Abstract
In this study, silver nanoparticles (AgNPs) were biosynthesised by using acidophilic actinobacterial SH11 strain isolated from pine forest soil. Isolate SH11 was identified based on 16S rRNA gene sequence to Streptomyces kasugaensis M338‐M1T and S. celluloflavus NRRL B‐2493T (99.8% similarity, both). Biosynthesised AgNPs were analysed by UV–visible spectroscopy, which revealed specific peak at λ = 420 nm. Transmission electron microscopy analyses showed polydispersed, spherical nanoparticles with a mean size of 13.2 nm, while Fourier transform infrared spectroscopy confirmed the presence of proteins as the capping agents over the surface of AgNPs. The zeta potential was found to be −16.6 mV, which indicated stability of AgNPs. The antibacterial activity of AgNPs from SH11 strain against gram‐positive (Staphylococcus aureus and Bacillus subtilis) and gram‐negative (Escherichia coli) bacteria was estimated using disc diffusion, minimum inhibitory concentration and live/dead analyses. The AgNPs showed the maximum antimicrobial activity against E. coli, followed by B. subtilis and S. aureus. Further, the synergistic effect of AgNPs in combination with commercial antibiotics (kanamycin, ampicillin, tetracycline) was also evaluated against bacterial isolates. The antimicrobial efficacy of antibiotics was found to be enhanced in the presence of AgNPs.
Inspec keywords: antibacterial activity, silver, nanoparticles, electrokinetic effects, Fourier transform infrared spectra, microorganisms, nanofabrication
Other keywords: actinobacterial mediated synthesis, silver nanoparticles, pathogenic bacteria, biosynthesis, acidophilic actinobacterial SH11 strain, pine forest soil, 16S rRNA gene sequence, Streptomyces kasugaensis M338‐M1T, S. celluloflavus NRRL B‐2493T, UV–visible spectroscopy, transmission electron microscopy, Fourier transform infrared spectroscopy, zeta potential, gram positive bacteria, Staphylococcus aureus, Bacillus subtilis, gram negative bacteria, Escherichia coli, disc diffusion, wavelength 420 nm, Ag
1 Introduction
Resistance of clinical pathogens to commercial antibiotics is increasing problem, which is intensified by a lack of new therapeutic agents [1]. Hence, there is search for novel antimicrobial compounds by many researchers and pharmaceutical companies [2]. Silver nanoparticles (AgNPs) provide an alternative to commercial antibiotics by developing novel applications in the pharmaceuticals and medicine [3].
Nanoscience and nanotechnology are emerging fields, which involve the synthesis and application of nanoscale materials and structures usually in the range of 1–100 nm [4]. One of the most important aspects of nanotechnology is synthesis of metal nanoparticles. The chemical procedures for synthesis of AgNPs are expensive and use toxic solvents extensively [5, 6]. Unlike chemical procedures, the biological systems for synthesis of AgNPs such as bacteria, fungi and plants are cheap, easy and eco‐friendly [7, 8, 9, 10].
Microorganisms play an important role in remediation of toxic metals through reduction of metal ions and consequently has been considered as nanofactories [11]. One of the most promising organisms for synthesis of metal nanoparticles is actinobacteria, which is known as main producer of diverse secondary metabolites, such as antibiotics and many others biologically active compounds [6]. Since the last decade these organisms have being intensively isolated from extreme environments and screened for novel bioactive compounds [12, 13]. However, the synthesis of AgNPs by actinobacteria from extreme habitats such as acidophilic soils (pH < 4.0) has been not well studied.
Currently, the AgNPs are explored and extensively investigated as potential antimicrobials [14, 15, 16]. The small size and the high surface area to volume ratio of nanoparticles enhance their interaction with the microbial cells to carry out a broad range of potential antimicrobial activities [17]. AgNPs have been found to be active against gram‐positive as well as gram‐negative bacteria, including multidrug resistant strains as well as biofilm formation [18, 19, 20].
Moreover, some authors have studied the synergism of AgNPs in combination with antibiotics such as amoxicillin and polymyxin B and found higher bactericidal activity by binding of AgNPs with antibiotics [21, 22, 23, 24].
The aim of this study was to synthesise AgNPs using acidophilic actinobacterium and to evaluate their activity against human pathogenic bacteria.
2 Materials and methods
2.1 Isolation of actinobacterial strain
The acidophilic actinobacterial SH11 strain was isolated from humic (H) layer of pine forest soil collected from southern slope of inland dune, near Torun, Poland (52°55′ 37″N, 18° 42′11″E) by the dilution plate procedure on the acidified starch–casein agar [25] as described previously by Golinska et al. [26]. The isolate was maintained on acidified yeast extract–malt extract (ISP2) agar slants [27] at room temperature and as suspension of spores and mycelial fragments in 20% glycerol (v/v) at −80 °C.
2.2 Identification and phylogenetic analysis of actinobacterial SH11 strain
Biomass for the molecular study was prepared by growing of SH11 strain in yeast extract–malt extract broth (ISP2; pH 5.5) at 28 °C for 2 weeks. Cells were harvested by centrifugation and washed twice in distilled water. The extraction of genomic DNA and amplification of 16S rRNA gene from SH11 strain were carried out according to the method described by Golinska et al. [28]. The amplified product was purified by GenEluateTM PCR Clean‐Up Kit (Sigma) and sent for sequencing at the Institute of Biochemistry and Biophysics Polish Academy of Sciences, Warsaw, Poland. The closest phylogenetic neighbours of isolate SH11 based on 16S rRNA gene sequences were found using the EzTaxon server (http://eztaxon‐e.ezbiocloud.net/) [29]. The resultant gene sequences were aligned with corresponding sequences of the type strains of the genera Streptomyces using ClustalW. Phylogenetic analyses were carried out using MEGA6 software [30]. The phylogenetic tree was inferred by using the neighbour‐joining [31], the maximum‐likelihood [32] and maximum‐parsimony [33] tree‐making algorithms. Further, the tree topology was evaluated by a bootstrap analysis [32] of the neighbour‐joining data based on 1000 resampling using MEGA6 software [30]. The root position of the unrooted tree was estimated using the 16S rRNA gene sequence of Streptomyces albus subsp. albus DSM 40313T (GenBank accession number AJ621602).
