Abstract
Spirochetes are emerging pathogens for which culture and identification are partly unresolved. In fact, 16S rRNA-based sequencing is by far the most widely used PCR methodology that is able to detect such uncultivable pathogens. However, this assay actually has some limitations linked to potential problems of contamination, which hampers diagnosis. To circumvent this, we have devised a simple PCR strategy involving targeting of the gene encoding the RNA polymerase beta subunit (rpoB), a highly conserved enzyme. The complete sequence of the Leptospira biflexa (serovar patoc) rpoB gene was determined and compared with the published sequences for Borrelia burgdorferi and Treponema pallidum. From the resulting analysis, degenerate nucleotide primers were designed and tested for their ability to amplify a portion of the rpoB gene from various spirochetes. Using two different pairs of these primers, we succeeded in obtaining specific rpoB-amplified fragments for all members of the genera Leptospira, Treponema, and Borrelia tested and no other bacteria. Our findings may have significant implications for the development of a new tool for the identification of spirochetes, especially if clinical samples are contaminated or when the infecting strain is uncultivable.
In 1999, and contrary to the predictions of the middle of this century, infectious diseases remain the primary cause of death worldwide (17). In fact, over recent decades, many demographic, social, environmental, and economical factors have contributed to the emergence of infectious diseases. These emerging pathogens correspond to infectious agents that are responsible for diseases which have increased over the last 20 years or whose incidence will most probably be enhanced in the near future. Consequently, and in the light of the dramatic rise in the numbers of such pathological states, the diagnosis of infectious pathogens was one of the main priorities announced by the Centers for Disease Control and Prevention and the World Health Organization (3).
The need for rapid and specific characterization of such infectious agents has stimulated the development of new molecular biological tests. Of these, PCR has emerged as a major technique for the detection and identification of specific microbial genes (6, 16). Identification of the pathogen responsible may not always be possible, because cultures can take from days to weeks to identify by conventional morphological methods. It is of note that, like other fastidious bacteria, spirochetes can be difficult or even impossible to grow (19). Observation of these non-Gram stain-reactive bacteria requires dark-field microscopy, and their identification has long relied on serotyping (19). Diagnosis of human spirochaetal infections caused by Leptospira sp., Borrelia sp., and Treponema sp., has been problematic (2, 4, 7, 13), and, as yet, no reliable methods have been widely adopted. Results obtained from comparison of the sequences of the 16S rRNA-encoding gene (24), which is a widely used method for identifying pathogens (5, 25), are unsatisfactory. Indeed, the sensitivity of this approach has been questioned (9, 22). In this study, we have devised a simple PCR strategy to specifically detect Leptospira, Borrelia, or Treponema. This was achieved by using consensus primers targeting a fragment of the gene encoding the β-subunit of the RNA polymerase (rpoB). Comparison of sequences derived from this gene has previously been used for taxonomic analyses of a variety of species of Archaea and Proteobacteria (11, 14, 18).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Spirochete strains (Borrelia burgdorferi, Borrelia recurrentis, Treponema pallidum, Leptospira biflexa serovar patoc, Leptospira interrogans serovars icterohaemmorragiae and australis) were obtained from the American Type Culture Collection (ATCC). Borrelia and Leptospira strains were grown at 30°C on BSK-H (Sigma Chemical Corp., St. Louis, Mo.) or EMJH (DIFCO, Detroit, Mich.) medium, respectively. Treponema pallidum was propagated by intratesticular injections in rabbits. Leptospira interrogans (serovar pomona), Leptospira borgpetersenii (serovars sejroë and tarassovi), Leptospira kirschneri (serovars cynopteri and grippotyphosa), Leptospira noguchii (serovars panama and louisiana), Leptospira santarosai (serovar bataviae), Borrelia garinii, Borrelia afzelii, Borrelia valaisiana, and Borrelia hermsii were provided by bioMérieux, Marcy l'Etoile, France. Other bacterial strains tested as negative controls (Campylobacter jejuni, Chlamydia pneumoniae, Chlamydia trachomatis, Citrobacter kasei, Corynebacterium jeikeium, Coxiella burnetii, Escherichia coli (three isolates), Francisela tularensis, Enterobacter aerogenes, Enterobacter cloacae, Haemophilus influenzae, human granulocytic Ehrlichia, Klebsiella pneumoniae, Legionella pneumophila, Listeria ivanovii, Listeria monocytogenes, Mycobacterium tuberculosis, Neisseria meningitidis, Pasteurella spp., Pseudomonas aeruginosa (two isolates), Rickettsia prowazekii, Rickettsia rhipicephali, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Streptococcus B agalactiae, Streptococcus salivarius, and Yersinia enterocolitica were either purchased from ATCC or were clinical isolates obtained from patients hospitalized in Marseille.
