Abstract
The last decade has observed a rapid advancement in utilising biological system towards bioremediation of metal ions in the form of respective metal nanostructures or microstructures. The process may also be adopted for respective metal nanoparticle biofabrication. Among different biological methods, bacteria‐mediated method is gaining great attention for nanoparticle fabrication due to their eco‐friendly and cost‐effective process. In the present study, silver nanoparticle (AgNP) was synthesised via continuous biofabrication using Aeromonas veronii, isolated from swamp wetland of Sunderban, West Bengal, India. The biofabricated AgNP was further purified to remove non‐conjugated biomolecules using size exclusion chromatography, and the purified AgNPs were characterised using UV–visible spectroscopy, X‐ray diffraction, field emission scanning electron microscopy and transmission electron microscopy (TEM). Additionally, the presence of proteins as capping and stabilising agents was confirmed by the amide‐I and amide‐II peaks in the spectra obtained using attenuated total reflection Fourier transform infrared spectroscopy. The size of biofabricated AgNP was 10–20 nm, as observed using TEM. Additionally, biofabricated AgNP shows significant antibacterial potential against E. coli and S. aureus. Hence, biofabricated AgNP using Aeromonas veronii, which found resistant to a significant concentration of Ag ion, showed enhanced antimicrobial activity compared to commercially available AgNP.
Inspec keywords: silver, nanoparticles, microorganisms, nanofabrication, purification, chromatography, ultraviolet spectra, visible spectra, X‐ray diffraction, field emission scanning electron microscopy, transmission electron microscopy, proteins, molecular biophysics, biochemistry, molecular configurations, attenuated total reflection, Fourier transform infrared spectra, particle size, antibacterial activity, biomedical materials, nanomedicine
Other keywords: capping agents, stabilising agents, amide‐I peaks, amide‐II peaks, attenuated total reflection Fourier transform infrared spectroscopy, antibacterial potential, E. coli, S. aureus, Aeromonas veronii, antimicrobial activity, size 10 nm to 20 nm, Ag, proteins, TEM, transmission electron microscopy, field emission scanning electron microscopy, X‐ray diffraction, UV‐visible spectroscopy, size exclusion chromatography, nonconjugated biomolecules, purification, swamp wetland, Aeromonas veronii, cost‐effective process, eco‐friendly, bacteria‐mediated method, biological methods, metal nanoparticle biofabrication, microstructures, metal nanostructures, metal ions, bioremediation, biological system, mangrove swamp, bacteria, silver nanoparticles
1 Introduction
Nanoparticles are the microscopic particles that bridge the gap between atomic scale and bulk particles [1]. The diverse physico‐chemical properties of nanoparticles make them popular for many applications. These physico‐chemical properties are mainly because of surface functionalities and high surface to the volume ratios [2, 3, 4, 5]. Nanoparticles play important roles in textile, material, semiconductor, sensor, cosmetics and therapeutic industries [6, 7, 8, 9]. Although, chemical and physical processes are huge success for nanoparticles synthesis, green synthesis is required to unload the energy consumption and the pressure on ecosystem. Green synthesis is an easy, eco‐friendly, reliable and non‐toxic process than other commercial processes. There are a number of biological resources reported for the green synthesis of nanoparticles including, plant extract, bacteria and fungi [10, 11]. The advantages of bacteria over other biological resources are mainly easy fabrication, low‐cost culture maintenance and re‐inoculations, and also can be adopted for relatively large‐scale production through fermentation [10].
In metal and microbe interaction, for the same reason, the molecular mechanism of metal ion invagination and clearance has now become the current issues of environmental toxicology. Silver is one of the most popular elements in the periodic table with potential biological applications [12]. Additionally, modifying the properties of transition element like silver (Ag) into functional materials attracts the process as an emerging class in analytical chemistry. Silver nanoparticle (AgNP) has shown very different physico‐chemical properties upon decreasing the size near to Fermi wavelength, because of the strong quantum confinement of free electrons [13]. Additionally, the chemiluminescence property of AgNP has the potential to expand the chemiluminescence‐mediated analytical applications [14]. Apart from therapeutic applications, AgNP is also used in photographic reaction and catalysis due to its size‐dependent optical and electrical properties [15]. Following green chemistry approach, ionic silver (Ag+) is reduced to elemental silver (Ag), which further self‐assemble and stabilise in silver nano‐crystal structure on interaction with microbial biomolecules [16]. Some researchers reported the involvement of metabolic process, like the use of nitrate or organic compounds by cyanobacteria as one of the mechanism for AgNP synthesis [1]. Published works have shown the involvement of proteins in the reduction of metals during biological synthesis of metal nanoparticle [17, 18, 19]. Later, Xie et al. [11] have reported that reduction kinetics of silver ion and subsequent anisotropic growth of nanocrystal are driven by different amino acids, rather than proteins during biological synthesis. Proteins, DNA, lipopolysaccharides and phospholipids are the predominant biomolecules involved in reduction and capping of nanoparticles in microbial green synthesis [15]. Besides the biomolecules, the presence of phytochemicals and secondary metabolites are involved in stabilisation of the nanoparticles [20]. Biological moieties capped nanoparticles are found thermodynamically stable than non‐capped nanoparticles, due to their prominent biocompatible surface functionality and hydrophilicity [21]. In addition to stability, biogenic AgNP has proven as an excellent antimicrobial agent when capped with antibodies, compared to antibiotic and chemically synthesised AgNP, independently [22]. The enhanced antimicrobial potential was due to increased reactive oxygen species generation in pathogenic bacteria because of synergistic interaction of AgNP and the capped moieties with pathogenic bacteria. Green synthesis of nanoparticles mostly exploits bacteria over fungi and plants, owing to their easy availability and inexpensive culture maintenance. In addition to that, diverse metabolism and efficient strategies to overcome the metal toxicity gives them additional advantage for metal nanoparticle synthesis [23]. Various studies have been reported for extracellular as well as intracellular synthesis of AgNPs from bacteria like Bacillus thuringiensis [10], Pseudomonas hibiscicola [24], Deinococcus radiodurans [25] and so on. Despite the significant progress in nanoparticle synthesis from bacteria, very few articles have reported live bacteria‐mediated AgNP synthesis, which can further be adopted in fermentation for large‐scale production [10].