2.3 Actinobacterial synthesis of AgNPs
For synthesis of AgNPs, the 100 ml of ISP2 broth (pH 5.5) in 250 ml Erlenmeyer flask was inoculated with SH11 strain and incubated at 28 °C in the orbital shaker (150 rpm) for 14 days. The actinobacterial biomass was then centrifuged at 6000g for 15 min and washed three times with sterile distilled water. Subsequently, the cell pellet was suspended in 100 ml sterilised distilled water and incubated at 28 °C for 24 h. After that the cell biomas was separated by centrifugation at 6000g for 20 min, and obtained supernatant was challenged with AgNO3 (1 mM final concentration) and kept at room temperature for 2–3 days. The supernatant without AgNO3 was used as a control.
2.4 Characterisation of AgNPs
2.4.1 Visual detection and spectroscopy analysis
The biosynthesis of Ag+ ions into AgNPs in the reaction solution was observed by colour change from light‐yellow to dark‐brown. The presence of biosynthesised AgNPs was confirmed by UV–visible spectroscopy analysis in the wavelength range of 200–800 nm using Nano Drop ND 2000 (Thermo Scientific, USA).
To determine the biomolecules responsible for the reduction of silver ions and stabilisation of AgNPs in the solution, the Fourier transform infrared spectroscopy (FTIR) analysis was carried out. For FTIR analysis the powder of biosynthesised AgNPs were combined with dry KBr in the ratio of 1:100. The FTIR spectrum of sample was recorded by FTIR instrument (Spectrum 2000, Perkin‐Elmer, USA). The measurements were carried out in the range of 4000–400 cm−1 at a resolution of 4 cm−1.
2.4.2 Transmission electron microscopy (TEM) analysis
The TEM analysis was performed to determine the size and morphology of biosynthesised AgNPs by using FEI Tecnai F20 X‐Twintool Electron Microscope (USA) operated at an accelerating voltage of 100 kV. For analysis the biosynthesised AgNPs solution was dropped on copper grids coated with carbon (400 µm mesh size) and kept at room temperature to dry. TEM measurements were then performed and the obtained data were evaluated by Statistica Software (StatSoft, USA).
2.4.3 Zeta potential analysis
To determine the stability of AgNPs, the zeta potential analysis was conducted by using the Zetasizer Nano ZS 90 (Malvern Instrument Ltd, UK). For the analysis the 25 µl of AgNPs solution was diluted ten times and sonicated for 15 min at 20 Hz. Then sample was filtered through of 0.22 µm millipore filter and used for study.
2.5 Determination of minimum inhibitory concentration (MIC) of antibiotics against tested bacteria
The MIC is defined as the lowest concentration of chemicals that inhibits the growth of the organism. The MIC assay of antibiotics (ampicillin, kanamycin or tetracycline) against gram‐positive (Staphylococcus aureus ATCC6338 and Bacillus subtilis PCM2021) and gram‐negative (Escherichia coli ATCC8739) bacteria was performed using eTest strips (Biomerieux) in the range of 0.016–256 µg/ml of antibiotic. Briefly, 100 µl of bacterial inoculum (1 × 106 CFU/ml) was spread on to the surface of the tripticase soy agar (Becton Dickinson) medium with sterile spreader. Subsequently, eTest stripe was placed onto the surface of inoculated medium and incubated for 24 h at 37 °C. The assay was performed in triplicate.
2.6 Determination of the MIC and minimum bactericidal concentration (MBC) of AgNPs
The synthesised AgNPs were tested (in triplicate) for determination of MIC by microtiter broth dilution method. Trypticase soy broth (TSB, Becton Dickinson) was used as diluents for bacterial strains. The final concentration of bacteria in each well was 1 × 106 CFU/ml, and various concentrations (from 5.0 to 200 µg/ml) of AgNPs were used. The positive and negative controls were also maintained. The microtiter plates were incubated at 37 °C for 24 h and then read at 450 nm using BIOLOG multimode reader to determine the MIC and MBC values. The MIC was defined as the lowest concentration of AgNPs that inhibited the growth of microorganisms, while the MBC end point was defined as the lowest concentration of AgNPs that killed ≥90% of bacterial cells.
2.7 Assessment of antibacterial activity of AgNPs individually and in combination with antibiotics
The MIC values of AgNPs from actinobacterial strain SH11 and commercial antibiotic were used to estimate accurate synergistic effect of AgNPs on antibiotic (kanamycin, ampicillin and tetracycline) activity. The assay was performed using 96‐well culture plates in triplicate. The final concentration of bacteria in each well was 1 × 106 CFU/ml. The TSB (Becton Dickinson) was used as diluent for bacterial strains, antibiotics and AgNPs. The positive and negative controls were maintained. The microtiter plates were read at 450 nm on multimode reader (BIOLOG, USA) after incubation at 37 °C for 24 h to determine the bacterial growth inhibition (%).
2.8 Bacterial viability assay after treatment with biosynthesised AgNPs
The viability assays of bacterial pathogens after treatment with AgNPs were carried out by using Live/Dead baclight kit (Invitrogen). To determine viability of bacterial cells, 1 ml of bacterial suspension (E. coli, B. subtilis or S. aureus) in TSB (1 × 106 CFU/ml) was treated with biosynthesised AgNPs (at final concentration of MIC of AgNPs for each tested bacteria) and incubated for 24 h in the orbital shaker (100 rpm) at 37 °C. The bacterial cells in TSB (1 × 106 CFU/ml) were used as a control. After incubation time the SYTO 9 (green‐fluorescent nucleic acid stain) and propidium iodide (the red‐fluorescent nucleic acid stain) dyes were added (both 1.5 µl/ml) into each experimental sample and controls. Then all samples were mixed thoroughly and incubated at room temperature for 30 min in the darkness. Subsequently, the samples were 1000 times diluted with sterile distilled water and the 100 µl of each stained sample was filtered through 0.2 μm black polycarbonate filters (Isopore; Millipore). Each filter, containing labelled bacteria, was placed on a glass slide with two drops of BacLight mounting oil and covered with a clear glass cover slip. The sample counting was performed with an E800 epifluorescence microscope (Nikon Eclipse E200, Melville, New York, USA) with a 100 W mercury light source. Images were captured with a Cool Cam camera (Cool Camera, Decatur, Georgia, USA) and associated software (MultiScanBase v.18.03, Image Pro Plus V.4.5; Media Cybernetic, Silver Spring, Maryland, USA). The numbers of dead and live cells were estimated from a count of five or more randomly chosen microscopic fields (×1000). At least 400 bacteria were counted per sample. The SYTO 9 and propidium iodide dyes differ both in their spectral characteristic and in their ability to penetrate healthy bacterial cells. Thus, with an appropriate mixture of the SYTO 9 and propidium iodide stains, bacteria with intact cell membranes stain fluorescent green, whereas bacteria with damaged membranes strain fluorescent red.
