Abstract
Recently, the authors reported newly synthesised polyethylene glycol (PEG)ylated silver (9%)‐doped zinc oxide nanoparticle (doped semiconductor nanoparticle (DSN)) which has high potency for killing Leishmania tropica by producing reactive oxygen species on exposure to sunlight. The current report is focused on Leishmania DNA interaction and damage caused by the DSN. Here, we showed that the damage to Leishmania DNA was indirect, as the DSN was unable to interact with the DNA in intact Leishmania cell, indicating the incapability of PEGylated DSN to cross the nucleus barrier. The DNA damage was the result of high production of singlet oxygen on exposure to sunlight. The DNA damage was successfully prevented by singlet oxygen scavenger (sodium azide) confirming involvement of the highly energetic singlet oxygen in the DNA degradation process.
Inspec keywords: silver, zinc compounds, nanoparticles, nanomedicine, DNA, microorganisms, cellular biophysics, biomedical engineering
Other keywords: photo‐induced Leishmania DNA degradation, PEGylated silver‐doped zinc oxide nanoparticle, Leishmania tropica, reactive oxygen species, sunlight, Leishmania DNA interaction, Leishmania cell, DNA damage, singlet oxygen scavenger, sodium azide, DNA degradation process, ZnO:Ag
1 Introduction
Continuous demand for anti‐parasitic drugs has urged researchers to explore metal nanoparticles for being less toxic and more capable to exhibit anti‐parasitic activity against parasitic diseases. Nanoparticles have the ability to penetrate the cell membrane, affecting the metabolic and genomic activities acting as a drug to control or cure a disease. However, there is an indispensable need of understanding the interactions of metal nanoparticles with cellular molecules [1]. Previously, metal nanoparticles were used for DNA interactions and degradation studies [2]. Metal nanoparticles attack the DNA and possibly affect its synthesis, replication and structural integrity thus impeding the DNA function [3]. The large surface to volume ratio makes metal nanoparticles suitable candidates to produce reactive oxygen species (ROS) especially singlet oxygen in the immediate vicinity [4]. The singlet oxygen basically causes degradation of DNA by promoting DNA cleavage with base modification or direct strand scission and leading to cell death [5].
Previously, gold, silver, platinum, copper and zinc oxide (ZnO) nanoparticles have been tested against cancer and bacterial DNA degradation [6, 7, 8, 9]. However, work on ZnO nanoparticles for DNA degradation toward parasitic organism especially Leishmania has not been reported. Leishmania causes a severe clinical manifestation called Leishmaniasis; a complex disease ranging from skin lesions to chronic systemic infection in the liver and spleen [10]. The current work was aimed to delineate the interaction mechanism of silver‐doped ZnO nanoparticles doped semiconductor nanoparticle (DSN) with both the intact Leishmania DNA and purified DNA using ultraviolet‐visible (UV–vis) absorption spectroscopy. Moreover, comet assay and gel electrophoresis were performed to study the degradation of Leishmania DNA by the singlet oxygen produced by the illumination of DSN with sunlight. Overall, the current report gives a prediction of the oxidative stress on the Leishmania DNA produced by DSN and provides an insight into the potential of this engineered nanoparticle as anti‐leishmanial agents.
2 Material and methods
2.1 Nanoparticle and Leishmania culture
In the current paper, we used our previously synthesised ZnO nanoparticle doped with 9% silver (DSN) by co‐precipitation method. Briefly, triton X‐100 (5%) was dissolved in ethanol (Sol A). Zinc acetate dihydrate and silver nitrate (9 mol% of Zn precursor) were dissolved in ethanol (final concentration of 50 mM of precursors) (Sol B). After mixing Sol B with A, the final solution was titrated against 100 mM sodium hydroxide (NaOH). The resulting material was subjected to the argon atmosphere in a tube furnace at 30 sccm, at 4°C/min and finally maintaining at 100°C for 4 h. Later, the nanoparticle was capped with polyethylene glycol (PEG)‐400. The particle size was measured by dynamic light scattering ( DLS) (Brookhaven Instruments, Inc., New York, USA), after dissolving 100 µg/ml of DSN in de‐ionised water and sonicating for 15 min (3 min on and 1 min off) and performed on particle size analyser operating at an angle of 90°, with a solid‐state laser (15 mW, 659 nm) as light source. Dust particles were removed through Millipore Millex‐HV filters with a pore size of 0.8 µm before DLS measurements.