Taking this into consideration, we studied the continuous synthesis of intracellular AgNPs and its biofabrication using live bacteria isolated from mangrove swamp. Swamp areas, like Sunderbans in West Bengal, India, are floodplain formed from major downstream rivers. These areas are having high moisture and hydric soil with minerals, and supposed to have bacterial species with bioactive compounds to withstand stressful habitat including metal resistance. Thus, the bacteria were initially screened and cultured in nutrient broth medium. Later on, bacteria with the maximum metal resistance were selected from other bacteria on the basis of minimum inhibitory concentration (MIC) assay. The selected metal resistant bacterium was further identified through 16s rRNA sequencing. Biofabricated AgNP was subjected to size exclusion chromatography (SEC) to remove the loosely bound non‐conjugated biomolecules from the AgNP. UV–visible spectroscopy, field emission scanning electron microscopy (FESEM), transmission electron microscopy (TEM) and X‐ray diffraction (XRD) were used to characterise the fabricated AgNP. Additionally, attenuated total reflection Fourier transform infrared (ATR‐FTIR) spectroscopy was also used to determine the possible functional groups adsorbed over biofabricated AgNP surface, and the backlight assay to compare the antibacterial property of biofabricated AgNP with commercial AgNP.
2 Materials and methods
2.1 Media and chemicals
Silver nitrate (AgNO3) and sephadex (G‐100) were purchased from Sigma‐Aldrich Chemicals, USA. Nutrient broth, tannic acid and sodium hexametaphosphate (SHMP) were purchased from HiMedia Laboratories Pvt Ltd, India. Glutaraldehyde was purchased from Merck, India. All other reagents used were of analytical grade. Bacterial strains like Escherichia coli (MTCC 443) and Staphylococcus aureus (MTCC 737) were purchased from Institute of Microbial Technology (IMTECH) Chandigarh, India.
2.2 MIC of AgNO3 against isolated bacteria
The water sample was collected from swamp formed near Hooghly River, West Bengal, India, in a sterile bottle, and stored at 4°C. The water sample was then serially diluted, and plated on nutrient agar. The plates were incubated at 37°C for 24 h, and culture from single colony was again streaked on agar plates, in order to get a pure culture. The isolated culture was identified by 16s rRNA sequencing at SciGenom Laboratorsy (Kochi, India). The contig sequence thus obtained was aligned using BLAST to identify the closest bacterial genome. The phylogenetic tree was also constructed using MEGA (version 5.0) software. Additionally, AgNO3 MIC against the cultured bacteria was observed for studying the silver resistance in bacteria. For MIC, 20 μl of overnight grown bacterium culture (bacterial population) was inoculated with different concentrations of AgNO3 in a 96‐well plate, with a final culture volume of 300 μl for 24 h. The growth was observed by optical density (OD) at 600 nm at 37°C using a micro‐plate reader (Synergy H1 hybrid reader, Biotek, Singapore).
2.3 Biofabrication and purification of AgNP
To synthesise AgNP during the bacterial growth in the culture medium, 0.312 mM AgNO3 (< MIC value for the bacteria) was added into the culture medium after the log phase was reached. The biosynthesis and purification of AgNP was obtained as reported in published protocol [10]. Briefly, AgNP was synthesised in 500 ml nutrient broth flask containing 0.312 mM AgNO3, and inoculated with 1 ml overnight culture of the isolated bacterial strain. The bacterial culture was allowed to grow for 24 h, and then was centrifuged to separate the bacterial biomass (pellet) from the media (supernatant). The bacterial pellet was then re‐suspended in deionised water, and lysed with lysozyme and incubated for 1 h at 37°C. Following the incubation, sonication at 90% amplitude for 10 min was done to complete the bacterial membrane disruption. Thus, the intracellular AgNP was released into the aqueous media upon cell lysis. AgNP containing cell lysate was then centrifuged at 10,000 rpm for 30 min, and the resulting supernatant containing AgNP was filtered through 0.22 μm membranes. The filtered cell suspension was subjected to G‐100 sephadex resin to separate the AgNP from non‐conjugated moieties using SEC and 1 mM SHMP in deionised water as mobile phase.
2.4 Characterisation of biofabricated AgNP
2.4.1 UV–visible spectroscopy
The biofabrication of AgNP was initially monitored using UV–visible spectroscopy (Cary 100, Agilent Technology, Singapore). The cell suspensions containing biofabricated AgNP were scanned at a resolution of 2 nm within the range of 200–800 nm. The UV–visible spectra of AgNP were examined before and after SEC to check AgNP presence in different elution fractions.
