azo dye‐decolorising species reported are able to degrade dyes at pH values near neutrality. Klebsiella pneumoniae RS‐13 was found to completely degrade Methyl red at pH of 6.0–8.0; pH between 6.0 and 7.5 was also optimal for the decolorisation of Reactive red 195 by B. cereus strain M [30]. In our case, a high decolorisation rate was obtained at different pH values.
Fig. 8.
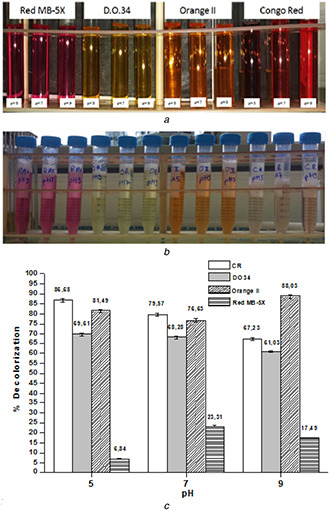
Decolorisation test of the azo dyes using free B. subtilis ATCC6633
(a) T 0, (b) After 24 h of incubation at 37°C, (c) Results of decolorisation rate measured by UV‐Vis spectroscopy
Data points indicate the mean of three independent replicates; ±SEM is indicated by error bars
The disappearance of the colour may be due to the adsorption of the dye onto the surface of the bacteria following the mechanism of biosorption [31] or due to an enzymatic degradation following the secretion of azoreductase, since this enzyme can be present in the intra‐ and extra‐cellular bacterial compartments [26]. Indeed, it has been reported that azoreductases are involved in enhancing survival and the acceleration of azo dye decolorisation [32, 33]. This confirmed that azoreductase 1 could function to decolorise azo dyes. Therefore, we searched the B. Subtilis ATCC 6633 genome sequence in the GenBank by a BLAST search using the entire protein sequence deduced from a main azoreductase 1 gene of B. subtilis ATCC6633 as the query (see Fig. 9). The search identified a single gene encoding a 208 amino acid residue, which displayed moderate primary structure identity (96%) and was similar in size to B. subtilis azoreductase. The high percentage of homology between the bacterial sequence and the enzymatic sequence can be responsible for the secretion of azoreductase 1 in the decolorisation medium thus leading to the decolorisation of azo dyes.
Fig. 9.

Amino acid sequences of B. subtilis ATCC6633 azoreductase 1
In parallel, we assessed the impact of azo dyes on the growth of B. subtilis by measuring the optical density following the cells growth in contact with azo dyes. No inhibition of cell growth was observed during 24 h of incubation under aerobic conditions, whatever the dye tested. We noticed that the optical density changed slightly according to the pH for each dye. After 24 h of incubation, the maximum optical density was detected in CR and Orange II solutions (Fig. 10). These results are in agreement with previous decolorisation tests indicating that the decolorisation rate was related to the growth rate of B. subtilis in azo dyes solutions.
Fig. 10.
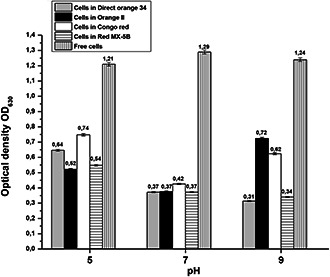
Optical density of decolorisation solutions at 630 nm after 24 h of incubation
Data points indicate the mean of three independent replicates; ±SEM is indicated by error bars
3.3.2 CR biodegradation mechanism
B. subtilis ATCC6633 exhibited the best decolorisation rates with CR and Orange II. As a result, we selected CR for further investigation regarding its enzymatic degradation and biosorption mechanism. Biosorption using different microorganisms is known as an efficient way to remove CR from industrial liquid effluents. In [31], the authors investigated the biosorption performance of nonviable Penicillium YW 01 biomass for removal of CR in solutions. In our case, the biosorption of CR by B. subtilis ATCC6633 was evidenced after 24 h of incubation (azo dye + cells) at 37°C and 10 min of centrifugation of the reaction mixture. This operation gave rise to an intense coloration of the deposited pellets and a degradation of the supernatant coloration (Fig. 11 a and Fig. 6b). These results suggest that CR is transported across the cell membrane, thereby, enhancing the decolorisation performance.
Fig. 11.
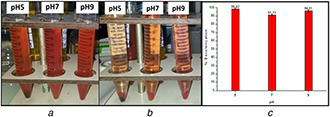
Biosorption test and Extracellular decolorisation test on CR
(a) Before, (b) After decolorisation and centrifugation, (c) Decolorisation of CR using extracellular supernatant of B. subtilis
Data points indicate the mean of three independent replicates; ±SEM is indicated by error bars
The decolorisation assay performed with the supernatant of a B. subtilis culture showed decolorisation rates of up to 90% under aerobic conditions (Fig. 11 c). B. subtilis showed high degradation efficiency under aerobic conditions after 24 h without any pretreatment, contrary to some results described in the literature [34, 35]. Moreover, in [36], it is reported that CR biodegradation alone under aerobic conditions is time consuming and a pretreatment is proposed by sonication to accelerate the process.
