Abstract
In the present study, an efficient biosurfactant producing bacterial strain Pseudomonas aeruginosa MKVIT3 was isolated from an oil logging area in Vellore district of Tamil Nadu, India. Liquid chromatography–mass spectrometry (LC‐MS/MS) analysis was performed for the identification of different congeners present in the extracted biosurfactant. The column purified biosurfactant was used to stabilise the formation of silver nanoparticles (NP) using borohydrate reduction in reverse micelles. The silver NP were characterised using UV‐vis absorption spectroscopy, Powder‐XRD TEM analysis and zeta potential. A comparative study of the antimicrobial activity and cytotoxic efficacy was done for the extracted purified biosurfactant and the silver NP. The LC‐MS/MS analysis of the biosurfactant revealed the presence of five rhamnolipid congeners. The synthesised silver NP showed the characteristic absorption peak in UV‐vis at 440 nm. Powder‐XRD and TEM analysis revealed the average particle size of the NP as 17.89 ± 8.74 nm as well as their cubic structure. Zeta potential value of −30.9 mV suggested that the silver NPs are stable in the suspension. Comparative study of the antimicrobial activity revealed that the silver NP are more potent than the biosurfactant in inhibiting the growth of microbes. Cytotoxic activity revealed that the biosurfactant are more effective than the synthesised silver NP.
Inspec keywords: microemulsions, silver, nanoparticles, nanomedicine, cellular biophysics, surfactants, microorganisms, chromatography, mass spectroscopic chemical analysis, ultraviolet spectra, visible spectra, antibacterial activity
Other keywords: Ag, microemulsion synthesis, silver nanoparticles, Pseudomonas aeruginosa, MKVIT3 strain, antimicrobial activities, cytotoxic activities, biosurfactant producing bacterial strain, oil logging area, Vellore district, Tamil Nadu, India, liquid chromatography–mass spectrometry, LC‐MS/MS analysis, extracted biosurfactant, column purified biosurfactant, borohydrate reduction, reverse micelles, UV‐vis absorption spectroscopy, powder‐XRD TEM analysis, zeta potential, antimicrobial activity, cytotoxic efficacy, extracted purified biosurfactant, rhamnolipid congeners
1 Introduction
In recent times, biosynthesis of metal nanoparticles (NPs) using plants [1] and microbes [2, 3] have emerged as a simple, eco‐friendly and cost‐effective substitute for the synthesis of metal NPs. Metal NPs are usually prepared from silver, gold, platinum, palladium [4], titanium, copper, zinc etc. [5] However, silver NPs proved to be the most effectual as they are effective against various microorganisms and are safe for humans [6]. Recently, the reverse micelle technique for synthesis of silver NPs have been widely used [7]. However, most of the surfactants that are used were chemical surfactants, which are toxic, economically nonviable and pollute the environment. Therefore, researchers have started looking into the synthesis of silver NPs by using biosurfactants produced by the microorganisms as the capping agent for better aggregation and stabilisation [8]. Biosurfactants or green surfactants are surface active microbial metabolites comprising a vast range of chemical structures including glycolipids, phospholipids, lipopeptides, fatty acids, lipopolysaccharides and neutral lipids [9]. They have bulky and complicated structures, lower toxicity, higher biodegradability with superior environmental compatibility [10, 11, 12] and they easily form a variety of liquid crystals in the aqueous solutions [13]. Biosurfactants have been reported to show antimicrobial [14], antitumor [15], antiviral [16], anti‐mycoplasmal [17], anti‐adhesive activity [18] etc. Rhamnolipid, a type of glycolipid biosurfactant produced by Pseudomonas aeruginosa, have unique potential for application in various injuries, organ repairs, skin problems along with antibacterial properties [19, 20] Rhamnolipid matrices were also used for delivering drugs effectively by overcoming the problem of poor solubility of the drugs in blood brain barrier [21] and therefore, a suitable candidate for the synthesis of silver NPs by microemulsion technique.
In light of the above information, an improved attempt was made to isolate the rhamnolipid biosurfactant produced by P. aeruginosa MKVIT3 strain with an improved technique. Silver NPs were synthesised with the extracted biosurfactant using microemulsion method and were characterised using UV‐vis spectroscopy, Powder‐XRD, TEM and zeta potential. The rhamnolipid and its silver NPs were evaluated antimicrobial and cytotoxic activities.
2 Experimental
2.1 Isolation of microorganism
Soil sample was obtained from an oil logging area of Shree Ganapathi garage in Vellore, Tamil Nadu, India. The collected soil sample (1 g) was added to a 500 ml Erlenmeyer flask which contained 100 ml of sterilised nutrient broth (NB) and mineral salt solution in 1:1 ratio. To this 2% (v/v) used engine oil was also added as the sole carbon source [22]. The flasks were incubated at 35°C in a rotary shaker (Orbitek LJEIL; Scigenics Biotech Pvt. Ltd., Chennai, India) at 150 rpm. After three days, 5 ml of the culture broth was collected from each flask and moved into a second set of flasks containing fresh medium and incubated under the same conditions as above to reduce the unwanted microbial load. The process was reiterated for three times. For the development of bacterial colonies, serial dilutions of the culture broth was done from the former set of flasks and were inoculated onto nutrient agar (NA) plates and incubated at 35°C. The morphologically different bacterial colonies that developed on the plates were streaked onto NA plates to attain pure cultures of the isolates. The pure cultures were then upheld on NA slants and kept at 4°C in the refrigerator.
2.2 Screening for biosurfactant producing bacteria
The isolated bacterial strains were grown in NB at 35°C for 24 h with shaking at 150 rpm and were used as mother inoculums. The mother inoculum (5 ml) of each isolates were moved to an Erlenmeyer flasks (500 ml) containing 100 ml of sterilised mineral salt medium (MSM) with glucose (2% w/v) as the carbon source and incubated at 35°C with shaking. The composition of the MSM used was as follows (g/l): K2 HPO4 (1.0), KH2 PO4 (0.5), NH4 NO3 (4.0), KCl (0.1), MgSO4. 7H2 O (0.5), FeSO4. 7H2 O (0.01), CaCl2 (0.01), yeast extract (0.1) and trace element solution (10 ml) containing (g/l): ZnSO4. 7H2 O (0.7), MnSO4. 7H2 O (0.5), CuSO4. 5H2 O (0.5), H3 BO3 (0.26) and (NH4)6 Mo7 O2. 4H2 O (0.06) [23]. The pH was adjusted to 7.0 ± 0.2. The biosurfactant production by the bacterial isolates was studied by drop collapse assay and surface tension (ST) reduction.
2.2.1 Drop collapse assay
A drop of 48 hold culture broth was dropped on a drop of crude oil placed on a glass slide and the drop collapse activity was monitored [24].