DNA amplification and Leptospira biflexa rpoB gene sequencing.
Genomic DNA was extracted by standard procedures (20). PCR amplifications were performed with 3.2 pmol of each degenerate SEB primer (Eurogentec, Seraing, Belgium) (Fig. 1), 10 mM deoxynucleoside triphosphates (dNTPs), and 1 U of Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.) in 50 μl of 1× PCR buffer. Following a first denaturation step (95°C for 1.5 min), a three-step cycle of 95°C for 20 s, 50°C for 30 s, and 72°C for 1 min was repeated 35 times. The final stage of the PCR program was a single 3-min extension at 72°C (Peltier thermal cycler model PTC 200; MJ Research, Watertown, Mass.). The amplicons obtained were then resolved by 1% agarose gel electrophoresis and visualized by staining with ethidium bromide. For sequencing, samples were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and then incorporated into the dRhodamine Terminator Cycle Sequencing Ready Reaction buffer (DNA sequencing kit; Perkin-Elmer). Reaction products were resolved and translated into sequence data by using an Applied Biosystems model ABI 310 automatic DNA sequencer (Perkin-Elmer). The sequence of the 5′ extremity of the rpoB gene was obtained by using the Universal Genome Walker kit (Clontech, Palo Alto, Calif.) according to the manufacturer's instructions. The complete rpoB sequence was collected by aligning and then combining gene fragments obtained in individual sequencing reactions.
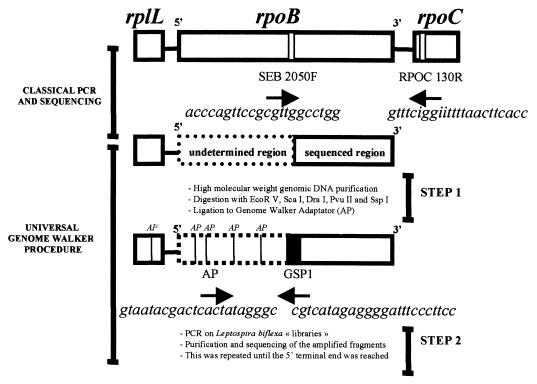
FIG. 1.
Strategy of Leptospira biflexa gene sequencing. Primers SEB 2050F and RPOC 130R were determined after alignment of the rpoB and rpoC genes of Salmonella enterica serovar Typhimurium, Escherichia coli, and Bacillus subtilis and definition of a consensus sequence. AP, Genome Walker Adaptator primer provided in the kit; GSP, gene-specific primer that corresponds to the 5′ end of the known sequence.
PCR protocols.
Bacterial DNA was purified with a QIAamp tissue kit (Qiagen). Oligonucleotide primers were synthesized in our laboratory (392 DNA/RNA Synthesizer; Perkin-Elmer). Amplification of rpoB fragments was carried out by using the LTB primers listed in Table 1 (3.2 pmol each) added to a 50-μl PCR mix containing 10 mM dNTP, 1 U of Taq polymerase (Gibco BRL), and 2 μl of DNA extract. The thermal cycle comprised a first denaturation step (94°C for 2 min) and then a three-step cycle of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min was repeated 35 times. The final stage of the PCR program was a single 3-min extension step at 72°C. In addition, for each bacterial DNA extract, a 16S rRNA PCR was performed in order to ensure the quality of the DNA extraction. This PCR was performed with the same PCR program as that used for rpoB gene amplification, but with oligonucleotide primers FD1 and RP2 (26). All amplicons obtained from spirochetal DNA were purified and sequenced as described above.
TABLE 1.
Oligonucleotide primers tested for PCR amplification of the rpoB gene
| LTB primer |
Nucleotide sequence (5′→3′) |
|---|---|
| 740F | TGGITIGAATTIGAIATIGA |
| 1130F | AATTGICTIGAAIIAGA |
| 1730F | CTTGGICCIGGIGGACTTTC |
| 1730R | GAAAGTCCICCIGGICCAAG |
| 1920F | GAAGGICCAAAIATIGG |
| 1920R | CCIATITTTGGTCCTTC |
| 2900F | ATGCIATITTIATTTC |
| 2900R | AGAAATIAAIATIGCATCCTC |
| 3700R | GCGTGIATTTTITCATCIAC |
| 3850R | GCTTCIAGIGCCCTIAC |
Data analysis.
