Abstract
There are conflicting reports on the effects of HIV on COVID-19. Here, we analyzed disease severity and immune cell changes during and after SARS-CoV-2 infection in 236 participants from South Africa, of which 39% were people living with HIV (PLWH), during the first and second (Beta dominated) infection waves. The second wave had more PLWH requiring supplemental oxygen relative to HIV-negative participants. Higher disease severity was associated with low CD4 T cell counts and higher neutrophil to lymphocyte ratios (NLR). Yet, CD4 counts recovered and NLR stabilized after SARS-CoV-2 clearance in wave 2 infected PLWH, arguing for an interaction between SARS-CoV-2 and HIV infection leading to low CD4 and high NLR. The first infection wave, where severity in HIV negative and PLWH was similar, still showed some HIV modulation of SARS-CoV-2 immune responses. Therefore, HIV infection can synergize with the SARS-CoV-2 variant to change COVID-19 outcomes.
Research organism: Virus
Introduction
HIV is a prevalent infection in KwaZulu-Natal, South Africa (Kharsany et al., 2018) which also has a high SARS-CoV-2 attack rate (Tegally et al., 2021a; Tegally et al., 2021b). HIV depletes CD4 T helper cells (Dalgleish et al., 1984) which are a critical part of the adaptive immune response and are also the main target of HIV infection. CD4 T cell death occurs after cellular infection with HIV (Westendorp et al., 1995), or in bystander or incompletely infected cells due to activation of cellular defense programs (Doitsh et al., 2010; Doitsh et al., 2014), and is halted and, to some extent, reversed by antiretroviral therapy (ART), even sub-optimal therapy (Jackson et al., 2018).
The loss of CD4 T cells leads to dysregulation of many aspects of the immune response, including germinal center formation and antibody affinity maturation, which requires help from the highly HIV susceptible CD4 T follicular helper cells (Okoye and Picker, 2013; Pallikkuth et al., 2012; Perreau et al., 2013). In association with this, HIV also causes B cell dysregulation and dysfunction (Moir and Fauci, 2013). Moreover, T cell trafficking, activation, and exhaustion profiles of both CD4 and CD8 subsets are also modulated by HIV infection (Day et al., 2006; Deeks et al., 2004; Mavigner et al., 2012).
Both antibody and T cell responses are critical for effective control and clearance of SARS-CoV-2. More severe COVID-19 disease correlates with lymphopenia and low T cell concentrations (Lucas et al., 2020; Sekine et al., 2020; Chen et al., 2020a), whilst mild disease correlates with a robust T cell response to SARS-CoV-2 (Grifoni et al., 2020; Sekine et al., 2020; Rydyznski Moderbacher et al., 2020; Mathew et al., 2020; Mateus et al., 2020; Liao et al., 2020; Chen et al., 2020b). Neutralizing antibodies and associated expansion of antibody secreting B cells (ASC) are elicited in most SARS-CoV-2 infected individuals (Woodruff et al., 2020; Robbiani et al., 2020; Quinlan et al., 2020), and neutralizing antibody titers strongly correlate with vaccine efficacy (Khoury et al., 2021; Earle et al., 2021), indicating their key role in the response to SARS-CoV-2 infection. In contrast, high neutrophil numbers are associated with more severe disease and an elevated neutrophil to lymphocyte ratio (NLR) is often considered a risk factor for a more severe COVID-19 outcome (Liu et al., 2020a; Liu et al., 2020b; Zhang et al., 2020).
Results from epidemiological studies of the interaction between HIV and SARS-CoV-2 from other locations are mixed. Several large studies observed that disease severity and/or mortality risk is increased with HIV infection (Western Cape Department of Health in collaboration with the National Institute for Communicable Diseases, South Africa et al., 2021; Geretti et al., 2021; Bhaskaran et al., 2021; Tesoriero et al., 2021; Braunstein et al., 2021; Jassat et al., 2021a) while others found no statistically significant differences in clinical presentation, adverse outcomes, or mortality (Huang et al., 2021; Sigel et al., 2020; Shalev et al., 2020; Vizcarra et al., 2020; Stoeckle et al., 2020; Dandachi et al., 2021; Härter et al., 2020; Karmen-Tuohy et al., 2020; the Northwell COVID-19 Research Consortium et al., 2020; Inciarte et al., 2020; Hadi et al., 2020). Worse outcomes for PLWH tended to be in patients with low CD4 (Hoffmann et al., 2021a; Dandachi et al., 2021; Braunstein et al., 2021) and low absolute CD4 count was a risk factor for more severe disease (Western Cape Department of Health in collaboration with the National Institute for Communicable Diseases, South Africa et al., 2021).
HIV is known to interfere with protective vaccination against multiple pathogens (Avelino-Silva et al., 2016; Carson et al., 1995; Cooper et al., 2011; Fuster et al., 2016), typically as a consequence of sub-optimal antibody responses. In line with this, results from a South-African phase IIb trial of the Novavax NVX-CoV2373 vaccine, which uses a stabilised prefusion spike protein, showed 60% efficacy in HIV-uninfected individuals. However, overall efficacy dropped to 49% upon inclusion of PLWH (Shinde et al., 2021), although it is important to note that the numbers of PLWH in the study were very small. Nonetheless, there were more breakthrough cases in PLWH in the vaccine arm than the placebo arm.
An important consideration in infections in South Africa is the infecting variant, which in the second infection wave peaking January 2021 was predominantly the B.1.351 variant of concern (VOC) now designated as the Beta variant. In the current third infection wave it is predominantly the B.1.617.2 Delta variant. We and others have shown that the Beta variant has evolved the ability to escape neutralization by antibody responses elicited by earlier strains of SARS-CoV-2 or by vaccines based on those strains (Cele et al., 2021; Wibmer et al., 2021; Garcia-Beltran et al., 2021; Hoffmann et al., 2021b). Loss of vaccine efficacy of the AstraZeneca ChAdOx vaccine in South Africa was associated with this drop in neutralization capacity (Madhi et al., 2021). The second infection wave driven by Beta infections also showed increased mortality of hospitalized cases relative to the first infection wave (Jassat et al., 2021b).
What factors contributed to the evolution of the Beta variant in South Africa is yet unclear. One possibility is intra-host evolution in immunosuppressed PLWH with advanced HIV who are unable to clear SARS-CoV-2 (Karim et al., 2021). There is also evidence that variants evolved other adaptations to the host in addition to those in the spike glycoprotein which lead to antibody escape and enhanced transmission. These include evolution of resistance to the host interferon response (Guo et al., 2021; Thorne et al., 2021), as well as enhanced cell-to-cell transmission (Rajah et al., 2021). Changes in the virus may make infection with some variants substantially different in disease course, transmission dynamics, and effect on PLWH relative to ancestral SARS-CoV-2 strains or other variants.
Here, we aimed to determine the effects of HIV on the immune response to SARS-CoV-2 infection in KwaZulu-Natal, South Africa. This is important because we need to better understand COVID-19 disease course and vaccine efficacy in this population, as well as the possible reasons for the emergence of variants which lead to immune escape from neutralizing antibodies. Our results indicate that infections in the Beta variant infection wave led to more severe disease in PLWH relative to HIV-negative participants. Higher severity was associated with a lower CD4 T cell count. Yet, the CD4 count recovered, indicating that these participants may not have had a low CD4 count when first exposed to SARS-CoV-2. In addition, there were changes in the response of immune cell subsets associated with SARS-CoV-2 infection in PLWH relative to HIV-negative participants in the first infection wave, even in the absence of a statistically significant increase in disease severity, indicating that HIV infection may modulate the immune response to SARS-CoV-2.
Results
HIV infection is associated with higher disease severity in the Beta variant infection wave
We initiated a longitudinal observational cohort study to enroll and track patients with a positive COVID-19 qPCR test presenting at three hospitals in Durban, South Africa. Patients presented due to either COVID-19 symptoms or because they were known contacts of a confirmed COVID-19 case.
All participants were initially admitted to a hospital facility, then discharged after varying periods and followed up as outpatients. Enrollment was between June 2020 and May 2021. Participants were followed up weekly for the first month post-enrollment, and at 3-month intervals thereafter. At each study visit, a blood sample and a combined nasopharyngeal and oropharyngeal swab was taken. The purpose of a combined swab was to maximize the detection probability by qPCR of SARS-CoV-2 in the upper respiratory tract. Blood was used to determine HIV status, HIV viral load, and cellular parameters such as the concentration of CD4 T cells and the NLR. We also tested the frequencies of more specific immune cell subsets by flow cytometry (only available for infection wave 1 samples).
Up to May 2021, 236 participants were enrolled in the study, for a total of 986 study visits (Supplementary file 1). All participants are assumed to be vaccinated with BCG in infancy in accordance with South African national guidelines. The majority of participants were female, possibly reflecting better linkage to care. Enrollment was a median 11 days post-symptom onset (Supplementary file 2). De-identified participant data used here are available as Source Data included in the supplementary materials.
Out of 236 study participants, 93 (39%) were PLWH (Table 1) and 89% of study participants were of African descent. PLWH were significantly younger than HIV uninfected participants. Hypertension, diabetes and obesity, known risk factors for more severe COVID-19 disease (Zhou et al., 2020; the Northwell COVID-19 Research Consortium et al., 2020), were common: Hypertension and obesity were present in 24%, and 42% of study participants respectively, a similar prevalence to that reported in the province of KwaZulu-Natal where this study was performed (van Heerden et al., 2017; Malaza et al., 2012). Diabetes prevalence in our study was 18%, compared to 13% reported for South Africa (Federation, 2019). Hypertension and diabetes were significantly lower in the PLWH group (Table 1). 28 or 30% of PLWH were HIV viremic at any point in the study. For individuals on ART, median ART duration was 9 years. ART regimen was determined by liquid chromatography with tandem mass spectrometry (LC-MS/MS) and was predominately efavirenz (EFV) based, with some participants transitioning to a dolutegravir (DTG) based regimen. In addition, there was a small subset of PLWH on a ritonavir boosted lopinavir (LPV/r) as well as other ART combinations. About 12% of PLWH had no detectable ART despite a clinical record of ART, or were ART naive (Supplementary file 3). The absolute CD4 T cell count and the CD4 to CD8 T cell ratio was significantly lower in PLWH relative to HIV negative participants at enrollment. The incidence of active TB and the fraction of participants with a history of TB were much higher in the PLWH group (Table 1).