2.2.2 ST measurement
ST reduction was evaluated at every 24 h up to 120th h (5th day) with a digital tensiometer (K11, Kruss, Germany). The values stated are the mean of five observations. The isolates that reduced the ST of the culture medium below 45 mN m−1 were chosen as effective ones for further experiments.
2.3 Identification of bacterial strain
The genomic DNA of the selected bacteria was extracted using standard protocol. The 16S rDNA was PCR amplified using universal primer pair, 968F (AACGCGAAGAACCTTAC) and 1541R (AAGGAGGTGATCCAGCCGCA) [25]. PCR was performed in a 25 µl volume in thermal cycler (Mastercycler Nexus gradient, Eppendorf, Germany) with a final concentration of 1X standard buffer, 1.5 mmol l−1 MgCl2, 0.2 μmol l−1 of each primer, 0.2 mmol l−1 dNTPs, 0.25 U Taq DNA polymerase (Sigma Aldrich, USA) and 25 ng of template DNA. The PCR reaction conditions consisted of initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 45 s and a final extension at 72°C for 7 min. PCR products were analysed on 1.2% agarose gel and visualised under Bio Doc‐It Imaging system (UVP, USA). PCR products were purified with GenEluteTM PCR Clean‐up kit (Sigma Aldrich, USA). PCR products were sequenced bidirectionally using an automated sequencer by Beckman coulter (GenomeLab GeXP, Genetic Analysis System, USA). Sequence data was aligned and evaluated to find the adjacent homolog of the isolated bacterial strain. A phylogenetic tree was built with aligned 16S rDNA sequences using 1000 bootstrap replication on Mega 6 [26].
2.4 Extraction of biosurfactant
For the extraction of biosurfactant, 48 h grown culture broth was centrifuged for 20 min at 10,000 rpm at 4°C to obtain a cell‐free supernatant which served as the source of crude biosurfactant. 6 N HCl was added to the clear supernatant to amend the pH at 2. The acidified supernatant was then stored overnight at 4°C. Biosurfactant was continuously extracted with ethyl acetate at room temperature. A 1:1 mixer of ethyl acetate and supernatant was agitated vigorously and left stationary for phase separation. The organic layer was separated and evaporated in a rotary evaporator to obtain a viscous solid product (crude biosurfactant) [27]. The crude biosurfactant was dried and the amount was determined gravimetrically.
2.5 Purification of biosurfactant
Column purification of the crude extract was performed in a 26 × 3.3 cm2 column containing 50 g of activated silica gel (60–120 mesh) (Sigma‐Aldrich, Bengaluru, India) CHCl3 slurry. The column was loaded with 1 g of sample prepared in 5 ml of CHCl3. Different ratio of CHCl3 /CH3 OH: 50:3 v/v (300 ml), 50:5 v/v (200 ml) and 50:50 v/v (100 ml) was used as the mobile phase at a flow rate of 1 ml min−1 and 20 ml of fractions were collected. Biosurfactant containing fractions were combined and dried under vacuum to get the pure biosurfactant.
2.6 Characterisation of biosurfactant
2.6.1 Biochemical
Presence of various bio‐molecules in the extracted biosurfactant was verified by different biochemical tests [28] like ninhydrin test for amino acids and proteins, anthrone test for carbohydrate and saponification test for lipid.
2.6.2 Fourier transform infrared spectroscopy (FT‐IR)
The functional groups and the bond type present in the column‐purified compound were determined with a FTIR spectrometer (Perkin Elmer, Waltham, MA, USA) in a range of 4000–500 cm−1.
2.6.3 Liquid chromatography–mass spectrometry (LC‐MS/MS)
Identification of different congeners present in biosurfactant was analysed using LC‐MS/MS (Thermo Scientific Exactive plus LC‐MS/MS mass spectrometer, Waltham, MA, USA). The column purified biosurfactant sample was dissolved in methanol and 1 µl of it was injected into Hypersil Gold C18 column (2.1 × 50 mm2). The LC flow rate was 0.5 ml min−1. An acetonitrile/water gradient with 0.01% formic acid was used (95–5%) as the mobile phase. ESI‐MS was performed in positive ion mode. Full scan data were obtained by scanning from m/z 150–2000 and fragmentor voltage used was 135.0 V.
2.7 Synthesis of silver NP
Silver NPs were synthesised by modified in situ in water‐in‐oil microemulsion phase [29]. Briefly, 1 ml of 0.05 mol l−1 aqueous AgNO3 was mixed with 6.25 ml of n ‐heptane and 0.1 g of column purified biosurfactant and stirred for 10 min at room temperature. Then, 1 ml of 0.1 mol l−1 aqueous NaBH4 solution was added to the mixture and stirred for 30 min. To break the reverse micelles, 10 ml of ethanol was added to the mixture which resulted in precipitation. The precipitate was separated by centrifugation at 10,000 rpm for 30 min at room temperature and 10 ml of n‐ butanol was added to attain a suspension by sonication.
2.8 Characterisation of the silver NPs
The UV‐Vis analysis was performed on a Hitachi U‐2910 spectrophotometer (Tokyo, Japan) from 300–800 nm wavelength range. Powder‐XRD was done with a Ni filter, Cu anode (2.2 KW) and a Lynx eye detector on the Bruker D8 Advance Powder XRD (Karlsruhe, Germany), operated in θ :2θ goniometer. TEM measurements were performed on a Philips CM200 TEM instrument (Amsterdam, Netherlands) operated at an accelerating voltage of 200 kV and 2.4 Å resolutions. The NP charge quantified as zeta potential was analysed using NanoPartica SZ‐100 series (HORIBA, Kyoto, Japan). To determine the theoretical average number of silver atoms present in one silver NP (1) and the molar concentration of the silver NP solution (2) the following equations are used [30]
| (1) |
| (2) |
Where, N is the average number of silver atoms per NP, ρ is the density of face centred cubic (fcc) silver (1.05 × 10−20 g nm−3), D is the average diameter of NPs, M is the atomic mass of silver (107.86 g mol−1), C is the molar concentration of the NP solution, N T is the total number of silver atoms added to the solution, V is the volume of the reaction solution in litres and N A is Avogadro's number.
2.9 Antimicrobial activity
The antimicrobial screening of the biosurfactant and its silver NPs was carried out against four strains of bacteria (Staphylococcus aureus, Bacillus subtilis, Escherichia coli and Klebsiella pneumoniae) and two strains of fungi (Aspergillus niger and Aspergillus flavus) using agar well diffusion assay method [31]. 50 µg of biosurfactant and 0.1 ml of the silver NPs solution was used for the study. Ciprofloxacin (10 µg/disc) was used as a positive control for bacterial culture and Fluconazole (1.0 mg/disc) for the fungal culture. The antimicrobial activity was assessed on the basis of diameter of zone of inhibition which was measured at cross angles.