3.3.3 Extraction and analysis of dye CR biotransformation metabolites
GC‐MS analysis data were used to determine the probable metabolites produced and also to propose a decolorisation pathway of CR dye by B. subtilis. Result of the mass spectrum analysis revealed the presence of one intermediate CR dye degradation metabolites, viz. biphenyl‐1,4′‐diamine with the retention time of 15.78 min as shown in Fig. 12. The proposed biodegradation pathway of CR dye by B. subtilis is depicted in Fig. 13. The possible mineralisation of CR dye starts with the initial conversion of the dye to form biphenyl‐1,4′‐diamine and unidentified intermediate catalysed by azoreductase following a cleavage of electrophilic azo bonds which leads to their decolorisation [37].
Fig. 12.
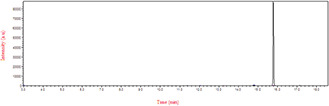
GC/MS Chromatogram of CR metabolite after decolorisation by B. subtilis
Fig. 13.
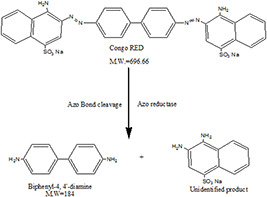
Proposed biodecolorisation pathway of CR dye by B. subtilis
3.3.4 Cyclic CR degradation using B. subtilis ATCC6633 immobilised by iron–oxide nanoparticles
We aim to ensure the recovery and the reuse of bacteria in a continuous and cyclic decolorisation process. At the end of each decolorisation batch of 24 h, the coated cells were collected by application of a magnetic field and then reused for another decolorisation cycle. The immobilised bacteria were successfully reused up to seven decolorisation cycles and we achieved a decolorisation rate exceeding 80% at different pH values. We noticed a high and stable decolorisation rate of ∼95% at pH = 7, which is the usual growth pH of B. subtilis (Fig. 14). Both the coated cells and free cells had a similar decolorisation activity, suggesting that the coated cells did not experience a mass transfer problem. The presence of LB media may have helped the cells to maintain their viability by providing necessary cofactors such as reducing equivalents (NADH).
Fig. 14.
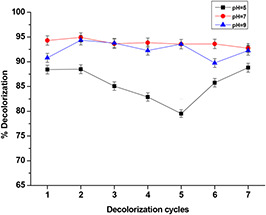
Reusability test of immobilised B. subtilis in decolorisation process of CR
Magnetically separated/immobilised cells could thus be repeatedly used for CR decolorisation and retained high catalytic activity throughout several cycles.
4 Conclusion
In the current work, we have synthesised iron–oxide nanoparticles by reverse co‐precipitation, which were used to develop a novel cell immobilisation and separation system based on the deposition of nanoparticles on the surface of bacteria. We showed that the immobilisation rate increased with the concentration of nanoparticles. This system was targeted for the decolorisation and degradation of azo dyes. B. subtilis cells coated with nanoparticles showed a strong ability to decolorise CR at different pH values following different mechanisms. The magnetic recovery offers the possibility to reuse the bacteria for several cycles of decolorisation at a high rate. These results confirmed that the immobilisation of bacteria by magnetic nanoparticles for the treatment of effluents is a simple, low cost and efficient method. This system could be used in many biological processes to limit the dependence on conventional processes and offers new opportunities for the development of bacterial bioreactors applicable in continuous flux for the treatment of industrial pollutant effluents such as biological treatment of leather, degradation of textile dyes and toxic aromatic molecules present in the effluents of textile industries.
5 Acknowledgments
The authors gratefully acknowledge Campus France for the financial support through PHC Toubkal project No. 32506VM.