2.4.2 Zeta analyser and ATR‐FTIR spectroscopy
Colloidal particles when dispersed in solution obtain a charge on its surface, which indirectly contributes in the particle dispersivity. Particles with highly negative (> −30 mV) or highly positive (> + 30 mV) zeta potential are considered as very stable nanoparticles [20]. Zeta analyser is used to determine the surface potential of the fabricated nanoparticle. Zeta potential of biofabricated AgNP was examined before and after SEC of elutions 3 and 5, relatively pure fractions.
In Fourier transform infrared (FTIR), chemical bond vibration specific characterisation was done for qualitative analysis of the chemical bonds adsorbed onto the surface, since bonds vibrate only at specific infrared wavenumbers. Hence, the bond level characterisation of AgNP and corona were done by ATR‐FTIR (Alpha FTIR Bruker, Germany) between 2000 and 500 cm−1, averaged over 24 scans with 4 cm−1 resolution.
2.4.3 TEM and FE‐SEM
The morphology and size distribution of biofabricated AgNP was analysed using TEM (Technai F30, FEI, Netherland). Biofabricated AgNP suspension was sonicated, and 50 μl of sonicated sample was placed on carbon‐coated copper grid. The grid was placed in front of lamp to evaporate the solvent prior to TEM analysis. Further, the FE‐SEM (Nova Nano SEM 450, FEI, Netherland) was also used for observing the morphology of fabricated AgNP. For both TEM and FE‐SEM, samples were taken from SEC elution fractions. For analysing the morphological changes in bacteria upon AgNP synthesis, bacterial sample (with and without AgNO3) from early stationary phase was fixed by glutaraldehyde and tannic acid, prior to their analysis using FE‐SEM. Both the samples were gold coated, and then scanned at 10 kV.
2.4.4 X‐ray diffraction
Crystal structure and particle size were determined using XRD. AgNP suspension was air dried in hot air oven at 50°C, and the dried samples were used for analysis. Structural signature of biofabricated AgNP spectrum was determined by X‐ray diffractograms (Ultima IV model Rigaku, Tokyo, Japan) recorded with Cu K‐alpha radiation at a current of 40 mA and voltage of 40 kV with a scan rate of 10°/min. The diffracted X‐ray intensities were measured from 20° to 80°, 2θ angle. The size of the crystal was measured using Scherrer's equation as follows:
| (1) |
where λ is the wavelength of X‐ray (1.540 × 10−10 m), K = 0.9, proportionality coefficient (shape factor), θ is the Braggs angle and β is the full width at half maximum in radians.
2.4.5 Antibacterial activity of biofabricated AgNP
The antibacterial activity of biofabricated AgNP was evaluated on gram‐negative (E. coli) and gram‐positive bacteria (S. aureus). The antibacterial property was evaluated through LIVE/DEAD BacLight viability assay. The effect of biofabricated AgNP on E. coli and S. aureus cells was studied using a LIVE/DEAD BacLight viability fluorescence kit (L7007, Molecular probes, Invitrogen, USA) using a fluorescence microscope with a 40X objective lens. The samples were prepared for image acquisition at fluorescence microscope, following the protocol as described in our previous article [10]. For a comparative analysis, we took commercial Sigma‐Aldrich AgNP suspension (product code‐730785, Sigma‐Aldrich, USA) as a standard. The bacteria were treated with 10 μg biofabricated and commercial AgNP, and stained with BacLight viability kit, before imaging at fluorescence microscope.
3 Results and discussion
3.1 Isolation, identification and determination of MIC of bacterial strain
Initially, the isolated bacterium was characterised through gram staining. Gram staining is a differential staining technique, which utilises crystal violet and a counter stain to differentiate gram‐positive from gram‐negative bacteria [26]. Fig. 1 a shows pink colour bacteria, gram‐negative bacteria only give pink colour upon staining with safranin as a counter stain. Thus, the isolated bacterium was gram‐negative bacteria. To determine the MIC value of Ag+ for the bacterium, bacterial growth was monitored by observing change in OD600 nm of the culture with time in presence of varying Ag+ concentration. From the stationary phase of growth kinetics, OD at 600 nm was plotted against AgNO3 concentration (Fig. 1 b). The data directly represent the MIC of Ag+ against the bacterial isolate. The MIC concentration of 0.312 mM AgNO3 was obtained for ≥50% bacterial growth, than other higher concentrations. After optimising MIC concentration, the bacterial strain was identified from contig sequence obtained from 16s rRNA sequencing. The contig sequence of 16s rRNA of the isolated bacterium was found to be 99% identical with Aeromonas veronii. Fig. 1 c shows the maximum neighbourhood tree, indicating Aeromonas veronii TH0426 the most identical strain to our isolated bacterium, as analysed using clustalW multiple sequence alignment [27]. Aeromonas veronii is a rod‐shaped gram‐negative bacteria, predominantly found in freshwater and sewage [28, 29].
Fig. 1.