2.10 Cytotoxic activity
2.10.1 Preparation of 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) solution
MTT was dissolved in Dulbecco's phosphate buffer saline of pH 7.4 to achieve the concentration of 5 mg/ml. The MTT solution was then filter‐sterilised through a 0.2 µM filter into a sterile, light protected container and stored at −20°C.
2.10.2 Preparation of solubilisation solution
40% (v/v) N,N’‐ dimethylformamide was prepared in 2% (v/v) glacial acetic acid. To this, 16% (w/v) sodium dodecyl sulphate (SDS) was added and the pH was adjusted to 4.7. To avoid precipitation of SDS, the solution was stored at room temperature.
2.10.3 MTT assay
To determine the cytotoxic activity of the biosurfactant and its silver NPs, cell viability study was done using conventional MTT reduction assay with some modifications [32]. Briefly, Chang liver cells (5 × 103 cells) and test compounds of different concentrations (10–250 µg) were seeded in 96 well plates containing 100 µl/well and incubated for desired period of exposure. To the above solution, 10 µl MTT solution was added to attain a final concentration of 0.45 mg ml−1. It was then incubated for 24 h at 37°C. After 24 h, 100 µl solubilisation solution was added to each well to dissolve formazan crystals and the absorbance of the solution were recorded at 570 nm using a Hitachi U‐2910 spectrophotometer. The experiment was performed in triplicate.
2.11 Statistical analysis
Data are stated as mean ± standard deviation (SD) and were analysed using Graph Pad Prism 5.0 software.
3 Results and discussion
3.1 Isolation of biosurfactant producing microorganism
From the collected soil samples, eight morphologically different bacterial colonies were isolated and each bacterial isolate was screened for production of biosurfactant.
3.2 Screening for biosurfactant producing bacteria
3.2.1 Drop collapse assay
Among the eight bacterial isolates, culture broth of two isolates collapsed the drop of crude oil immediately within 1 min, implying the presence of biosurfactant in the culture media. The remaining six bacterial cultures could not collapse the crude oil drop.
3.2.2 ST measurement
The two bacterial isolates showing positive result in drop collapse assay could reduce the ST of the medium below 45 mN m−1 (Table 1). The isolates which can reduce the ST of the medium below 45 mN m−1, are considered to be biosurfactant‐producing microbes [33]. Strain MKVIT3 was selected for biosurfactant production for the preparation of NPs, since the ST reduction was highest in case of this strain.
Table 1.
ST of the bacterial isolates at different time intervals
| Bacterial isolates | Surface tension (mN m−1) at | |||||
|---|---|---|---|---|---|---|
| 0th h | 24th h | 48th h | 72th h | 96th h | 120th h | |
| Ca | 71.1 ± 0.30b | 71.0 ± 0.24 | 71.0 ± 0.40 | 69.9 ± 0.30 | 69.9 ± 0.23 | 69.8 ± 0.27 |
| MKVIT1 | 61.9 ± 0.40 | 54.4 ± 0.44 | 53.5 ± 0.43 | 50.0 ± 0.31 | 54.0 ± 0.33 | 55.2 ± 0.23 |
| MKVIT2 | 59.1 ± 0.30 | 50.5 ± 0.25 | 46.5 ± 0.30 | 47.2 ± 0.25 | 49.7 ± 0.15 | 52.7 ± 0.25 |
| MKVIT3c | 60.8 ± 0.30 | 45.8 ± 0.44 | 27.4 ± 0.22 | 31.5 ± 0.23 | 33.9 ± 0.24 | 38.8 ± 0.28 |
| MKVIT4 | 69.8 ± 0.12 | 68.5 ± 0.17 | 60.1 ± 0.24 | 60.0 ± 0.43 | 62.7 ± 0.41 | 62.8 ± 0.31 |
| MKVIT5 | 60.8 ± 0.15 | 57.7 ± 0.21 | 54.4 ± 0.16 | 57.8 ± 0.23 | 57.0 ± 0.16 | 57.3 ± 0.26 |
| MKVIT6 | 69.4 ± 0.16 | 59.2 ± 0.24 | 58.6 ± 0.22 | 57.1 ± 0.35 | 56.2 ± 0.40 | 56.1 ± 0.30 |
| MKVIT7c | 63.9 ± 0.40 | 49.4 ± 0.21 | 36.4 ± 0.41 | 38.3 ± 0.12 | 40.2 ± 0.31 | 43.3 ± 0.22 |
| MKVIT8 | 68.8 ± 0.12 | 60.5 ± 0.17 | 54.1 ± 0.24 | 57.8 ± 0.43 | 60.7 ± 0.41 | 62.8 ± 0.31 |
a Abiotic control
b Data expressed as mean ± SD (n = 5)
c Denotes the biosurfactant producing bacterial strains
3.3 Identification of the efficient isolate
The sequences of bacterial strain MKVIT3 was submitted to NCBI GenBank data base (Accession Number KT725781) and compared by blasting with the existing sequences. Results showed that the strain MKVIT3 had complete similarity (100%) with different P. aeruginosa strains that are included in the phylogenetic tree (Fig. 1). Thus, strain MKVIT3 was identified as P. aeruginosa MKVIT3.
Fig. 1.

Phylogenetic tree of strain MKVIT3 based on the 16S rDNA sequences and closest relatives
3.4 Extraction of crude biosurfactant
The yield of biosurfactant from strain MKVIT3, when grown in 2% (w/v) glucose containing MSM, was 3.24 g L−1. The colour of the crude biosurfactant was dark honey in appearance.
3.5 Characterisation of biosurfactant
3.5.1 Biochemical
In the ninhydrin test, Ruhemann's purple complex formation was absent implying the absence of amino acid or protein in the isolated biosurfactant. In the anthrone test, a bluish green colour formation was noted, indicating the presence of carbohydrates in the biosurfactant. In the saponification test, NaOH saponifies the lipids existing in the biosurfactant, indicating the presence of lipids in the biosurfactant. These results imply that the biosurfactant produced by bacterial strain MKVIT3 contains sugar and lipid molecules.