The obtained sequences, which were performed at least in duplicate, were analyzed, corrected, and assembled with the Auto-Assembler program of the ABI PRISM 310 Genetic Analyzer package (Perkin-Elmer). Comparisons between sequences from various spirochete strains were performed by using the PC Gene program (Intelligenetics). Percentages of similarity between the rpoB gene of Leptospira biflexa and those of Treponema pallidum (gbAE001205.1) and Borrelia burgdorferi (gbAE001144.1) were determined by using the FASTA program. Alignment of these three sequences was carried out with CLUSTAL algorithm (10), and potential consensus primers (LTB primers listed in the Table 1) were chosen to hybridize to highly conserved regions within this alignment.
Nucleotide sequence accession number.
The complete sequence of the rpoB gene has been submitted to the GenBank database under accession no. AF150880.
RESULTS AND DISCUSSION
Sequencing of the rpoB gene for Leptospira biflexa.
Oligonucleotide primers targeting highly conserved regions of the rpoB gene (15), deduced by reference to the alignment of the rpoB and rpoC sequences of Staphylococcus aureus, Escherichia coli, and Bacillus subtilis, permitted amplification of the 3′ terminal end of the rpoB gene of Leptospira biflexa (Fig. 1). The sequence of the 5′ end of the gene was obtained with the Universal Genome Walker kit. To our knowledge, this method, which allows determination of unknown genomic DNA sequences adjacent to known ones without molecular cloning (21), has not previously been used for elucidation of bacterial genome sequence. Briefly, genomic DNA is digested with five different restriction enzymes, and following purification of the DNA fragments, each is ligated to a specific Genome Walker Adaptator primer. PCR is then performed with the Adaptator primer supplied by the manufacturer and a gene-specific primer, the sequence of which is complementary to a region as close as possible to the known 5′ extremity of the gene. Amplification and subsequent sequence determination then permit recovery of the intervening sequence containing at least part of the missing 5′ gene fragment. This procedure was found to be a convenient means of amplifying the 5′-terminal end of the Leptospira biflexa rpoB gene.
Analysis.
A full-length Leptospira biflexa rpoB gene of 3,687 bp encoding a protein of 1,229 amino acids was obtained. Comparative analysis of this sequence with those of Borrelia burgdorferi (1) and Treponema pallidum (23) was performed and indicated sequence similarity values of approximately 60%. The alignment obtained meant that the most conserved regions could be determined. Consequently, it was possible to design oligonucleotide sequences that could be used as specific primers for spirochetes belonging to these three distinct genera. Thus, 10 LTB primers in which inosine was introduced in place of ambiguous bases (12) were chosen (Table 1).
PCR assays.
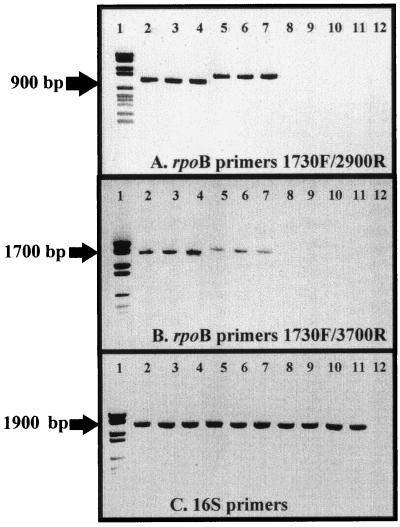
To assess whether degenerated primers permitted amplification of rpoB gene fragments, all possible combinations were tested in PCR experiments carried out at various annealing temperatures. Our results, presented in part in Fig. 2, illustrate the fact that amplification products of approximately 900 and 1,700 bp were obtained for all spirochete strains analyzed. Amplification was observed with primers LTB 1730F and either LTB 2900R or LTB 3700R, with an annealing temperature of 52°C. In contrast, all other pairs of LTB primers tested failed to yield amplification products. In a second set of experiments, amplification of the rpoB gene fragments was also observed with both pairs of primers by using four other strains of Borrelia and eight other strains of Leptospira, respectively (listed in Materials and Methods [data not shown]). The successful amplification reaction was specific to spirochetes. Among the 26 other bacterial strains tested, no rpoB amplicon was observed. In contrast, and as expected, positive responses were obtained from all strains with the 16S rRNA primers, demonstrating the quality of the DNA samples tested (Fig. 2C). All amplified fragments derived from spirochetes were then sequenced and compared with those in either the laboratory or the public domain databases, confirming their nature. Interestingly, rpoB amplicons were larger for Leptospira species than for members of the genera Borrelia and Treponema. This difference corresponds to a high number of nucleotide insertions inside the Leptospira rpoB sequence compared with those of other genera.