Table 1. Participant characteristics.
| All (n=236) | HIV- (n = 143, 60.6%) | HIV+ (n=93, 39.4%) | Odds ratio (95% CI) | p-value | |
|---|---|---|---|---|---|
| Demographics | |||||
| Age years, median (IQR) | 45 (35–57) | 49 (35–62) | 41 (35–50) | - | 0.003* |
| Male sex, n (%) | 82 (34.7) | 48 (33.6) | 34 (36.6) | 1.1 (0.7–2.0) | 0.68 |
| Current smoker, n (%) | 13 (5.5) | 4 (2.8) | 9 (9.7) | 3.7 (1.2 – gt10) | 0.038 |
| Comorbidity, n (%) | |||||
| Hypertension#, n=235 | 57 (24.1) | 42 (29.4) | 15 (16.1) | 0.5 (0.2–0.9) | 0.023 |
| Diabetes | 42 (17.8) | 32 (22.4) | 10 (10.8) | 0.4 (0.2–0.9) | 0.024 |
| Obesity#, n=221 | 91 (42.3) | 64 (47.1) | 27 (29.0) | 0.6 (0.3–1.0) | 0.086 |
| Active TB | 10 (4.2) | 1 (0.7) | 9 (9.7) | >10 | 0.001 |
| History TB | 32 (13.6) | 3 (2.1) | 29 (31.2) | >10 | <0.0001 |
| HIV associated parameters | |||||
| HIV viremic, n (% of all HIV) | - | - | 28 (30.1) | - | - |
| Years ART, median (IQR) | - | - | 9.4 (3.9–13.2) | - | - |
| CD4 cells/μL median (IQR) n=221 | 633 (326–974) | 887 (534–1148) | 464 (200–702) | - | <0.0001* |
| CD4/CD8 | 1.2 (0.8–1.7) | 1.6 (1.2–2.1) | 0.8 (0.4–1.1) | - | <0.0001* |
| Disease severity, n (%) | |||||
| Asymptomatic | 33 (14.0) | 25 (17.5) | 8 (8.6) | 0.4 (0.2–1.0) | 0.058 |
| Ambulatory with symptoms | 128 (54.2) | 80 (55.9) | 48 (51.6) | 0.8 (0.5–1.4) | 0.59 |
| Supplemental oxygen | 62 (26.3) | 30 (21.0) | 32 (34.4) | 2.0 (1.1–3.5) | 0.024 |
| Death | 13 (5.5) | 8 (5.6) | 5 (5.4) | 1.0 (0.3–2.9) | >0.99 |
| COVID-19 treatment, n (%) | |||||
| Corticosteroids | 74 (31.2) | 47 (32.9) | 27 (29.0) | 0.8 (0.5–1.5) | 0.57 |
| Anticoagulants | 53 (22.5) | 35 (24.5) | 18 (19.4) | 0.7 (0.4–1.4) | 0.43 |
| Symptom, n (%) | |||||
| Sore throat | 88 (37.3) | 55 (38.5) | 33 (35.5) | 0.9 (0.5–1.5) | 0.68 |
| Runny nose | 53 (22.5) | 30 (21.0) | 23 (24.7) | 1.2 (0.7–2.3) | 0.53 |
| Cough | 153 (64.8) | 91 (63.6) | 62 (66.7) | 1.1 (0.7–2.0) | 0.68 |
| History of fever#, n=235 | 58 (24.7) | 29 (20.3) | 29 (31.2) | 1.8 (1.0–3.3) | 0.063 |
| Shortness of breath | 148 (62.7) | 87 (60.8) | 61 (65.6) | 1.2 (0.7–2.1) | 0.49 |
p-value calculated via 2-sided Fisher’s Exact test, except for * which was calculated via Mann-Whitney U test. # Not including pregnancy or unable to be measured.
A minority of study participants (14%) were asymptomatic and presented at the hospital because of a close contact with a confirmed COVID-19 case. To include the asymptomatic participants in our analysis, we used time from diagnostic swab as our timescale, which was tightly distributed for symptomatic participants relative to symptom onset at a median of 3 to 4 days apart (Supplementary file 2).
The majority of participants in the study (54%) had symptoms but did not progress beyond mild disease, defined here as not requiring supplemental oxygen during the course of disease and convalescence. Twenty-six percent of participants required supplemental oxygen but did not die and 6% of participants died. Our cohort design did not specifically enroll critical SARS-CoV-2 cases. The requirement for supplemental oxygen, as opposed to death, was therefore our primary measure for disease severity.
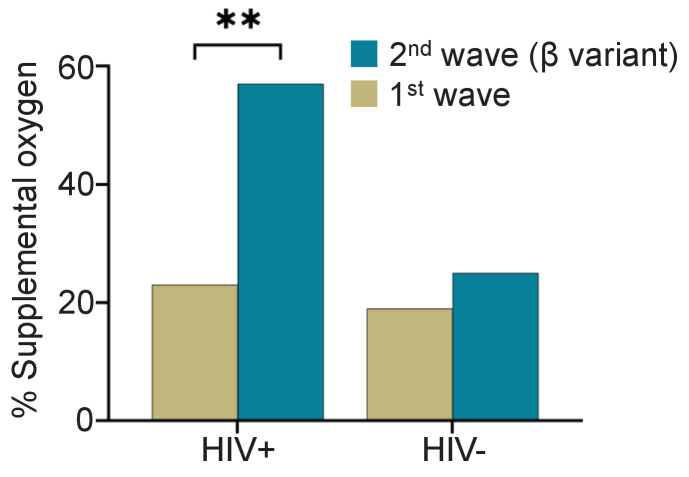
There was a significant difference in the frequency of participants requiring supplemental oxygen (without subsequent death) between HIV-negative participants and PLWH (21% versus 34% respectively, odds ratio of 2.0 with 95% confidence intervals of 1.1–3.5, Table 1).
To determine if the fraction of participants requiring supplemental oxygen differed between the first infection wave and the Beta variant dominated second infection wave, we compared disease severity between the first infection wave (Figure 1, Supplementary file 4), and the second infection wave (Figure 1, Supplementary file 5). In the first infection wave, there was no significant difference in the fraction of participants requiring supplemental oxygen between HIV-negative and PLWH participants (Supplementary file 4, p=0.5). However, significantly more PLWH required supplemental oxygen in the second wave (Supplementary file 5, odds ratio of 4.0 with 95% CI of 1.6–10.4, p=0.005). Comparing within the HIV-negative and PLWH groups, there was only a moderate increase in the fraction of participants requiring supplemental oxygen between SARS-CoV-2 infection wave 1 and infection wave 2 in HIV-negative participants (19% to 25%) which was not significant (Figure 1). In contrast, the number of PLWH participants requiring supplemental oxygen more than doubled from 24% to 57% (p=0.0025, Figure 1).
Figure 1. Fraction of PLWH and HIV-negative participants requiring supplemental oxygen during the first and the Beta variant dominated second infection waves.
p=0.0025 by Fisher’s Exact test.
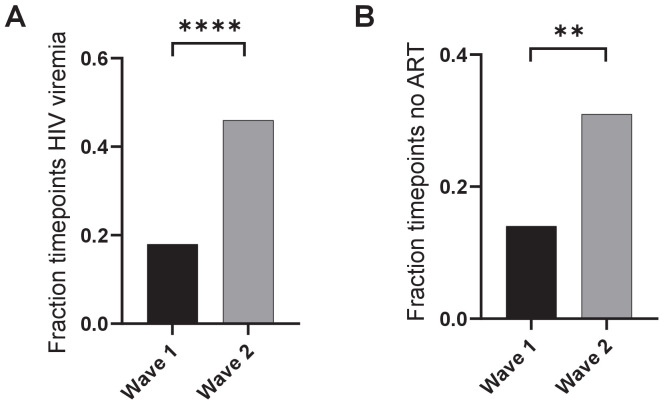
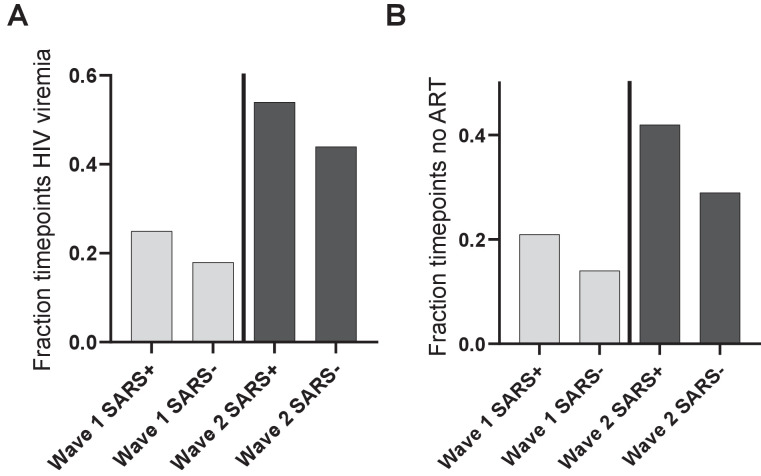
Figure 1—figure supplement 1. Viremia and ART in PLWH in wave 1 versus wave 2.
Figure 1—figure supplement 2. Effect of ART regimen on disease severity.
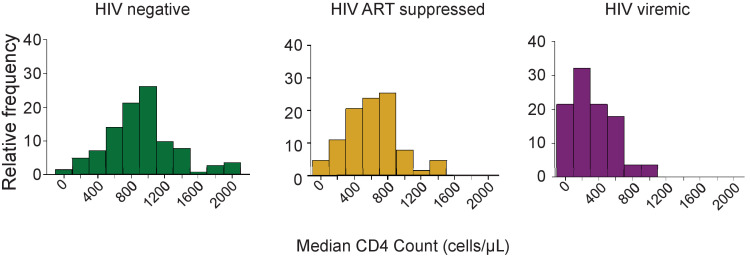
Figure 1—figure supplement 3. Distribution of CD4 counts by HIV status.