3.5.2 Fourier transform infrared spectroscopy
The FT‐IR spectra of the column purified biosurfactant is shown in Fig. 2. Comparison of the FT‐IR spectra with literature revealed the presence of different functional groups in the purified biosurfactant sample [34, 35]. The strong band at 3275.13 cm−1 indicate to hydroxyl (–OH) group stretching. The deformation vibrations at 1413.82 and 1365.6 cm−1 verified the presence of alkyl groups. Existence of ester carbonyl groups (1737.86 cm−1) and glycosidic bond (C–O–C) (1031.92 cm−1) was also confirmed. The peak at 2933.73 cm−1 indicated the presence of –CH aliphatic stretching vibrations of hydrocarbon chain of alkyl (CH2 ‐CH3) groups. The pattern of the absorption bands seen in the FT‐IR spectra indicated similarity with that of polysaccharide or polysaccharide‐like substances. Hence, from the above analysis, the purified biosurfactant is expected to be a glycolipid.
Fig. 2.
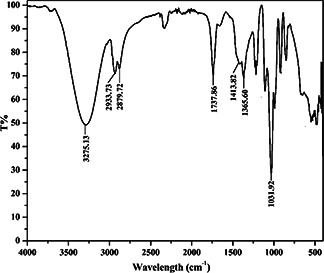
FT‐IR spectra of the column‐purified biosurfactant
3.5.3 Liquid chromatography–mass spectrometry
The column purified biosurfactant was further analysed in LC‐MS/MS from the total ion chromatogram in positive mode [36] to identify the compounds present in it (Fig. S1, supplementary figure). Five rhamnolipid congeners with the pseudo‐molecular ion peaks between 150 and 2000m/z were identified using LC‐MS/MS (Fig. S2, supplementary figure). The assignment of the five pseudo‐molecular ions was based on the comparison of congeners from the literature [37, 38]. Mono‐rhamnolipid homologues were found to be major (61.20%) constituents in the mixture. The molecular ion peak at m/z 661 corresponding to the [M‐H+Na2]+ sodium adduct ion of [Rha‐C14 ‐C14] was the predominant component.. A di‐rhamnolipid corresponding to the molecular [M + K]+ potassium adduct with m/z of 717 was the major isoform (12.69%) within the di‐rhamnolipid homologues and was recognised as [Rha‐Rha‐C10 ‐C12]/[Rha‐Rha‐C12 ‐C10]. The m/z value of 477 corresponded to [M + H]+ of the mono‐rhamnolipid [Rha‐C8 ‐C10]/[Rha‐C10 ‐C8]. The peak at m/z 623 and 667 has been assigned to [M + H]+ and molecular [M‐H + Na2]+ sodium adduct ion of di‐rhamnolipid [Rha‐Rha‐C10 ‐C8]/[Rha‐Rha‐C8 ‐C10], respectively. The m/z at 479 corresponded to [M‐H]− of the di‐rhamnolipid [Rha‐Rha‐C10]. Thus, the biosurfactant obtained from the P. aeruginosa MKVIT3 strain was a mixture of both mono and di‐rhamnolipid. The naturally occurring rhamnolipid are always found as a mixture of different rhamnolipid congeners, as witnessed with different strains of P. aeruginosa [39, 40, 41, 42].
3.6 Characterisation of silver NPs
To monitor the formation and stability of the synthesised silver NPs the UV‐Vis absorption spectra was recorded. It was seen that the surface plasmon resonance (SPR) of silver NPs was visualised at 440 nm (Fig. 3 a) which shows the UV‐visible spectra of silver NPs formation using 0.05 mol l−1 AgNO3 solution [43]. This result proved that the silver NPs can be synthesised in reverse micelles using biosurfactant as the stabiliser.
Fig. 3.
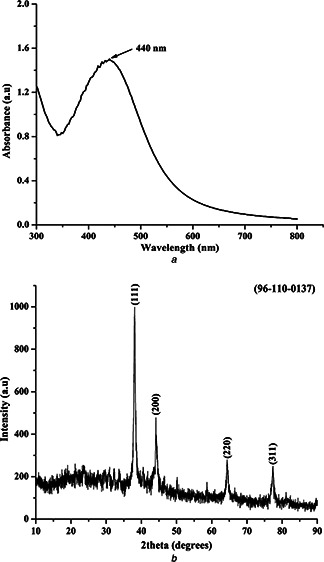
SPR of silver NPs was visualised at 440 nm and X‐ray diffraction pattern of the silver NPs matched with the standard JCPDS Ag pattern
a UV‐vis absorption spectra of the synthesised silver NPs
b XRD pattern spectra of the synthesised silver NPs
The X‐ray diffraction pattern of the silver NPs (Fig. 3 b) matched with the standard JCPDS Ag pattern (96‐110‐0137). Four sharp peaks were observed at 2θ values of 38.088, 44.168, 64.471 and 77.448 which were indexed as (111), (200), (220) and (311) bands of cubic structures of silver NPs. Using the Debye–Scherrer equation, Kλ /β cosθ, where K is the Scherrer constant with value from 0.9 to 1, λ the wavelength of the X‐ray, β full width at half maximum and θ the Bragg angle in radians, the average crystallite size of silver NPs was found to be about 20 nm [44, 45].
The size, shape and morphology of the silver NPs were elucidated using transmission electron microscopy. Fig. 4 a shows the representative TEM micrograph of the synthesised silver NPs. From the TEM image it was confirmed that the NPs were spherical in shape, largely uniform with a narrow size distribution. The silver NPs formed were in the size range of 2–30 nm with an average particle size of 17.89 ± 8.74 nm. The typical selected area electron beam diffraction (SAED) pattern (Fig. 4 b) of silver NPs showed four Debye–Scherrer rings corresponding to (111), (200), (220) and (311) planes of a fcc crystalline lattice [45, 46]. The above mentioned structural characteristic confirmed that the synthesised silver NPs have anisotropic crystalline structure and it also corresponded to the Powder‐XRD of the synthesised silver NPs. The particle size distribution histogram plot constructed from the TEM micrograph (ImageJ 1.49v, USA) is shown in Fig. 4 c. These results are in fine tuning with the observed optical properties of silver NPs.
Fig. 4.
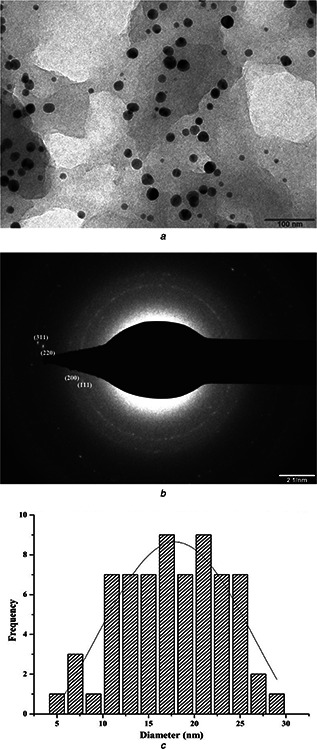
Representative TEM micrograph of the synthesised silver NPs and typical SAED pattern of silver NPs
a TEM of the synthesised silver NPs. Scale bar corresponds to 100 nm
b SAED pattern of silver NPs. Scale bar corresponds to 2 nm
c Histogram showing particle size distribution of the silver NPs
Zeta potential is a crucial parameter for the characterisation of stability in silver NPs suspensions. A minimum of ±30 mV zeta potential value is required for implication of stable nano‐suspension [47]. Whereas, the nano‐suspensions are least stable at isoelectric point, where the zeta potential value is zero. A zeta potential value of −30.9 mV was recorded for the synthesised silver NPs (Fig. 5) indicating an anionic charge on the NP surface. Thus, the result confirmed the high stability of the freshly prepared nano‐suspension.