3 Statistical analysis
To determine whether there are any significant differences among the activities of AgNPs, antibiotics and the combination of AgNPs and antibiotics, we applied ‘one way analysis of variance’ for the relative variability within above parameters.
4 Results
4.1 Identification and phylogenetic analysis of actinobacterial SH11 strain
Almost complete 16S rRNA gene sequence (1423 nucleotides [nt]) of the isolate SH11 was determined (GenBank accession number: KU587966). Isolate SH11 formed a distinct subclade in the Streptomyces 16S rRNA gene tree together with the type strains of S. kasugaensis M338‐M1T and S. celluloflavus NRRL B‐2493T, the taxons which were supported by N‐J tree‐making algorithm and by a bootstrap value of 100% (Fig. 1). The isolate shared its highest 16S rRNA similarity with the type strains of S. kasugaensis M338‐M1T and S. celluloflavus NRRL B‐2493T, namely 99.8%, a value which corresponded to three nucleotide differences at 1421 locations.
Fig. 1.

Neighbour‐joining tree based on nearly complete 16S rRNA gene sequences (1374–1485 nucleotides) showing relationships between the isolate SH11 and the type strains of Streptomyces sp. Numbers at the nodes are percentage bootstrap values based on 1000 resampled datasets, only values above 30% are given. T‐type strain. Bar 0.005 substitutions per nucleotide position. The root position of the tree was determined using S. albus subsp. albus DSM 40313T as outgroup
4.2 Synthesis and characterisation of AgNPs
The AgNPs were successfully synthesised from SH11 actinobacterial strain which was observed by colour change of the cell free filtrate from light‐yellow to dark‐brown after challenging with 1 mM silver nitrate (Fig. 2). Furthermore, the UV–visible spectroscopy analysis of AgNPs revealed sharp narrow peak at λ = 420 nm and confirmed presence of biosynthesised AgNPs (Fig. 2).
Fig. 2.
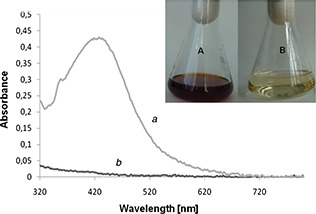
Detection of AgNPs from SH11 strain by UV–visible spectrophotometer – experimental (a), control (b). Visual detection of synthesised AgNPs – (A) experimental sample after treatment with AgNO3 after 72 h of incubation, (B) control before treatment with AgNO3
The FTIR spectroscopy analysis showed intense absorption bands at 3440.7, 2927.4, 2858.6, 1631.9, 1437.2, 1387.6 and 1030.1 cm−1 which corresponded to different functional groups on nanoparticles and indicated the presence of stabilising protein molecules on the surface of AgNPs (Figs. 3 a and b).
Fig. 3.
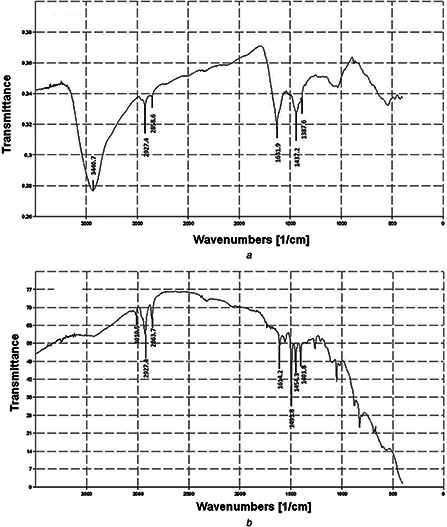
FTIR analysis of AgNPs synthesised from SH11
(a) Experimental, (b) Control
The TEM analysis indicated that the biosynthesised AgNPs from SH11 strain were spherical in shape and polydispersed. However, few aggregates of AgNPs were also found at some places. The AgNPs showed the size in the range of 4–24 nm, with a mean size of 13.2 (±2.9) nm (Fig. 4).
Fig. 4.
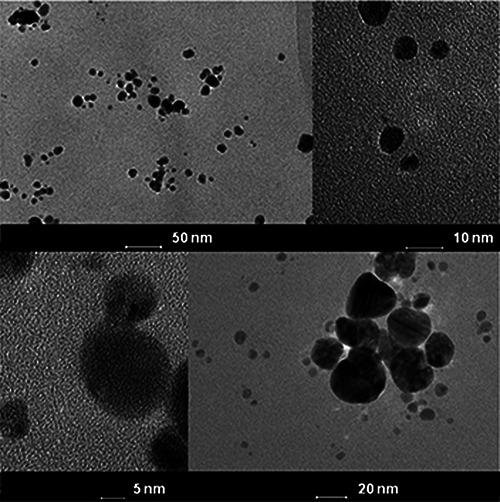
TEM micrograph and selected area diffraction pattern of AgNPs synthesised from SH11 strain
The zeta potential of biosynthesised AgNPs was found to be −16.6 mV, which indicated stability of Ag nanoparticles (Fig. 5).
Fig. 5.