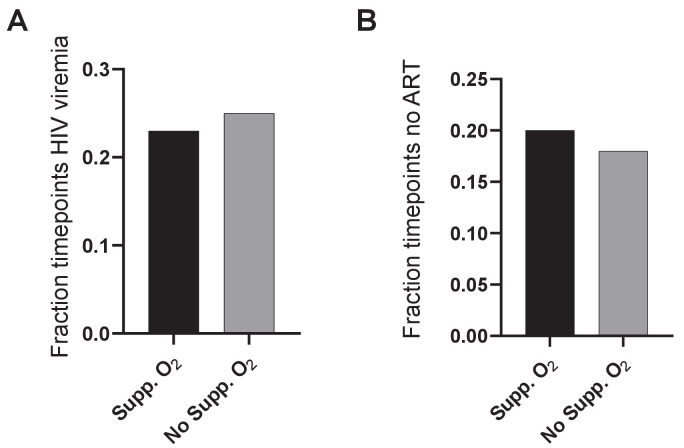
Figure 1—figure supplement 4. Viremia and ART in PLWH requiring versus not requiring supplemental oxygen.
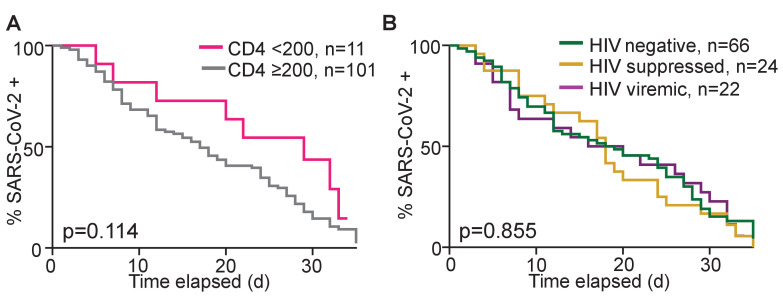
Figure 1—figure supplement 5. Dependence of time to SARS-CoV-2 clearance on CD4 count and HIV status.
To examine whether the differences in the requirement for supplemental oxygen in PLWH were because of differences in the level of HIV control between waves, we examined the fraction of timepoints where participants showed HIV viremia. We excluded low level viremia and set the threshold at VL > 200 HIV RNA copies/mL (Ryscavage et al., 2014). Furthermore, we determined whether ART was detectable in the blood by LC-MS/MS. Second wave participants had approximately twofold higher fraction of timepoints where HIV viremia was detected (Figure 1—figure supplement 1A). In agreement with this, the fraction of participants with no detectable ART in the blood was also about twofold higher (Figure 1—figure supplement 1B). These observations are consistent with diminished suppression of HIV in second wave PLWH enrolled in this study. The specific HIV regimen had no discernible effect on disease severity (Figure 1—figure supplement 2).
We compared comorbidities and other characteristics between the PLWH and HIV negative participants on supplemental oxygen (Table 2). Strikingly, the median age of PLWH on supplemental oxygen was 21 years younger relative to HIV negative (41 versus 62, p=0.003). PLWH had significantly lower frequency of comorbidities which are usually associated with more severe COVID-19 disease: both hypertension (p=0.03) and diabetes (p=0.03) were lower. In contrast, the median CD4 T cell count across all study visits was lower in PLWH (277 versus 339), although this difference did not reach statistical significance (p=0.07). There was no significant difference in the fraction of participants treated with corticosteroids (p=0.2).
Table 2. Characteristics by HIV status of participants requiring supplemental oxygen.
| All (n=68) | HIV- (n = 35, 51.5%) | HIV+ (n=33, 48.5%) | Odds ratio (95% CI) | p-value | |
|---|---|---|---|---|---|
| Demographics | |||||
| Age years, median (IQR) | 51 (38–64) | 62 (47–66) | 41 (36–56) | - | 0.003* |
| Male sex, n (%) | 25 (36.8) | 12 (34.3) | 13 (39.4) | 1.2 (0.5–3.3) | 0.80 |
| Current smoker, n (%) | 2 (2.9) | 1 (2.9) | 1 (3.0) | 1.1 (<0.1 – >10) | gt 0.99 |
| Comorbidity, n (%) | |||||
| Hypertension | 26 (38.2) | 18 (51.4) | 8 (24.2) | 0.3 (0.1–0.8) | 0.026 |
| Diabetes | 17 (25.0) | 13 (37.1) | 4 (12.1) | 0.2 (0.1–0.8) | 0.025 |
| Obesity#, n=57 | 23 (40.4) | 11 (31.4) | 12 (36.4) | 1.8 (0.6–5.1) | 0.42 |
| Active TB | 6 (8.8) | 1 (2.9) | 5 (15.2) | 6.1 (0.9 – >10) | 0.10 |
| History TB | 16 (23.5) | 2 (5.7) | 14 (42.4) | 12.2 (2.7 – >10) | lt 0.001 |
| HIV associated parameters | |||||
| HIV viremic, n (% of all HIV) | - | - | 9 (27.3) | - | - |
| Years ART, median (IQR) | - | - | 11.6 (6.1–13.3) | - | - |
| CD4 cells/μL median (IQR) n=65 | 309 (170–545) | 339 (227–592) | 277 (134–461) | - | 0.072* |
| COVID-19 treatment, n (%) | |||||
| Corticosteroids | 43 (63.2) | 25 (71.4) | 18 (54.5) | 0.5 (0.2–1.3) | 0.21 |
| Anticoagulants | 31 (45.6) | 18 (51.4) | 13 (39.4) | 0.6 (0.2–1.6) | 0.34 |
p-value calculated via two-sided Fisher’s Exact test, except for * which was calculated via Mann-Whitney U test. # Not including pregnancy or unable to be measured.
Interestingly, when comparing HIV-negative participants requiring supplemental oxygen to those not requiring supplemental oxygen (Supplementary file 6), those on supplemental oxygen were significantly older (62 versus 47 years, p=0.002), and had significantly higher frequency of hypertension (p=0.002) and diabetes (p=0.02). This differed from PLWH, where differences in age and comorbidities were not significant between PLWH requiring supplemental oxygen and those not (Supplementary file 7), although there was a trend to a higher frequency for hypertension (p=0.1).
HIV viremic participants showed lower CD4 counts relative to HIV suppressed or HIV negative participants (Figure 1—figure supplement 3). Surprisingly, there was no difference in either the fraction of HIV viremic timepoints or fraction of timepoints where ART was not detected in the blood between the group of PLWH requiring supplemental oxygen and the no supplemental oxygen group (Figure 1—figure supplement 4). We also analyzed the time of SARS-CoV-2 clearance as a function of CD4 count and HIV status and found that while participants with a low CD4 count (<200) showed a trend of longer time to SARS-CoV-2 clearance (p=0.11), HIV viremia had no effect (Figure 1—figure supplement 5). Hence, while the PLWH enrolled in the second wave had both worse control of HIV infection and had a higher fraction requiring supplemental oxygen, we did not observe that the PLWH requiring supplemental oxygen had a higher frequency of HIV viremia.
SARS-CoV-2 has differential effects on CD4 count and the neutrophil to lymphocyte ratio between infection waves in PLWH
We next determined whether the increased disease severity in PLWH in infection wave two was reflected in the cellular immune response to SARS-CoV-2 infection. We therefore examined the CD4 count and NLR, both known to be strongly associated with disease severity. We used a three-point scale for disease severity, where 1: asymptomatic, 2: mild, and 3: supplemental oxygen (at any point in the study) or death. Death was merged with supplemental oxygen because of the small number of participants who died, and was not excluded in any of the subsequent analyses.
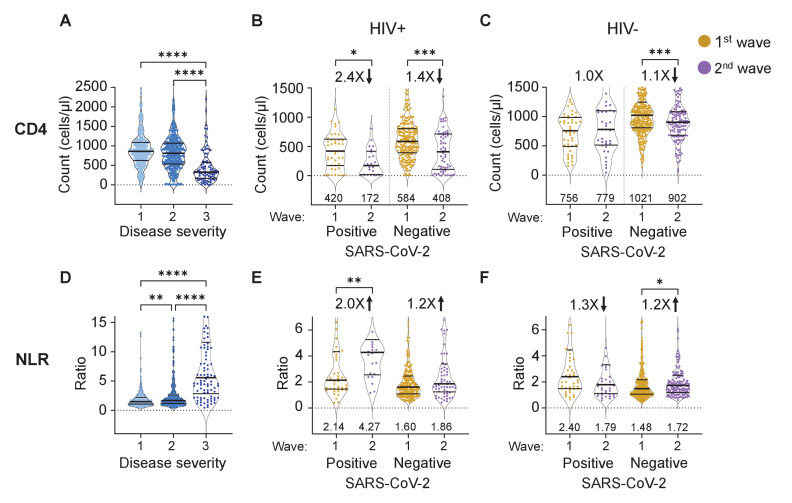
As expected, we observed a significant decrease in CD4 T cell count at the highest severity which included disease that required administration of supplemental oxygen and/or resulted in death (Figure 2A, see Figure 2—figure supplement 1 for all data points and number of data points per graph).
Figure 2. The differential effect of HIV on the CD4 count and neutrophil to lymphocyte ratio between waves.
(A) The concentration of CD4 T cells in the blood in all participants in all infection waves and at all timepoints as a function of disease severity. Disease severity was scored as 1: asymptomatic, 2: mild, and 3: on supplemental oxygen or death. CD4 counts in PLWH (B) and HIV negative (C) participants in wave 1 versus wave 2 during active SARS-CoV-2 infection and after SARS-CoV-2 clearance. (D) Neutrophil to lymphocyte ratio (NLR) in the blood in all participants in all infection waves and at all timepoints as a function of disease severity. NLR in PLWH (E) and HIV negative (F) participants in wave 1 versus wave 2 during active SARS-CoV-2 infection and after SARS-CoV-2 clearance. SARS-CoV-2 positive indicates a timepoint where SARS-CoV-2 RNA was detected. Data shown as violin plots with median and IQR, with the median denoted below each plot. Fold-change in the second wave versus first wave is indicated, with arrow denoting direction of change. p-values are * <0.05; ** <0.01; *** < 0.001, **** < 0.0001 as determined by Kruskal-Wallis test with Dunn's multiple comparison correction or by Mann-Whitney U test. Plots scales were restricted to highlight changes close to the median.