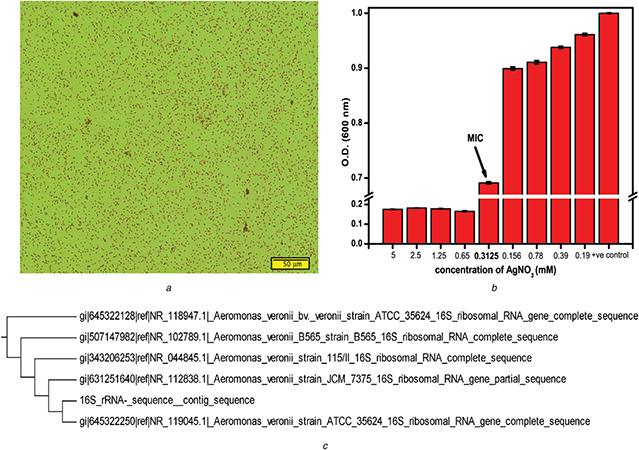
Screening and identification of bacterial strain
(a) Gram staining of bacteria isolated from mangrove swamp (scale bar is 50 μm), (b) MIC of AgNO3 against the isolated gram‐negative bacteria, error bar represent SEM, (c) 16s rRNA sequence based phylogenetic tree
3.2 UV–visible spectroscopic analysis
Biofabricated AgNP was continuously synthesised during the growth of Aeromonas veronii. The reduction of silver ion into AgNP was confirmed by the change in colour of culture media from 0 to 24 h of growth (Fig. 2 a). The difference in colour before and after synthesis is due to the surface plasmon resonance (SPR) property of AgNP [30]. Fig. 2 b shows the UV–visible spectra of AgNP suspension obtained during continuous bacterial growth. SPR peak at 413 nm was observed in UV–visible spectra, which confirmed the presence of biofabricated AgNP in supernatant of cell lysate, a characteristic SPR peak of AgNP [31, 32]. Interestingly, before cell lysis, the nanoparticle was observed in pellet containing intact cells and not in the media supernatant, indicating the intracellular fabrication of AgNP. The SPR peak for before and after size exclusion purification was found to be similar, confirming the purification of biofabricated AgNP from non‐conjugated biolmolecules did not affect the SPR property (Fig. 2 c). Fabrication of different sizes and shapes nanoparticle using bacteria were obtained by varying conditions like pH, temperature, incubation period and concentration of metal ion [25]. Although, different studies have reported to fabricate in vitro AgNP, i.e. using bacterial biomass [20, 33], we fabricated AgNP from live bacteria, which can be upgraded for industrial‐scale production using biofermentation. In contrast, the live bacterial cell found at the end of the batch can be further used as inoculums for next fed‐batch culture. Since A. veronii can sustain significant metal resistance, it may offer advantages over other freshwater bacteria where nanoparticles synthesis is an alternative for bioremediation. Our results are consistent with previous studies on intracellular synthesis of nanoparicle [34].
Fig. 2.
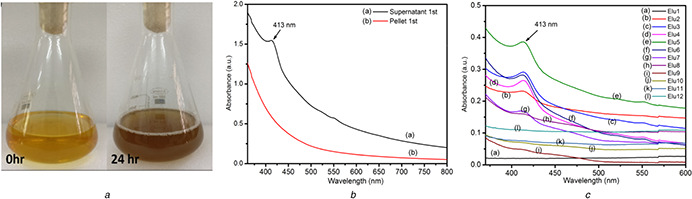
Analysis of AgNP biofabrication
(a) Visual observation of AgNP synthesis by colour change, (b) Absorption spectrum of biofabricated AgNP in resuspended cell lysate (pellet first) and bacterial growth medium (supernatant first), (c) Absorption spectrum of all the eluted fractions from size‐exclusion column
3.3 Zeta potential analysis
The potential difference between the dispersion medium and the accessible surface of dispersed particle is known as surface potential of the particle, or zeta potential. Fig. 3 represents the zeta potential of fabricated AgNP before and after the purification. Zeta potential of biofabricated AgNP before SEC was found to be −16.0 mV, which is due to the adsorption of negatively charged biomolecules (Fig. 3 a). Figs. 3 b and c show the zeta potentials of two eluents (eluents 3 and 5) with high AgNP specific SPR peak. The surface potential of biofabricated AgNP was more negative (−30.5 and −23.6 mV for eluents 3 and 5, respectively) after purification, compared to before purification. The increase in negative potential of biofabricated AgNP is likely because of negatively charged SHMP adsorption onto the AgNP surface, in addition to the biomolecules already adsorbed onto the surface. SHMP is reported for its protective and stabilising tendency for nanoparticles [35]. Thus, increase in negative surface potential of the purified AgNP further increases the particle monodispersity and stability.
Fig. 3.