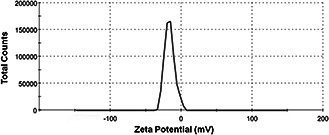
Zeta potential graph of the AgNPs synthesised from SH11 strain (−16.6 mV) at pH7
4.3 Antibacterial activity of biosynthesised AgNPs individually and in combination with antibiotics
The AgNPs exhibited the lowest MIC against S. aureus and B. subtilis, both at 40 μg/ml, followed by E. coli at 70 μg/ml. The MBC was estimated for all tested bacteria at 140 µg/ml (Table 1).
Table 1.
MIC of antibiotics and AgNPs synthesised from actinobacterial strain SH11 and synergistic effect of AgNPs combined with antibiotics against bacterial isolates by dilution plate method. Bacterial growth inhibition (%)
| Bacteria | MIC of AgNPs, µg/ml | MIC of antibiotics, µg/ml | AgNPsa | Kanamycina | Ampicillina | Tetracyclinea | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kan | Amp | Tet | A | A + AgNPs | A | A + AgNPs | A | A + AgNPs | |||
| E. coli ATCC8739 | 70 ±0.06 | 1.5 ±0.5 | 2.0 ±0.0 | 0.79 ±0.35 | 71.9 ±0.01 | 39.62 ±0.01 | 72.65 ±0.03 | 73.94 ±0.06 | 72.31 ±0.02 | 23.13±0.02 | 74.3 ±0.03 |
| S. aureus ATCC6338 | 40 ±0.05 | 1.16 ±0.28 | 0.1 ±0.03 | 0.25 ±0.0 | 15.8 ±0.05 | 39.54 ±0.01 | 86.55 ±0.02 | 47.4 ±0.04 | 86.05 ±0.005 | 44.2 ±0.03 | 87.04 ±0.004 |
| B. subtilis PCM2021 | 40 ±0.15 | 29.3 ±0.17 | 0.1 ±0.01 | 0.15 ±0.08 | 39.4 ±0.01 | 85.9 ±0.01 | 86.13 ±0.01 | 34.62 ±0.22 | 51.9 ±0.02 | 17.14±0.04 | 51.4 ±0.004 |
Mean value are significantly different at p ≤ 0.05; values are expressed in Mean ± SD.
MIC, minimal inhibitory concentration; Kan, kanamycin; Amp, ampicillin; Tet, tetracycline; A, antibiotic; AgNPs, silver nanoparticles.
a Growth inhibition (%) in the presence of MICs of AgNPs or/and antibiotic.
The results of antibiotic sensitivity test for bacterial strains were presented in Table 1. All the bacterial pathogens exhibited the highest sensitivity to ampicillin, followed by kanamycin and tetracycline.
The enhanced antibacterial activity of antibiotics in the presence of AgNPs was observed against all tested pathogens. However, antibacterial activity of tetracycline was most enhanced. The highest synergistic activity of AgNPs was found against S. aureus, followed by B. subtilis and E. coli (Table 1).
4.4 Bacterial viability assay after treatment with AgNPs
Live/Dead bacLight kit was used to estimate the per cent survival rate of bacterial cells after treatment with AgNPs. The ratio of live to dead bacterial cells of analysed control samples (without AgNPs) was found to be 91.9 and 8.1% for S. aureus, 90.2 and 9.8% for B. subtilis then 90.1 and 9.9% for E. coli. While, in experimental samples after treatment with AgNPs the ratio of live/dead bacterial cells was recorded as follows 85, 7 and 14.3% for S. aureus, 60.0 and 40.0% for B. subtilis and 30.8 and 69.2% for E. coli (Fig. 6).
Fig. 6.
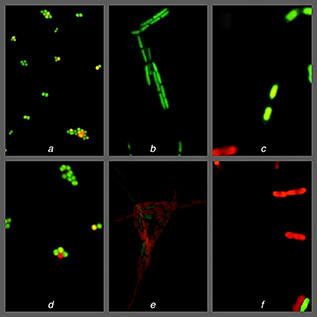
Live/dead analysis of effective activity of AgNPs on bacterial pathogens
Control – (a) S. aureus, (b) B. subtilis, (c) E. coli; Experimental – (d) S. aureus, (e) B. subtilis, (f) E. coli
5 Discussion
Antibiotic resistance of the pathogenic bacteria has been a great problem since the last decade [23]. Hence, there is a need for the development of new antibacterial agents. The researchers are moving towards nanoparticles especially AgNPs to solve the problem of emerging pathogens including multi‐drug resistant bacteria [34]. Hence, the AgNPs have appeared as a promising antibacterial candidate in the medical field [23, 34, 35].
In this study, SH11 actinobacterial strain was studied for synthesis of AgNPs by using a simple biological synthesis process.
The colour change from pale‐yellow to dark‐brown indicates the formation of AgNPs. The brown colour is the result of excitation of surface plasmon vibration in the metal nanoparticles and is typical of the AgNPs presence [36]. The visible observation of synthesis of AgNPs by colour change were also reported by Zarina and Nanda [2], and Golinska et al. [37] with a cell filtrate of Streptomyces ‐MS 26 and Pilimelia columellifera subsp. pallida after treatment with 1 mM AgNO3. Such observation clearly indicates that SH11 strain had the potential to reduce the silver ions to AgNPs.
The UV–visible spectrum in the form of sharp peak, at around 420 nm is specific for the synthesis of AgNPs [2, 38]. Our results are similar to findings of Otari et al. [38] and Zarina and Nanda [2]. Authors reported the absorption peak at about 420 nm when studied Rhodococcus sp. and Streptomyces sp., respectively.
FTIR spectroscopy studies reveal the presence of specific functional groups constituting organic molecules [39, 40]. The band at 3440.7 cm−1 is characteristic for vibrations of functional groups such as amino groups (N–H) [41]. The peaks in the range of 2850–2960 cm−1 are associated with stretching vibration of C–H and the absorbance peak at 1387.6 cm−1 can be assigned to CH3 sym. bending which is characteristic for alkanes group [4]. The absorption band at 1631.9 cm−1 resulted from the amide I vibration, which originated mainly from stretching of carbonyl groups (C = O) [41]. Moreover, the peak at 1437.2 cm−1 can be assigned to the C–H deformation vibration, which represents the presence of alcoholic bonded [19]. The peak at around 1030.1 cm−1 can be assigned as C–N stretching vibrations and these results showed similarity with findings of Zarina and Nanda [4] and Chauhan et al. [42]. The FTIR spectroscopy study confirmed the presence of proteins on the surface of AgNP.