Leishmania tropica KWH23 promastigotes were grown in Medium 199 (M199) (pH 7.2) with 25 mM HEPES (4‐(2‐hydroxyethyl)‐l‐piper‐azi‐neethanesulfonic acid), 10% foetal bovine serum and antibiotics at 24°C. For axenic amastigotes, Leishmania promastigotes were cultured in M199 at pH 5 and maintained at 33°C for 6 days [11]. The Leishmania cells were taken at a concentration of 2 × 108 cells/ml in all experiments. The cells were treated with 10 µg/ml of DSN. The experiments were carried out in two conditions, that is, one in dark and other in sunlight for 15 min. After that they were incubated at 24°C in full dark condition for 24 h. Full mortality (100% killing) was achieved after 24 h by checking the motility of Leishmania under microscope on an Neubauer chamber. The same protocol was repeated in all experiments.
2.2 DNA extraction
DNA was extracted from Leishmania (both treated and untreated) by adding 100 µl of lysis buffer (0.3 M sucrose, 10 mM Tris‐hydrochloride (HCl) (pH 7.5), 2 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 1% triton X‐100, 10 mM sodium dodecyl sulphate and 8.5 µl proteinase K solution (20 µl/ml) and then incubated in a water bath at 60°C for 2 h. Equal quantity of phenol: chloroform (1:1) was added to the above solution and centrifuged at 10,000 rpm for 10 min. After taking the aqueous layer with the DNA, 1 mL of chilled isopropanol was added and centrifuged again at 10,000 rpm for 10 min. After precipitation, DNA was washed with 70% ethanol and suspended in Tris‐EDTA (TE) buffer (1 M Tris‐HCl, 0.5 M EDTA).
2.3 DNA interaction by UV–vis spectroscopy
Leishmania DNA was dissolved by overnight stirring in double de‐ionised water (pH = 7.0) and kept at 4°C. De‐ionised water was used to prepare buffer (20 mM phosphate buffer (monosodium phosphate (NaH2 PO4)) (disodium phosphate (Na2 HPO4)), pH = 7.2). The DNA concentration was determined via absorption spectroscopy using the molar absorption coefficient of 6600 M−1 cm−1 (270 nm) for Leishmania DNA [12, 13] and it was found to be 1.8 × 10−4 M. The DSN nanoparticle was dissolved in de‐ionised water. The UV absorption titrations were performed in the absence and presence of DNA. Same concentration of DNA was added both to the DSN and reference solutions to eliminate the absorbance of DNA itself. DSN‐DNA solutions were allowed to incubate for 10 min at room temperature before measurements were made. Absorption spectra were recorded using cuvettes of 1 cm path length at room temperature (25 ± 1°C).
2.4 Comet assay
The comet assay of promastigotes and amastigotes was carried out according to Singh et al. [14] protocol with the following modifications. A 10 µl of each fresh and DSN treated Leishmania cell suspension (10,000 cells) were mixed with 100 µl of 0.7% low‐melting agarose in phosphate buffer at 24°C. Subsequently, 100 µl of this mixture was layered onto two slides (one for each sample) already pre‐coated with thin layer of 1% agarose and afterwards covered with a cover‐slip. After solidifying the agarose at 4°C, the slides were immersed in freshly prepared lysing solution consisted of 0.1 M EDTA, 10 mM Tris base, 2.5 M sodium chloride, 1% of triton X‐100 and 10% dimethyl sulfoxide (DMSO) for 30 min. For unwinding, the slides were submerged in alkaline electrophoresis buffer consisting of 10 N NaOH and 0.2 M EDTA (pH 13) for 1 h and then electrophoresed at 24 V for 20 min. Afterwards, slides were further immersed in neutralisation buffer for 3–5 min and were stained with ethidium bromide (1 µg/ml). The slides were observed under fluorescence microscope (Nikon, Tokyo, Japan) provided with epi‐flourescence and equipped with a rhodamine filter (excitation wavelength, 546 nm and barrier filter, 580 nm).
2.5 ROS scavenging and DNA degradation
To the treated Leishmania (dark and sunlight) and to isolated Leishmania DNA (treated with 10 µg/ml of DSN), 0.1 mM of sodium azide and 1 mM of mannitol were added as singlet oxygen and hydroxyl radical scavengers. From the treated Leishmania, DNA was isolated by the above method. All DNA samples were electrophoresed on 1% agarose gel at 100 V. The DNA was visualised by using 0.5 µg/ml of ethidium bromide.
