ABSTRACT
The mitochondrial genome of the pathogenic yeast Candida albicans displays a typical organization of several (eight) primary transcription units separated by noncoding regions. Presence of genes encoding Complex I subunits and the stability of its mtDNA sequence make it an attractive model to study organellar genome expression using transcriptomic approaches. The main activity responsible for RNA degradation in mitochondria is a two-component complex (mtEXO) consisting of a 3ʹ-5ʹ exoribonuclease, in yeasts encoded by the DSS1 gene, and a conserved Suv3p helicase. In C. albicans, deletion of either DSS1 or SUV3 gene results in multiple defects in mitochondrial genome expression leading to the loss of respiratory competence. Transcriptomic analysis reveals pervasive transcription in mutants lacking the mtEXO activity, with evidence of the entire genome being transcribed, whereas in wild-type strains no RNAs corresponding to a significant fraction of the noncoding genome can be detected. Antisense (‘mirror’) transcripts, absent from normal mitochondria are also prominent in the mutants. The expression of multiple mature transcripts, particularly those translated from bicistronic mRNAs, as well as those that contain introns is affected in the mutants, resulting in a decreased level of proteins and reduced respiratory complex activity. The phenotype is most severe in the case of Complex IV, where a decrease of mature COX1 mRNA level to ~5% results in a complete loss of activity. These results show that RNA degradation by mtEXO is essential for shaping the mitochondrial transcriptome and is required to maintain the functional demarcation between transcription units and non-coding genome segments.
KEYWORDS: Mitochondria, RNA degradation, pervasive transcription, mtEXO, exoribonuclease, Candida albicans
Introduction
The transcriptome consists of multiple coding and noncoding RNAs that originate through complex and evolutionarily diverse multi-step interconnected processes of synthesis, maturation, and degradation. Recent advances in RNA characterization techniques revealed unexpected widespread transcriptional activity, extending beyond annotated genes to the majority of the genome, and occurring both in eukaryotic and prokaryotic cells [1–4]. Prevalence of pervasive transcription is most evident in the study of mutants deficient in RNA degradation [5]. About 75% of the human nuclear genome is covered by identified primary transcripts, even though actual coding sequences constitute only a small fraction thereof [6,7], and evolutionary conservation indicates sequence-level functionality only for about 10% of its length [8]. Therefore RNA degradation has to play an important role in shaping the final, functional transcriptome.
The mitochondrial genetic system, despite its evident endosymbiotic origins [9], shows many unique derived characteristics that set it apart from the genomes of prokaryotes and the eukaryotic nucleus [10]. With the exception of Jakobid protists that still retain a bacteria-like transcriptional apparatus in their mitochondria [11], synthesis of polycistronic transcription units depends on a single-subunit polymerase related to T-odd bacteriophage enzymes [12]. Mitochondrial transcription is initiated from simple promoters, and its regulation is limited. Primary mitochondrial transcripts encompassing protein-coding, tRNA and rRNA sequences undergo extensive processing by various mechanisms, including tRNA excision (punctuation) [13], intron splicing, and other processing events [14,15]. Comprehensive studies of the mitochondrial transcriptomes in yeasts S. cerevisiae [14], S. pombe [16] and C. albicans [15] demonstrated that the final abundance of RNAs derived from the same primary transcript can differ by several orders of magnitude, similar variation was also observed between RNAs derived from the major heavy strand transcript of human mitochondria [17]. It is thus clear that RNA processing and degradation play a key role in the expression of mitochondrial genes.
Enzymatic complexes exhibiting processive exoribonucleolytic activity are the main actors in RNA degradation [18]. The main mitochondrial exoribonuclease complex, known as mtEXO or the mitochondrial degradosome [19] exhibits 3ʹ-to-5ʹ directionality and is composed of two subunits: the Suv3 ATP dependent helicase and an exoribonuclease. Whereas the Suv3 helicase is conserved in the mitochondrial degradosome of all studied eukaryotic lineages, including fungi [20,21], plants [22], protists [23], and humans [24], the exoribonucleolytic activity can be assured by two different enzymes: the phosphorolytic polynucleotide phosphorylase (PNPase) in animals [25] and plants [26], and the hydrolytic Dss1 RNase in fungi [21,27] and in trypanosomes [28].
In mammalian cells, degradosome-dependent RNA degradation, turnover, and surveillance play a key role in the maintenance of organellar gene expression. Dysfunction of the mammalian mitochondrial RNA degradosome results in a plethora of phenotypes, including accumulation of noncoding antisense transcripts and dsRNA molecules, with significant physiological consequences [29–32].
As an example of a minimal 3ʹ-to-5ʹ RNA decay enzymatic system, the S. cerevisiae mtEXO complex has been extensively studied using enzymology [33], structural biology [34], and genetic [35–37] approaches. Work on S. cerevisiae mutants deficient in the mtEXO function clearly identified RNA degradation as the key factor in mitochondrial gene expression [27,35]. Both mtEXO subunits are essential for mitochondrial respiration in S. cerevisiae, and deletion of SUV3 or DSS1 results in multiple RNA metabolism defects, including accumulation of introns and precursors, as well as destabilization of mature transcripts [27,35].
Studies in S. cerevisiae are, however, hindered by the mitochondrial genome instability that is often associated with defects in organellar gene expression [38]. This phenomenon is also observed in mtEXO mutants, and their dysfunction results in mtDNA instability (generation of irreversible rho−/rho0 cytoplasmic petites) through impaired translation [19,35,39,40]. As RNA processing is inevitably compromised in such petites, in the S. cerevisiae system it is difficult to separate the primary phenotypes of SUV3 or DSS1 deletion from the secondary effects related to mtDNA instability.
In addition to the mitochondrial genome instability typical in petite-positive species, significant sequence divergence between different laboratory strains, and extremely AT-rich low complexity intergenic regions of S. cerevisiae mtDNA, present challenges for transcriptomic studies, in particular by making unambiguous mapping of short sequence reads to intergenic sequences difficult. A petite-negative species, like C. albicans is thus more suited for functional analysis of factors affecting mitochondrial RNA processing. Additionally, mtDNA of petite-negative yeasts is also generally more stable and less prone to the accumulation of point mutations [41].