6 References
- 1. Pandey A. Singh P. Iyengar L.: ‘Bacterial decolorization and degradation of azo dyes’, Int. Biodeterior. Biodegrad., 2007, 59, (2), pp. 73 –84 [Google Scholar]
- 2. Hao O.J. Kim H. Chiang P.‐C.: ‘Decolorization of wastewater’, Crit. Rev. Environ. Sci. Technol., 2000, 30, (4), pp. 449 –505 [Google Scholar]
- 3. Gupta V.: ‘Application of low‐cost adsorbents for dye removal–a review’, J. Environ. Manag., 2009, 90, (8), pp. 2313 –2342 [DOI] [PubMed] [Google Scholar]
- 4. Saratale R.G. Saratale G.D. Chang et al.: ‘Bacterial decolorization and degradation of azo dyes: a review’, J. Taiwan Inst. Chem. Eng., 2011, 42, (1), pp. 138 –157 [Google Scholar]
- 5. Vandevivere P.C. Bianchi R. Verstraete W.: ‘Treatment and reuse of wastewater from the textile wet‐processing industry: review of emerging technologies’, J. Chem. Technol. Biotechnol., 1998, 72, (4), pp. 289 –302 [Google Scholar]
- 6. Sudha M. Saranya A. Selvakumar G. et al.: ‘Microbial degradation of azo dyes: a review’, Int. J. Cur. Microbiol. Appl. Sci., 2014, 3, (2), pp. 670 –690 [Google Scholar]
- 7. Chang J.‐S. Chen B.‐Y. Lin Y.S.: ‘Stimulation of bacterial decolorization of an azo dye by extracellular metabolites from Escherichia coli strain NO3 ’, Bioresour. Technol., 2004, 91, (3), pp. 243 –248 [DOI] [PubMed] [Google Scholar]
- 8. Saratale R.G. Saratale G.D. Kalyani D.C. et al.: ‘Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium‐GR’, Bioresour. Technol., 2009, 100, (9), pp. 2493 –2500 [DOI] [PubMed] [Google Scholar]
- 9. Anjaneyulu Y. Chary N.S. Raj D.S.S.: ‘Decolourization of industrial effluents–available methods and emerging technologies–a review’, Rev. Environ. Sci. Bio/Technol., 2005, 4, (4), pp. 245 –273 [Google Scholar]
- 10. Yao Z. Wang L. Qi J.: ‘Biosorption of methylene blue from aqueous solution using a bioenergy forest waste: Xanthoceras sorbifolia seed coat’, CLEAN – Soil, Air, Water, 2009, 37, (8), pp. 642 –648 [Google Scholar]
- 11. Lade H. Kadam A. Paul D. et al.: ‘Biodegradation and detoxification of textile azo dyes by bacterial consortium under sequential microaerophilic/aerobic processes’, EXCLI J., 2015, 14, pp. 158 –174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rafii F. Hall J. Cerniglia C.: ‘Mutagenicity of azo dyes used in foods, drugs and cosmetics before and after reduction by Clostridium species from the human intestinal tract’, Food Chem. Toxicol., 1997, 35, (9), pp. 897 –901 [DOI] [PubMed] [Google Scholar]
- 13. Chacko J.T. Subramaniam K.: ‘Enzymatic degradation of azo dyes‐a review’, Int. J. Environ. Sci., 2011, 1, (6), p. 1250 [Google Scholar]
- 14. Shanmugam B.K. Mahadevan S.: ‘Metabolism and biotransformation of azo dye by bacterial consortium studied in a bioreaction calorimeter’, Bioresour. Technol., 2015, 196, pp. 500 –508 [DOI] [PubMed] [Google Scholar]
- 15. Le T.H. Kim S.J. Bang S.H. et al.: ‘Phenol degradation activity and reusability of Corynebacterium glutamicum coated with NH(2)‐functionalized silica‐encapsulated Fe3 O4 nanoparticles’, Bioresour. Technol., 2012, 104, pp. 795 –798 [DOI] [PubMed] [Google Scholar]
- 16. Gao X.‐L. Shao M.F. Xu Y.S. et al.: ‘Non‐selective separation of bacterial cells with magnetic nanoparticles facilitated by varying surface charge’, Front. Microbiol., 2016, 7, pp. 1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lewinski N. Colvin V. Drezek R.: ‘Cytotoxicity of nanoparticles’, Small, 2008, 4, (1), pp. 26 –49 [DOI] [PubMed] [Google Scholar]
- 18. Hoskins C. Cuschieri A. Wang L.: ‘The cytotoxicity of polycationic iron oxide nanoparticles: common endpoint assays and alternative approaches for improved understanding of cellular response mechanism’, J. Nanobiotechnol., 2012, 10, (1), p. 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheng‐Nan S. Chao W. Zan‐Zan et al.: ‘Magnetic iron oxide nanoparticles: synthesis and surface coating techniques for biomedical applications’, Chin. Phys. B., 2014, 23, (3), p. 037503 [Google Scholar]