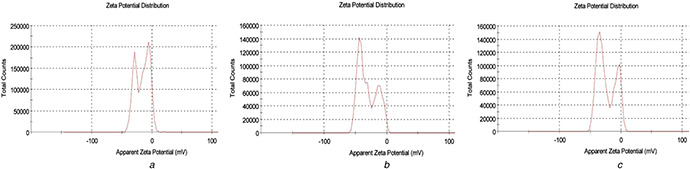
Zeta potential of biofabricated AgNP
(a) Before purification and after purification using SEC of, (b) Third elution fraction, (c) Fifth elution fraction
3.4 ATR‐FTIR analysis
ATR‐FTIR analysis was performed for the biofabricated AgNP to identify the functional groups of possible biomolecules responsible for capping and stabilising the nanoparticle. The bond vibration level study of the samples, pellet and supernatant collected upon cell lysis were analysed (Fig. 4 a). Absorption peaks at 534, 585, 520 and 579 cm−1 for both ATR‐FTIR spectra of pellet and supernatant confirm the presence of AgNP in the samples [36]. Nevertheless, the presence of 534 cm−1 peak in supernatant indicated major fraction of AgNP population partition into supernatant, upon centrifugation of cell lysate. Apart from this peak, the figure showed another two peaks around 1640 cm−1 and 1545, 1549 cm−1 for spectra of pellet and supernatant, which indicated the presence of amide‐I bond corresponding to C = O stretching, and amide‐II corresponding to N–H bending and C–N stretching vibrations, respectively. These two amide peaks in the FTIR analysis supported the presence of protein in cell‐free filtrate, and protein interaction with AgNP [37]. It was also reported that many enzymes like NADH‐dependent reductase and nitrate‐dependent reductase of the electron transport chain possibly take part in the reduction of Ag+ to Ag0. The presence of biomolecules like proteins, peptides and DNA on the interface of nanoparticles has shown to bestow novel physico‐chemical and physiological properties on them, which can have many biological applications [38]. Thus, the presence of proteins and peptides in our biofabricated AgNP can be used as an alternative biocompatible agent for therapeutic and analytical applications.
Fig. 4.
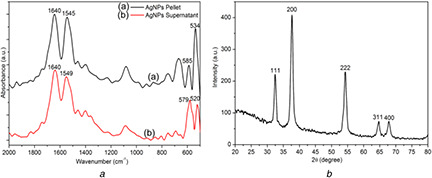
Characterisation of functional groups and crystalline structure
(a) FTIR spectra of biofabricated AgNP from A. veronii, (b) XRD pattern of biofabricated AgNP after purification
3.5 XRD analysis
XRD technique was established to identify the phase, orientation and grain size of nanoparticles. The XRD pattern in Fig. 4 b indicated the crystalline nature of biofabricated AgNP, after purification. The Bragg's diffractions at 2θ angle of 32.364, 37.6648, 54.185, 64.7219 and 67.832 correspond to different Miller indices of (111), (200), (222), (311) and (400), respectively. These indices were well indexed to tetrahedral face‐centred cubic crystal structure of biofabricated AgNP, which match with the reported reference value of joint committee on power diffraction standards. The similar type of crystalline structure of AgNP was previously reported in published studies [3, 39]. The average particle sizes of AgNP were determined using Scherrer's equation (1)
where λ is the wavelength of X‐ray (1.540 × 10−10 m), K is 0.9, a proportionality coefficient (shape factor), θ is the Braggs angle and β is the full width at half maximum in radians. According to above equation, size of fabricated AgNP was calculated to 15.13 nm.
3.6 FE‐SEM and TEM analysis
For bacterial morphology, FE‐SEM micrographs were obtained for intact and AgNO3 treated bacteria, as shown in Fig. 5. The FE‐SEM micrograph indicated that the intact bacterium is rod shape (Fig. 5 a). The live rod‐shaped bacteria with intact membrane in FE‐SEM images further supported our 16s rRNA sequencing for bacterial identification and MIC assay. The spherical molecules attached over the cell membrane of bacteria in Fig. 5 b show the presence of fabricated AgNP on their surface. The morphology of biofabricated AgNP was found to be spherical in shape with sizes <50 nm.
Fig. 5.
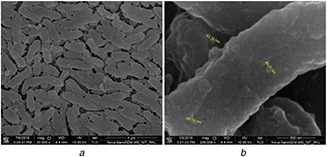
FE‐SEM micrographs of Aeromonas veronii
(a) Surface morphology of bacterium, (b) Bacterial surface showing fabricated AgNP
To obtain a better resolution of biofabricated AgNPs after purification, elutions 3 and 5 were scanned using TEM. Figs. 6 a and b show the spherical shape of fabricated AgNP in TEM micrographs. The sizes of biofabricated AgNP were found in a range of 5–15 nm. The irregular shape and size of AgNP are commonly found in green synthesis, but our biofabicated AgNP shapes were found consistently spherical before and after the purification, as shown in FE‐SEM (Fig. 5 b) and TEM (Fig. 6 a) micrographs [34, 40]. Earlier reports suggested fabrication of larger size nanoparticles of 200, 80.98 and 300 nm from bacteria like Pseudomonas stutzeri, Bacillus megaterium and Verticillum sp, respectively [25, 34].
Fig. 6.