Candida albicans is a yeast species known primarily as the most important infectious fungal pathogen affecting immunocompromised individuals [42]. It has also emerged as a promising model organism to study various aspects of fungal cell biology and evolution [15,43–48]. It belongs to a distinct hemiascomycete clade with a conspicuous departure from the universal genetic code (the ‘CTG clade’), where the CUG codon is read as serine instead of leucine [49,50].
The genome of Candida albicans, like all known yeast genomes, contains genes encoding the two-subunit mitochondrial degradosome (mtEXO), composed of the Suv3 helicase (conserved throughout the entire eukaryotic domain), and the exoribonuclease which like in other fungi is a 3ʹ-to-5ʹ hydrolytic exoribonuclease belonging to the RNR (RNase II-like) family, orthologous to the Dss1 protein of S. cerevisiae [33–35,37,39].
Previous studies on the function of mitochondrial degradosome components in C. albicans were limited to the observation that a suv3/suv3 mutant was unable to produce chlamydospores and hyphae, grew poorly on acetate as carbon source [51], and was deficient in biofilm formation [52]. The involvement of Suv3, or any other nuclear-encoded protein, in mitochondrial RNA metabolism had not been studied in this species.
Even though the mitochondrial genome of C. albicans encodes more proteins (14, not counting intron-encoded ORFs) than that of S. cerevisiae (nine protein coding genes), it is more compact (33.6 kb of unique sequence, compared to 75–85 kb), but still contains large (up to 5.6 kb) noncoding stretches between the eight primary transcripts [15], in contrast to the very compact mtDNAs of mammals (~16.5 kb), and S. pombe (~19 kb). As C. albicans is a petite-negative yeast that does not tolerate loss of mtDNA [41], its mitochondrial genome is essentially stable. In addition, common laboratory strains of this species have identical mtDNA sequence and do not show any differences in the expression of mitochondrial transcripts [15]. These traits make C. albicans a convenient system to study the mitochondrial transcriptome using RNA-seq methodology.
Previously we described the major features of the transcriptome in wild-type C. albicans mitochondria, identifying eight primary transcription units and significant differences in steady-state levels of mature transcripts [15]. Describing the transcriptome of wild-type mitochondria presents a snapshot of the steady-state levels of mature RNAs. As evidenced by multiple studies mentioned above, studying the transcriptome of mutants deficient in RNA degradation is required to reveal the true extent and specificity of transcription. In this work we analysed the changes in the organellar transcriptomic landscape resulting from the inactivation of the mitochondrial degradosome (mtEXO) through genomic deletion of genes encoding orthologs of the Suv3 helicase and the Dss1 exoribonuclease in C. albicans. We also investigated the effects of impaired RNA degradation on the expression of mature transcripts, the proteins they encode, and the activity of respiratory complexes in order to obtain a more comprehensive picture of the role of RNA degradation in mitochondrial gene expression.
Materials and methods
Strains and media
Candida albicans strain BWP17 (arg4::hisG/arg4::hisG, his1::hisG/his1::hisG, ura3::imm434/ura3::imm434, iro1::imm434 /iro1::imm434) [53] was used as the wild-type control and the starting point for construction of mutants.
The ΔCadss1/ ΔCadss1 strain (arg4/arg4, his1::hisG/his1::hisG, ura3::imm434 /ura3::imm434, iro1::imm434 /iro1::imm434, dss1::HIS1/dss1::SAT) was constructed using PCR based targeting [54]. The first allele was disrupted by integration of the CaSAT1 gene with ~40 nt flanks homologous to the upstream and downstream sequence of CaDSS1, amplified by PCR on the template of pFASAT1 [54]. In the second round, a deletion cassette was constructed by yeast recombinational cloning [55]. The CaHIS1 gene from the plasmid pFAHIS1 [54], together with ~1 kb flanks upstream and downstream of CaDSS1, all obtained by PCR, were used for in vivo recombination into the pRS426 vector in the S. cerevisiae CW252 strain. Deletion cassettes were amplified on the template of recombined plasmids by PCR and introduced into C. albicans heterozygous strains by electroporation [56]. Genotypes of the obtained knockouts were confirmed by PCR.
The ΔCasuv3/ ΔCasuv3 strain (arg4/arg4, his1::hisG/his1::hisG, ura3::imm434 /ura3::imm434, iro1::imm434 /iro1::imm434, suv3::HIS1/suv3::HIS1, eno1::CaCAS9-SAT/ENO1) was constructed using the C. albicans CRISPR/Cas9 Solo system as described previously [57]. The guide sequence started 14 nt downstream of the CaSUV3 ATG codon.
For the reconstitution of the wild-type gene, DSS1 or SUV3 genes together with upstream (~1 kb) and downstream (~0.5 kb) fragments were cloned into the pFAURA3 vector using SLIC [58]. The construct was subsequently linearized and transformed into C. albicans homozygous knockout strains. Reconstituted strains were verified by PCR, confirming the presence of the gene in the target locus.
For the isolation of mitochondria, strains were grown in liquid YPGal medium (1% yeast extract, 2% peptone and 2% galactose) containing 80 g/l uridine at 37°C until logarithmic growth phase. Respiratory growth was tested on agar plates with either YPD (1% yeast extract, 2% peptone and 2% glucose) or YPG (1% yeast extract, 2% peptone and 2% glycerol). Salicylhydroxamic acid (SHAM) was optionally added at 5 mM. Growth curves were measured using the Bioscreen C Automated Microbiology Growth Curve Analysis System (Thermo) in liquid YPD or YPG media, with the optional addition of SHAM at 5 mM. Numerical analysis of growth curves was performed using the Growthcurver R package [59]. Synthetic complete media were 0.67% Yeast Nitrogen Base (Bifco), 0.2% CSM-His-Leu-Trp-Ura mix (MP Biomedicals) and 2% glucose, supplemented with histidine (20 mg/l), tryptophan (20 mg/l), leucine (60 g/l), and uracil (20 g/l) as appropriate.
Sequences of all the primers used in the construction and verification of strains are listed in the Supplementary Table S1.