- 20. Hola K. Markova Z. Zoppellaro G. et al.: ‘Tailored functionalization of iron oxide nanoparticles for MRI, drug delivery, magnetic separation and immobilization of biosubstances’, Biotechnol. Adv., 2015, 33, (6), pp. 1162 –1176 [DOI] [PubMed] [Google Scholar]
- 21. Sugiura W. Yoda T. Matsuba T. et al.: ‘Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus stearothermophilus ’, Biosci. Biotechnol. Biochem., 2006, 70, (7), pp. 1655 –1665 [DOI] [PubMed] [Google Scholar]
- 22. Mikhaylova M. Kim D.K. Bobrysheva N. et al.: ‘Superparamagnetism of magnetite nanoparticles: dependence on surface modification’, Langmuir, 2004, 20, (6), pp. 2472 –2477 [DOI] [PubMed] [Google Scholar]
- 23. Aono H. Nagamachi T. Naohara T. et al.: ‘Synthesis conditions of nano‐sized magnetite powder using reverse coprecipitation method for thermal coagulation therapy’, J. Ceram. Soc. Jpn., 2016, 124, (1), pp. 23 –28 [Google Scholar]
- 24. Fedlheim D.L. Foss C.A.: ‘Metal nanoparticles: synthesis, characterization, and applications’ (CRC Press, Marcel Dekker Inc. Publisher, 2001) [Google Scholar]
- 25. Mahmed N. Heczko O. Lancok A. et al.: ‘The magnetic and oxidation behavior of bare and silica‐coated iron oxide nanoparticles synthesized by reverse co‐precipitation of ferrous ion (Fe2+) in ambient atmosphere’, J. Magn. Magn. Mater., 2014, 353, pp. 15 –22 [Google Scholar]
- 26. Leelakriangsak M. Borisut S.: ‘Characterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1’, Songklanakarin J. Sci. Technol., 2012, 34, (5), pp. 509 –516 [Google Scholar]
- 27. Lu A.H. Salabas E.L. Schüth F.: ‘Magnetic nanoparticles: synthesis, protection, functionalization, and application’, Angew. Chem., Int. Ed., 2007, 46, (8), pp. 1222 –1244 [DOI] [PubMed] [Google Scholar]
- 28. He Y.P. Miao Y.M. Li C.R. et al.: ‘Size and structure effect on optical transitions of iron oxide nanocrystals’, Phys. Rev. B, 2005, 71, (12), p. 125411 [Google Scholar]
- 29. Li Y.‐G. Gao H.S. Li W.L. et al.: ‘In situ magnetic separation and immobilization of dibenzothiophene‐desulfurizing bacteria’, Bioresour. Technol., 2009, 100, (21), pp. 5092 –5096 [DOI] [PubMed] [Google Scholar]
- 30. Cui D. Li G. Zhao M. et al.: ‘Decolourization of azo dyes by a newly isolated Klebsiella sp. strain Y3, and effects of various factors on biodegradation’, Biotechnol. Biotechnol. Equip., 2014, 28, (3), pp. 478 –486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Y. Wang G. Wang B. et al.: ‘Biosorption of Acid Black 172 and Congo Red from aqueous solution by nonviable Penicillium YW 01: kinetic study, equilibrium isotherm and artificial neural network modeling’, Bioresour. Technol., 2011, 102, (2), pp. 828 –834 [DOI] [PubMed] [Google Scholar]
- 32. Liu G. Zhou J. Wang J. et al.: ‘Acceleration of azo dye decolorization by using quinone reductase activity of azoreductase and quinone redox mediator’, Bioresour. Technol., 2009, 100, (11), pp. 2791 –2795 [DOI] [PubMed] [Google Scholar]
- 33. Liu G. Zhou J. Jin R. et al.: ‘Enhancing survival of Escherichia coli by expression of azoreductase AZR possessing quinone reductase activity’, Appl. Microbiol. Biotechnol., 2008, 80, (3), p. 409 [DOI] [PubMed] [Google Scholar]
- 34. Telke A.A. Joshi S.M. Jadhav S.U. et al.: ‘Decolorization and detoxification of Congo red and textile industry effluent by an isolated bacterium Pseudomonas sp. SU‐EBT’, Biodegradation, 2010, 21, (2), pp. 283 –296 [DOI] [PubMed] [Google Scholar]
- 35. Li R. Ning X.A. Sun J. et al.: ‘Decolorization and biodegradation of the Congo red by Acinetobacter baumannii YNWH 226 and its polymer production's flocculation and dewatering potential’, Bioresour. Technol., 2015, 194, pp. 233 –239 [DOI] [PubMed] [Google Scholar]
- 36. Gopinath K.P. Murugesan S. Abraham J. et al.: ‘ Bacillus sp. mutant for improved biodegradation of Congo red: random mutagenesis approach’, Bioresour. Technol., 2009, 100, (24), pp. 6295 –6300 [DOI] [PubMed] [Google Scholar]
- 37. Chung K.‐T. Stevens S.E. Cerniglia C.E.: ‘The reduction of azo dyes by the intestinal microflora’, Crit. Rev. Microbiol., 1992, 18, (3), pp. 175 –190 [DOI] [PubMed] [Google Scholar]