In another group, Leishmania cells were exposed to serial concentrations (0.01–10 mM) of sodium azide and mannitol without any DSN for any inhibitory effect of these scavengers. These cells were incubated at 24°C for 24 h. Viable cells were counted by both trypan blue and motility of Leishmania cells on Neubauer chamber.
3 Results and discussion
3.1 Particle size
The present paper revealed the effect of PEGylated silver‐doped ZnO nanoparticle on L. tropica DNA, that is, on the DNA of Leishmania cell pre‐treated with the nanoparticle and also on isolated Leishmania DNA. First, the size of the synthesised nanoparticle was measured on DLS and it was found that the mean hydrodynamic diameter was 45 nm (Fig. 1). This size was not in accordance with the scanning electron microscopy in our previous findings, which showed an average size of 23 nm [15]. The results of different size determination methods are based on the applied principles, for example, DLS gives mean hydrodynamic diameter of all the particles in solution. Moreover, the agglomeration of particles playa an important role, and thus on polydispersity it gives a weighted hydrodynamic diameter of collection of particles with larger size [16].
Fig. 1.
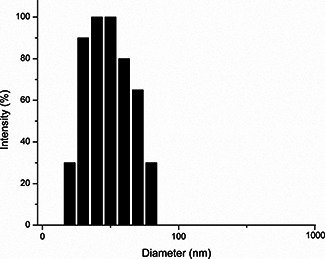
DLS of the synthesised nanoparticle
3.2 DNA interaction by UV–vis spectroscopy
Electronic absorption spectrum was initially used to examine the interaction between DSN and Leishmania DNA. Fig. 2 shows the UV–vis spectrum of free Leishmania DNA, free DSN and DSN + DNA. Fig. 3 shows the absorption peak of treated DNA (DNA extracted from Leishmania cell pre‐treated with DSN). It was found that in the presence of DNA, hypochromism (decrease in absorbance) occurred. As shown in Fig. 3, there was no shift in the original peak of the DNA on the addition of DSN. The hypochromic shift for DSN + DNA peak was probably due to the dilution effect of solvent. Any kind of interaction (either intercalation or electrostatic interaction) between the DSN and DNA results in a shift. For instance, if there was an intercalative interaction than there must be hypochromic shift along with red shift. As intercalative mode of interaction involves the π –π stacking interaction in which energy for the said transition reduces, which results in a red shift [17, 18, 19]. Interestingly, there was no interaction of DSN with the Leishmania cell DNA. The results were confirmed using both UV–vis spectrometry and gel electrophoresis during dark exposure. This might have been resulted due to the incompetence of PEGylated DSN delivery to nucleus that is highly selective and is the most significant barrier to efficient delivery systems [20].
Fig. 2.
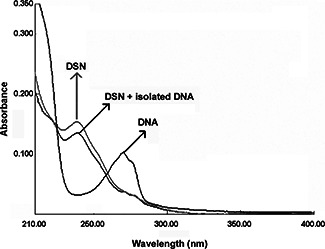
UV–vis of isolated Leishmania DNA without exposure to sunlight: (i) free DNA, (ii) treated with DSN, and (iii) DSN alone
Fig. 3.
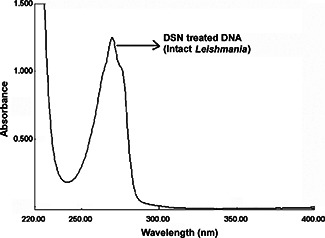
UV–vis of Leishmania DNA without exposure to sunlight, treated with DSN and showing no interaction with intact cell DNA
3.3 Gel electrophoresis
In our previous paper, we found that DSN were capable of producing a good quantum yield of ROS (0.27) with more than 80% of singlet oxygen and remaining hydroxyl radical on exposure to sunlight for 15 min [15]. The intensity of light was adjusted to control the singlet oxygen generation because more exposure to light causes photosensitiser photobleaching and oxygen depletion [4]. In the current paper, 0.1 mM sodium azide and 1 mM mannitol were used to scavenge both singlet oxygen and hydroxyl radicals [17, 18]. Low concentrations of both the scavengers were used because higher concentrations caused Leishmania cell death (Fig. 4). Sodium azide scavenged singlet oxygen and completely inhibited the DNA degradation in both Leishmania cell pre‐treated with DSN and also on isolated Leishmania DNA (Fig. 5). However, mannitol was unable to prevent DNA degradation. Thus, this can be elucidated that singlet oxygen was involved in the degradation of Leishmania DNA. Furthermore, experiments in the dark showed that no nuclease activity of the current nanoparticles was found. However, the current results were in accordance with the previously used ZnO and copper nanoparticles for the DNA degradation by producing ROS especially singlet oxygen [6, 8, 18].
