Abstract
A clinical evaluation of an automated quantitative PCR assay, the COBAS AMPLICOR HCV MONITOR test, version 2.0 (v2.0), was carried out to assess the performance of this test in comparison with that of the previous, manual version, the AMPLICOR HCV MONITOR test, and with that of nested PCR. Serial dilutions of serum samples infected with genotype 1b, 2a, or 3, as well as synthetic RNA transcripts and serum samples derived from 87 patients with chronic hepatitis C and infected with genotype 1a, 1b, 2a, 2b, 3a, 3b, 4, or 5, were analyzed to determine the ability of the system to efficiently quantify various hepatitis C virus (HCV) genotypes. These experiments showed that the COBAS AMPLICOR HCV MONITOR test, v2.0, has mean intra-assay, interassay, and interoperator coefficients of variation that range from 22 to 34.5% and a 3-logarithm dynamic range, which spans from 103 to 106 copies/ml. Compared to the previous, manual version of the test, the COBAS AMPLICOR HCV MONITOR test, v2.0, showed an improved efficacy for all genotypes, especially genotypes 2, 3, and 4, whose estimated concentrations were on average 1 logarithm higher. When used to monitor patients under treatment, however, both versions showed the same patterns of viremia, indicating that the COBAS AMPLICOR HCV MONITOR test, v2.0, and the AMPLICOR HCV MONITOR test were equally effective at detecting relative viremia changes in serial samples. As expected, the automated test was less sensitive than nested PCR; among specimens from a cohort of patients treated with interferon, nested PCR identified three more viremic specimens, which probably contained very low concentrations of HCV RNA.
The broad potential diagnostic and therapeutic monitoring applications of PCR have prompted a significant effort toward improving the performance and reliability of hepatitis C virus (HCV) quantitation assays. The occurrence of false-positive results due to amplicon carryover contamination, false-negative results due to enzymatic inhibition, and poor reproducibility of PCR, shown previously by several proficiency studies, has always been a concern (2, 4, 12, 29). However, technical advances marked by the introduction of uracil-N-glycosylase and internal controls have successfully addressed most of these issues and have allowed an increased use of PCR in clinical laboratories (5, 17, 19, 23). Standardization and accuracy of quantitation have been fundamental in the case of HCV, a pathogen that cannot be directly detected by conventional techniques and whose viremia levels have important clinical implications (13, 21, 26). Indeed, the inclusion of HCV RNA levels in the diagnostic algorithm for chronic hepatitis C has been made easier by the partial automation of the procedure achieved with the development of dedicated laboratory instrumentation (3, 9, 28). The recent development of an automated PCR system for the quantitation of HCV RNA may further support the clinical use of viral load monitoring in the management of chronically infected individuals (1, 15).
In this respect, we have evaluated the analytical and clinical performance of a new automated test, the COBAS AMPLICOR HCV MONITOR test, version 2.0 (v2.0), compared to those of the first-generation manual version (AMPLICOR HCV MONITOR) and nested PCR. The COBAS AMPLICOR HCV MONITOR test, v2.0, incorporates changes in the formulation of the amplification solution that are likely to improve its ability to efficiently quantify all major viral subtypes, whereas the first-generation AMPLICOR HCV MONITOR test was reported to underestimate the quantities of genotypes other than genotype 1.
MATERIALS AND METHODS
The COBAS AMPLICOR HCV MONITOR test, v2.0, was evaluated in two phases. The first phase focused on intra-assay, interassay, and interoperator reproducibility, and the second phase focused on the efficiency of quantification of different HCV genotypes and a comparison to nested PCR.
Analytical evaluation.
To assess the precision of the automated test, coded samples, obtained by diluting high-titer serum specimens previously quantified by the COBAS AMPLICOR HCV MONITOR test, v2.0, were provided to each of the participating laboratories by Roche Molecular Systems. Two specimens each infected with genotypes 1, 2, and 3 were diluted to approximately 103, 104, 105, and 106 copies/ml and were tested in six replicates per run for intra-assay reproducibility, two replicates per run for 6 subsequent days for interassay reproducibility, and two replicates per run performed by two operators for interoperator reproducibility.