Figure 2—figure supplement 1. The differential effect of HIV on the CD4 count and neutrophil to lymphocyte ratio between waves - full dataset and number of data points per plot.
Figure 2—figure supplement 2. No significant increase in control of HIV infection at convalescence relative to active SARS-CoV-2 infection.
We then asked whether PLWH in infection wave two showed different CD4 T cell responses to SARS-CoV-2. Since decreased CD4 count could be due to HIV infection alone, we separated the data into timepoints when SARS-CoV-2 was detectable by qPCR and after SARS-CoV-2 was cleared. Upon SARS-CoV-2 clearance, the immune response of convalescent participants should start the return to baseline, and differences due to SARS-CoV-2 should decrease and reflect HIV mediated effects only.
The CD4 counts in PLWH in infection wave 2 were lower during active SARS-CoV-2 infection relative to wave 1 (Figure 2B, median 172 versus 420 cells/μL, a decrease of 2.4-fold) and were below the 200 cells/μL clinically used threshold indicating a low CD4 count. However, CD4 counts for PLWH for both wave 2 and wave 1 recovered post-SARS-CoV-2 clearance (408 for wave 2 versus 584 cells/μL for wave 1), consistent with the low CD4 count in PLWH in wave 2 being SARS-CoV-2 induced. CD4 counts for both groups were substantially above the 200 cells/μL threshold after SARS-CoV-2 clearance. HIV-negative participants showed no or minor differences in CD4 counts between waves, although these minor differences showed significance due to the large number of participant timepoints for this group (Figure 2C).
The NLR had a remarkably similar pattern. An elevated NLR associated strongly with higher disease severity (Figure 2D). PLWH with active SARS-CoV-2 infection in wave 2 showed a twofold increase in the NLR relative to PLWH with active SARS-CoV-2 infection in wave 1 (Figure 2E). This difference declined to 1.2-fold once SARS-CoV-2 was cleared, consistent with differences in NLR being SARS-CoV-2 driven and not a result of other pathology in PLWH in wave 2. In contrast, the NLR was lower in HIV negative participants in wave 2 relative to wave 1 in the presence of SARS-CoV-2 (Figure 2F).
The observed recovery of the CD4 count may result from improved access to ART due to the hospital visit in wave 2. We therefore checked whether the fraction of HIV viremic participants decreased upon convalescence and whether there was an associated decrease in the number of PLWH with undetectable ART. We observed no significant differences in either viremia or fraction of PLWH with undetectable ART in either wave between timepoints which were SARS-CoV-2 positive and those that were negative (Figure 2—figure supplement 1). This indicates that the increase in the CD4 was not due to better linkage to care after the hospital visit but rather due to SARS-CoV-2 clearance.
Differences in the frequencies and associations of immune cell subsets in PLWH and HIV-negative participants
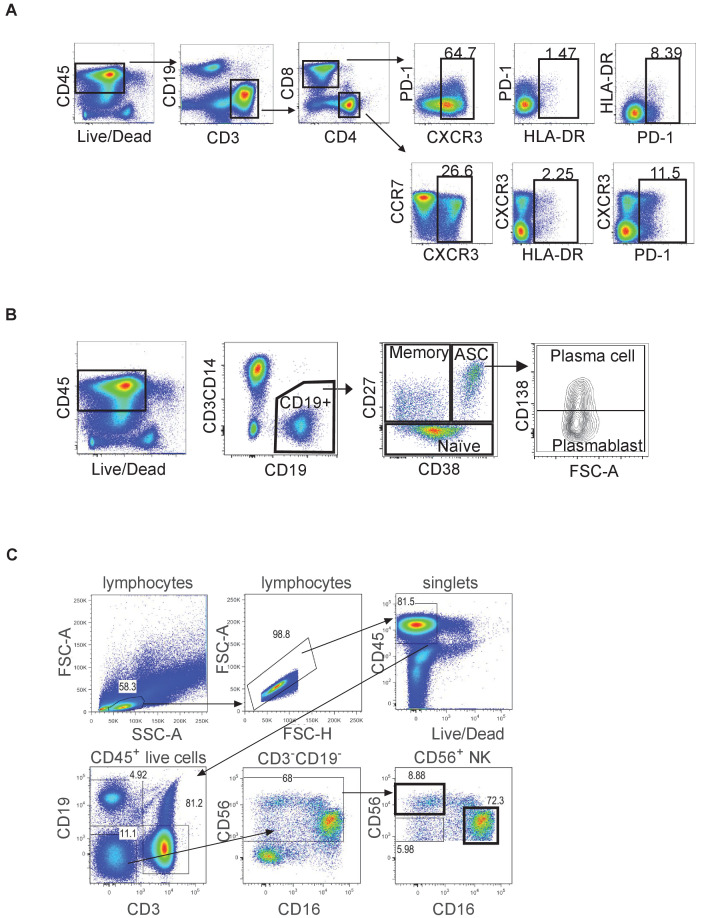
To examine differences in immune cell subset associations between HIV-negative and PLWH participant groups, we conducted detailed phenotyping of immune cells using longitudinal fresh PBMC samples and correlated these to measured phenotypes and clinical parameters in both HIV-negative and PLWH groups (Figure 3; see Figure 3—figure supplement 1 for gating strategies). We used established approaches for gating of cell subsets (Sanz et al., 2019; Khodadadi et al., 2019). This was only performed for the first wave participants, where cells were available for additional phenotyping by flow cytometry.
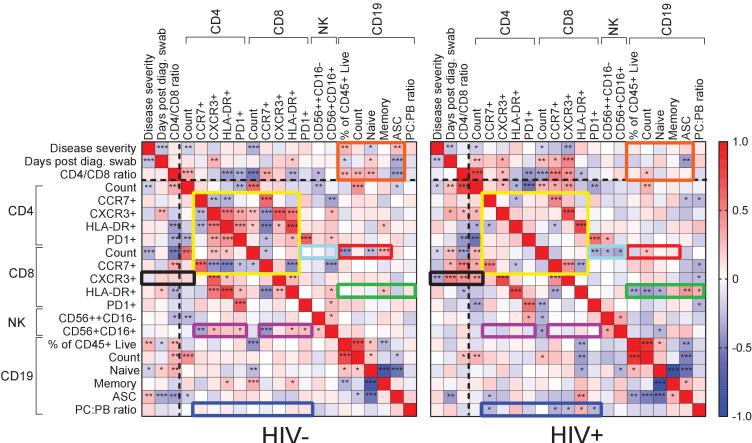
Figure 3. Immune cell and clinical correlates in HIV negative and PLWH groups.
Spearman rank correlation values () are shown from red (1.0) to blue (−1.0). p-values per correlation are *< 0.5; **< 0.01; ***< 0.001. The number of matched pairs for HIV negative participants ranged from 77 to 229 and for PLWH from 48 to 164. Rectangles represent regions where a set of correlations is present in one group and absent in the other. Black dashed lines represent the divide between clinical and cellular parameters.
Figure 3—figure supplement 1. Gating strategy.
For HIV-negative participants, there were significant negative and positive correlations between CD4 T cell parameters, and between these and the CD8 T cell count and phenotypes (Figure 3, yellow box). There were negative correlations between CD4 and the CD8 CCR7+ T cell phenotype and CD56+CD16+ NK cells (purple box). The fraction of NK cells positively correlated with the CXCR3 fraction of CD4 T cells, with HLA-DR on CD8 T cells, and with PD-1 on both cell types (purple box). In addition, there were correlations between CD8 T cell count and CD19 B cell parameters, such as fractions of naÏve and memory B cells (red box). Interestingly, disease severity as well as the CD4/CD8 ratio showed correlations with B cell parameters, including the frequency of antibody secreting cells (ASC), which were lost in PLWH (orange box).
New correlations arose in PLWH, particularly involving CD8 T cells: CXCR3+ CD8 T cells were negatively correlated with disease severity but positively correlated with the CD4/CD8 ratio and the CD4 T cell count (Figure 3, black box). CD8 T cell activation (HLA-DR+) was correlated with several CD19+ B cell phenotypes (green box), and the plasma cell to plasmablast ratio, determined by CD138 expression, correlated with both CD4 and CD8 T cell phenotypes (blue box). In addition, CD8 T cell count showed negative correlations with CD8 PD-1 and NK cell phenotypes only in PLWH (turquoise box).
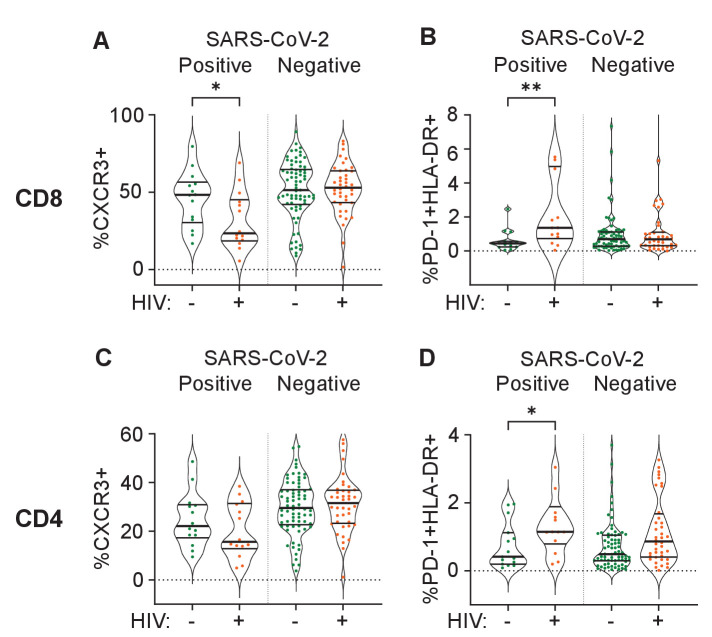
Out of the set of markers examined, the combination of PD-1 and HLA-DR expression is linked to T cell activation (Sauce et al., 2007; Vollbrecht et al., 2010), while CXCR3 expression is essential to recruitment of T cells to tissues (Groom and Luster, 2011). We therefore asked whether these markers showed differences between HIV negative and PLWH in the first infection wave during the time participants were positive for SARS-CoV-2, despite there being no significant differences in disease severity in this wave. In CD8 T cells, we observed a significant decrease in the fraction of CXCR3 expressing cells in the blood compartment in PLWH relative to HIV-negative participants (Figure 4A). We also observed an increase in the fraction of PD-1+HLA-DR+ cells (Figure 4B). For CD4 cells, there was no significant decrease in the fraction of CXCR3+ cells although a decrease was apparent (Figure 4C). Similarly to CD8 T cells, there was an increase in PD-1+HLA-DR+ CD4 T cells in PLWH (Figure 4D). There was no difference between PLWH and HIV-negative participants in any cell/marker combination after SARS-CoV-2 clearance.