Fig. 4.
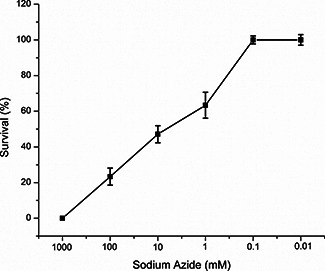
Dose‐dependent response of sodium azide against L. tropica
Fig. 5.
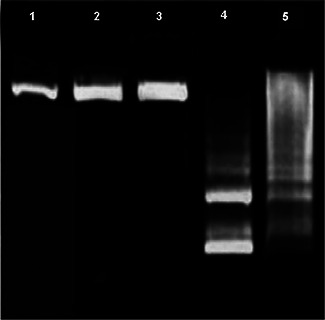
Gel electrophoresis of Leishmania DNA. Lane 1 represents the untreated (control) Leishmania DNA, Lane 2 shows DSN treated Leishmania in dark and its DNA, Lane 3 shows DNA isolated from treated Leishmania with DSN on exposure to light in the presence of sodium azide (singlet oxygen scavenger), Lane 4 shows DNA isolated from treated Leishmania with DSN on exposure to light in the presence of mannitol and DMSO (hydroxyl radical scavenger) and lane 5 shows DNA isolated from Leishmania and later on treated with DSN and exposed to sunlight
3.4 Comet assay
There are many assays available to measure the DNA damage by oxidative stress but the powerful and sensitive method is comet assay or single cell gel electrophoresis [19, 21]. Currently, this method was used to measure the oxidative stress generated by the ROS especially singlet oxygen by the DSN on Leishmania cells in the presence of sunlight. Comet assay was performed on both promastigotes and amastigotes because Leishmania exists in two morphological forms, that is, extracellular form (promastigote) and intracellular form (amastigote). Leishmania (promastigotes and amastigotes) cells were treated accordingly and were lysed in high salt concentrations for unwinding and electrophoresis at alkaline pH and neutral pH for the detection of both single and double stranded breaks. In the presence of electric field, the DNA migrated to anode and both amastigotes and promastigote cells were showing a comet like appearance indicating the DNA damage (Fig. 6). Interestingly, there was no clear difference in the comet formation by both amastigote and promastigote forms. It was confirmed that the DSN was capable of causing DNA degradation in both forms. The comets were only observed in the light treated promastigote and amastigote cells in the presence of DSN (average DNA tail of 32.78% and tail moment of 5.94 µm), whereas no comets were present in case of dark treated cells. Furthermore, the comet tail was unidirectional on rotation of electric field, showing the attachment of DNA with the head [22]. However, the UV–vis spectroscopy has shown no interaction of Leishmania DNA pre‐treated with DSN, thus the comets produced currently might be because of the entry of nanoparticles to the cytoplasm, and thus on activation with light it produced the singlet oxygen which caused indirect damage to DNA [6]. However, ZnO nanoparticles have the capability to interact directly or indirectly with DNA causing DNA damage independent of the cell type [23]. Moreover, our paper is in accordance with the previous reports of oxidative stress produced by the ZnO in the intracellular environment which leads to cell death [24].
Fig. 6.
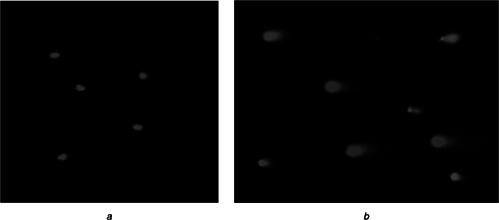
Comet assay of Leishmania cells
a Comets of axenic amastigotes
b Comets of promastigotes
Appearance of comets with different unwinding and electrophoresis conditions subsequently treating with DSN on exposure in sunlight
4 Conclusion
It can be concluded that the results were in accordance with our previous findings that silver (9%)‐doped ZnO nanoparticle was responsible for the killing of Leishmania by producing ROS especially singlet oxygen on excitation in sunlight. Singlet oxygen degraded Leishmania DNA on production in sunlight. Furthermore, the UV–vis spectroscopy showed that the nanoparticles in dark were not affecting the DNA confirming the involvement of the major role of light inducing singlet oxygen production which was further confirmed both by comet formation and gel electrophoresis. However, this PEGylated DSN was incapable of reaching the nucleus but it was able to degrade DNA by indirect mechanism. In summary, the ability of DSN to degrade DNA and killing Leishmania makes it a suitable anti‐leishmanial candidate. Furthermore, the use of this nanoparticle as a therapeutic agent is advantageous because human body has the capability to metabolise both silver and zinc since they are major micronutrients. However, the incapability of the DSN to reach nucleus can be utilised in designing better and more active anti‐leishmanial drug by chemically modifying the DSN with a particular carrier macromolecule. Currently, our focus is on using such nanoparticles and designing new kinds of carrier systems and molecules, which can be helpful in delivering such nanoparticles to the targeted site.