Cloned, synthetic transcripts representing genotypes 1 to 5 were quantified spectrophotometrically at 260 nm. These transcripts were serially diluted and analyzed by the COBAS AMPLICOR HCV MONITOR test, v2.0, to determine the linearity of the test by using different HCV genotype targets.
Clinical evaluation.
A total of 211 serum samples, which contained genotype 1a, 1b, 2a, 2b, 3a, 3b, 4, or 5, were obtained from 87 patients during interferon (IFN) therapy for type C chronic hepatitis. The diagnosis was based on serological, biochemical, and histological parameters, and the presence of HCV RNA was determined by an in-house nested PCR or a single-step reverse transcription-PCR (AMPLICOR HCV test; Roche Diagnostic Systems, Branchburg, N.J.) (28). HCV RNA levels had also been previously quantified in 101 specimens by the AMPLICOR HCV MONITOR test (Roche Diagnostic Systems) (3).
Automated HCV RNA quantitation.
The concentrations of HCV RNA in both the coded and the clinical samples were determined by the COBAS AMPLICOR HCV MONITOR test, v2.0, by following the manufacturer's instructions. In brief, 100 μl of serum was subjected to chaotropic lysis in the presence of known amounts of an internal quantitation standard (QS). The QS is a synthetic RNA transcript with primer binding regions identical to those of the HCV target sequence, a randomized internal sequence of similar length and base composition as the HCV target sequence, and a unique probe binding region that differentiates the QS from the target amplicon. After isopropanol precipitation and an ethanol wash, the target viral RNA and the QS were resuspended in the Specimen Diluent (Roche), and this mixture was mixed with an equal volume of the amplification ready solution (Master Mix; Roche) containing the primers KY78 (biotinylated) and KY80 (nonbiotinylated), deoxynucleoside triphosphates, AmpErase, and rTth DNA polymerase.
Amplification, amplicon dilution, detection, and quantitation were automatically performed by the COBAS AMPLICOR analyzer (1, 6, 15).
Compared to the AMPLICOR HCV MONITOR test, the COBAS AMPLICOR HCV MONITOR test, v2.0, has a reformulated master mixture that includes a cosolvent that enables a more complete denaturation of the target and a more efficient annealing and extension of the primers. In addition, manganese acetate is provided as a separate reagent dispensed in the amplification solution immediately prior to amplification.
Statistical analysis included calculation of average copy number, standard deviation (SD), coefficient of variation (CV), regression, and Pearson's correlation.
RESULTS
The performance of the COBAS AMPLICOR HCV MONITOR test, v2.0, for the quantitation of HCV RNA was assessed by the two participating laboratories, laboratories A and B, with coded, mock-infected specimen panels and then with clinical specimens.
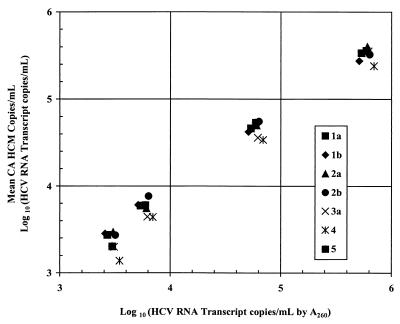
As shown in Tables 1 and 2, the interassay, intra-assay, and interoperator reproducibilities of the assay were analyzed by testing several replicates of samples that each contained genotype 1b, 2b, or 3a at expected HCV RNA concentrations of approximately 103, 104, 105, and 106 copies/ml. The variations between the mean and the observed concentrations for the intra-assay, interassay, and interoperator analyses were 25.7, 34.5, and 25%, respectively. Additionally, the mean CVs for the individual genotypes were similar. Several dilutions of a high-titer, coded specimen containing genotype 1b were also used to analyze the dynamic range of the assay. In these experiments, performed in triplicate, the COBAS AMPLICOR HCV MONITOR test, v2.0, showed a linear range that spanned 3 logarithms, from 103 to 106 copies/ml (data not shown). Similar results were also obtained with cloned, synthetic transcripts of genotypes 1 to 5 quantified spectrophotometrically at 260 nm (Fig. 1).