Fig. 5.
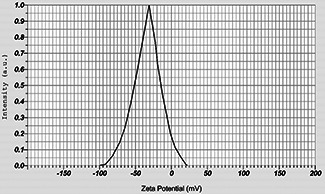
Zeta potential of the synthesised silver NPs
By substituting the values in (1) and (2), the average number of silver atoms present in one NP was found to be 8784 and the molar concentration of the silver NPs solution was found to be 6.88 × 10−4 mole L−1.
3.7 Antimicrobial activity
The biosurfactant and the silver NPs displayed antimicrobial activity towards the tested pathogenic strains of both bacterial and fungal culture (Table 2). The susceptibility of S. aureus, B. subtilis, E. coli and K. pneumoniae showed that both the biosurfactant and silver NPs possessed antibacterial properties that inhibited both Gram +ve and Gram –ve strains. Moreover, they also inhibited the growth of both the fungal strains. As seen in the Table 2, silver NPs have antimicrobial activity against all the strains of the microbes. Whereas, the biosurfactant have shown less activity against all the microbial strains as compared to the silver NPs. The silver NPs produced higher zone of inhibition, may be because of their larger specific surface area, smaller size and spherical shape which helps it to attach to the microbial cell wall and disquieting the functions of the microbes [48, 49].
Table 2.
Antimicrobial activity of the biosurfactant and silver NPs
| Diameter of zone of inhibition, in mm | ||||
|---|---|---|---|---|
| Test organisms | Biosurfactant | Silver NPs | Positive control | Negative control |
| S. aureus | 5.42 ± 1.0a | 10.8 ± 0.12 | 13.66 ± 1.52 | 0 |
| B. subtilis | 6.84 ± 0.54 | 9.45 ± 0.89 | 12.45 ± 1.57 | 0 |
| E. coli | 1.21 ± 0.89 | 8.73 ± 0.35 | 15.33 ± 1.15 | 0 |
| K. pneumoniae | 2.74 ± 0.5 | 10.32 ± 1.7 | 15.66 ± 0.57 | 0 |
| A. niger | 3.45 ± 2.1 | 9.96 ± 1.2 | 13.0 ± 2.0 | 0 |
| A. flavus | 5.67 ± 3.2 | 8.76 ± 0.72 | 13.66 ± 2.30 | 0 |
Values are expressed as mean ± SD (n = 3); Zone of inhibition not include the diameter of the well (7 mm). Positive control: Ciprofloxacin (10 µg/ disc), Fluconazole (1.0 mg/disc)
3.8 Cytotoxic activity
The effect of the biosurfactant and its silver NPs on cell viability was assessed using MTT assay as indicator of cell cytotoxicity. The MTT assay demonstrated the cytotoxicity indices as a measure of percentage cell mortality, which is calculated in Chang liver cell in a dose dependent manner at the end of 24 h incubation (Figs. 6 a and b). According to the findings of this study, cytotoxic level of biosurfactant in 10, 25, 50, 100, and 250 µg and control showed the inhibition of 89.02 ± 0.13%, 76.14 ± 0.78%, 50.23 ± 0.86%, 40.09 ± 0.79%, 20.32 ± 0.15% and 99.18 ± 0.05%, respectively. However, the silver NPs showed the inhibition level of 96.76 ± 0.03%, 90.54 ± 0.67%, 74.89 ± 0.14%, 62.01 ± 0.13% and 50.13 ± 0.86% for the same concentration. The IC50 value of the biosurfactant was found to be 50 µg, which was lower than that specified by silver NPs (250 µg).
Fig. 6.
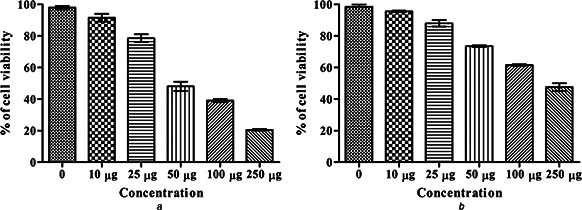
Cytotoxic activity
a Biosurfactant
b Silver NPs. Values are expressed as mean ± SD (n = 3)
The cytotoxic effect of the biosurfactant was probably due to the prevalence of a detergent‐like effect leading to the disruption of cell membranes [50, 51]. Reed, reported that margination and cell condensation, nuclear fragmentation and presence of apoptotic bodies are the main cause of the cytotoxicity of the biosurfactant [52]. The biosurfactant toxicity indexed by MTT reduction indicates for intracellular mitochondrial activities and could imply that the mitochondria simultaneously lead to the damage of cell membrane [53]. Based on this, we can deduce that the cell membranes could be the actionable target of biosurfactant [54, 55, 56].
Numerous possible mechanism of action for anticancer activity of silver NPs can be proposed. Cytotoxic activity of the silver NPs might be due to its physiochemical interaction with the intracellular DNA and proteins. Reports have shown that the cytotoxicity might also be due to initiation of apoptosis activated by the caspase‐3 enzyme [57]. Yuan et al. [58] reported that oxidative stress is one of the crucial mechanisms of cytotoxicity induced by silver NPs. The shape and size are important properties that influence the toxicity of silver NPs by elevating reactive oxygen species [59]. Because of the physicochemical differences, some silver NPs are broken‐down in the lysosomes and then the release of silver ions result in oxidative stress. In addition, oxidative stress can also lead to genotoxic stress and even upregulation of p53 gene [60, 61], which is an important lead for the application of nanomaterial as anticancer nano‐medicine, as upregulation of p53 gene initiates apoptosis [62]. Moreover, the presence of biosurfactant may be one of the reasons for the silver NPs to exhibit cytotoxic activity. Additional, studies are needed to determine the exact mechanism behind the cytotoxicity of the synthesised silver NPs.