5 Acknowledgments
Financial support from the Higher Education Commission (HEC) of Pakistan and the Pakistan Science Foundation (PSF) are gratefully acknowledged.
6 References
- 1. An H. Jin B.: ‘Prospects of nanoparticle–DNA binding and its implications in medical biotechnology’, Biotechnol. Adv., 2012, 30, (6), pp. 1721 –1732 (doi: 10.1016/j.biotechadv.2012.03.007) [DOI] [PubMed] [Google Scholar]
- 2. Shen Q. Nie Z. Guo M. et al.: ‘Simple and rapid colorimetric sensing of enzymatic cleavage and oxidative damage of single‐stranded DNA with unmodified gold nanoparticles as indicator’, Chem. Commun., 2009, 8, (8), pp. 929 –931 (doi: 10.1039/b818081d) [DOI] [PubMed] [Google Scholar]
- 3. Basu S. Jana S. Pande S. et al.: ‘Interaction of DNA bases with silver nanoparticles: assembly quantified through SPRS and SERS’, J. Colloid Interface Sci., 2008, 321, (2), pp. 288 –293 (doi: 10.1016/j.jcis.2008.02.015) [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y. Aslan K. Previte M.J. et al.: ‘Plasmonic engineering of singlet oxygen generation’, Proc. Natl. Acad. Sci., 2008, 105, (6), pp. 1798 –1802 (doi: 10.1073/pnas.0709501105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burrows C.J. Muller J.G.: ‘Oxidative nucleobase modifications leading to strand scission’, Chem. Rev., 1998, 98, (3), pp. 1109 –1152 (doi: 10.1021/cr960421s) [DOI] [PubMed] [Google Scholar]
- 6. Sharma V. Shukla R.K. Saxena N. et al.: ‘DNA damaging potential of zinc oxide nanoparticles in human epidermal cells’, Toxicol. Lett., 2009, 185, (3), pp. 211 –218 (doi: 10.1016/j.toxlet.2009.01.008) [DOI] [PubMed] [Google Scholar]
- 7. Midander K. Cronholm P. Karlsson H.L. et al.: ‘Surface characteristics, copper release, and toxicity of nano‐ and micrometer‐sized copper and copper(II) oxide particles: a cross‐disciplinary study’, Small, 2009, 5, (3), pp. 389 –399 (doi: 10.1002/smll.200801220) [DOI] [PubMed] [Google Scholar]
- 8. Hackenberg S. Zimmermann F.Z. Scherzed A. et al.: ‘Repetitive exposure to zinc oxide nanoparticles induces DNA damage in human nasal mucosa mini organ cultures’, Environ. Mol. Mutagenesis, 2011, 52, (7), pp. 582 –589 (doi: 10.1002/em.20661) [DOI] [PubMed] [Google Scholar]
- 9. Chatterjee A.K. Chakraborty R. Basu T.: ‘Mechanism of antibacterial activity of copper nanoparticles’, Nanotechnology, 2014, 25, (13), p. 135101 (doi: 10.1088/0957-4484/25/13/135101) [DOI] [PubMed] [Google Scholar]
- 10. Yasinzai M. Khan M. Nadhman A. et al.: ‘Drug resistance in leishmaniasis: Current drug‐delivery systems and future perspectives’, Future Med. Chem., 2013, 5, (15), pp. 1877 –1888 (doi: 10.4155/fmc.13.143) [DOI] [PubMed] [Google Scholar]
- 11. Bates P.A.: ‘Axenic culture of Leishmania amastigotes’, Parasitol. Today, 1993, 9, (4), pp. 143 –146 (doi: 10.1016/0169-4758(93)90181-E) [DOI] [PubMed] [Google Scholar]
- 12. Sirajuddin M. Ali S. Shah N.A. et al.: ‘Synthesis, characterization, biological screenings and interaction with calf thymus DNA of a novel azomethine 3‐((3, 5‐dimethylphenylimino) methyl) benzene‐1, 2‐diol’, Spectrochim. Acta A, Mol. Biomol. Spectrosc., 2012, 94, pp. 134 –142 (doi: 10.1016/j.saa.2012.03.068) [DOI] [PubMed] [Google Scholar]