TABLE 1.
COBAS AMPLICOR HCV MONITOR test, v2.0, interassay precisiona
| Specimen | Laboratory | Genotype | Sample dilution | No. of HCV RNA copies/ml
|
||
|---|---|---|---|---|---|---|
| Mean (n = 6) | SD | % CV | ||||
| 71815620 | A | 1b | Undiluted | 776,000 | 173,000 | 22 |
| 1:10 | 151,000 | 22,900 | 15 | |||
| 1:100 | 18,600 | 3,530 | 19 | |||
| 71823667 | A | 2b | Undiluted | 602,000 | 50,800 | 8 |
| 1:10 | 99,700 | 13,700 | 14 | |||
| 1:100 | 11,100 | 2,330 | 21 | |||
| 72237713 | A | 3a | Undiluted | 619,000 | 41,600 | 7 |
| 1:10 | 107,000 | 19,100 | 18 | |||
| 1:100 | 13,400 | 1,140 | 9 | |||
| 71815620 | B | 1b | 1:10 | 146,000 | 27,100 | 19 |
| 1:100 | 16,500 | 3,210 | 19 | |||
| 1:1,000 | 2,370 | 336 | 14 | |||
| 71823667 | B | 2b | 1:10 | 84,800 | 20,600 | 24 |
| 1:100 | 13,700 | 12,400 | 91 | |||
| 1:1,000 | 1,270 | 529 | 42 | |||
| 72237713 | B | 3a | 1:10 | 91,000 | 14,300 | 16 |
| 1:100 | 9,560 | 753 | 8 | |||
| 1:1,000 | 2,210 | 1,900 | 86 | |||
Three specimens containing HCV genotype 1b, 2b, or 3a were serially diluted and analyzed in duplicate over 6 consecutive days by the two participating laboratories. Means, SDs, and CVs were then calculated for each sample.
TABLE 2.
COBAS AMPLICOR HCV MONITOR test, v2.0, intra-assay precisiona
| Specimen | Genotype | Sample dilution | No. of HCV RNA copies/ml
|
||
|---|---|---|---|---|---|
| Mean (n = 6) | SD | % CV | |||
| 71824305 | 1a | Undiluted | 824,000 | 105,000 | 13 |
| 1:10 | 116,000 | 10,200 | 9 | ||
| 1:100 | 18,000 | 3,190 | 18 | ||
| 1:1,000 | 1,890 | 959 | 51 | ||
| 71815620 | 1b | Undiluted | 1,220,000 | 492,000 | 40 |
| 1:10 | 89,600 | 10,600 | 12 | ||
| 1:100 | 14,200 | 1,320 | 9 | ||
| 1:1,000 | 2,980 | 781 | 26 | ||
| 71823667 | 2b | Undiluted | 821,000 | 200,000 | 24 |
| 1:10 | 60,700 | 7,020 | 12 | ||
| 1:100 | 7,630 | 881 | 12 | ||
| 1:1,000 | 1,320 | 408 | 31 | ||
| 72249021 | 2c | Undiluted | 744,000 | 210,000 | 28 |
| 1:10 | 91,200 | 20,500 | 22 | ||
| 1:100 | 11,800 | 2,480 | 21 | ||
| 1:1,000 | 1,470 | 279 | 19 | ||
| 72237713 | 3a | Undiluted | 797,000 | 202,000 | 25 |
| 1:10 | 78,600 | 15,200 | 19 | ||
| 1:100 | 10,900 | 2,710 | 25 | ||
| 1:1,000 | 1,740 | 969 | 56 | ||
| 72249864 | 3a | Undiluted | 574,000 | 63,000 | 11 |
| 1:10 | 75,400 | 5,410 | 7 | ||
| 1:100 | 10,100 | 3,220 | 32 | ||
| 1:1,000 | 1,080 | 78 | 7 | ||
Six specimens containing HCV genotype 1a, 1b, 2b, 2c, or 3a were serially diluted and analyzed in six replicates by the two participating laboratories. Means, SDs, and CVs were then calculated for each sample.