Enzymatic assays
Mitochondrial proteins were extracted by 5% DDM treatment. Protein concentration measurement was performed by the BCA protein assay (Pierce). 100 μg of mitochondrial protein in 1x loading buffer (Invitrogen) was loaded onto a BN-PAGE gradient 3–12% bis TRIS gel (Invitrogen). Electrophoresis was performed in the light blue cathode buffer variant, in the X Cell Sure Lock® Mini Cell system according to the manufactures instruction (Invitrogen). In-gel activity assays were performed as described previously [60,61]. For complex I activity staining, gels were pre-washed with reaction buffer (0.1 M Tris–HCl pH 7.4) for 20 min, followed by the addition of fresh buffer supplemented by 0.2 mM NADH (Bioshop) and 0.2% nitrotetrazolium blue (NTB) (SIGMA Aldrich) and further incubated for 1 h with agitation. The reaction was stopped by the addition of fixing solution (45% methanol, 10% acetic acid). Gels were scanned after overnight destaining in the fixing buffer. For ATPase activity staining, gels were incubated in the reaction buffer (270 mM glycine, 35 mM Tris, 14 mM MgSO4) for 30 min at RT. Gels were transferred into fresh buffer with 0.2% Pb(NO3)2 and 8 mM ATP (pH 8.3) (Bioshop). The gel was subsequently scanned for quantification of the white lead phosphate precipitate correlating with ATP hydrolysis. The lead phosphate precipitate was later completely dissolved by adding fixing solution (50% methanol, 10% acetic acid) and the gels were stained by Coomassie dye [61].
In vitro colorimetric assays for Complex I, Complex III, and Complex IV activity were performed as described previously [62]. 10 μM rotenone, 10 μg/ml antimycin and 10 nM NaN3 were used as inhibitors for Complex I, Complex III and Complex IV activity, respectively. For the mitochondrial ATPase activity assays mitochondria stored at −80°C were thawed and ATP hydrolysis was measured without osmotic protection at pH 8.4 in the presence of a saturating amount of ATP as described previously [63], with the optional addition of 10 μg/ml oligomycin as the inhibitor of the Fo fragment.
Northern blot analysis
Northern hybridization was performed essentially as described previously [15,64]. The probes and 32P labelling protocols used to detect each transcript were described previously [15]. Quantitative analysis of blots scanned on the Typhoon FLA 9000 (GE) biomolecular imager was performed in Fiji [65].
Sequence analysis of C. albicans SUV3 and DSS1 genes
DNA and amino acid sequences of SUV3 and DSS1 from C. albicans were obtained from the Candida Genome Database [66]. Amino acid sequence identity and similarity was calculated from Needleman-Wunsch global pairwise alignments obtained using STRETCHER from the EMBOSS suite [67].
Isolation of mitochondria and RNA extraction
RNA was obtained from mitochondria isolated from the log-phase liquid cultures of C. albicans grown in YPGal as described previously [15].
Transcriptome sequencing, mapping and analysis
RNA prepared from purified mitochondria was used to construct mitochondrial RNA-seq libraries. RNA-seq libraries Ion Total RNA-Seq Kit v2 (ThermoFisher Scientific) were prepared starting with 400–500 ng of mitochondrial RNA, according to the manufacturer’s protocol. The RNA fragmentation step was shortened to 4 min, preserving intact tRNAs. RNA quality and library construction was monitored using BioAnalyzer 2100 (Agilent Technologies) according to the manufacturer’s protocol.
The libraries were sequenced on the Ion Torrent Proton™ NGS System according to the manufacturer’s instructions. Raw sequencing data were processed using the Torrent Suite™ Software (Life Technologies). Barcode removal and quality trimming were performed in Torrent Suite™ using default parameters (30% QC threshold, reads <25 nt rejected). The resulting reads had a mean length of about 120 nt, and median length of about 110 nt. The processed reads were exported as FASTQ files.
The complete mtDNA sequence of C. albicans strain SC5314 (GenBank:AF285261.1), with one of two identical copies of the inverted repeat region removed and additional annotations [15] was used as a reference. Reads were mapped to the reference sequence using BWA-mem [68] which was found to be the best performing aligner for Ion Torrent data [69]. SAMtools [70] was used to manipulate the resulting alignments and to calculate coverage depth for each position in the reference sequence. The vioplot package in R was used to visualize coverage depth histograms. Coverage graphs were obtained by visualizing BWA files obtained using bamCoverage from the deepTools2 package [71] in pyGenomeTracks [72]. Reads mapping to the annotated transcription units in the sense and antisense orientation were quantified using featureCounts [73] from the Rsubread package [74].
Mass spectrometry
Mass spectrometry on proteins from isolated mitochondria was preceded by filter-aided sample preparation (FASP) [75,76], using sequencing grade trypsin (Promega), Microcon 30 K spin columns (Milipore) and 30 µl of protein extract (3 µg/µl) per digestion. The LC-MS-MS/MS experiment was performed on an Orbitrap Elite spectrometer (Thermo). Protein concentrations, relative to the concentration of mitochondrial porin, were estimated based on MS spectra, with the use of MaxQuant v1.6.17.0 software and built-in LFQ algorithm [77,78]. Alternatively, when the number of detected peptide fragments was not sufficient (as in the case of Nad1p, represented by three peptides in each sample), relative abundance was estimated based on peak intensities [79].
Results
Both mtEXO components are essential for the functioning of the C. albicans mitochondrial respiratory system
The Suv3 protein in C. albicans is encoded by the C2_04350C_A gene (orf19.4519) which is predicted to encode a protein of 720 amino acids, sharing 46% sequence identity (64% similarity) with its S. cerevisiae ortholog. The product of the C. albicans C2_08550C_A (orf19.3624) gene encodes a protein of 1146 amino acids with 22% sequence identity (41% similarity) to Dss1p of S. cerevisiae.
As C. albicans is an obligate diploid, we constructed homozygous deletion mutants in the background of the BWP17 wild-type strain [53]. The ΔCadss1/ ΔCadss1 strain was constructed in two rounds of transformation using PCR cassettes with the SAT1 and HIS1 markers. The ΔCasuv3/ ΔCasuv3 strain was obtained using the CRISPR-Cas9 system adapted to C. albicans [57]. Additionally, wild-type CaSUV3 and CaDSS1 genes were reconstituted in their native genomic loci by recombination following transformation of the respective homozygous deletants with a linearized pFAURA3 plasmid [80] containing a PCR-amplified wild-type gene sequence. All DNA constructs were verified by sequencing, and the genotypes of all strains were confirmed by PCR. For each genotype two independently obtained strains were constructed and tested.