It is claimed that the carbonyl group of amino acid residues and peptides of proteins has the stronger ability to bind to metal ions [7, 43, 44]. Therefore, proteins are responsible for formation of AgNPs, form a coat which covers the surface of metal nanoparticles (named capping agent of the AgNPs) and effect on its stabilisation by preventing the agglomeration of nanoparticles [34, 43, 45].
The TEM study clearly image the shape and size of biosynthesised nanoparticles [46]. The AgNPs synthesised from SH11 actinobacterial strain were spherical and small sized (26 nm). The obtained results are similar to findings of Sadhasivam and coworkers [47], who reported nanoparticles size range of 20–30 nm synthesised from Streptomyces sp. culture. Sukanya et al. [48] and Abdeen et al. [49] also reported spherical and small sized Ag nanoparticles from actinobacterial strains, which were found to be in size range of 5–40 and 10–20 nm, respectively. Moreover, the small amount of aggregations of biosynthesised AgNPs from SH11 strain suggests the inherent stability of nanoparticles in the reaction mixture, which might be due to presence of capping agents formed by proteins or other biomolecules, as mentioned previously [36].
The zeta potential is one of the main factors affecting the interaction between the particles in the liquid. Zetasizer analysis revealed negative zeta potential of AgNPs from SH11 strain (−16.6 mV). It has been reported that the particles with higher negative or positive zeta potential possess a force to repel each other and do not form aggregates. It has been reported that the particles with higher negative or positive zeta potential possess a force to repel each other and do not form aggregates. The low zeta potential minimize the electrostatic repulsive force and therefore maximize aggregation [40, 50]. The AgNPs biosynthesised in this study were found to be more stable than AgNPs synthesised by the marine isolate of S. albidoflavus studied by Prakasham et al. [51]. The zeta potential of obtained AgNPs were found to be −8.5 mV [51].
Among the different nanosized antibacterial agents, AgNPs have proved to be the most effective because of its broad‐spectrum activity against bacteria, viruses and eukaryotic microorganisms [35, 52, 53]. The antibacterial efficacy of the nanoparticles depends on the shape and size of AgNPs [54, 55]. Moreover, the nanoparticles smaller than 10 nm interact with bacteria and produce electronic effects, which enhance the reactivity of nanoparticles. Thus, it is proven that the bactericidal effect of AgNPs is size dependent [56].
The highest antibacterial activity of AgNPs was noticed against E. coli followed by B. subtilis and S. aureus. These findings were compared with studies of Selvakumar et al. [57]. Authors also examined the antimicrobial activity of AgNPs synthesised from actinobacteria against either gram‐negative (Pseudomonas aeruginosa, E. coli, K. pneumoniae, Enterobacter faecalis) or gram‐positive (S. aureus) human pathogenic bacteria. However, they recorded the maximum antimicrobial activity of synthesised AgNPs against S. aureus followed by P. aeruginosa, E. faecalis and E. coli, and the least against Klebsiella pneumoniae. Similarly Prakasham et al. [51] studied the antimicrobial activity of AgNPs from S. albidoflavus against gram‐positive (B. subtilis and Micrococcus luteus) and gram‐negative (E. coli and K. pneumoniae) bacterial strains. The maximum antimicrobial activity of AgNPs was found against K. pneumoniae, followed by M. luteus, B. subtilis and the least E. coli.
Biogenic AgNPs are intensively tested for their antimicrobial activity against a broad range of microorganisms including gram‐positive and gram‐negative bacteria, fungi, yeasts and microbial biofilms [10]. It is believed that DNA loses its replication ability and cellular proteins become inactivated upon silver ion treatment [4].
The results of minimal inhibitory and bactericidal concentrations confirmed that AgNPs biosynthesised from actinobacteria SH11 strain were efficient against tested bacteria. However, both gram‐positive bacterial pathogens were found to be more sensitive than gram‐negative strain of E. coli. Similarly, live/dead analysis displayed effective activity of AgNPs on bacterial pathogens.
Singh et al. [58] studied MIC of silver NPs synthesised from Acinetobacter calcoaceticus and reported activity of AgNPs against human pathogenic bacteria in much higher concentration range of 150–600 μg/ml when compared with our findings.
However, Manivasagan et al. [6] assessed the MIC of AgNPs synthesised from Nocardiopsis sp. MBRC‐1 against various pathogenic bacteria and fungi. The authors determined lowest MIC against B. subtilis at 7 µg/ml, B. subtilis at 10 μg/ml and Candida albicans at 10 µg/ml, suggesting the broad‐spectrum nature of their MIC. It seems that, the higher sensitivity of pathogens could be affected by chemical nature of biosynthesised nanoparticles or nature of microbial cells.
The combination of antibiotics with biosynthesised AgNPs against clinical pathogens offers valuable contribution to nanomedicine [10]. Enhanced effect of bacterial AgNPs on antimicrobial activity of antibiotics has been also observed in studies of Manikprabhu and Lingappa [59]. It is claimed that synergistic effect against bacteria depends on the interaction of AgNPs with a chemical structure of antibiotic [10, 60]. Chauhan et al. [61] suggested that the increase in synergistic effect may be caused by the bonding reaction between antibiotic and nanomaterials. The antibiotic molecules contain many active groups such as hydroxyl and amide groups, which react easily with nanosilver by chelation.
Combination of AgNPs with antibiotics reduces the toxicity of both toward human cells by decreasing the required dosage with enhanced microbicidal properties. Furthermore, such combination restores the ability of the drug to kill bacteria that have acquired resistance to them [10, 58, 62].