- 13. Sirajuddin M. Ali S. McKee V. et al.: ‘Synthesis, spectroscopic characterization, crystal structure, DNA interaction study and in‐vitro biological screenings of 4‐(5‐chloro‐2‐hydroxyphenylamino)‐4‐oxobut‐2‐enoic acid’, Spectrochim. Acta A, Mol. Biomol. Spectrosc., 2015, 134, pp. 244 –250 (doi: 10.1016/j.saa.2014.06.099) [DOI] [PubMed] [Google Scholar]
- 14. Singh N.P. McCoy M.T. Tice R.R. et al.: ‘A simple technique for quantitation of low levels of DNA damage in individual cells’, Exp. Cell Res., 1988, 175, (1), pp. 184 –191 (doi: 10.1016/0014-4827(88)90265-0) [DOI] [PubMed] [Google Scholar]
- 15. Nadhman A. Nazir S. Khan M.I. et al.: ‘PEGylated silver doped zinc oxide nanoparticles as novel photosensitizers for photodynamic therapy against Leishmania’, Free Radical Biol. Med., 2014, 77, pp. 230 –238 (doi: 10.1016/j.freeradbiomed.2014.09.005) [DOI] [PubMed] [Google Scholar]
- 16. Sharma V. Anderson D. Dhawan A.: ‘Zinc oxide nanoparticles induce oxidative DNA damage and ROS‐triggered mitochondria mediated apoptosis in human liver cells (HepG2)’, Apoptosis, 2012, 17, (8), pp. 852 –870 (doi: 10.1007/s10495-012-0705-6) [DOI] [PubMed] [Google Scholar]
- 17. Zhang X. Rosenstein B.S. Wang Y. et al.: ‘Identification of possible reactive oxygen species involved in ultraviolet radiation‐induced oxidative DNA damage’, Free Radical Biol. Med., 1997, 23, (7), pp. 980 –985 (doi: 10.1016/S0891-5849(97)00126-3) [DOI] [PubMed] [Google Scholar]
- 18. Jose G.P. Santra S. Mandal S.K. et al.: ‘Singlet oxygen mediated DNA degradation by copper nanoparticles: potential towards cytotoxic effect on cancer cells’, J. Nanobiotechnol., 2011, 9, (9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moller P. Knudsen L.E. Loft S. et al.: ‘The comet assay as a rapid test in biomonitoring occupational exposure to DNA‐damaging agents and effect of confounding factors’, Cancer Epidemiol. Biomarkers Prevention, 2000, 9, (10), pp. 1005 –1015 [PubMed] [Google Scholar]
- 20. Bonner D.K. Leung C. Chen‐Liang J. et al.: ‘Intracellular trafficking of polyamidoamine–poly (ethylene glycol) block copolymers in DNA delivery’, Bioconjugate Chem., 2011, 22, (8), pp. 1519 –1525 (doi: 10.1021/bc200059v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nandhakumar S. Parasuraman S. Shanmugam M. et al.: ‘Evaluation of DNA damage using single‐cell gel electrophoresis (comet assay)’, J. Pharmacol. Pharmacotherapeutics, 2011, 2, (2), p. 107 (doi: 10.4103/0976-500X.81903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klaude M. Eriksson S. Nygren J. et al.: ‘The comet assay: mechanisms and technical considerations’, Mutation Res,/DNA Repair, 1996, 363, (2), pp. 89 –96 (doi: 10.1016/0921-8777(95)00063-1) [DOI] [PubMed] [Google Scholar]
- 23. Demir E. Akça H. Kaya B. et al.: ‘Zinc oxide nanoparticles: genotoxicity, interactions with UV‐light and cell‐transforming potential’, J. Hazardous Mater., 2014, 264, pp. 420 –429 (doi: 10.1016/j.jhazmat.2013.11.043) [DOI] [PubMed] [Google Scholar]
- 24. Kocbek P. Teskač K. Kreft M.E. et al.: ‘Toxicological aspects of long‐term treatment of keratinocytes with ZnO and TiO2 nanoparticles’, Small, 2010, 6, (17), pp. 1908 –1917 (doi: 10.1002/smll.201000032) [DOI] [PubMed] [Google Scholar]