4 Conclusion
In the present study, a new biosurfactant producing bacterial strain, namely P. aeruginosa MKVIT3, was isolated from an oil logging area in Vellore district of Tamil Nadu, India. The extracted biosurfactant was found to be glycolipid in nature. LC‐MS/MS analysis revealed the presence of five rhamnolipid congeners in the column‐purified biosurfactant. Spherical silver NPs were successfully synthesised using the column‐purified biosurfactant by microemulsion technique. The synthesised silver NPs were characterised using UV‐vis spectroscopy, Powder‐XRD, TEM and zeta potential. The characterisation study confirmed the formation of silver NP, their spherical shape and average diameter of 17.89 ± 8.74 nm. Zeta potential value of −30.9 mV suggested that the silver NPs are stable in the suspension. Comparative study of the antimicrobial activity revealed that the silver NPs are more potent than the biosurfactant in inhibiting the growth of microbes. Cytotoxic activity revealed that the biosurfactant are more effective than the silver NP with an IC50 value of 50 µg.
5 Acknowledgments
We are grateful to VIT University, Vellore, for support of this research, SAIF/IIT‐Bombay, Mumbai, for providing the TEM analysis.
6 References
- 1. Kouvaris P. Delimitis A. Zaspalis V. et al.: ‘Green synthesis and characterization of silver nanoparticles produced using Arbutus unedo leaf extract’, Mater. Lett., 2012, 76, pp. 18 –20 (doi: 10.1016/j.matlet.2012.02.025) [DOI] [Google Scholar]
- 2. Duran N. Marcato P.D. Alves O.L. et al.: ‘Mechanistic aspects of biosynthesis of silver nanoparticles by several fusarium oxysporum strains’, J. Nanobiotechnol., 2005, 3, p. 8 (doi: 10.1186/1477-3155-3-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Husseiny M.I. El‐Aziz M.A. Badr Y. et al.: ‘Biosynthesis of gold nanoparticles using Pseudomonas aeruginosa ’, Spectrochim. Acta A, 2007, 67, pp. 1003 –1006 (doi: 10.1016/j.saa.2006.09.028) [DOI] [PubMed] [Google Scholar]
- 4. Leela A. Vivekanandan M.: ‘Tapping the unexploited plant resources for the synthesis of silver nanoparticles’, African J. Biotechnol., 2008, 7, pp. 3162 –3165 [Google Scholar]
- 5. Retchkiman‐Schabe P.S. Canizal G. Becerra‐Herrera R. et al.: ‘Biosynthesis and characterization of Ti/Ni bimetallic nanoparticles’, Opt. Mater., 2006, 29, pp. 95 –99 (doi: 10.1016/j.optmat.2006.03.014) [DOI] [Google Scholar]
- 6. Jeong S.H. Yeo S.Y. Yi S.C.: ‘The effect of filler particle size on the antibacterial properties of compounded polymer/ silver fibers’, J. Mat. Sci., 2005, 40, pp. 5407 –5411 (doi: 10.1007/s10853-005-4339-8) [DOI] [Google Scholar]
- 7. Lin J. Zhou W. O'Connor C.J.: ‘Formation of ordered arrays of gold nanoparticles from CTAB reverse micelles’, Mater. Lett., 2001, 49, pp. 282 –286 (doi: 10.1016/S0167-577X(00)00385-2) [DOI] [Google Scholar]
- 8. Reddy A.S. Chen C.Y. Baker S.C. et al.: ‘Synthesis of silver nanoparticles using surfactin: a biosurfactant as stabilizing agent’, Mater. Lett., 2009, 63, pp. 1227 –1230 (doi: 10.1016/j.matlet.2009.02.028) [DOI] [Google Scholar]
- 9. Desai J.D. Banat I.M.: ‘Microbial production of surfactants and their commercial potential’, Microbiol. Mol. Biol. Rev., 1997, 61, pp. 47 –64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banat I.M. Franzetti A. Gandolfi I. et al.: ‘Microbial biosurfactants production, applications and future potential’, Appl. Microbiol. Biotechnol., 2010, 87, pp. 427 –444 (doi: 10.1007/s00253-010-2589-0) [DOI] [PubMed] [Google Scholar]
- 11. Makkar R.S. Cameotra S.S. Banat I.M.: ‘Advances in utilization of renewable substrates for biosurfactant production’, AMB Express, 2011, 1, p. 5 (doi: 10.1186/2191-0855-1-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poremba K. Gunkel W. Lang S. et al.: ‘Marine biosurfactants III: toxicity testing with marine microorganisms and comparison with synthetic surfactants’, Z. Naturforsch C: J. Biosci., 1991, 46, pp. 210 –216 [DOI] [PubMed] [Google Scholar]
- 13. Kiran G.S. Sabu A. Selvin J.: ‘Synthesis of silver nanoparticles by glycolipid biosurfactant produced from marine brevibacterium casei MSA19’, J. Biotechnol., 2010, 148, pp. 221 –225 (doi: 10.1016/j.jbiotec.2010.06.012) [DOI] [PubMed] [Google Scholar]
- 14. Gottenbos B. Grijpma D. Vander Mei H.C. et al.: ‘Antimicrobial effects of positively charged surfaces on adhering gram‐positive and gram negative bacteria’, J. Antimicrob. Chemother., 2001, 48, pp. 7 –13 (doi: 10.1093/jac/48.1.7) [DOI] [PubMed] [Google Scholar]
- 15. Bernheimer A. Avigad L.: ‘Nature and properties of a cytolytic agent produced by Bacillus subtilis ’, J. Gen. Microbiol., 1970, 61, pp. 361 –369 (doi: 10.1099/00221287-61-3-361) [DOI] [PubMed] [Google Scholar]
- 16. Vollenbroich D. Ozel M. Vater J. et al.: ‘Mechanism of inactivation of enveloped viruses by the biosurfactant surfactin from Bacillus subtilis ’, Biologicals, 1997, 25, pp. 289 –297 (doi: 10.1006/biol.1997.0099) [DOI] [PubMed] [Google Scholar]
- 17. Vollenbroich D. Pauli G. Ozel M. et al.: ‘Antimycoplasma properties and applications in cell culture of surfactin, a lipopeptide antibiotic from Bacillus subtilis ’, Appl. Environ. Microbiol., 1997, 63, pp. 44 –49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Hoogmoed C.G. Van der Kuijl‐Booij M. Van der Mei H.C. et al.: ‘Inhibition of streptococcus mutans NS adhesion to glass with and without a salivary conditioning film by biosurfactant‐ releasing streptococcus mitis strains’, Appl. Environ. Microbiol., 2000, 66, pp. 659 –663 (doi: 10.1128/AEM.66.2.659-663.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bharali P. Saikia J.P. Ray A. et al.: ‘Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent’, Colloids Surf. B Biointerfaces, 2013, 103, pp. 502 –509 (doi: 10.1016/j.colsurfb.2012.10.064) [DOI] [PubMed] [Google Scholar]
- 20. Desanto K.: US Patent, US20110123623, 2011.