Respiratory competence of the mutant strains was assessed by comparing growth on a non-fermentable carbon source – glycerol with the fermentable glucose (Fig 1(a)). As some respiratory defects in C. albicans can be partially compensated by the activity of an alternative oxidase (AOX) pathway [81,82], we included a known specific AOX inhibitor – salicylhydroxamic acid (SHAM) in the assay.
Figure 1.

The homozygous ΔCadss1 and ΔCasuv3 strains are respiratory-deficient. (a). Growth on solid agar media containing glucose (YPD) or glycerol (YPG) as the carbon source. Alternative oxidase inhibitor – salicylhydroxamic acid (SHAM) was optionally added at 5 mM. A series of 10x dilutions from an overnight YPD starter culture were spotted on plates and incubated for 48 h at 30°C. (b). Growth curves based on turbidometric measurements of 1 ml liquid cultures (Bioscreen C Automated Microbiology Growth Curve Analysis System) were determined at 30°C for 30 h. Three technical replicates for wild-type (WT) and reconstituted (rec) strains, and three technical replicates of two independent biological replicates for deletant (Δ) strains were used to calculate plotted mean ± SD values
The homozygous ΔCadss1 and ΔCasuv3 strains exhibit significantly impaired growth on glycerol media, regardless of the presence of SHAM. On glucose the mutants continue to grow, even in the presence of SHAM, albeit at a visibly reduced rate. The respiratory defect in the ΔCasuv3 strain seems to be slightly more pronounced than that of the ΔCadss1 mutant. Reconstitution of a wild-type gene in either deletant strain restored their ability to grow on non-fermentable media to wild-type levels.
In order to confirm the results of the growth test we also performed growth curve measurements in liquid media (Fig 1(b)). On glucose media both mutant strains grow at a rate that is similar to that of the wild-type, but they show a longer lag phase and have a lower carrying capacity (maximum density at plateau, estimated at 75% of wild-type in YPD, and 40–60% in YPD + SHAM). On liquid glycerol media the growth defect is very clear, and more pronounced in the case of the ΔCasuv3 strain, which essentially fails to exhibit measurable growth. The addition of SHAM seems to only slightly aggravate the phenotype of both mutants in this assay. Reconstitution of a single functional allele of CaSUV3 or CaDSS1 restores the carrying capacity and doubling-time in respiratory media to near wild-type levels.
These results indicate that both DSS1 and SUV3 are required to maintain respiratory competence in C. albicans.
Pervasive transcription of the mitochondrial genome in C. albicans mtEXO mutants
In order to gain insight into the role of mtEXO in C. albicans cells we performed RNA-seq analysis of mitochondrial transcriptomes in the homozygous ΔCasuv3 and ΔCadss1 strains, comparing them with the wild-type BWP17 strain. RNA sequencing libraries were prepared from isolated mitochondria and sequenced using the Ion Torrent Proton™ NGS System. This NGS technology was previously used to describe the mitochondrial transcriptome of wild-type C. albicans [15], and is known to perform well in gene expression studies using reads mapping to a known template [83], in spite of the slightly higher rate of read errors in homopolymer sequences [84] compared to other short-read methods.
Reads were mapped to the mtDNA sequence of C. albicans strain SC5314 (GenBank:AF285261.1) with one of the two identical 6.8 kb repeat regions removed. Previous RNA-seq analysis showed that the sequence of the mitochondrial genome of BWP17 and SN148 laboratory strains is identical to this reference [15].
Two independent cultures of the wild-type strain, and two independent homozygous deletants of each gene were used for RNA-seq. As there was no significant variation in mapped read coverage between the replicates, reads from the two cultures of the same genotype were analysed together. Table 1 presents the sequencing and mapping statistics. The percentage of mapped reads in the wild-type strain (48%) was comparable to that obtained in our previous study on the wild-type C. albicans mitochondrial transcriptome [15]. Mitochondrial RNA preparations from respiratory deficient strains showed a higher percentage of reads mapping to the mtDNA reference, consistent with our observations that the amount of cytoplasmic material co-purifying with mitochondria is higher in respiratory competent wild-type strains.
Table 1.
Summary of the RNA-seq mapping in the wild-type (BWP17) and homozygous ΔCadss1 and ΔCasuv3 strains
| BWP17 | ΔCadss1 | ΔCasuv3 | |
|---|---|---|---|
| Total reads | 23007600 | 13282788 | 13301880 |
| Mapped to mtDNA (%) | 11105411 (48%) | 11747820 (84%) | 11667218 (88%) |
| Mapped to transcription units (TUs) (% of mapped) | 11014996 (99%) | 10792218 (92%) | 10307551 (88%) |
| Sense (% of mapped to TUs) | 11003471 (99.9%) | 9807730 (91%) | 8851904 (86%) |
Further analysis and visualization of the results obtained in the wild-type control strain (Fig 2) also follows the previously published observations. The majority (99%) of reads map to the identified primary transcription units in the sense orientation (Fig 2(a,b)). Only 1% of reads map to regions not annotated as the primary transcripts, and this activity is limited to a few short fragments, previously identified in the major noncoding stretch in the repeat region. Few, if any reads map to regions separating the primary transcription units. The amount of reads mapping to the transcription units in antisense orientation in wild-type strains is negligible (<1%). Distribution of coverage depth shows that in the wild-type strain 3685 positions in the reference sequence have no RNA-seq reads mapping to them, and the regions with highest coverage correspond to a small subset of the genome (Fig 2(c)).
Figure 2.

Statistics of RNA-seq reads mapping to the mtDNA reference sequence in the wild-type (BWP17) and homozygous ΔCadss1 and ΔCasuv3 strains. (a). Percentage of reads mapping to the annotated transcription units (TU [15]), and intergenic regions. (b). Percentage of reads mapping to the annotated transcription units in sense and antisense orientation. Data for a. and b. were obtained using featureCounts [73] from the Rsubread package [74]. (c). Violin plots showing the distribution of sites in the the mtDNA reference sequence with varying coverage depth. Width of the plot corresponds to the frequency of sites covered by the number of reads shown on the X axis. White circles and dark bars correspond to the median and the interquartile range (IQR), respectively. Coverage depth was calculated using the -depth option of SAMtools [70] and visualized in R using the vioplot package
These results conform to the expected landscape of the mature transcriptome, with relatively few highly expressed short transcripts, mostly corresponding to tRNAs and, to a lesser extent, rRNAs, mRNAs present at significantly lower steady-state levels, and the noncoding and intronic sequences efficiently removed.