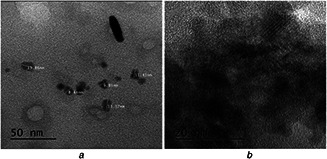
TEM analysis of biofabricated AgNPs
(a) Spherical shape biofabricated AgNP, (b) High‐resolution image of small size fabricated AgNP
3.7 Antibacterial activity of biofabricated AgNP
The antibacterial activity of biofabricated AgNP was evaluated using LIVE/DEAD Baclight bacterial viability kit. This LIVE/DEAD commercial kit uses the formulation of two dyes, Syto9 and propidium iodide (PI) dyes, to determine the bacterial viability through fluorescence imaging. The PI dye is a cell impermeable dye, which gives red fluorescence upon binding to nucleic acids, whereas Syto9 is a permeable dye but having less affinity towards nucleic acid. Upon the treatment, when cell membrane is compromised or deformed, PI dye enters the cell and intercalate to nucleic acids, hence give red colour to dead/deformed cells. Otherwise, Syto9 permeate and intercalate to nucleic acids where the membrane remains intact upon the treatment, and show fluoresce green colour. Hence, the deformed/non‐viable cells appear red and viable cells appear green on staining with the BacLight viability kit. As shown in Figs. 7 a ‐i and b ‐i, untreated E. coli and S. aureus exhibit only green fluorescence, indicating 100% cell viability. However, Figs. 7 a ‐ii and 7b ‐ii show biofabricated AgNP treated E. coli and S. aureus with 38 and 35% live cells, confirming cell death more than 60%. On the other hand, 10 µg/ml commercial AgNPs from sigma did not show significant antibacterial potential, and treated culture showed 70% E. coli and 64% S. aureus viable cells (Figs. 7 a ‐iii and 7b ‐iii). The cell viability percentage was calculated from fluorescence image of bacteria using imageJ software [41]. The Baclight result indicates the loss of membrane integrity is the one of the main mechanism for antibacterial activity of biofabricated AgNP. The relatively higher fraction of non‐viable cells in biofabricated AgNP treated bacterial culture indicated higher efficacy than commercial AgNP. The stronger antibacterial activity of biofabricated AgNP can be attributed to the synchronised antimicrobial activity of adsorbed biomolecule of nanoparticle corona and nanoparticle itself. Although the exact mechanism of bactericidal action of biofabricated AgNP is not well defined. However, most accepted hypothesis attributed to Ag+ ‐mediated perturbation of cytoskeleton assembly, resulting in deformed cell membrane/texture, as also observed in our studies [42].
Fig. 7.
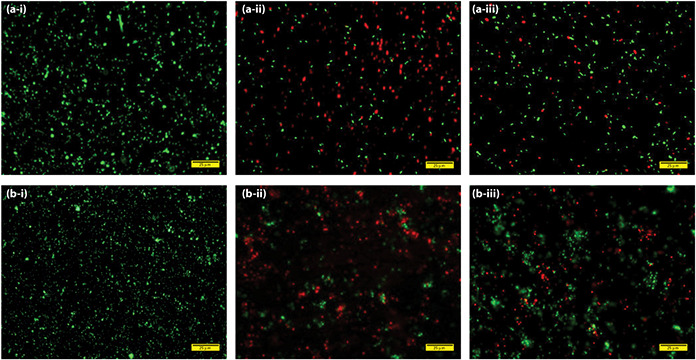
Fluorescence microscopy images of E. coli and S. aureus cells stained with LIVE/DEAD Backlight viability kit
(a‐i, b‐i) Intact E. coli and S. aureus cells, (a‐ii, b‐ii) Bacteria treated with 10 μg/ml biofabricated AgNP, (a‐iii, b‐iii) Bacteria treated with 10 μg/ml commercial AgNP from Sigma‐Aldrich (scale bar is 25 μm)
4 Conclusion
In this report, we optimised the biofabrication of homogenous AgNP using live bacteria isolated from mangrove swamp area. The bacterial strain was identified as a freshwater bacterium, Aeromonas veronii, through 16s rRNA sequencing, and the bacteria were screened based on its ability to resist high metal concentration, specifically Ag+ ion. MIC of AgNO3 against the Aeromonas veronii was 0.321 mM, which is relatively very high concentration considering the antibacterial property of the ion, and was used for the AgNP fabrication. The active biomolecules, responsible for capping and stabilisation of AgNP, were comprised of mostly proteins, as evident from ATR‐FTIR spectra. The bioreduction of AgNP was initially monitored by UV–visible spectroscopy, and later XRD technique further confirmed the synthesis of the tetrahedral face‐centred cubic packed AgNP. Additionally, FE‐SEM micrographs indicated the spherical fabricated AgNP on bacterial cell, but relatively in very less quantity compared to microorganism doing the extracellular fabrication. The size of biofabricated AgNP was found to be <15 nm, as depicted in TEM micrographs. Hence, unlike other bacteria‐mediated fabrication where the fabrication was done using cell mass, we successfully fabricated significant concentrations of AgNP using live bacteria. However, the fabrication took relatively more time, i.e. 24 h, compared to when cell mass is used, which can be compensated by adopting the live bacteria‐mediated nanoparticle fabrication in biofermentation. Moreover, the nanoparticle fabricated here showed enhanced antibacterial potentials compared to chemically synthesised commercial counter part, which furhter enhanced the novelty of the work.
5 Acknowledgment
The authors are thankful to the Department of Ceramic Engineering, Department of Chemical Engineering and Department of Physics, National Institute of Technology, Rourkela, Odisha, India, for providing the FE‐SEM/SEM, TEM and XRD facilities, respectively, for fulfilment of the research work. For the financial support, we acknowledge Department of Science and Technology (IFA‐13, CH91), Department of Biotechnology (MED/2015/102) and Ministry of Human Resource and Development, Govt. of India, India.