FIG. 1.
Quantitation of different HCV genotype transcripts by the COBAS AMPLICOR HCV MONITOR test, v2.0 (CA HCM). Serial dilutions of synthetic, genotype-specific HCV transcripts of known concentration were analyzed by the COBAS AMPLICOR HCV MONITOR test, v2.0.
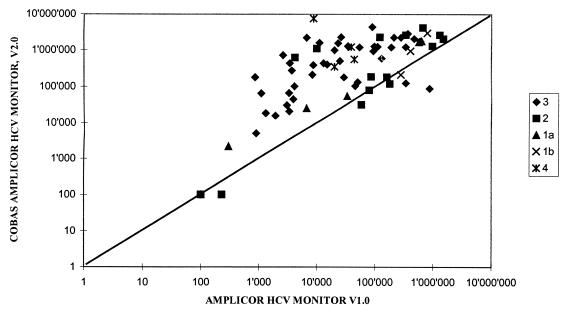
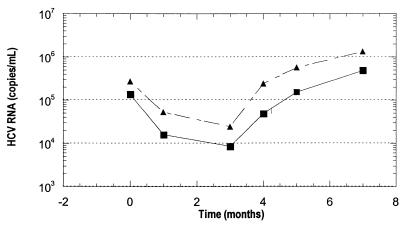
The efficiency of the automated system in quantifying HCV RNA independent of genotype was further evaluated by testing specimens obtained from 72 patients during IFN therapy for chronic type C hepatitis. Compared with the first-generation AMPLICOR HCV MONITOR test, the COBAS AMPLICOR HCV MONITOR test, v2.0, produced a one- to fivefold increase in HCV titers with all genotypes tested. With synthetic transcripts this observation was most marked for genotypes 2, 3, and 4; however, the two tests showed a significant correlation (Fig. 2). The differences in the absolute copy number determined by each test method showed the same profile and did not affect the ability to monitor the viral load changes over the time course of viremia observed in patients under antiviral treatment (Fig. 3).
FIG. 2.
Comparison of COBAS AMPLICOR HCV MONITOR test, v2.0, and AMPLICOR HCV MONITOR test in the quantitation of different HCV genotypes. Specimens containing different concentrations of genotype-specific HCV RNA were analyzed by the two tests.
FIG. 3.
Viral load profile of a patient infected with genotype 1b, determined by using the COBAS AMPLICOR HCV MONITOR test, v2.0 (▴), and the first-generation AMPLICOR HCV MONITOR test (■).
The viral loads were measured by the COBAS AMPLICOR HCV MONITOR test, v2.0, in a subset of clinical samples taken from 24 patients during IFN therapy and were compared to the nested PCR results. The nested PCR test was able to detect virus in 45 of 64 (70%) specimens infected with HCV genotype 1, 2, or 3. The COBAS AMPLICOR HCV MONITOR test, v2.0, was able to detect and quantify virus in 42 of 64 (66%) of the same set of specimens. The three specimens in which virus was not detected by the COBAS AMPLICOR HCV MONITOR test, v2.0, were infected with genotypes 1a, 1b, and 2. These three specimens were sequential samples with low titers of virus taken from patients who had shown primary biochemical and virological responses to IFN therapy.
DISCUSSION
The low level of efficiency of IFN therapy in the management of chronic hepatitis C has prompted the search for predictive markers that refine the timing of treatment and improve treatment regimens (3, 6). Viral load appears to be one of the most informative markers both before and during therapy, but its application in clinical practice is still controversial (7, 8, 10, 11, 16, 18, 22, 24, 25, 27). One of the factors that has prevented its widespread use is the less than optimal reproducibility, the variable dynamic range, and the HCV genotype-dependent sensitivities of the available assays (14, 20, 21, 30).
In the present study we have evaluated the performance of an automated, quantitative PCR assay, the COBAS AMPLICOR HCV MONITOR test, v2.0, that has recently been developed to overcome some of these issues.