The RNA-seq results obtained in the mtEXO mutants are in stark contrast to those observed in the wild-type control. In total, 12% and 8% of reads map to the regions outside the annotated transcription units in the ΔCasuv3 and ΔCadss1 strains, respectively, compared to 1% observed in the wild-type control (Fig 2(a)). Additionally, reads mapping to the primary transcripts in the antisense orientation, virtually absent from the wild-type transcriptome, also show a marked increase (14% and 9% in the ΔCasuv3 and ΔCadss1 strains, respectively) (Fig 2(b)). Quantitative analysis of the distribution of coverage depth in the mtEXO mutants also shows a drastically different picture compared to control (Fig 2(c)) There are no sites in the reference genome without any reads mapped to them (the lowest coverage values are 576 and 85 in in the ΔCasuv3 and ΔCadss1 strains, respectively), and the majority of genome has very high RNA-seq read coverage.
A cursory observation of the coverage by reads mapping to the mitochondrial genome sequence (Fig 3) shows nearly uniform coverage of the entire genome in the mutant strains. In particular, RNA-seq reads cover the noncoding regions, including fragments separating different transcription units, and the main non-coding region. Introns, particularly those in the COX1 gene, also show a much higher coverage.
Figure 3.

Coverage of the C. albicans mtDNA reference sequence with one of two identical copies of the inverted repeat region removed by forward and reverse RNA-seq reads in wild-type (BWP17) and homozygous ΔCadss1 and ΔCasuv3 strains. Transcription units and gene annotations are according to [15]. BWA files obtained using bamCompare [71] were visualized in pyGenomeTracks [72]. The depth coverage axis was set at the maximum value of 5000 reads to better visualize low-coverage regions, truncating the highest values. Note that for TU02 and TU04 the sense strand is the reverse strand
These results indicate that in the absence of mtEXO function, transcriptional activity covering the entire mitochondrial genome is revealed, including a significant amount of antisense (‘mirror’) transcripts. Removal of excised introns is also visibly impaired. The effect is apparent in both homozygous mutants, but it is somewhat more pronounced in the ΔCasuv3 homozygous mutant, reflecting the slightly more pronounced respiratory phenotype in that strain.
Steady state levels of multiple mitochondrial transcripts are affected in C. albicans mtEXO mutants
In order to verify how the dysfunction of mtEXO affects the processing and steady-state levels of mature mitochondrial transcripts, we performed a series of Northern blot experiments on RNA isolated from mitochondria of the homozygous ΔCadss1 mutant, with the wild-type BWP17 strain used as control. The deletant with one wild-type CaDSS1 allele reconstituted by recombination was also included in the analysis. The results (Fig 4(a–d) and Table 2) show that the steady-state levels of several mature mRNAs are significantly affected by the deletion of CaDSS1.
Figure 4.
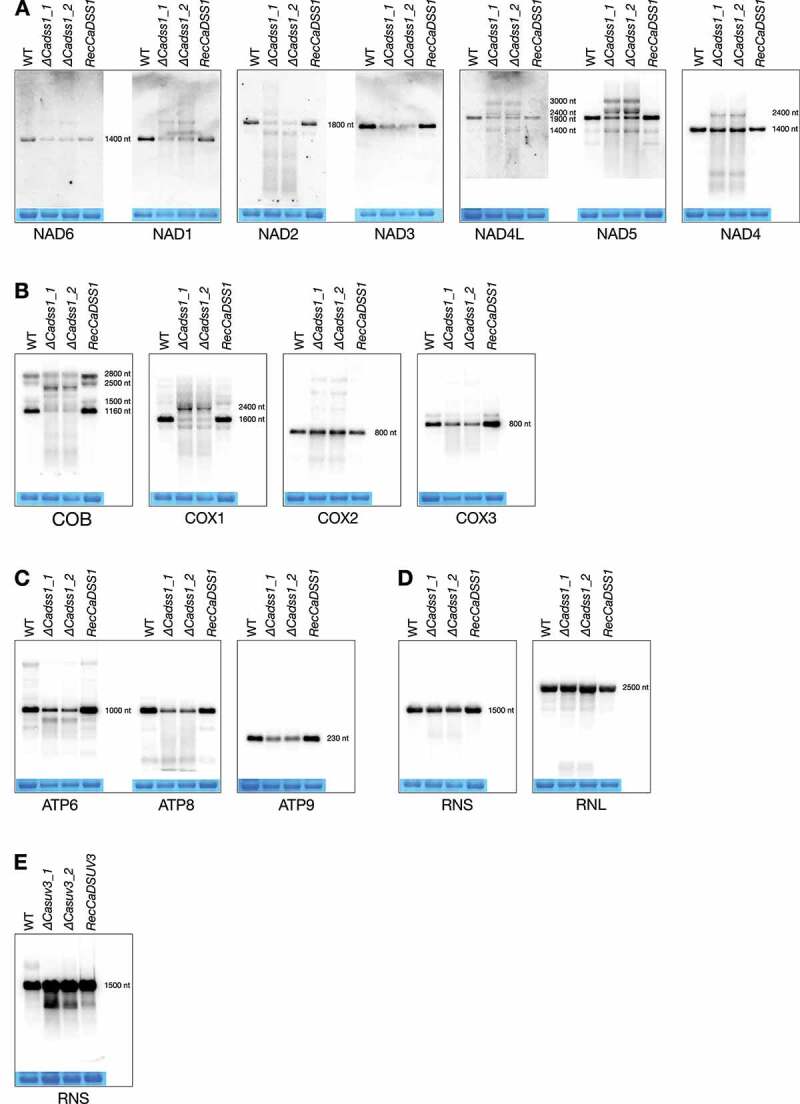
Northern blot analysis of mitochondrial mRNA and rRNA transcripts from wild-type (WT, BWP17), homozygous ΔCadss1 mutants (a–d, two independent strains), a strain with a single wild-type DSS1 allele reintroduced by recombination (a–d, recCaDSS1) of C. albicans, homozygous ΔCasuv3 mutants (e, two independent strains), and a strain with a single wild-type SUV3 allele reintroduced by recombination (e, recCaSUV3). (a). mRNAs encoding subunits of Complex I. (b). mRNAs encoding subunits of Complex III (COB) and Complex IV (COX). (c). mRNAs encoding subunits of the ATP synthase (Complex V). (d). rRNAs of the small (RNS) and large (RNL) subunits of the mitoribosome. (e). Northern blot analysis of mitochondrial rRNA of the small (RNS) subunit in ΔCasuv3 mutants. RNAs were prepared from purified mitochondria and separated by agarose/formaldehyde gel electrophoresis in denaturing conditions. Methylene blue staining of the small subunit cytoplasmic rRNA in the blot is shown below each autoradiogram as a loading control. In panels a–d blot series [NAD2, COB, COX1, RNS]; [NAD4, COX2, RNL]; [NAD4L, ATP9]; [COX3, ATP6]; [NAD5, ATP8] were prepared by stripping and re-hybridizing the same membrane, hence the same loading controls. Two blots within a single frame indicate that the probes hybridize to two ORFs contained in the same bicistronic mature transcript. Approximate sizes of mature transcripts and most prominent secondary species are indicated
Table 2.