6 References
- 1. Thakkar K.N. Mhatre S.S. Parikh R.Y.: ‘Biological synthesis of metallic nanoparticles’, Nanomed. Nanotechnol. Biol. Med., 2010, 6, (2), pp. 257 –262 [DOI] [PubMed] [Google Scholar]
- 2. Panda S. Yadav K.K. Nayk P.S. et al.: ‘Screening of metal‐resistant coal mine bacteria for biofabrication of elemental silver nanoparticle’, Bull. Mater. Sci., 2016, 39, (2), pp. 397 –404 [Google Scholar]
- 3. Arakha M. Borah S.M. Saleem M. et al.: ‘Interfacial assembly at silver nanoparticle enhances the antibacterial efficacy of nisin’, Free Radical Biol. Med., 2016, 101, pp. 434 –445 [DOI] [PubMed] [Google Scholar]
- 4. Arakha M. Pal S. Samantarrai D. et al.: ‘Antimicrobial activity of iron oxide nanoparticle upon modulation of nanoparticle‐bacteria interface’, Sci. Rep., 2015, 5, p. 14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arakha M. Saleem M. Mallick B.C. et al.: ‘The effects of interfacial potential on antimicrobial propensity of ZnO nanoparticle’, Sci. Rep., 2015, 5, 09578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iravani S.: ‘Bacteria in nanoparticle synthesis: current status and future prospects’ Int Sch Res Notices 2014, pp. 1 –18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gil P.R. Parak W.J.: ‘Composite nanoparticles take aim at cancer’, ACS Nano, 2008, 2, (11), pp. 2200 –2205 [DOI] [PubMed] [Google Scholar]
- 8. Duncan T.V.: ‘Applications of nanotechnology in food packaging and food safety: barrier materials, antimicrobials and sensors’, J. Colloid Interface Sci., 2011, 363, (1), pp. 1 –24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arakha M. Roy J. Nayak P.S. et al.: ‘Zinc oxide nanoparticle energy band gap reduction triggers the oxidative stress resulting into autophagy‐mediated apoptotic cell death’, Free Radical Biol. Med., 2017. [DOI] [PubMed] [Google Scholar]
- 10. Nayak P.S. Arakha M. Kumar A. et al.: ‘An approach towards continuous production of silver nanoparticles using bacillus thuringiensis’, RSC Adv., 2016, 6, (10), pp. 8232 –8242 [Google Scholar]
- 11. Xie J. Lee J.Y. Wang D.I. et al.: ‘Silver nanoplates: from biological to biomimetic synthesis’, ACS Nano, 2007, 1, (5), pp. 429 –439 [DOI] [PubMed] [Google Scholar]
- 12. Sondi I. Salopek‐Sondi B.: ‘Silver nanoparticles as antimicrobial agent: a case study on E. Coli as a model for gram‐negative bacteria’, J. Colloid Interface Sci., 2004, 275, (1), pp. 177 –182 [DOI] [PubMed] [Google Scholar]
- 13. Zheng K. Yuan X. Goswami N. et al.: ‘Recent advances in the synthesis, characterization, and biomedical applications of ultrasmall thiolated silver nanoclusters’, RSC Adv., 2014, 4, (105), pp. 60581 –60596 [Google Scholar]
- 14. Iranifam M.: ‘Chemiluminescence reactions enhanced by silver nanoparticles and silver alloy nanoparticles: applications in analytical chemistry’, Trends Anal. Chem., 2016, 82, pp. 126 –142 [Google Scholar]
- 15. Lengke M.F. Fleet M.E. Southam G.: ‘Biosynthesis of silver nanoparticles by filamentous cyanobacteria from a silver (I) nitrate complex’, Langmuir, 2007, 23, (5), pp. 2694 –2699 [DOI] [PubMed] [Google Scholar]
- 16. Basavaraja S. Balaji S.D. Lagashetty A. et al.: ‘Extracellular biosynthesis of silver nanoparticles using the fungus fusarium semitectum’, Mater. Res. Bull., 2008, 43, (5), pp. 1164 –1170 [Google Scholar]
- 17. Xie J. Lee J.Y. Wang D.I. et al.: ‘Identification of active biomolecules in the high‐yield synthesis of single‐crystalline gold nanoplates in algal solutions’, Small, 2007, 3, (4), pp. 672 –682 [DOI] [PubMed] [Google Scholar]
- 18. Makarov V.V. Love A.J. Sinitsyna O.V. et al.: ‘‘Green’ nanotechnologies: synthesis of metal nanoparticles using plants’, Acta Naturae, 2014, 6, (1), pp. 35 –44 [PMC free article] [PubMed] [Google Scholar]
- 19. Siddiqi K.S. Husen A.: ‘Fabrication of metal nanoparticles from fungi and metal salts: scope and application’, Nanoscale Res. Lett., 2016, 11, (1), p. 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Das V.L. Thomas R. Varghese R.T. et al.: ‘Extracellular synthesis of silver nanoparticles by the bacillus strain CS 11 isolated from industrialized area’, 3 Biotech, 2014, 4, (2), pp. 121 –126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan J.‐K. Wang Y.Y. Zhu L. et al.: ‘Green synthesis and characterization of zinc oxide nanoparticles using carboxylic curdlan and their interaction with bovine serum albumin’, RSC Adv., 2016, 6, (81), pp. 77752 –77759 [Google Scholar]