Among tested antibiotics, tetracycline showed best synergistic activity against gram‐positive and gram‐negative bacteria when combined with biosynthesised AgNPs. The effective enhanced activity of ampicillin was noticed against S. aureus and B. subtilis, and kanamycin against S. aureus. The synergetic effect of AgNPs biosynthesised from Streptomyces ‐MS26 in combination with ampicillin, azithromycin, cefotaxime, erythromycin and ofloxacin against bacterial pathogens was reported by Zarina and Nanda [4]. The maximum synergistic effect was observed for amoxicillin, cefixime, amoxicillin and erythromycin against M. luteus, Pseudomonas sp., S. aureus and multi‐drug resistant pathogen, respectively.
In this study, the actinobacteria SH11 strain isolated from acid soil has shown potential for efficient synthesis of AgNPs by eco‐friendly and simple process. The spherical, small sized and stable AgNPs showed significant antibacterial activity. The efficacy of nanosilver increased when used in combination with commercially used antibiotics. Therefore, such AgNPs can be used as antimicrobial agent alone or in combination with antibiotics after more trials in experimental animals.
6 Acknowledgments
This study was supported by Grant Symphony 1, No. 2013/08/W/NZ8/00701 from The Polish National Science Centre.
7 References
- 1. Shirley A. Dayanand B. Sreedhar S. et al.: ‘Antimicrobial activity of silver nanoparticle synthesized from novel Streptomyces sp.’, Dig. J. Nanomater. Biostr., 2010, 5, pp. 447 –451. [Google Scholar]
- 2. Zarina A. Nanda A.: ‘Green approach for synthesis of silver nanoparticles from marine Streptomyces ‐ MS 26 and their antibiotic efficacy’, J. Pharm. Sci. Res., 2014, 6, pp. 321 –327 [Google Scholar]
- 3. Selvarani M. Prema P.: ‘Synergistic antibacterial evaluation of commercial antibiotics combined with nanoiron against human pathogens’, Int. J. Pharm. Sci. Rev. Res., 2013, 27, pp. 183 –190 [Google Scholar]
- 4. Zarina A. Nanda A.: ‘Combined efficacy of antibiotics and biosynthesized silver nanoparticles from Streptomyces albaduncus ’, Int. J. PharmTech Res., 2014b, 6, pp. 1862 –1869 [Google Scholar]
- 5. Probhu S. Poulose E.K.: ‘Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects’, Int. Nano Lett., 2012, 2, p. 32 [Google Scholar]
- 6. Manivasagan P. Venkatesan J. Senthilkumar K. et al.: ‘Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC‐1’, Bio. Med. Res. Int., 2013, 2013, pp. 1 –9 10.1155/2013/287638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Narasimha G. Janardhan A. Alzohairy M. et al.: ‘Extracellular synthesis, characterization and antibacterial activity of silver nanoparticles by Actinomycetes isolative’, Int. J. Nano Dimens., 2013, 4, pp. 77 –83 [Google Scholar]
- 8. Golinska P. Wypij M. Ingle A.P. et al.: ‘Biogenic synthesis of metal nanoparticles from actinomycetes: biomedical applications and cytotoxicity’, Appl. Microbiol. Biotechnol., 2014, 98, pp. 8083 –8097 [DOI] [PubMed] [Google Scholar]
- 9. Salunkhe G.R. Ghosh S. Santosh Kumar R.J. et al.: ‘Rapid efficient synthesis and characterization of silver, gold, and bimetallic nanoparticles from the medicinal plant Plumbago zeylanica and their application in biofilm control’, Int. J. Nanomed., 2014, 27, pp. 2635 –53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singh R. Shedbalkar U.U. Wadhwani S.W. et al.: ‘Bacteriagenic silver nanoparticles: synthesis, mechanism, and applications’, Appl. Microbiol. Biotechnol., 2015, 99, pp. 4579 –4593 [DOI] [PubMed] [Google Scholar]
- 11. Saminathan K.: ‘Biosynthesis of silver nanoparticles using soil Actinomycetes Streptomyces sp.’, Int. J. Curr. Microbiol. Appl. Sci., 2015, 4, pp. 1073 –1083 [Google Scholar]
- 12. Bull A.T.: ‘Actinobacteria from the extremobiosphere’, in Horikoshi K. Antranikian G. Bull A.T. Robb F. Stetter K.O. (eds.): ‘Extremophiles handbook’ (Springer, New York, 2010), pp. 3 –15 [Google Scholar]
- 13. Pan T. He H. Li C. et al.: ‘ Streptomyces daqingensis sp. nov., isolated from saline‐alkaline soil’, Int. J. Syst. Evol. Microbiol., 2016, 66, pp. 1358 –1363, doi: 10.1099/ijsem.0.000887 [DOI] [PubMed] [Google Scholar]
- 14. Gutierrez F.M. Olive P.L. Banuelos A. et al.: ‘Synthesis, characterization, and evaluation of antimicrobial and cytotoxic effect of silver and titanium nanoparticles’, Nanomedicine, 2010, 6, pp. 681 –688 [DOI] [PubMed] [Google Scholar]
- 15. Potara M. Bawaskar M. Simon T. et al.: ‘Biosynthesized silver nanoparticles performing as biogenic SERS‐nanotags for investigation of C26 colon carcinoma cells’, Colloids Surf. B, Biointerfaces, 2015, 133, pp. 296 –303, doi: 10.1016/j.colsurfb.2015.06.024 [DOI] [PubMed] [Google Scholar]
- 16. Franci G. Falanga A. Galdiero S. et al.: ‘Silver nanoparticles as potential antibacterial agents’, Molecules, 2015, 20, pp. 8856 –8874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rai M. Kon K. Ingle A. et al.: ‘Broad‐spectrum bioactivities of silver nanoparticles: the emerging trends and future prospects’, Appl. Microbiol. Biotechnol., 2014, 98, pp. 1951 –1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banu A. Rathod V.: ‘Biosynthesis of monodispersed silver nanoparticles and their activity against Mycobacterium tuberculosis ’, J. Nanomed. Biotherapeutic Discov., 2013, 3, p. 110 [Google Scholar]