Steady-state levels of mature mitochondrial mRNAs determined by Northern blots (normalized to small subunit cytoplasmic rRNA), levels of selected mitochondrially-encoded proteins determined by MS (normalized to mitochondrial porin), and enzymatic complex activities in the homozygous ΔCadss1 strain, expressed as % of the wild-type (BWP17) value. n/a – not available (peptides not detected by MS)
| Complex | Subunit | mRNA in ΔCadss1 | Protein in ΔCadss1 | Activity in ΔCadss1 |
|---|---|---|---|---|
| I | NAD1 | 13% | 26% | 40% |
| NAD2 | 22% | n/a | ||
| NAD3 | 29% | 44% | ||
| NAD4 | 78% | n/a | ||
| NAD4L | 18% | n/a | ||
| NAD5 | 35% | 42% | ||
| NAD6 | 24% | n/a | ||
| III | COB | 19% | 12% | 77% |
| IV | COX1 | 5% | 2% | 5% |
| COX2 | 120% | 9% | ||
| COX3 | 58% | n/a | ||
| V | ATP6 | 28% | n/a | 55% (total) 53% (oligomycin-sensitive) |
| ATP8 | 24% | n/a | ||
| ATP9 | 60% | n/a |
Of the mitochondrial mRNAs encoding Complex I subunits, three bicistronic transcripts (cf. Fig 3 for the organization of transcription units): NAD6-NAD1, NAD2-NAD3, and NAD4L-NAD5 all show decreased steady-state levels of mature transcripts (13%-29% of wild-type, Table 2), and, to a varying degree, accumulation of high molecular weight precursors, particularly apparent for NAD4L-NAD5 (Fig 4(a)). The monocistronic NAD4 mRNA also shows precursor accumulation, but without significantly decreased mature transcript level (78% of wild-type). Among transcripts encoding subunits of Complexes III and IV, expression of intron-containing transcripts of COB and COX1 is severely affected (Fig 4(b) and Table 2), with the accumulation of unspliced precursors and loss of the mature mRNA. The COX1 transcript is by far the most severely affected, with only ~5% of mature mRNA remaining (Table 2). The level of the COX3 transcript is only moderately affected, and COX2 shows no significant decrease in the mutants. The bicistronic ATP6-ATP8 transcript shows decreased steady-state level of the mature mRNA to about 24%-28%, whereas the ATP9 mRNA is only moderately decreased (60% of wild-type, Fig 4(c) and Table 2). No significant reduction in both rRNAs (RNS and RNL) was observed (Fig 4(d)).
In addition to these effects on mature transcripts, a more subtle phenotype is also apparent in the majority of Northern blots (Fig 4(a–d)). Hybridization with samples from the homozygous ΔCadss1 mutants shows an increased level of background ‘smear’ signal, both larger and smaller than the probe target. This is particularly apparent with the NAD4 (Fig 4(a)) and COB (Fig 4(b)) probes, but can be also observed for other samples. Wild-type and reconstituted samples show significantly lower background. This is likely to be the effect of the presence of low-levels of multiple RNA fragments covering the entire genome, which was revealed by RNA-seq.
Northern blot analysis of mitochondrial RNA from the homozygous ΔCasuv3 mutant showed broadly similar results to the ones obtained in ΔCadss1 (not shown), with one notable exception. Hybridization with the small subunit rRNA probe (RNS) shows a very significant background signal, with apparent distinct fragments shorter than the mature rRNA, which disappear upon reconstitution of one wild-type allele (Fig 4(e)).
Respiratory complex synthesis and activity is affected in C. albicans mtEXO mutants
In order to assess the effect of transcriptome changes on the synthesis of mitochondrial oxidative phosphorylation system complexes, we performed a mass spectrometry analysis of the mitochondrial proteome, followed by enzymatic visualization of the activity of Complex I and ATPase (Complex V) on mitochondrial protein extracts separated in native polyacrylamide gels (Fig 5(a)), and in vitro quantitative colorimetric assays [62] for the activity of Complex I, Complex III, and Complex IV (Fig 5(b–d)). The function of Complex V was assessed using a relatively simple (compared to the ATP synthesis measurement) assay of ATPase activity sensitive to oligomycin (a specific Fo inhibitor) (Fig 5(e)) [63].
Figure 5.

Respiratory complex activity in the homozygous ΔCadss1 and ΔCasuv3 strains, compared to wild-type (WT, BWP17), and strains with a single wild-type allele reintroduced by recombination (recCaDSS1 and recCaSUV3). (a). Native polyacrylamide gels with the activity of Complex I and Complex V visualized in situ. Coomassie blue staining (Co.) shows complex assembly. (b). Time-course curves of colorimetric assays for Complex I activity measured as decreasing OD340. Reaction inhibited with rotenone served as the negative control. (c). Time-course curves of colorimetric assays for Complex III activity measured as increasing OD550. Reaction inhibited with antimycin served as the negative control. (d). Time-course curves of colorimetric assays for Complex IV activity measured as decreasing OD550. Reaction inhibited with NaN3 served as the negative control. In b, c, and d, curves show mean ± SD values for three replicates. (e). Mitochondrial ATPase activity. Bars show mean ± SD values for three replicates for total ATPase activity in mitochondrial protein extracts, with the activity remaining after inhibition of the Fo fragment by oligomycin shown in blue
Mass spectrometry analysis (normalized to the levels of mitochondrial porin) indicates that the levels of detected mitochondrially encoded proteins Cob, Cox1, Cox2, Nad1, Nad3, and Nad5 are markedly decreased in the mutant strains, particularly in ΔCadss1 (Table 2, Supplementary Table S2). This effect is particularly apparent for the Complex IV subunits Cox1 and Cox 2 (2% and 9% of wild-type in ΔCadss1, respectively). The decrease in detected protein levels mostly follows that of the respective mRNAs, with the notable exception of Cox2, where the protein is present at a significantly lowered amount despite the mRNA being detected at normal (or even slightly increased) level, suggesting that translation and/or protein stability is affected. Not all mitochondrially-encoded proteins were detected by MS even in the wild-type strain, which is to be expected based on similar results obtained in human cells [76].