In a first phase of the evaluation we analyzed the interassay, intra-assay, and interoperator precisions of the COBAS AMPLICOR HCV MONITOR test, v2.0, using standard specimens diluted to several concentrations. Overall, the reproducibility was characterized by mean CVs of 26 to 35%. This is an improvement over the 20 to 60% CVs shown by the first-generation manual version of the test (3). Automation of amplicon dilution and detection postamplification is probably the step that has contributed the most to the increased precision of the COBAS AMPLICOR HCV MONITOR test, v2.0. Although the CVs for some concentrations are quite large, the associated SDs of less than or equal to 0.33 log are not clinically significant.
The assay showed a dynamic range that spanned 3 logarithms, between 103 and 106 copies/ml. This range appears to be suitable for the monitoring of viremia changes during antiviral therapy because it can discriminate patients with high versus low viremia levels and has adequate sensitivity to assess primary virological responses (8, 10, 11, 16, 25, 27). The extent of viral clearance during the first 4 weeks of treatment is considered a valid marker of the response to treatment by IFN monotherapy. The dynamic range may be useful for determination of the timely modification of treatment for the patients who do not show any significant viral load changes compared to those at the baseline (8, 16, 23, 30). The clinical performance of the COBAS AMPLICOR HCV MONITOR test, v2.0, was also evaluated with samples that have recently been assessed by an in-house, nested PCR assay, previously reported to have a detection limit of 102 copies/ml (22). Only three specimens with viruses that were detectable by nested PCR had levels of viremia below the limit of detection of the COBAS AMPLICOR HCV MONITOR test, v2.0. These samples, obtained from patients who had shown a primary response to IFN therapy, probably contained very low levels of HCV RNA, as often seen in sustained responders prior to viral clearance. Another aspect of test performance investigated was the influence of genotype on quantitation, recently pointed out as one of the drawbacks of the first-generation manual assay (14, 20). Several changes in the formulations of the test reagents have been introduced to improve performance, including a buffer change, the addition of a cosolvent to the amplification solution, and the packaging of Mn2+ solution as a separate reagent. It is probable that these modifications have resulted in a more efficient hybridization of the primers to the stem-loop structures of the 5′ noncoding region target sequence and in an increase in overall reagent stability. These changes have produced equivalent efficiencies of amplification of genotypes 1 through 5, demonstrated with synthetic transcripts and clinical specimens. Although these improvements have resulted in a difference between the results generated by the two versions of the test, there is no impact on the monitoring of treatment responses, as the kinetics of viremia follow the same pattern. Additionally, the COBAS AMPLICOR HCV MONITOR test, v2.0, is being calibrated to the World Health Organization International Standard for HCV. This will provide a mechanism for easier comparison of results between future versions and other technologies.
Even though the relative change in copy number between serial samples rather than absolute copy number is most important when one is monitoring therapies, the improved genotype-specific performance of this assay will make the data obtained by different tests more easily interpretable and comparable. In addition, the workload reduction and increased accuracy granted by automation may expand the use of viral load monitoring in the management of chronic type C hepatitis and help in better defining its benefits and limitations.
ACKNOWLEDGMENTS
We thank Roche Molecular Systems, Inc., the manufacturer of the COBAS AMPLICOR HCV test, v2.0, for providing the test reagents and the HCV RNA standards.