- 21. Pornsunthorntawee O. Chavadej S. Rujiravanit R.: ‘Characterization and encapsulation efficiency of rhamnolipid vesicles with cholesterol addition’, J. Biosci. Bioeng., 2011, 112, pp. 102 –106 (doi: 10.1016/j.jbiosc.2011.03.009) [DOI] [PubMed] [Google Scholar]
- 22. Francy D.S. Thomas J.M. Raymond R.L. et al.: ‘Emulsification of hydrocarbons by subsurface bacteria’, J. Ind. Microbiol., 1991, 8, pp. 237 –246 (doi: 10.1007/BF01576061) [DOI] [Google Scholar]
- 23. Patowary K. Saikia R.R. Kalita M.C. et al.: ‘Degradation of polyaromatic hydrocarbons employing biosurfactant producing Bacillus pumilus KS2’, Ann. Microbiol., 2015, 65, pp. 225 –234 (doi: 10.1007/s13213-014-0854-7) [DOI] [Google Scholar]
- 24. Bodour A.A. Miller‐Maier R.M.: ‘Application of a modified drop‐collapse technique for surfactant quantitation and screening of biosurfactant‐producing microorganisms’, J. Microbiol. Methods, 1998, 32, pp. 273 –280 (doi: 10.1016/S0167-7012(98)00031-1) [DOI] [Google Scholar]
- 25. White T.J. Bruns T. Lee S. et al.: ‘PCR Protocols: A guide to methods and applications’, in Innis M.A. Gelfand D.H. Sninsky J.J. White T.J. (Eds.): ‘Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics’ (Academic Press, Inc., New York, 1990), pp. 315 –322 [Google Scholar]
- 26. Tamura K. Peterson D. Peterson N. et al.: ‘MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods’, Mol. Biol. Evol., 2011, 28, pp. 2731 –2739 (doi: 10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. George S. Jayachandran K.: ‘Analysis of rhamnolipid biosurfactants produced through submerged fermentation using orange fruit peelings as sole carbon source’, Appl. Biochem. Biotechnol., 2009, 158, pp. 694 –705 (doi: 10.1007/s12010-008-8337-6) [DOI] [PubMed] [Google Scholar]
- 28. Sawhney S.K. Singh R.: ‘Introductory practical biochemistry’ (Narosa Publishing House, India, 2000), pp. 16 –17 [Google Scholar]
- 29. Palanisamy P. Raichur A.M.: ‘Synthesis of spherical NiO nanoparticles through a novel biosurfactant mediated emulsion technique’, Mater. Sci. Eng. C, 2009, 29, pp. 199 –204 (doi: 10.1016/j.msec.2008.06.008) [DOI] [Google Scholar]
- 30. Liu X. Atwater M. Wang J. et al.: ‘Extinction coefficient of gold nanoparticles with different sizes and different capping ligands’, Colloids Surf. B Biointerfaces, 2007, 58, pp. 3 –7 (doi: 10.1016/j.colsurfb.2006.08.005) [DOI] [PubMed] [Google Scholar]
- 31. Bharali P. Saikia J.P. Paul S. et al.: ‘Colloidal silver nanoparticles/rhamnolipid (SNPRL) composite as novel chemotactic antibacterial agent’, Int. J. Biol. Macromol., 2013, 61, pp. 238 –242 (doi: 10.1016/j.ijbiomac.2013.07.006) [DOI] [PubMed] [Google Scholar]
- 32. Kotha A. Sekharam M. Cilenti L. et al.: ‘Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein’, Mol. Cancer Ther., 2006, 5, pp. 621 –629 (doi: 10.1158/1535-7163.MCT-05-0268) [DOI] [PubMed] [Google Scholar]
- 33. Viramontes‐Ramos S. Portillo‐Ruiz M.C. Ballinas‐Casarrubias M.L. et al.: ‘Selection of biosurfactan/bioemulsifier‐producing bacteria from hydrocarbon‐contaminated soil’, Brazil. J. Microbiol., 2010, 41, pp. 668 –675 (doi: 10.1590/S1517-83822010000300017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bordoloi N.K. Konwar B.K.: ‘Bacterial biosurfactant in enhancing solubility and metabolism of petroleum hydrocarbons’, J. Hazard. Mater., 2009, 170, pp. 495 –505 (doi: 10.1016/j.jhazmat.2009.04.136) [DOI] [PubMed] [Google Scholar]
- 35. Thenmozhi R. Sornalaksmi A.N. Nagasathya A. et al.: ‘Characterisation of biosurfactant produced by bacterial isolates from engine oil contaminated soil’, Adv. Environ. Biol., 2011, 5, pp. 2402 –2408 [Google Scholar]
- 36. Hultberg M. Alsberg T. Khalil S. et al.: ‘Suppression of disease in tomato infected by Pythium ultimum with a biosurfactant produced by Pseudomonas koreensis ’, BioControl, 2010, 55, pp. 435 –444 (doi: 10.1007/s10526-009-9261-6) [DOI] [Google Scholar]
- 37. Abdel‐Mawgoud A.M. Lepine F. Deziel E.: ‘Rhamnolipids: diversity of structures, microbial origins and roles’, Appl. Microbiol. Biotechnol., 2010, 86, pp. 1323 –1336 (doi: 10.1007/s00253-010-2498-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pantazaki A.A. Papaneophytou C.P. Lambropoulou D.A.: ‘Simultaneous polyhydroxyalkanoates and rhamnolipids production by Thermus thermophilus HB8’, AMB Express, 2011, 1, pp. 1 –13 (doi: 10.1186/2191-0855-1-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abalos A. Pinazo A. Infante M.R. et al.: ‘Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes’, Langmuir, 2001, 17, pp. 1367 –1371 (doi: 10.1021/la0011735) [DOI] [Google Scholar]
- 40. Benincasa M. Abalos A. Oliveira I. et al.: ‘Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa LBI from soapstock’, Antonie Van Leeuwenhoek, 2004, 85, pp. 1 –8 (doi: 10.1023/B:ANTO.0000020148.45523.41) [DOI] [PubMed] [Google Scholar]
- 41. Haba E., Abalos A. Jauregu O. et al.: ‘Use of liquid chromatography‐mass spectroscopy for studying the composition and properties of rhamnolipids produced by different strains of Pseudomonas aeruginosa ’, J. Surfactants Deterg., 2003, 6, pp. 155 –161 (doi: 10.1007/s11743-003-0260-7) [DOI] [Google Scholar]