Visualization of Complex I and Complex V on native gels shows only a moderate decrease in the levels of assembled complexes (Fig 5(a)). The levels of unassembled Fo and Fc fragments of Complex V seem to be slightly increased in the deletant strains. Synthesis and assembly of these complexes is thus at least partially preserved, consistent with the presence (albeit at a reduced level) of the respective mRNAs. As native gel assays can detect gross abnormalities in the respiratory complex synthesis, but will not necessarily reveal more subtle defects, we performed quantitative assays of the respiratory complex enzymatic activity. The results show a partial, but significant reduction for Complexes I, III and V, and a complete loss of Complex IV activity in both homozygous deletants (Fig 5(b–e)). The activity of Complex I in both mutants is markedly reduced to about 40% of the wild-type, and is largely consistent with the observed decrease in mature mRNA and protein levels (Table 2). This remaining activity is, however, still higher than that of a control with the CI activity inhibited by rotenone (27%). Similarly, the activity of Complex III in the mutants is moderately decreased (77% and 80% of wild-type activity for ΔCadss1 and ΔCasuv3, respectively), albeit not as severely, as in the control extract treated with antimycin (6% of wild-type activity). On the other hand, Complex IV activity in both ΔCadss1 and ΔCasuv3 strains is severely reduced (5% and 22% of wild-type activity for ΔCadss1 and ΔCasuv3, respectively), comparable to the control extract treated with NaN3 (13% of wild-type activity). The stark difference between the effect on Complexes III and IV is indicative of a threshold effect, with 19% of remaining COB mRNA being still sufficient to maintain the enzymatic activity of the complex at about 77% level; whereas the decrease of mature COX1 mRNA to 5% results in a nearly complete loss of the activity of the complex. Both total (55% and 74% of wild-type activity for ΔCadss1 and ΔCasuv3, respectively) and oligomycin-sensitive (53% and 90% for ΔCadss1 and ΔCasuv3, respectively) ATP hydrolysis activity of Complex V shows a moderate, but significant reduction in the mutant strains, particularly apparent in ΔCadss1, in line with the observed reduction in mature mRNAs and proteins (Table 2).
Such pronounced deficiency in Complex IV, coupled with a reduced activity of the remaining complexes, would be sufficient to account for the respiratory negative phenotype of the mutants.
Discussion
Pervasive transcription is revealed in the absence of mtEXO activity
In order to gain insight into the role of the mtEXO complex in C. albicans, we analysed the transcriptome of homozygous ΔCadss1 and ΔCasuv3 mutant strains using the same methodology we previously applied to wild-type mitochondria in this species. Like in that previous study [15], in wild-type cells there are no detectable RNA molecules corresponding to the intergenic noncoding sequences, with the notable exception of two short transcripts of unknown function originating from the region between transcription units TU1 and TU2. Eight clearly separate primary transcription units, each with its own promoter initiating transcription, are apparent. Correspondingly, about 10% of the reference mtDNA sequence has zero coverage by RNA-seq reads in normally functioning mitochondria.
In stark contrast, in the transcriptomes of mtEXO deficient mutants, there is evidence of RNA-seq reads covering the entire mitochondrial genome sequence in both directions. The levels of different transcripts still vary significantly (although visibly less than in wild-type), but the separation of different transcription units is less apparent, as there are reads covering all the noncoding segments. There are no sites in the reference genome that are not covered by some RNA-seq reads. In total, from 8% to 12% of reads map to noncoding regions outside the primary transcription units, compared to 1% in the wild-type.
RNA-seq, like most transcriptomic analysis methods, provides information on the steady state of identified RNAs. The observed increase in RNA-seq reads corresponding to intergenic regions and antisense transcripts could thus, in principle, result from either increased transcription of these noncoding sequences, or from deficient degradation of these transcripts. The latter explanation is, however, much more likely in view of what is known about the enzymatic activity of Suv3 and Dss1 orthologs in other organisms. Studies in S. cerevisiae [33] and C. glabrata [34] show that Dss1 is a 3ʹ-to-5ʹ exoribonuclease that is responsible for the RNase activity of the complex, and the Suv3 helicase is required for efficient feeding of the substrate to its active site. When RNA species absent from normal cells appear in a mutant deficient in a known RNase activity, impaired RNA degradation is the most likely explanation, in line with interpretation of studies performed in RNase deficient mammalian cells [85], as well as human mitochondria [29]. In contrast, proteins with an RNase activity have never been shown to be participate in transcription initiation in yeast mitochondria.
The loss of mtEXO activity reveals the existence of transcripts covering the majority of the genome, consistent with pervasive transcription demonstrated in the Eukaryotic nucleus [3,5–7,86,87], and in Prokaryotes [4,88,89]. Significant amounts of noncoding RNAs mapping outside annotated genes were also identified in plant organelle transcriptomes, mostly in chloroplasts [90–93]. Mitochondrial transcriptome studies of RNA degradation deficient mutants were mostly limited to human cells [29,30,32], where the extremely compact genome is transcribed in its entirety [17,94]. Our results in C. albicans demonstrate that pervasive mitochondrial transcription occurs in fungal mitochondria that contain longer noncoding sequences, and is thus likely a universal phenomenon. As in other systems, the effects of pervasive mitochondrial transcription in wild-type C. albicans are undetectable, most likely due to efficient degradation, and become apparent only in mtEXO deficient mutants.
Antisense transcripts accumulate in the absence of mtEXO activity
A significant fraction of the abnormal noncoding RNAs that we identified in the mtEXO deficient mutants corresponds to transcripts mapping to the sequences of transcription units, but in an antisense orientation. These ‘mirror RNAs’ are virtually undetectable in wild-type C. albicans mitochondria, and in the mutants constitute up to 14% of all reads mapping to the transcription units.