- 19. Shanmugaiah V. Harikrishnan H. Al‐Harbi N.S. et al.: ‘facile synthesis of silver nanoparticles using Streptomyces sp.VSMGT1014 and their antimicrobial efficiency’, Dig. J. Nanomater. Biostr., 2015, 10, pp. 179 –187 [Google Scholar]
- 20. Mallevre F. Fernandes T.F. Aspray T.J.: ‘ Pseudomonas putida biofilm dynamics following a single pulse of silver nanoparticles’, Chemosphere, 2016, 153, pp. 356 –364 [DOI] [PubMed] [Google Scholar]
- 21. Li P. Li J. Wu C. et al.: ‘Synergistic antibacterial effects of β‐lactam antibiotic combined with silver nanoparticles’, Nanotechnology, 2005, 16, pp. 1912 –1917 [Google Scholar]
- 22. Ruden S. Hilpert K. Berditsch M. et al.: ‘Synergistic interaction between silver nanoparticles and membrane‐permeabilizing antimicrobial peptides’, Antimicrob. Agents Chemother., 2009, 53, pp. 3538 –3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dar M.A. Ingle A. Rai M.: ‘Enhanced antimicrobial activity of silver nanoparticles synthesized by Cryphonectria sp. evaluated singly and in combination with antibiotics’, Nanomed. Nanotechnol. Biol. Med., 2013, 9, pp. 105 –110 [DOI] [PubMed] [Google Scholar]
- 24. McShan D. Zhang Y. Deng H. et al.: ‘Synergistic antibacterial effect of silver nanoparticles combined with ineffective antibiotics on drug resistant Salmonella typhimurium DT104’, J. Environ. Sci. Health C. Environ. Carcinog. Ecotoxicol. Rev., 2015, 33, pp. 369 –384 [DOI] [PubMed] [Google Scholar]
- 25. Küster E. Williams S.T.: ‘Selection of media for isolation of Streptomycetes ’, Nature, 1964, 202, pp. 928 –929 [DOI] [PubMed] [Google Scholar]
- 26. Golinska P. Ahmed L. Wang D. et al.: ‘ Streptacidiphilus durhamensis sp. nov., isolated from a spruce forest soil’, Anton. Van Leeuw., 2013, 104, pp. 199 –206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shirling E.B. Gottlieb D.: ‘Methods for characterization of Streptomyces sp.’, Int. J. Syst. Bacteriol., 1966, 16, pp. 313 –340 [Google Scholar]
- 28. Golinska P. Kim B.‐Y. Dahm H. et al.: ‘Streptacidiphilus hamsterleyensis sp. nov., isolated from a spruce forest soil’, Anton. Van Leeuw., 2013b, 104, pp. 965 –972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim O.S. Cho Y.J. Lee K. et al.: ‘Introducing EzTaxon‐e: a prokaryotic 16S rRNA gene sequence data‐base with phylotypes that represent uncultured species’, Int. J. Syst. Evol. Microbiol., 2012, 62, pp. 716 –721 [DOI] [PubMed] [Google Scholar]
- 30. Tamura K. Stecher G. Peterson D. et al.: ‘MEGA6: molecular evolutionary genetics analysis version 6.0’, Mol. Biol. Evol., 2013, 30, pp. 2725 –2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saitou N. Nei M.: ‘The neighbor‐joining method: a new method for constructing phylogenetic trees’, Mol. Biol. Evol., 1987, 4, pp. 406 –425 [DOI] [PubMed] [Google Scholar]
- 32. Felsenstein J.: ‘Confidence limits on phylogenies: an approach using the bootstrap’, Evol., 1985, 39, pp. 783 –791 [DOI] [PubMed] [Google Scholar]
- 33. Fitch W.M.: ‘Toward defining the course of evolution: minimum change for a specific tree topology’, Syst. Zool., 1971, 20, pp. 406 –416 [Google Scholar]
- 34. Raheman F. Shivaji D. Ingle A. et al.: ‘Silver nanoparticles: novel antimicrobial agent synthesized from an endophytic fungus Pestalotia sp. isolated from leaves of Syzygium cumini (L)’, Nano Biomed. Eng., 2011, 3, pp. 174 –178 [Google Scholar]
- 35. Birla S.S. Tiwari V.V. Gade A.K. et al.: ‘Fabrication of silver nanoparticles by Phoma glomerata and its combined effect against Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus ’, Lett. Appl. Microbiol., 2009, 48, pp. 173 –179 [DOI] [PubMed] [Google Scholar]
- 36. Alani F. Murray M.Y. William A.: ‘Biosynthesis of silver nanoparticles by a new strain of Streptomyces sp. compared with Aspergillus fumigates ’, World J. Microbiol. Biotechnol., 2012, 28, pp. 1081 –1086 [DOI] [PubMed] [Google Scholar]
- 37. Golinska P. Wypij M. Rathod D. et al.: ‘Synthesis of silver nanoparticles from two acidophilic strains of Pilimelia columellifera subsp. pallida and their antibacterial activities’, J. Basic Microbiol., 2015, 55, pp. 1 –16 [DOI] [PubMed] [Google Scholar]
- 38. Otari S.V. Patil R.M. Nadaf N.H. et al.: ‘Green biosynthesis of silver nanoparticles from an actinobacteria Rhodococcus sp.’, Mater. Lett., 2012, 72, pp. 92 –94 [Google Scholar]
- 39. Duran N. Marcato P.D. Duran M. et al.: ‘Mechanistic aspects in the biogenic synthesis of extracellular metal nanoparticles by peptides, bacteria, fungi and plants’, Appl. Microbiol. Biotechnol., 2011, 90, pp. 1609 –1624 [DOI] [PubMed] [Google Scholar]
- 40. Rai M. Ingle A. Gade A. et al.: ‘Synthesis of silver nanoparticles by Phoma gardeniae and in vitro evaluation of their efficacy against human disease‐causing bacteria and fungi’, IET Nanobiotech., 2015a, 9, pp. 71 –75 [DOI] [PubMed] [Google Scholar]