FIG. 2.
Specific PCR amplification of the spirochetal rpoB gene. The PCR assay was performed by using Taq polymerase with an annealing temperature of 52°C. Lanes: 1, molecular mass markers (DNA molecular weight marker VI; Boehringer); 2, Borrelia burgdorferi; 3, Borrelia recurrentis; 4, Treponema pallidum; 5, Leptospira biflexa serovar patoc; 6, Leptospira interrogans, serovar australis; 7, Leptospira interrogans, serovar icterohaemmorragiae; 8, Escherichia coli; 9, Staphylococcus aureus; 10, Streptococcus salivarius; 11, Pseudomonas aeruginosa; 12, negative control without DNA. (A) rpoB gene primers LTB 1730F and LTB 2900R. (B) rpoB gene primers LTB 1730F and LTB 3700R. (C) 16S rRNA gene primers FD1 and RP2.
Possible applications.
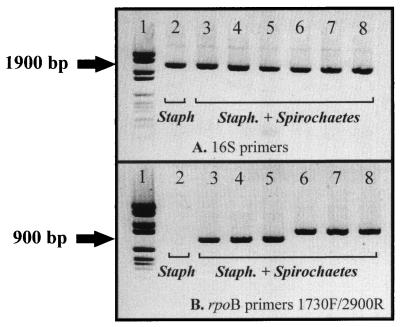
The procedure described in this paper offers a rapid, convenient, and specific tool to identify Leptospira, Treponema, and Borrelia, which are all potential agents for human spirochetal infections (19). These results are of importance in light of the increased incidence of meningoencephalitis caused by spirochetes (8). In this respect, the proposed PCR methodology is a powerful molecular biological technique which should allow, for example, the examination of cerebrospinal fluid samples under routine conditions, without a requirement for axenic cultivation of the pathogens involved. The proven specificity of the rpoB gene primers is also of importance. These oligonucleotides will enable spirochetes to be detected even when a sample is contaminated by other bacteria. Thus, when suspensions of spirochetes and Staphylococcus aureus are mixed together before DNA extraction, the observed 16S rRNA PCR bands most probably correspond to the concomitant amplification of both pathogens. This point clearly illustrates the limitation of the 16S rRNA-based PCR protocols. Indeed, under such experimental conditions, identification was impossible, since the mixed amplicons obtained could not be sequenced under routine conditions (personal observation). In contrast, by using the rpoB-based PCR assay, the presence of spirochetes can be specifically demonstrated. Indeed, as illustrated in Fig. 3, the presence of contaminating Staphylococcus aureus DNA was shown not to be an impediment to the detection of spirochete DNA. This is an important point when considering the fact that Staphylococcus aureus is the first contaminant agent in cerebrospinal fluids.
FIG. 3.
Detection of spirochetes from a mixed bacterial suspension using specific primers for the rpoB gene. The PCR assay was performed under the same conditions as described in the legend to Fig. 2, but with spirochetal DNA previously mixed with a Staphylococcus extract. Lanes: 1, molecular mass markers (DNA molecular weight marker VI; Boehringer); 2, S. aureus DNA alone; 3, Borrelia burgdorferi; 4, Borrelia recurrentis; 5, Treponema pallidum; 6, Leptospira biflexa serovar patoc; 7, Leptospira interrogans serovar australis; 8, Leptospira interrogans, serovar icterohaemmorragiae. (A) 16S rRNA primers FD1 and RP2. (B) rpoB primers LTB 1730F and LTB 2900R.
In summary, we believe that the proposed rpoB-based PCR assay should provide a new means to detect spirochetes. Moreover, when coupled with sequence analysis of amplified fragments, this technique would permit further epidemiologic investigation and thus provide an opportunity to perceive new patterns of diseases caused by Leptospira, Borrelia, and Treponema, most particularly by the strains that we investigated here.