- 42. Pornsunthorntawee O. Wongpanit P. Chavadej S. et al.: ‘Structural and physicochemical characterization of crude biosurfactant produced by Pseudomonas aeruginosa SP4 isolated from petroleum‐contaminated soil’, Bioresour. Technol., 2008, 99, pp. 1589 –1595 (doi: 10.1016/j.biortech.2007.04.020) [DOI] [PubMed] [Google Scholar]
- 43. Dipankar C. Murugan S.: ‘The green synthesis, characterization and evaluation of the biological activities of silver nanoparticles synthesized from Iresine herbstii leaf aqueous extract’, Colloids Surf. B: Biointerfaces, 2012, 98, pp. 112 –119 (doi: 10.1016/j.colsurfb.2012.04.006) [DOI] [PubMed] [Google Scholar]
- 44. Spreadborough J. Christian J.W.: ‘High temperature X‐Ray diffractometer’, J. Sci. Instrum., 1959, 36, pp. 116 –118 (doi: 10.1088/0950-7671/36/3/302) [DOI] [Google Scholar]
- 45. Kumar C.G. Mamidyala S.K.: ‘Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa ’, Colloids Surf. B: Biointerfaces, 2011, 84, pp. 462 –466 (doi: 10.1016/j.colsurfb.2011.01.042) [DOI] [PubMed] [Google Scholar]
- 46. Cullity B.D.: ‘Elements of X‐ray diffraction’ (Addison‐Wesley Publishing Co Inc., MA, 1978), pp. 233 –247 [Google Scholar]
- 47. Jacobs C. Muller R.H.: ‘Production and characterization of a budesonide nanosuspension for pulmonary administration’, Pharma. Res., 2002, 19, pp. 189 –194 (doi: 10.1023/A:1014276917363) [DOI] [PubMed] [Google Scholar]
- 48. Baker C. Pradhan A. Pakstis L. et al.: ‘Synthesis and antibacterial properties of silver nanoparticles’, J. Nanosci. Nanotechnol., 2005, 5, pp. 244 –249 (doi: 10.1166/jnn.2005.034) [DOI] [PubMed] [Google Scholar]
- 49. Guzman M.G. Dille J. Godet S.: ‘Synthesis of silver nanoparticles by chemical reduction method and their antibacterial activity’, Int. J. Chem. Bio. Eng., 2009, 2, pp. 104 –111 [Google Scholar]
- 50. Kim S.Y. Kim J.Y. Kim S.H. et al.: ‘Surfactin from Bacillus subtilis displays anti‐proliferative effect via apoptosis induction, cell cycle arrest and survival signaling suppression’, FEBS Lett., 2007, 581, pp. 865 –871 (doi: 10.1016/j.febslet.2007.01.059) [DOI] [PubMed] [Google Scholar]
- 51. Lee J.H. Nam S.H. Seo W.T. et al.: ‘The production of surfactin during the fermentation of cheonggukjang by potential probiotic Bacillus subtilis CSY191 and the resultant growth suppression of MCF‐7 human breast cancer cells’, Food Chem., 2012, 131, pp. 1347 –1354 (doi: 10.1016/j.foodchem.2011.09.133) [DOI] [Google Scholar]
- 52. Reed J.C.: ‘Apoptosis‐targeted therapies for cancer’, Cancer Cells, 2003, 3, pp. 17 –22 (doi: 10.1016/S1535-6108(02)00241-6) [DOI] [PubMed] [Google Scholar]
- 53. Inacio A.S. Mesquita K.A. Baptista M. et al.: ‘ In vitro surfactant structure‐toxicity relationships: implications for surfactant use in sexually transmitted infection prophylaxis and contraception’, PLoS One, 2011, 6, p. e19850 (doi: 10.1371/journal.pone.0019850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Abbasi H. Noghabi K.A. Ortiz A.: ‘Interaction of a bacterial monorhamnolipid secreted by Pseudomonas aeruginosa MA01 with phosphatidylcholine model membranes’, Chem. Phys. Lipids, 2012, 165, pp. 745 –752 (doi: 10.1016/j.chemphyslip.2012.09.001) [DOI] [PubMed] [Google Scholar]
- 55. Ortiz A. Teruel J.A. Espuny M.J. et al.: ‘Effects of dirhamnolipid on the structural properties of phosphatidylcholine membranes’, Int. J. Pharm., 2006, 325, pp. 99 –107 (doi: 10.1016/j.ijpharm.2006.06.028) [DOI] [PubMed] [Google Scholar]
- 56. Pantazaki A.A. Choli‐Papadopoulou T.: ‘On the Thermus thermophilus HB8 potential pathogenicity triggered from rhamnolipids secretion: morphological alterations and cytotoxicity induced on fibroblastic cell line’, Amino Acids, 2012, 42, pp. 1913 –1926 (doi: 10.1007/s00726-011-0917-z) [DOI] [PubMed] [Google Scholar]
- 57. Sriram M.I. Kanth S.B.M. Kalishwaralal K. et al.: ‘Antitumor activity of silver nanoparticles in Dalton's lymphoma ascites tumor model’, Int. J. Nanomed., 2010, 5, pp. 753 –762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yuan X. Setyawati M.I. Tan A.S. et al.: ‘Highly luminescent silver nanoclusters with tunable emissions: cyclic reduction–decomposition synthesis and antimicrobial properties’, NPG Asia Mater., 2013, 5, p. e39 (doi: 10.1038/am.2013.3) [DOI] [Google Scholar]
- 59. Giovanni M. Tay C.Y. Setyawati M.I. et al.: ‘Toxicity profiling of water contextual zinc oxide, silver, and titanium dioxide nanoparticles in human oral and gastrointestinal cell systems’, Environ. Toxicol., 2014, 30, pp. 1459 –1469 (doi: 10.1002/tox.22015) [DOI] [PubMed] [Google Scholar]
- 60. Setyawati M.I. Tay C.Y. Leong D.T.: ‘Effect of zinc oxide nanomaterials‐induced oxidative stress on the p53 pathway’, Biomater., 2013, 34, pp. 10133 –10142 (doi: 10.1016/j.biomaterials.2013.09.024) [DOI] [PubMed] [Google Scholar]
- 61. Ng K.W. Khoo S.P. Heng B.C. et al.: ‘The role of the tumor suppressor p53 pathway in the cellular DNA damage response to zinc oxide nanoparticles’, Biomater., 2011, 32, pp. 8218 –8225 (doi: 10.1016/j.biomaterials.2011.07.036) [DOI] [PubMed] [Google Scholar]
- 62. Satapathy S.R. Mohapatra P. Preet R. et al.: ‘Silver‐based nanoparticles induce apoptosis in human colon cancer cells mediated through p53’, Nanomed., 2013, 8, pp. 1307 –1322 (doi: 10.2217/nnm.12.176) [DOI] [PubMed] [Google Scholar]