Figure 4. Differences between PLWH and HIV-negative participants in immune cell markers.
Percent of CD8 T cells positive for CXCR3 (A) or double positive for HLA-DR and PD-1 (B). Percent of CD4 T cells positive for CXCR3 (C) or double positive for HLA-DR and PD-1 (D). Data is composed of 15 participant timepoints which were SARS-CoV-2+HIV-, 14 SARS-CoV-2+HIV+, 40 SARS-CoV-2-HIV+, and 74 SARS-CoV-2-HIV-, where SARS-CoV-2+ indicates SARS-CoV-2 RNA was detected in the upper respiratory tract. p-values for differences between PLWH and HIV-negative participants are * <0.05; ** <0.01; *** < 0.001, **** < 0.0001 as determined by the Mann-Whitney U test.
Discussion
We observed that in our cohort, COVID-19 disease severity was higher in PLWH, consistent with some of the larger epidemiological studies (Western Cape Department of Health in collaboration with the National Institute for Communicable Diseases, South Africa et al., 2021; Geretti et al., 2021; Bhaskaran et al., 2021; Tesoriero et al., 2021; Braunstein et al., 2021; Jassat et al., 2021a), although in this study differences were detected in the frequency of participants requiring supplemental oxygen and not in mortality. Our cohort may not be a typical ’hospitalized cohort’ as the majority of participants did not require supplemental oxygen. We therefore cannot discern effects of HIV on critical SARS-CoV-2 cases since these numbers are too small in the cohort. However, focusing on lower disease severity enabled us to capture a broader range of outcomes which predominantly ranged from asymptomatic to requiring supplemental oxygen. Understanding this part of the disease spectrum could be important since it may indicate underlying changes in the immune response which affect long-term quality of life and response to vaccines.
We observed a higher fraction of PLWH requiring supplemental oxygen relative to HIV negative participants in the second, Beta variant dominated SARS-CoV-2 infection wave in KwaZulu-Natal, South Africa. The odds ratio for requiring supplemental oxygen in the second wave for PLWH was 4.0 relative to HIV negative participants. The 95% confidence intervals were wide at 1.6–10.4, reflecting the relatively small number of participants. However, confidence intervals did not overlap one.
Consistent with HIV infection leading to more severe SARS-CoV-2 infection outcomes in our study is the much younger age of PLWH requiring supplemental oxygen relative to HIV negative participants (41 versus 63 years). PLWH on supplemental oxygen also had lower frequencies of hypertension and diabetes. Age, hypertension, and diabetes are risk factors for more severe COVID-19 disease (Yang et al., 2020; Guan et al., 2020; Ambrosioni et al., 2021; Jassat et al., 2021a), and their absence may indicate that the more severe outcome is driven by another factor, with HIV infection being the simplest explanation.
The cause of the difference between waves in PLWH may be because PLWH enrolled in the second infection wave had worse suppression of HIV with ART: both the fraction of timepoints where viremia was detected and where ART was absent were about twofold higher and indeed were very high at about 40%. We therefore expected that this showed a direct link between HIV viremia and the requirement for supplemental oxygen during COVID-19 disease in PLWH. However, there was no difference in the frequency of viremia between those requiring supplemental oxygen and those not.
Furthermore, the substantial recovery of CD4 T cell counts in PLWH after SARS-CoV-2 clearance in wave 2 may be consistent with the Beta variant having more impact on the CD4 count relative to the ancestral SARS-CoV-2 strain infections in the first wave. A similar pattern was seen in the NLR, which was higher in wave two relative to wave 1 in PLWH with active SARS-CoV-2 infection, but then decreased to similar levels upon convalescence. The role of the Beta variant is supported by data showing extensive evolution, increasing the ability of Beta to escape the interferon response and result in more efficient viral cell-to-cell transmission (Guo et al., 2021; Thorne et al., 2021; Rajah et al., 2021). Beta variant hospitalizations also led to more deaths in South Africa (Jassat et al., 2021b). Therefore, the effect of the variant on PLWH in addition to HIV suppression status should be considered.
Our data detailing the SARS-CoV-2 response of more defined immune cell subsets in PLWH versus HIV negative participants is limited by the data only being available for the first infection wave. However, even in samples from that wave, there were multiple differences in correlations between cell subsets in PLWH relative to HIV negative participants, which may be another indication of differences in the immune response to SARS-CoV-2. We cannot deduce from these associations whether the differences could have an impact on disease severity. However, the fraction of CXCR3+ CD8 T cells decreased in the blood compartment and PD-1+HLA-DR+ CD8 and CD4 T cells increased. The increase in PD-1+HLA-DR+ T cells indicates T cell activation (Sauce et al., 2007; Vollbrecht et al., 2010) which associates with worse COVID-19 outcomes (Chen et al., 2020b). CXCR3 plays a key role in T cell homing to sites of inflammation and is activated by interferon-inducible ligands CXCL9, CXCL11, and CXCL10 (IP-10) (Groom and Luster, 2011; Rodda et al., 2021). A decrease in CXCR3 indicates either that T cells are less able to home to the site of infection, or that there is more inflammation in PLWH during SARS-CoV-2 infection and therefore more homing of the CXCR3+ CD8 T cells to tissues so that the fraction of CXCR3+ cells left in the blood decreases. Either way, the combination of these changes likely indicates either more pronounced SARS-CoV-2 infection or an impaired response in PLWH despite the similar infection outcomes in this wave.
In summary, PLWH showed increased disease severity mostly restricted to the second infection wave, where the Beta variant was dominant. Increased severity was associated with low CD4 T cell counts and high NLR which stabilized post-SARS-CoV-2 clearance in second wave infected PLWH to close to wave 1 PLWH values, arguing for a synergy between SARS-CoV-2 and HIV to decrease CD4 T cell numbers and increase the NLR rather than the status of HIV infection alone determining these parameters. More work is required to understand how these HIV related immune perturbations influence long-term immunity to SARS-CoV-2 infection and whether vaccine response will be affected.
Materials and methods
Ethical statement and study participants
The study protocol was approved by the University of KwaZulu-Natal Institutional Review Board (approval BREC/00001275/2020). Adult patients (>18 years old) presenting at King Edward VIII, Inkosi Albert Luthuli Central, or Clairwood Hospitals in Durban, South Africa, between June 2020 to May 2021, diagnosed to be SARS-CoV-2 positive as part of their clinical workup and able to provide informed consent were eligible for the study. Written informed consent was obtained for all enrolled participants.
Clinical laboratory testing
An HIV rapid test and viral load quantification was performed from a 4 ml EDTA tube of blood at an accredited diagnostic laboratory (Molecular Diagnostic Services, Durban, South Africa) using the RealTime HIV negative1 viral load test on an Abbott machine. CD4 count, CD8 count, and a full blood count panel were performed by an accredited diagnostic laboratory (Ampath, Durban, South Africa). Depending on the volume of blood which was drawn, the CD8, CD4, and full blood count was not available for every participant, and numbers performed are detailed in the figure legends.
qPCR detection of SARS-CoV-2
RNA was extracted from combined oropharyngeal and nasophryngeal swabs from 140 μl viral transport medium using the QIAamp Viral RNA Mini kit (cat. no. 52906, QIAGEN, Hilden, Germany) according to manufacturer’s instructions, and eluted into 100 μl AVE buffer. To detect SARS-CoV-2 RNA, 5 μl RNA was added to the TaqPath 1-step RT-qPCR mastermix. 3 SARS-CoV-2 genes (ORF1ab, S and N) were amplified using the TaqPath COVID-19 Combo Kit and TaqPath COVID-19 CE-IVD RT-PCR Kit (ThermoFisher Scientific, Massachusetts, United States) in a QuantStudio 7 Flex Real-Time PCR system (ThermoFisher Scientific). Data was analyzed using the Design and Analysis software (ThermoFisher Scientific). For positive samples, Ct values are represented as the average of the Ct values of all three genes. A sample was scored positive where at least two out of the three genes were detected, and inconclusive if only one of the genes was detected.