REFERENCES
- 1.Albadalejo J, Alonso R, Antinozzi R, Bogard M, Bourgault A-M, Colucci G, Fenner T, Petersen H, Sala E, Vincelette J, Young C. Multicenter evaluation of COBAS AMPLICOR HCV, an integrated PCR system for the rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J Clin Microbiol. 1998;36:862–865. doi: 10.1128/jcm.36.4.862-865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonino F, Brunetto M R, Negro F, Baldi M, Saracco G, Abate M L, Fabiano A, Verme G. Hepatitis C virus infection and disease. Diagnostic problems. J Hepatol. 1993;17(Suppl. 3):S78–S82. doi: 10.1016/s0168-8278(05)80429-5. [DOI] [PubMed] [Google Scholar]
- 3.Colucci G, Gutekunst K. Development of a quantitative PCR assay for monitoring HCV viraemia levels in patients with chronic hepatitis C. J Viral Hepatitis. 1997;4(Suppl. 1):75–78. doi: 10.1111/j.1365-2893.1997.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 4.Conry-Cantilena C. Hepatitis C virus diagnostics: technology, clinical applications and impacts. Trends Biotechnol. 1997;15:71–76. doi: 10.1016/S0167-7799(97)84206-0. [DOI] [PubMed] [Google Scholar]
- 5.Damen M, Cuypers H T M, Zaaijer H W, Reesink H W, Schaasberg W P, Gerlich W H, Niesters H G M, Lelie P N. International collaborative study on the second EUROHEP HCV-RNA reference panel. J Virol Methods. 1996;58:175–185. doi: 10.1016/0166-0934(96)02011-3. [DOI] [PubMed] [Google Scholar]
- 6.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 7.Dusheiko G M, Khakoo S, Soni P, Grellier L. A rational approach to the management of hepatitis C infection. Br Med J. 1996;312:357–364. doi: 10.1136/bmj.312.7027.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flichman D, Colombatto P, Randone A, Baldi M, Bellati G, Negro F, Oliveri F, Colucci G, Verme G, Bonino F, Brunetto M R. Quantitative detection of hepatitis C virus RNA in the serum of patients with chronic hepatitis C treated with interferon: a pilot study. Clin Diagn Virol. 1997;8:63–70. doi: 10.1016/s0928-0197(97)00013-5. [DOI] [PubMed] [Google Scholar]
- 9.Gerken G, Pontisso P, Roggendorf M, Rumi M G, Simmonds P, Trepo C, Zeuzem S, Colucci G. Clinical evaluation of a single reaction, diagnostic PCR assay for the detection of hepatitis C virus (HCV) RNA. J Hepatol. 1996;24:33–37. doi: 10.1016/s0168-8278(96)80183-8. [DOI] [PubMed] [Google Scholar]
- 10.Gerken G, Knolle P, Jacobs S, Meyer Zum Buschenfelde K H. Quantification and genotyping of serum HCV-RNA in patients with chronic hepatitis C undergoing interferon treatment. Arch Virol. 1997;142:459–464. doi: 10.1007/s007050050092. [DOI] [PubMed] [Google Scholar]
- 11.Gerken G, Knolle P. Molecular analysis of hepatitis C virus. In: Harrison T J, Zuckerman A J J, editors. The molecular medicine of viral hepatitis. London, United Kingdom: Wiley & Sons Ltd.; 1997. pp. 167–181. [Google Scholar]
- 12.Gretch D R. Diagnostic tests for hepatitis C. Hepatology. 1997;26:43S–47S. doi: 10.1002/hep.510260708. [DOI] [PubMed] [Google Scholar]
- 13.Gretch D R, dela Rosa C, Carithers R L, Willson R A, Williams B, Corey L. Assessment of hepatitis C viremia using molecular amplification technologies: correlation and clinical implications. Ann Intern Med. 1995;123:321–329. doi: 10.7326/0003-4819-123-5-199509010-00001. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus load among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by Quantiplex HCV RNA assay version 1 and 2, Roche Monitor assay, and in-house limiting dilution method. J Clin Microbiol. 1997;35:187–192. doi: 10.1128/jcm.35.1.187-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jungkind D, DiRienzo S, Beavis K G, Silverman N S. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karino Y, Toyota J, Sugawara M, Higashino K, Sato T, Ohmura T, Suga T, Okuuchi Y, Matsushima T. Early loss of serum hepatitis C virus RNA can predict a sustained response to interferon therapy in patients with chronic hepatitis C. Am J Gastroenterol. 1997;92:61–65. [PubMed] [Google Scholar]