- 22. Fayaz A.M. Balaji K. Girilal M. et al.: ‘Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram‐positive and gram‐negative bacteria’, Nanomed. Nanotech. Biol. Med., 2010, 6, (1), pp. 103 –109 [DOI] [PubMed] [Google Scholar]
- 23. Gallardo C. Monrás J.P. Plaza D.O. et al.: ‘Low‐temperature biosynthesis of fluorescent semiconductor nanoparticles (CdS) by oxidative stress resistant antarctic bacteria’, J. Biotechnol., 2014, 187, pp. 108 –115 [DOI] [PubMed] [Google Scholar]
- 24. Punjabi K. Yedurkar S. Doshi S. et al.: ‘Biosynthesis of silver nanoparticles by pseudomonas spp. Isolated from effluent of an electroplating industry’, IET Nanobiotechnol., 2017, 11, (5), pp. 584 –590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kulkarni R.R. Shaiwale N.S. Deobagkar D.N. et al.: ‘Synthesis and extracellular accumulation of silver nanoparticles by employing radiation‐resistant deinococcus radiodurans, their characterization, and determination of bioactivity’, Int. J. Nanomed., 2015, 10, p. 963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prophet E.B.: ‘Laboratory methods in histotechnology’ (Armed forces institute of pathology, Washington, 1992), pp. 1 –279 [Google Scholar]
- 27. Drancourt M. Bollet C. Carlioz A. et al.: ‘16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates’, J. Clin. Microbiol., 2000, 38, (10), pp. 3623 –3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Igbinosa I.H. Igumbor E.U. Aghdasi F. et al.: ‘Emerging aeromonas species infections and their significance in public health’, Sci. World J., 2012, 2012, p. 625023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minana‐Galbis D. Farfan M. Loren J.G. et al.: ‘Biochemical identification and numerical taxonomy of aeromonas spp. Isolated from environmental and clinical samples in Spain’, J. Appl. Microbiol., 2002, 93, (3), pp. 420 –430 [DOI] [PubMed] [Google Scholar]
- 30. Arokiyaraj S. Arasu M.V. Vincent S. et al.: ‘Rapid green synthesis of silver nanoparticles from chrysanthemum indicum L and its antibacterial and cytotoxic effects: an in vitro study’, Int. J. Nanomed., 2014, 9, p. 379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agnihotri S. Mukherji S. Mukherji S.: ‘Size‐controlled silver nanoparticles synthesized over the range 5–100 nm using the same protocol and their antibacterial efficacy’, RSC Adv., 2014, 4, (8), pp. 3974 –3983 [Google Scholar]
- 32. Naik R.R. Stringer S.J. Agarwal G. et al.: ‘Biomimetic synthesis and patterning of silver nanoparticles’, Nature Mater., 2002, 1, (3), pp. 169 –172 [DOI] [PubMed] [Google Scholar]
- 33. Peiris M.K. Gunasekara C.P. Jayaweera P.M. et al.: ‘Biosynthesized silver nanoparticles: are they effective antimicrobials?’, Memórias do Instituto Oswaldo Cruz, 2017, 112, (8), pp. 537 –543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Narayanan K.B. Sakthivel N.: ‘Biological synthesis of metal nanoparticles by microbes’, Adv. Colloid Interface Sci., 2010, 156, (1), pp. 1 –13 [DOI] [PubMed] [Google Scholar]
- 35. Parab H.J. Huang J.H. Lai T.C. et al.: ‘Biocompatible transferrin‐conjugated sodium hexametaphosphate‐stabilized gold nanoparticles: synthesis, characterization, cytotoxicity and cellular uptake’, Nanotechnology, 2011, 22, (39), p. 395706 [DOI] [PubMed] [Google Scholar]
- 36. Begam J.N.: ‘Biosynthesis and characterization of silver nanoparticles (AgNPs) using marine bacteria against certain human pathogens’, Int. J. Adv. Sci. Res., 2016, 2, (7), pp. 152 –156 [Google Scholar]
- 37. Selvi K.V. Sivakumar T.: ‘Isolation and characterization of silver nanoparticles from fusarium oxysporum’, Int. J. Curr. Microbiol. Appl. Sci, 2012, 1, (1), pp. 56 –62 [Google Scholar]
- 38. Goswami N. Zheng K. Xie J.: ‘Bio‐NCs–the marriage of ultrasmall metal nanoclusters with biomolecules’, Nanoscale, 2014, 6, (22), pp. 13328 –13347 [DOI] [PubMed] [Google Scholar]
- 39. Atta A.M. Allohedan H.A. Ezzat A.O. et al.: ‘Synthesis of dispersed and stabilized silver nanoparticles in acidic media’, Polym. Sci. Ser. B, 2014, 56, (6), pp. 762 –769 [Google Scholar]
- 40. Iravani S.: ‘Green synthesis of metal nanoparticles using plants’, Green Chem., 2011, 13, (10), pp. 2638 –2650 [Google Scholar]
- 41. Schneider C.A. Rasband W.S. Eliceiri K.W.: ‘NIH Image to ImageJ: 25 years of image analysis’, Nature Methods, 2012, 9, pp. 671 –675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dar M.A. Ingle A. Rai M.: ‘Enhanced antimicrobial activity of silver nanoparticles synthesized by cryphonectria sp. evaluated singly and in combination with antibiotics’, Nanomed. Nanotechnol. Biol. Med., 2013, 9, (1), pp. 105 –110 [DOI] [PubMed] [Google Scholar]