PBMC isolation and immune phenotyping by flow cytometry
PBMC were isolated by density gradient centrifugation using Histopaque 1077 (Sigma-Aldrich, St. Louis, Missouri, United States) and SepMate separation tubes (STEMCELL Technologies, Vancouver, Canada). For T cell and NK cell phenotyping, 106 fresh PBMCs were surface stained in 50 microliter antibody mix with the following antibodies from BD Biosciences (Franklin Lakes, NJ, USA): anti-CD45 Hv500 (1:100 dilution, clone HI30, cat. 560777); anti-CD8 BV395 (1:50 dilution, clone RPA-T8, cat. 563795); anti-CD4 BV496 (1:25 dilution, clone SK3, cat. 564651); anti-PD1 BV421 (1:50 dilution, clone EH12.1, cat. 562516); anti-CXCR3 PE-CF594 (1:25 dilution, clone 1C6/CXCR3, cat. 562451). The following antibodies were from BioLegend (San Diego, CA, USA): anti-CD19 Bv605 (1:100 dilution, clone HIB19, cat. 302244); anti-CD16 Bv650 (1:50 dilution, clone 3G8, cat. 302042); anti-CD56 Bv711 (1:50 dilution, clone HCD56, cat. 318336); anti-CD3 Bv785 (1:25 dilution, clone OKT3, cat. 317330); anti-CXCR5 FITC (1:25 dilution, clone J252D4, cat. 356914); anti-HLA-DR PE (1:50 dilution, clone L243, cat. 307606); anti-CCR7 PerCP-Cy5.5 (1:25 dilution, clone G043H7, cat. 353220); anti-CD38 PE-Cy7 (1:25 dilution, clone HIT2, cat. 303516); anti-ICOS APC (1:25 dilution, clone C398.4A, cat. 313510) and anti-CD45RA AF700 (1:25 dilution, clone HI100, cat. 304120). PBMCs were incubated with antibodies for 20 min at room temperature. For B-cell phenotyping, the following antibodies were used: (all from BioLegend) anti-CD45 APC (1:25 dilution, clone HI30, cat. 304012); anti-CD3 Bv711 (1:50 dilution, clone OKT3, cat. 317328), anti-CD14 Bv711 (1:25 dilution, clone M5E2, cat. 301838); anti-CD19 Bv605 (1:50 dilution, clone HIB19, cat. 302244); anti-CD27 Hv500 (1:50 dilution, clone O323, cat. 302836); anti-CD38 PE-Cy7 (1:25 dilution, clone HIT2, cat. 303516) and anti-CD138 BV785 (1:25 dilution, clone MI15, cat. 356538). Cells were then washed twice in PBS and fixed in 2% paraformaldehyde and stored at 4°C before acquisition on FACSAria Fusion III flow cytometer (BD) and analyzed with FlowJo software version 9.9.6 (Tree Star). Depending on the volume of blood which was drawn, full phenotyping was only available for participants where sufficient blood was available for the assay.
Statistical analysis
Data is described with the non-parametric measures of median and interquartile range, and significance determined using the non-parametric Mann-Whitney U test for pairwise comparisons, Fisher Exact test for pairwise comparisons of frequencies, and the Kruskal-Wallis test with multiple comparison correction by the Dunn Method for comparisons involved more than two populations. All tests were performed using Graphpad Prism eight or Stata software.
Acknowledgements
This work was supported by the Bill and Melinda Gates Investment INV-018944 to AS.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Alasdair Leslie, Email: Al.Leslie@ahri.org.
Henrik Kløverpris, Email: Henrik.Kloverpris@ahri.org.
Alex Sigal, Email: alex.sigal@ahri.org.
Miles P Davenport, University of New South Wales, Australia.
Lishomwa Ndhlovu, Weill Cornell Medical College, United States.
COMMIT-KZN Team:
Moherndran Archary, Kaylesh J Dullabh, Jennifer Giandhari, Philip Goulder, Guy Harling, Rohen Harrichandparsad, Kobus Herbst, Prakash Jeena, Thandeka Khoza, Nigel Klein, Rajhmun Madansein, Mohlopheni Marakalala, Mosa Moshabela, Kogie Naidoo, Zaza Ndhlovu, Kennedy Nyamande, Nesri Padayatchi, Vinod Patel, Theresa Smit, and Adrie Steyn
Funding Information
This paper was supported by the following grant:
Bill and Melinda Gates Foundation INV-018944 to Alex Sigal.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Formal analysis, Project administration.
Formal analysis, Methodology.
Data curation, Formal analysis.
Data curation.
Data curation, Formal analysis.
Data curation, Formal analysis.
Project administration.
Project administration.
Investigation.
Investigation.
Resources.
Data curation.
Project administration.
Methodology.
Resources.
Resources.
Writing - review and editing.
Writing - review and editing.
Conceptualization.
Resources.
Resources, Supervision.
Formal analysis.
Formal analysis, Supervision.
Formal analysis, Supervision, Investigation.
Conceptualization, Funding acquisition, Writing - review and editing.
Ethics
Human subjects: The study protocol was approved by the University of KwaZulu-Natal Institutional Review Board (approval BREC/00001275/2020). Adult patients (>18 years old) presenting either at King Edward VIII, Inkosi Albert Luthuli Central or Clairwood Hospitals in Durban, South Africa, between June 2020 to May 2021, diagnosed to be SARS-CoV-2 positive as part of their clinical workup and able to provide informed consent were eligible for the study. Written informed consent was obtained for all enrolled participants.
Additional files
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files.
References
- Ambrosioni J, Blanco JL, Reyes-Urueña JM, Davies MA, Sued O, Marcos MA, Martínez E, Bertagnolio S, Alcamí J, Miro JM, Investigators C-i, COVID-19 in HIV Investigators Overview of SARS-CoV-2 infection in adults living with HIV. The Lancet HIV. 2021;8:e294–e305. doi: 10.1016/S2352-3018(21)00070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelino-Silva VI, Miyaji KT, Mathias A, Costa DA, de Carvalho Dias JZ, Lima SB, Simoes M, Freire MS, Caiaffa-Filho HH, Hong MA, Lopes MH, Sartori AM, Kallas EG. CD4/CD8 ratio predicts yellow fever Vaccine-Induced antibody titers in Virologically suppressed HIV-Infected patients. JAIDS Journal of Acquired Immune Deficiency Syndromes. 2016;71:189–195. doi: 10.1097/QAI.0000000000000845. [DOI] [PubMed] [Google Scholar]
- Bhaskaran K, Rentsch CT, MacKenna B, Schultze A, Mehrkar A, Bates CJ, Eggo RM, Morton CE, Bacon SCJ, Inglesby P, Douglas IJ, Walker AJ, McDonald HI, Cockburn J, Williamson EJ, Evans D, Forbes HJ, Curtis HJ, Hulme WJ, Parry J, Hester F, Harper S, Evans SJW, Smeeth L, Goldacre B. HIV infection and COVID-19 death: a population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. The Lancet HIV. 2021;8:e24–e32. doi: 10.1016/S2352-3018(20)30305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein SL, Lazar R, Wahnich A, Daskalakis DC, Blackstock OJ. Coronavirus Disease 2019 (COVID-19) Infection Among People With Human Immunodeficiency Virus in New York City: A Population-Level Analysis of Linked Surveillance Data. Clinical Infectious Diseases. 2021;72:e1021–e1029. doi: 10.1093/cid/ciaa1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson PJ, Schut RL, Simpson ML, O'Brien J, Janoff EN. Antibody class and subclass responses to pneumococcal polysaccharides following immunization of human immunodeficiency virus-infected patients. Journal of Infectious Diseases. 1995;172:340–345. doi: 10.1093/infdis/172.2.340. [DOI] [PubMed] [Google Scholar]
- Cele S, Gazy I, Jackson L, Hwa SH, Tegally H, Lustig G, Giandhari J, Pillay S, Wilkinson E, Naidoo Y, Karim F, Ganga Y, Khan K, Bernstein M, Balazs AB, Gosnell BI, Hanekom W, Moosa MS, Lessells RJ, de Oliveira T, Sigal A, Network for Genomic Surveillance in South Africa. COMMIT-KZN Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, Zhang X, Zhang M, Wu S, Song J, Chen T, Han M, Li S, Luo X, Zhao J, Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. Journal of Clinical Investigation. 2020a;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, John Wherry E, Wherry EJ. T cell responses in patients with COVID-19. Nature Reviews Immunology. 2020b;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C, Thorne A, Klein M, Conway B, Boivin G, Haase D, Shafran S, Zubyk W, Singer J, Halperin S, Walmsley S, CIHR Canadian HIV Trials Network Influenza Vaccine Research Group Immunogenicity is not improved by increased antigen dose or booster dosing of seasonal influenza vaccine in a randomized trial of HIV infected adults. PLOS ONE. 2011;6:e17758. doi: 10.1371/journal.pone.0017758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Dandachi D, Geiger G, Montgomery MW, Karmen-Tuohy S, Golzy M, Antar AAR, Llibre JM, Camazine M, Díaz-De Santiago A, Carlucci PM, Zacharioudakis IM, Rahimian J, Wanjalla CN, Slim J, Arinze F, Kratz AMP, Jones JL, Patel SM, Kitchell E, Francis A, Ray M, Koren DE, Baddley JW, Hill B, Sax PE, Chow J. Characteristics, Comorbidities, and outcomes in a multicenter registry of patients with human immunodeficiency virus and coronavirus disease 2019. Clinical Infectious Diseases. 2021;73:e1964–e1972. doi: 10.1093/cid/ciaa1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- Deeks SG, Kitchen CM, Liu L, Guo H, Gascon R, Narváez AB, Hunt P, Martin JN, Kahn JO, Levy J, McGrath MS, Hecht FM. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, Hebbeler AM, Greene WC. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Muñoz-Arias I, Greene WC. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Ambrosino DM, Fiore-Gartland A, Goldblatt D, Gilbert PB, Siber GR, Dull P, Plotkin SA. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federation ID. Idf Diabetes Atlas. 2019 https://www.diabetesatlas.org/en/
- Fuster F, Vargas JI, Jensen D, Sarmiento V, Acuña P, Peirano F, Fuster F, Arab JP, Martínez F, Soto S, Core-HIV Study Group CD4/CD8 ratio as a predictor of the response to HBV vaccination in HIV-positive patients: A prospective cohort study. Vaccine. 2016;34:1889–1895. doi: 10.1016/j.vaccine.2016.02.055. [DOI] [PubMed] [Google Scholar]
- Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2523. doi: 10.1016/j.cell.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geretti AM, Stockdale AJ, Kelly SH, Cevik M, Collins S, Waters L, Villa G, Docherty A, Harrison EM, Turtle L, Openshaw PJM, Baillie JK, Sabin CA, Semple MG. Outcomes of coronavirus disease 2019 (COVID-19) Related hospitalization among people with human immunodeficiency virus (HIV) in the ISARIC world health organization (WHO) Clinical characterization protocol (UK): A prospective observational study. Clinical Infectious Diseases. 2021;73:e2095–e2106. doi: 10.1093/cid/ciaa1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom JR, Luster AD. CXCR3 in T cell function. Experimental Cell Research. 2011;317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen KY, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang J, Liang Z, Peng Y, Wei L, Liu Y, Hu Y, Peng P, Wang J, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N, Ou C, He J, Lei C. Clinical characteristics of coronavirus disease 2019 in China. New England Journal of Medicine. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K, Barrett BS, Mickens KL, Hasenkrug KJ, Santiago ML. Interferon resistance of emerging sars-cov-2 variants. bioRxiv. 2021 doi: 10.1101/2021.03.20.436257. [DOI] [PMC free article] [PubMed]
- Hadi YB, Naqvi SFZ, Kupec JT, Sarwari AR. Characteristics and outcomes of COVID-19 in patients with HIV: a multicentre research network study. AIDS. 2020;34:F3–F8. doi: 10.1097/QAD.0000000000002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härter G, Spinner CD, Roider J, Bickel M, Krznaric I, Grunwald S, Schabaz F, Gillor D, Postel N, Mueller MC, Müller M, Römer K, Schewe K, Hoffmann C. COVID-19 in people living with human immunodeficiency virus: a case series of 33 patients. Infection. 2020;48:681–686. doi: 10.1007/s15010-020-01438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Casado JL, Härter G, Vizcarra P, Moreno A, Cattaneo D, Meraviglia P, Spinner CD, Schabaz F, Grunwald S, Gervasoni C. Immune deficiency is a risk factor for severe COVID-19 in people living with HIV. HIV Medicine. 2021a;22:372–378. doi: 10.1111/hiv.13037. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS, Schulz S, Jäck HM, Jahrsdörfer B, Schrezenmeier H, Müller M, Kleger A, Münch J, Pöhlmann S, Groß R, Hörnich BF, Krüger N, Müller M, Münch J, Pöhlmann S. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021b;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Xie N, Hu X, Yan H, Ding J, Liu P, Ma H, Ruan L, Li G, He N, Wei S, Wang X. Epidemiological, virological and serological features of coronavirus disease 2019 (COVID-19) Cases in people living with human immunodeficiency virus in Wuhan: a Population-based cohort study. Clinical Infectious Diseases. 2021;73:e2086–e2094. doi: 10.1093/cid/ciaa1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inciarte A, Gonzalez-Cordon A, Rojas J, Torres B, de Lazzari E, de la Mora L, Martinez-Rebollar M, Laguno M, Callau P, Gonzalez-Navarro A, Leal L, Garcia F, Mallolas J, Mosquera M, Marcos MA, Ambrosioni J, Miro JM, Martinez E, Blanco JL, (on behalf the COVID-19 in HIV Investigators) Clinical characteristics, risk factors, and incidence of symptomatic coronavirus disease 2019 in a large cohort of adults living with HIV: a single-center, prospective observational study. AIDS. 2020;34:1775–1780. doi: 10.1097/QAD.0000000000002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L, Hunter J, Cele S, Ferreira IM, Young AC, Karim F, Madansein R, Dullabh KJ, Chen CY, Buckels NJ, Ganga Y, Khan K, Boulle M, Lustig G, Neher RA, Sigal A. Incomplete inhibition of HIV infection results in more HIV infected lymph node cells by reducing cell death. eLife. 2018;7:e30134. doi: 10.7554/eLife.30134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassat W, Cohen C, Tempia S, Masha M, Goldstein S, Kufa T, Murangandi P, Savulescu D, Walaza S, Bam JL, Davies MA, Prozesky HW, Naude J, Mnguni AT, Lawrence CA, Mathema HT, Zamparini J, Black J, Mehta R, Parker A, Chikobvu P, Dawood H, Muvhango N, Strydom R, Adelekan T, Mdlovu B, Moodley N, Namavhandu EL, Rheeder P, Venturas J, Magula N, Blumberg L, DATCOV author group Risk factors for COVID-19-related in-hospital mortality in a high HIV and tuberculosis prevalence setting in South Africa: a cohort study. The Lancet HIV. 2021a;8:e554–e567. doi: 10.1016/S2352-3018(21)00151-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassat W, Mudara C, Ozougwu L, Tempia S, Blumberg L, Davies MA, Pillay Y, Carter T, Morewane R, Wolmarans M, von Gottberg A, Bhiman JN, Walaza S, Cohen C, DATCOV author group Difference in mortality among individuals admitted to hospital with COVID-19 during the first and second waves in South Africa: a cohort study. The Lancet Global Health. 2021b;9:e1216–e1225. doi: 10.1016/S2214-109X(21)00289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F, Moosa M, Gosnell B, Cele S, Giandhari J, Pillay S, Tegally H, Wilkinson E, San J, Msomi N, Mlisana K, Khan K, Bernstein M, Manickchund N, Singh L, Ramphal U, Hanekom W, Lessells R. Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv. 2021 doi: 10.1101/2021.06.03.21258228. [DOI]
- Karmen-Tuohy S, Carlucci PM, Zervou FN, Zacharioudakis IM, Rebick G, Klein E, Reich J, Jones S, Rahimian J. Outcomes among HIV-Positive patients hospitalized with COVID-19. JAIDS Journal of Acquired Immune Deficiency Syndromes. 2020;85:6–10. doi: 10.1097/QAI.0000000000002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharsany ABM, Cawood C, Khanyile D, Lewis L, Grobler A, Puren A, Govender K, George G, Beckett S, Samsunder N, Madurai S, Toledo C, Chipeta Z, Glenshaw M, Hersey S, Abdool Karim Q. Community-based HIV prevalence in KwaZulu-Natal, South Africa: results of a cross-sectional household survey. The Lancet HIV. 2018;5:e427–e437. doi: 10.1016/S2352-3018(18)30104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadadi L, Cheng Q, Radbruch A, Hiepe F. The maintenance of memory plasma cells. Frontiers in Immunology. 2019;10:721. doi: 10.3389/fimmu.2019.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, Davenport MP. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nature Medicine. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nature Medicine. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Liu J, Li S, Liu J, Liang B, Wang X, Wang H, Li W, Tong Q, Yi J, Zhao L, Xiong L, Guo C, Tian J, Luo J, Yao J, Pang R, Shen H, Peng C, Liu T, Zhang Q, Wu J, Xu L, Lu S, Wang B, Weng Z, Han C, Zhu H, Zhou R, Zhou H, Chen X, Ye P, Zhu B, Wang L, Zhou W, He S, He Y, Jie S, Wei P, Zhang J, Lu Y, Wang W, Zhang L, Li L, Zhou F, Wang J, Dittmer U, Lu M, Hu Y, Yang D, Zheng X. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020a;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du X, Chen J, Jin Y, Peng L, Wang HHX, Luo M, Chen L, Zhao Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. Journal of Infection. 2020b;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A, Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, Padayachee SD, Dheda K, Barnabas SL, Bhorat QE, Briner C, Kwatra G, Ahmed K, Aley P, Bhikha S, Bhiman JN, Bhorat AE, du Plessis J, Esmail A, Groenewald M, Horne E, Hwa SH, Jose A, Lambe T, Laubscher M, Malahleha M, Masenya M, Masilela M, McKenzie S, Molapo K, Moultrie A, Oelofse S, Patel F, Pillay S, Rhead S, Rodel H, Rossouw L, Taoushanis C, Tegally H, Thombrayil A, van Eck S, Wibmer CK, Durham NM, Kelly EJ, Villafana TL, Gilbert S, Pollard AJ, de Oliveira T, Moore PL, Sigal A, Izu A, NGS-SA Group, Wits-VIDA COVID Group Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the b.1.351 variant. New England Journal of Medicine. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaza A, Mossong J, Bärnighausen T, Newell ML. Hypertension and obesity in adults living in a high HIV prevalence rural area in South Africa. PLOS ONE. 2012;7:e47761. doi: 10.1371/journal.pone.0047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, Weiskopf D. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D'Andrea K, Manne S, Chen Z, Huang YJ, Reilly JP, Weisman AR, Ittner CAG, Kuthuru O, Dougherty J, Nzingha K, Han N, Kim J, Pattekar A, Goodwin EC, Anderson EM, Weirick ME, Gouma S, Arevalo CP, Bolton MJ, Chen F, Lacey SF, Ramage H, Cherry S, Hensley SE, Apostolidis SA, Huang AC, Vella LA, Betts MR, Meyer NJ, Wherry EJ, UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavigner M, Cazabat M, Dubois M, L'Faqihi FE, Requena M, Pasquier C, Klopp P, Amar J, Alric L, Barange K, Vinel JP, Marchou B, Massip P, Izopet J, Delobel P. Altered CD4+ T cell homing to the gut impairs mucosal immune reconstitution in treated HIV-infected individuals. Journal of Clinical Investigation. 2012;122:62–69. doi: 10.1172/JCI59011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir S, Fauci AS. Insights into B cells and HIV-specific B-cell responses in HIV-infected individuals. Immunological Reviews. 2013;254:207–224. doi: 10.1111/imr.12067. [DOI] [PubMed] [Google Scholar]
- Okoye AA, Picker LJ. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunological Reviews. 2013;254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallikkuth S, Parmigiani A, Silva SY, George VK, Fischl M, Pahwa R, Pahwa S. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 2012;120:985–993. doi: 10.1182/blood-2011-12-396648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau M, Savoye A-L, De Crignis E, Corpataux J-M, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. Journal of Experimental Medicine. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan BD, Mou H, Zhang L, Guo Y, He W, Ojha A, Parcells MS, Luo G, Li W, Zhong G, Choe H, Farzan M. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv. 2020 doi: 10.1101/2020.04.10.036418. [DOI]