- 17.Longo M C, Berninger M S, Hartley H L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 18.Magrin S, Craxi A, Fabiano C, Simonetti R G, Fiorentino G, Marino L, Diquattro O, Di Marco V, Loiacono O, Volpes R, et al. Hepatitis C viremia in chronic liver disease: relationship to interferon-alpha or corticosteroid treatment. Hepatology. 1994;2:273–276. [PubMed] [Google Scholar]
- 19.Myers T W, Gelfand D H. Reverse transcription and DNA amplification by a Thermus thermophilus DNA polymerase. Biochemistry. 1991;30:7661–7666. doi: 10.1021/bi00245a001. [DOI] [PubMed] [Google Scholar]
- 20.Ohno T, Lau J Y N. The “gold standard,” accuracy, and the current concepts: hepatitis C virus genotype and viremia. Hepatology. 1996;24:1312–1315. doi: 10.1002/hep.510240552. [DOI] [PubMed] [Google Scholar]
- 21.Pawlotsky J M. Measuring hepatitis C virus viremia in clinical samples: can we trust the assay? Hepatology. 1997;26:1–4. doi: 10.1002/hep.510260131. [DOI] [PubMed] [Google Scholar]
- 22.Romeo R, Thiers V, Driss F, Berthelot P, Nalpas B, Brechot C. Hepatitis C virus RNA in serum of blood donors with or without elevated transaminase levels. Transfusion. 1993;33:629–633. doi: 10.1046/j.1537-2995.1993.33893342742.x. [DOI] [PubMed] [Google Scholar]
- 23.Rosenstraus M, Wang Z, Chang S Y, DeBoneville D, Spadoro S P. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J Clin Microbiol. 1997;36:191–197. doi: 10.1128/jcm.36.1.191-197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiratory Y, Kato N, Yokosuka O, Imazeki F, Hashimoto E, Hayashi N, Nakamura A, Asada M, Kuroda H, Tanaka N, et al. Predictor of efficacy of interferon therapy in chronic hepatitis C virus infection. Gastroenterology. 1997;113:558–566. doi: 10.1053/gast.1997.v113.pm9247476. [DOI] [PubMed] [Google Scholar]
- 25.Shiratory Y, Kato N, Yokosuka O, Hashimoto E, Hayashi N, Nakamura A, Asada M, Kuroda H, Ohkubo H, Arakawa Y, et al. Quantitative assays for hepatitis C virus in serum as predictor of the long-term response to interferon. J Hepatol. 1997;27:437–444. doi: 10.1016/s0168-8278(97)80346-7. [DOI] [PubMed] [Google Scholar]
- 26.Tedeschi V, Seeff L B. Diagnostic tests for hepatitis C: where are we now? Ann Intern Med. 1995;123:383–385. doi: 10.7326/0003-4819-123-5-199509010-00009. [DOI] [PubMed] [Google Scholar]
- 27.Trabaud M A, Bailly F, Si-Ahmed S N, Chevallier P, Sepetjan M, Colucci G, Trepo C. Comparison of HCV RNA assays for the detection and quantification of hepatitis C virus RNA levels in serum of patients with chronic hepatitis C treated with interferon. J Med Virol. 1997;52:105–112. [PubMed] [Google Scholar]
- 28.Young K K Y, Archer J J, Yokosuka O, Omata M, Resnick R M. Detection of hepatitis C virus RNA by a combined reverse transcription PCR assay: comparison with nested amplification and antibody testing. J Clin Microbiol. 1995;33:654–657. doi: 10.1128/jcm.33.3.654-657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaaijer H J, Cuypers H T M, Reesink H W. Reliability of HCV PCR results. Lancet. 1993;341:722–724. doi: 10.1016/0140-6736(93)90488-3. [DOI] [PubMed] [Google Scholar]
- 30.Zeuzem S, Lee J-H, Franke A, Rüster B, Prümmer D, Herrmann G, Roth W K. Quantification of the initial decline of serum hepatitis C virus RNA and response to interferon alfa. Hepatology. 1998;27:1149–1156. doi: 10.1002/hep.510270433. [DOI] [PubMed] [Google Scholar]