- 41. Kumar C.G. Mamidyala S.K.: ‘Extracellular synthesis of silver nano‐particles using culture supernatant of Pseudomonas aeruginosa ’, Colloids Surf. B, Biointerfaces, 2011, 84, pp. 462 –466 [DOI] [PubMed] [Google Scholar]
- 42. Chauhan R. Kumar A. Abraham J.: ‘A biological approach to the synthesis of silver nanoparticles with Streptomyces sp. JAR1 and its antimicrobial activity’, Sci. Pharm., 2013, 81, pp. 607 –621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gole A. Dash C. Ramakrishnan V. et al.: ‘Pepsin‐gold colloid conjugates: preparation, characterization and enzymatic activity’, Langmuir, 2001, 17, pp. 1674 –1679 [Google Scholar]
- 44. Balaji D.S. Basavaraja S. Deshpande R. et al.: ‘Extracellular biosynthesis of functionalized silver nanoparticles by strains of Cladosporium cladosporioides fungus’, Colloids Surf. B, Biointerfaces, 2001, 68, pp. 88 –92 [DOI] [PubMed] [Google Scholar]
- 45. Basavaraja S. Balaji S.D. Lagashetty A. et al.: ‘Extracellular biosynthesis of silver nanoparticle using the fungus Fusarium semitectum ’, Mater. Res. Bull., 2008, 45, pp. 1164 –1170 [Google Scholar]
- 46. Karthik L. Kumar A. Kirthi V.A. et al.: ‘ Streptomyces sp. LK3 mediated synthesis of silver nanoparticles and its biomedical application’, Bioprocess Biosyst. Eng., 2014, 37, pp. 261 –267 [DOI] [PubMed] [Google Scholar]
- 47. Sadhasivam S. Shanmugam P. Yun K.: ‘Biosynthesis of silver nanoparticles by Streptomyces hygroscopicus and antimicrobial activity against medically important pathogenic microorganisms’, Colloids Surf. B, Biointerfaces, 2010, 81, pp. 358 –362 [DOI] [PubMed] [Google Scholar]
- 48. Sukanya M.K. Saju K.A. Praseetha P.K. et al.: ‘Therapeutic potential of biologically reduced silver nanoparticles from actinomycete cultures’, J. Nanosci., 2013, 2013, pp. 1 –8, 10.1155/2013/940719 [DOI] [Google Scholar]
- 49. Abdeen S. Geo S. Sukanya S. et al.: ‘Biosynthesis of silver nanoparticles from Actinomycetes for therapeutic applications’, Int. J. Nano Dimens., 2014, 5, pp. 155 –162 [Google Scholar]
- 50. Rai M. Ingle A. Gade A. et al.: ‘Three‐Phoma spp. synthesized novel silver nanoparticles that possess excellent antimicrobial efficacy’, IET Nanobiotechnol., 2015, 9, pp. 280 –287, doi: 10.1049/iet‐nbt.2014.0068 [DOI] [PubMed] [Google Scholar]
- 51. Prakasham R.S. Buddana S.K. Yannam S.K. et al.: ‘Characterization of silver nanoparticles synthesized by using marine isolate Streptomyces albidoflavus ’, J. Microbiol. Biotech., 2012, 22, pp. 614 –621 [DOI] [PubMed] [Google Scholar]
- 52. Gaikwad S. Ingle A. Gade A. et al.: ‘Antiviral activity of mycosynthesized silver nanoparticles against herpes simplex virus and human parainfluenza virus type 3’, Int. J. Nanomed., 2013, 8, pp. 4303 –4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anasane N. Golinska P. Wypij M. et al.: ‘Acidophilic actinobacteria synthesised silver nanoparticles showed remarkable activity against fungi‐causing superficial mycoses in humans’, Mycoses, 2016, 59, pp. 157 –166 [DOI] [PubMed] [Google Scholar]
- 54. Rai M. Yadav A. Gade A.: ‘Silver nanoparticles as a new generation of antimicrobials’, Biotechnol. Adv., 2009, 27, pp. 76 –83 [DOI] [PubMed] [Google Scholar]
- 55. Rai M.K. Deshmukh S.D. Ingle A.P. et al.: ‘Silver nanoparticles: the powerful nanoweapon against multidrug‐resistant bacteria’, J. Appl. Microbiol., 2012, 112, pp. 841 –852 [DOI] [PubMed] [Google Scholar]
- 56. Morones J.R. Elechiguerra J.L. Camacho A. et al.: ‘The bactericidal effect of silver nanoparticles’, Nanotechnol., 2005, 16, pp. 2346 –2353 [DOI] [PubMed] [Google Scholar]
- 57. Selvakumar P. Viveka S. Prakash S. et al.: ‘Antimicrobial activity of extracellularly synthesized silver nanoparticles from marine derived Streptomyces rochei ’, Int. J. Pharm. Biol. Sci., 2012, 3, pp. 188 –197 [Google Scholar]
- 58. Singh R. Wagh P. Wadhwani S. et al.: ‘Synthesis, optimization, and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity when combined with antibiotics’, Int. J. Nanomed., 2013, 8, pp. 4277 –4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manikprabhu D. Lingappa K.: ‘Antibacterial activity of silver nanoparticles against methicillin‐resistant Staphylococcus aureus synthesized using model Streptomyces sp. pigment by photo‐irradiation method’, J. Pharm. Res., 2013, 6, pp. 255 –260 [Google Scholar]
- 60. Ghosh S. Patil S. Ahire M. et al.: ‘Synthesis of silver nanoparticles using Dioscorea bulbifera tuber extract and evaluation of its synergistic potential in combination with antimicrobial agents’, Int. J. Nanomed., 2012, 7, pp. 483 –496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chauhan R. Reddy A. Abraham J.: ‘Biosynthesis of silver and zinc oxide nanoparticles using Pichia fermentans JA2 and their antimicrobial property’, Appl. Nanosci., 2015, 5, pp. 63 –71 [Google Scholar]
- 62. Allahverdiyev A.M. Kon K.V. Abamor E.S. et al.: ‘Coping with antibiotic resistance: combining nanoparticles with antibiotics and other antimicrobial agents’, Expert Rev. Anti Infect. Ther., 2011, 9, pp. 1035 –1052 [DOI] [PubMed] [Google Scholar]