- Rajah MM, Hubert M, Bishop E, Saunders N, Robinot R, Grzelak L, Planas D, Dufloo J, Gellenoncourt S, Bongers A, Zivaljic M, Planchais C, Guivel‐Benhassine F, Porrot F, Mouquet H, Chakrabarti LA, Buchrieser J, Schwartz O. SARS-CoV-2 Alpha, Beta and Delta variants display enhanced Spike‐mediated syncytia formation. The EMBO Journal. 2021;10:e108944. doi: 10.15252/embj.2021108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, Hägglöf T, Oliveira TY, Viant C, Hurley A, Hoffmann HH, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP, Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, Nussenzweig MC. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, Takehara KK, Eggenberger J, Hemann EA, Waterman HR, Fahning ML, Chen Y, Hale M, Rathe J, Stokes C, Wrenn S, Fiala B, Carter L, Hamerman JA, King NP, Gale M, Campbell DJ, Rawlings DJ, Pepper M. Functional SARS-CoV-2-Specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, Crotty S. Antigen-Specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryscavage P, Kelly S, Li JZ, Harrigan PR, Taiwo B. Significance and clinical management of persistent low-level viremia and very-low-level viremia in HIV-1-infected patients. Antimicrobial Agents and Chemotherapy. 2014;58:3585–3598. doi: 10.1128/AAC.00076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz I, Wei C, Jenks SA, Cashman KS, Tipton C, Woodruff MC, Hom J, Lee FE. Challenges and opportunities for consistent classification of human B cell and plasma cell populations. Frontiers in Immunology. 2019;10:2458. doi: 10.3389/fimmu.2019.02458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauce D, Almeida JR, Larsen M, Haro L, Autran B, Freeman GJ, Appay V. PD-1 expression on human CD8 T cells depends on both state of differentiation and activation status. AIDS. 2007;21:2005–2013. doi: 10.1097/QAD.0b013e3282eee548. [DOI] [PubMed] [Google Scholar]
- Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A, Llewellyn-Lacey S, Kamal H, Bogdanovic G, Muschiol S, Wullimann DJ, Kammann T, Emgård J, Parrot T, Folkesson E, Rooyackers O, Eriksson LI, Henter JI, Sönnerborg A, Allander T, Albert J, Nielsen M, Klingström J, Gredmark-Russ S, Björkström NK, Sandberg JK, Price DA, Ljunggren HG, Aleman S, Buggert M, Karolinska COVID-19 Study Group Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev N, Scherer M, LaSota ED, Antoniou P, Yin MT, Zucker J, Sobieszczyk ME. Clinical characteristics and outcomes in people living with human immunodeficiency virus hospitalized for coronavirus disease 2019. Clinical Infectious Diseases. 2020;71:2294–2297. doi: 10.1093/cid/ciaa635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V, Bhikha S, Hoosain Z, Archary M, Bhorat Q, Fairlie L, Lalloo U, Masilela MSL, Moodley D, Hanley S, Fouche L, Louw C, Tameris M, Singh N, Goga A, Dheda K, Grobbelaar C, Kruger G, Carrim-Ganey N, Baillie V, de Oliveira T, Lombard Koen A, Lombaard JJ, Mngqibisa R, Bhorat A, Benadé G, Lalloo N, Pitsi A, Vollgraaff P-L, Luabeya A, Esmail A, Petrick FG, Oommen-Jose A, Foulkes S, Ahmed K, Thombrayil A, Fries L, Cloney-Clark S, Zhu M, Bennett C, Albert G, Faust E, Plested JS, Robertson A, Neal S, Cho I, Glenn GM, Dubovsky F, Madhi SA. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. New England Journal of Medicine. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigel K, Swartz T, Golden E, Paranjpe I, Somani S, Richter F, De Freitas JK, Miotto R, Zhao S, Polak P, Mutetwa T, Factor S, Mehandru S, Mullen M, Cossarini F, Bottinger E, Fayad Z, Merad M, Gnjatic S, Aberg J, Charney A, Nadkarni G, Glicksberg BS. Coronavirus 2019 and people living with human immunodeficiency virus: outcomes for hospitalized patients in New York city. Clinical Infectious Diseases. 2020;71:2933–2938. doi: 10.1093/cid/ciaa880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle K, Johnston CD, Jannat-Khah DP, Williams SC, Ellman TM, Vogler MA, Gulick RM, Glesby MJ, Choi JJ. In: Open Forum Infectious Diseases. Ellman T. M, editor. Oxford University Press; 2020. Covid-19 in hospitalized adults with hiv; pp. 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao NY, Korsman S, Davies MA, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021a;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Tegally H, Wilkinson E, Lessells RJ, Giandhari J, Pillay S, Msomi N, Mlisana K, Bhiman JN, von Gottberg A, Walaza S, Fonseca V, Allam M, Ismail A, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Williamson C, Petruccione F, Sigal A, Gazy I, Hardie D, Hsiao NY, Martin D, York D, Goedhals D, San EJ, Giovanetti M, Lourenço J, Alcantara LCJ, de Oliveira T. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nature Medicine. 2021b;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- Tesoriero JM, Swain CE, Pierce JL, Zamboni L, Wu M, Holtgrave DR, Gonzalez CJ, Udo T, Morne JE, Hart-Malloy R, Rajulu DT, Leung SJ, Rosenberg ES. COVID-19 outcomes among persons living with or without diagnosed HIV infection in New York state. JAMA Network Open. 2021;4:e2037069. doi: 10.1001/jamanetworkopen.2020.37069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- the Northwell COVID-19 Research Consortium. Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, Barnaby DP, Becker LB, Chelico JD, Cohen SL, Cookingham J, Coppa K, Diefenbach MA, Dominello AJ, Duer-Hefele J, Falzon L, Gitlin J, Hajizadeh N, Harvin TG, Hirschwerk DA, Kim EJ, Kozel ZM, Marrast LM, Mogavero JN, Osorio GA, Qiu M, Zanos TP. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne LG, Bouhaddou M, Reuschl AK, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MV, Ummadi M, Rojc A, Turner J, Obernier K, Braberg H, Soucheray M, Richards A, Chen KH, Harjai B, Memon D, Hosmillo M, Hiatt J, Jahun A, Goodfellow IG, Fabius JM, Shokat K, Jura N, Verba K, Noursadeghi M, Beltrao P, Swaney DL, Garcia-Sastre A, Jolly C, Towers GJ, Krogan NJ. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv. 2021 doi: 10.1101/2021.06.06.446826. [DOI]
- van Heerden A, Barnabas RV, Norris SA, Micklesfield LK, van Rooyen H, Celum C. High prevalence of HIV and non-communicable disease (NCD) risk factors in rural KwaZulu-Natal, South Africa. Journal of the International AIDS Society. 2017;20:e25012. doi: 10.1002/jia2.25012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra P, Pérez-Elías MJ, Quereda C, Moreno A, Vivancos MJ, Dronda F, Casado JL, COVID-19 ID Team Description of COVID-19 in HIV-infected individuals: a single-centre, prospective cohort. The Lancet HIV. 2020;7:e554–e564. doi: 10.1016/S2352-3018(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht T, Brackmann H, Henrich N, Roeling J, Seybold U, Bogner JR, Goebel FD, Draenert R. Impact of changes in antigen level on CD38/PD-1 co-expression on HIV-specific CD8 T cells in chronic, untreated HIV-1 infection. Journal of Medical Virology. 2010;82:358–370. doi: 10.1002/jmv.21723. [DOI] [PubMed] [Google Scholar]
- Westendorp MO, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin KM, Krammer PH. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature. 1995;375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- Western Cape Department of Health in collaboration with the National Institute for Communicable Diseases, South Africa. Boulle A, Davies MA, Hussey H, Ismail M, Morden E, Vundle Z, Zweigenthal V, Mahomed H, Paleker M, Pienaar D, Tembo Y, Lawrence C, Isaacs W, Mathema H, Allen D, Allie T, Bam JL, Buddiga K, Dane P, Heekes A, Matlapeng B, Mutemaringa T, Muzarabani L, Phelanyane F, Pienaar R, Rode C, Smith M, Tiffin N, Zinyakatira N, Cragg C, Marais F, Mudaly V, Voget J, Davids J, Roodt F, van Zyl Smit N, Vermeulen A, Adams K, Audley G, Bateman K, Beckwith P, Bernon M, Blom D, Boloko L, Botha J, Boutall A, Burmeister S, Cairncross L, Calligaro G, Coccia C, Corin C, Daroowala R, Dave JA, De Bruyn E, De Villiers M, Deetlefs M, Dlamini S, Du Toit T, Endres W, Europa T, Fieggan G, Figaji A, Frankenfeld P, Gatley E, Gina P, Govender E, Grobler R, Gule M, Hanekom C, Held M, Heynes A, Hlatswayo S, Hodkinson B, Holtzhausen J, Hoosain S, Jacobs A, Kahn M, Kahn T, Khamajeet A, Khan J, Khan R, Khwitshana A, Knight L, Kooverjee S, Krogscheepers R, Jacque Kruger J, Kuhn S, Laubscher K, Lazarus J, Le Roux J, Lee Jones S, Levin D, Maartens G, Majola T, Manganyi R, Marais D, Marais S, Maritz F, Maughan D, Mazondwa S. Risk factors for coronavirus disease 2019 (COVID-19) Death in a population cohort study from the western cape province, South Africa. Clinical Infectious Diseases . 2021;73:e2005–e2015. doi: 10.1093/cid/ciaa1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, de Oliveira T, Vermeulen M, van der Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nature Medicine. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, Ley AM, Kyu S, Howell JC, Ozturk T, Lee S, Suryadevara N, Case JB, Bugrovsky R, Chen W, Estrada J, Morrison-Porter A, Derrico A, Anam FA, Sharma M, Wu HM, Le SN, Jenks SA, Tipton CM, Staitieh B, Daiss JL, Ghosn E, Diamond MS, Carnahan RH, Crowe JE, Hu WT, Lee FE, Sanz I. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nature Immunology. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhou X, Zhu C, Song Y, Feng F, Qiu Y, Feng J, Jia Q, Song Q, Zhu B, Wang J. Immune Phenotyping Based on the Neutrophil-to-Lymphocyte Ratio and IgG Level Predicts Disease Severity and Outcome for Patients With COVID-19. Frontiers in Molecular Biosciences. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]