ACKNOWLEDGMENT
We thank Richard Birtles for critical review of the manuscript.
REFERENCES
- 1.Alekshun M, Kashlev M, Schwartz I. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene. 1997;186:227–235. doi: 10.1016/s0378-1119(96)00714-7. [DOI] [PubMed] [Google Scholar]
- 2.Baranton G, Marti Ras N, Postic D. Borrelia burgdorferi, taxonomy, pathogenicity and spread. Ann Med Interne. 1998;149:455–458. [PubMed] [Google Scholar]
- 3.Berkelman R L. Introduction. In: Hordsburgh C R Jr, Nelson A M, editors. Pathology of emerging infections. Washington, D.C.: American Society for Microbiology; 1997. pp. 1–5. [Google Scholar]
- 4.Binder W D, Mermel L A. Leptospirosis in an urban setting: case report and review of an emerging infectious disease. J Emerg Med. 1998;16:851–856. doi: 10.1016/s0736-4679(98)00097-3. [DOI] [PubMed] [Google Scholar]
- 5.Dobbins W O. The diagnosis of Whipples disease. N Engl J Med. 1995;132:390–392. doi: 10.1056/NEJM199502093320611. [DOI] [PubMed] [Google Scholar]
- 6.Ehrlich G D, Greenberg S J. PCR-based diagnostics in infectious disease. Boston, Mass: Blackwell Scientific Publications; 1994. [Google Scholar]
- 7.Engelkens H J, Niemel P L, van der Sluis J J, Meheus A, Stolz E. Endemic treponematoses. II. Pinta and endemic syphilis. Int J Dermatol. 1991;30:231–238. doi: 10.1111/j.1365-4362.1991.tb04626.x. [DOI] [PubMed] [Google Scholar]
- 8.Farr R W. Leptospirosis. Clin Infect Dis. 1995;21:1–8. doi: 10.1093/clinids/21.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S RNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 10.Higgins D G, Sharp P M. Clustal: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 11.Klenk H-P, Zillig W. DNA-dependent RNA polymerase subunit β as a tool for phylogenetic reconstructions: branching topology of the archeal domain. J Mol Evol. 1994;38:420–432. doi: 10.1007/BF00163158. [DOI] [PubMed] [Google Scholar]
- 12.Knoth K, Roberds S, Poteet C, Tamkun M. Highly degenerate, inosine-containing primers specifically amplify rare cDNA using the polymerase chain reaction. Nucleic Acids Res. 1988;16:10932–10937. doi: 10.1093/nar/16.22.10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koff A B, Rosen T. Nonvenereal treponematoses: yaws, endemic syphilis, and pinta. J Am Acad Dermatol. 1993;31:1075–1076. doi: 10.1016/0190-9622(93)70217-h. [DOI] [PubMed] [Google Scholar]
- 14.Mollet C, Drancourt M, Raoult D. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol. 1997;26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 15.Palenik B. Polymerase evolution and organism evolution. Curr Opin Genet Dev. 1992;2:931–936. doi: 10.1016/s0959-437x(05)80118-2. [DOI] [PubMed] [Google Scholar]
- 16.Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. [Google Scholar]
- 17.Pinner R W, Teutsch S M, Simonsen L, Klug L A, Graber J M, Clarke M J, Berkelman R L. Trends in infectious diseases mortality in the United States. JAMA. 1996;275:189–193. [PubMed] [Google Scholar]
- 18.Pülher G, Leffers H, Gropp F, Palm P, Klenk H P, Lottspeich F, Garrett R A, Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of eucaryotic nuclear genome. Proc Natl Acad Sci USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raoult D. Dictionnaire des Maladies Infectieuses. Paris, France: Editions Scientifiques et Médicales Elsevier; 1998. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Siebert P D, Chenchick A, Kellog D E, Lukyanov K A, Lukyanov S A. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S RNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 23.Weinstock G M, Hardham J M, McLeod M P, Sodergren E J, Norris S J. The genome of Treponema pallidum: new light on the agent of syphilis. FEMS Microbiol Rev. 1998;22:323–332. doi: 10.1111/j.1574-6976.1998.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 24.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson K H. Detection of culture-resistant bacterial pathogens by amplification and sequencing of ribosomal DNA. Clin Infect Dis. 1994;18:958–962. doi: 10.1093/clinids/18.6.958. [DOI] [PubMed] [Google Scholar]
- 26.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]