Accumulation of antisense transcripts was shown in human mitochondria with impaired degradosome function, where it contributes to the accumulation of double-stranded RNA (dsRNA), which is released from mitochondria and triggers interferon-dependent innate antiviral response [29,30,32]. In S. cerevisiae, a moderate increase in antisense transcripts corresponding to two genes was observed in mitochondria of a mutant lacking the mitochondrial targeting sequence of the Dis3 exoribonuclease [14].
Impaired mtEXO function results in aberrant mitochondrial gene expression and disruption of mitochondrial respiration
Northern blot analysis of homozygous ΔCadss1 and ΔCasuv3 mutants revealed some similarities to the phenotypes observed in S. cerevisiae. Notably, the expression and processing of intron-containing pre-mRNAs of COB and COX1 is severely affected. The visible increase in RNA-seq reads mapping to introns and intron-exon boundaries also points to a splicing defect. The link between RNA degradation and processing of intron-containing transcripts has been suggested by previous studies in S. cerevisiae [20,36] and S. pombe [21], and is clearly confirmed by our results. In the absence of mtEXO RNase activity introns accumulate, which in turn likely affects splicing, possibly by sequestering protein factors that assist this process. Also, introns in COX1 and COB genes contain open reading frames likely to encode maturase proteins [15] that need to be translated in order to enable efficient splicing. Generalized mitochondrial gene expression dysfunction observed in mtEXO mutants could thus affect splicing by interfering with maturase expression. Such interpretation is further strengthened by the observation that even though the introns of large subunit rRNA transcript (RNL) that lack ORFs do accumulate in the mutants, the expression of mature rRNA is not significantly affected.
The alterations in the mitochondrial transcriptome of ΔCadss1 and ΔCasuv3 mutants have a profound effect on their respiratory capability. Both strains exhibit a complete respiratory deficiency, independent of the alternative oxidase (AOX) activity. Analysis of the mitochondrial proteome and the activity of mitochondrial electron transfer chain complexes indicates defects that are generally consistent with the observed decreased levels of mature mRNAs. Activities of Complex I, III, and IV are clearly decreased in the mutants. Despite indications of a slight defect in the coordination of its assembly, evidenced by the presence of free Fo and Fc fragments, the mature Complex V is still present, its ATPase activity is, however, moderately, but significantly reduced. A significant decrease of mitochondrial ATP synthesis activity can occur in S. cerevisiae mutants that exhibit normal or only slightly decreased ATPase activity [95], but as the mutant strains are respiratory deficient, it is unclear whether this is also the case here.
Probably the most interesting aspect of the physiological phenotype of ΔCadss1 and ΔCasuv3 mutants is related to the remarkable difference in the effect on the activity of Complex III, which is only moderately (albeit significantly) reduced, and Complex IV, which is totally inactive in ΔCadss1 (and severely reduced in ΔCasuv3). This is consistent with the levels of mature mRNAs of COB and COX1, and indicates that even a large decrease in the mRNA level is well tolerated up to a certain threshold, below which the synthesis and activity of the respiratory complex collapses. This threshold appears to be quite low, as the activity of Complex III, Complex I, and Complex V ATPase is reduced only partially, and it takes the loss of almost all mature COX1 mRNA to completely disrupt Complex IV. Interestingly, the loss of COX1 mRNA results in a marked decrease of not only the Cox1 protein, but also of the Cox2 protein, despite normal levels of COX2 mRNA, corroborating that translation of mitochondrially encoded Complex IV subunits is tightly regulated and coordinated with the complex assembly [96].
Results from prior functional analyses performed in S. cerevisiae also indicate that the mitochondrial system is quite resilient against even significant decreases in mature mRNA levels. In strains exhibiting about five-fold decrease in the overall transcription rate, OXPHOS is still functional enough to enable respiratory growth [35], and mutants with about 10% of a COX1 mRNA were shown to still maintain minimal, but observable, respiratory function [97]. Varying degrees of reduction in multiple mature mRNA levels observed in this study allowed us to study this phenomenon of resilience in more detail. One could be tempted to seek analogy between the phenotype of disturbed mitochondrial gene expression revealed in this and other yeast studies, and the threshold effects observed when analysing the impact of pathological mtDNA mutations in humans on mitochondrial function, and on the physiological manifestation of the symptoms [98]. At this point, however, it remains speculative. General resilience to change and the presence of threshold effects could, however, be a fundamental property of mitochondrial systems.
The role of mtEXO RNase activity in the shaping of mitochondrial transcriptome
The results of this study revealed the role played by RNA degradation in shaping the transcriptome of mitochondria. In C. albicans mitochondria the functional distinction between coding and noncoding regions, i.e. traditionally identified transcription units and intergenic spacers is realized primarily on the post-transcriptional level, in spite of the presence of individual transcription start sites [15]. Homologs of mitochondrial transcription termination factors that that are present in Metazoa and plants have not been identified in Fungi, including yeasts [99], and mechanisms that might terminate mitochondrial transcription in this group remain unknown. In mtEXO mutants, primary transcripts extend to areas downstream of the 3ʹ terminus observed in wild-type mitochondria, suggesting that this 3ʹ-to-5ʹ RNase activity plays an important role in the generation of proper 3ʹ transcript ends. RNA-seq coverage of the noncoding intergenic regions is generally still lower than for transcription units, suggesting that additional mechanism may contribute to the generation of primary and mature transcripts. Whether this involves another RNase activity, like the ortholog of the PET127 gene [100] encoding a putative 5ʹ-3ʹ exoribonuclease, or yet another mechanism requires further investigation.
Since the organization of the mitochondrial genome into several polycistronic transcription units separated by noncoding regions of varying length is a feature shared by most (if not all) genomes of yeasts and other Fungi [101–103], it is reasonable to believe that the role of the mtEXO complex in shaping the functional mitochondrial transcriptome is also similar in other species of this group. The studies performed in S. cerevisiae dis3 mutant [14] did not, however, provide evidence for truly pervasive transcription encompassing the entirety of noncoding mtDNA regions, and the accumulation of mirror RNAs was relatively limited compared to what can be observed in C. albicans in our experiments (and in human mitochondria as well). This could be due to the fact that yeast Dis3 is primarily a cytoplasmic and nuclear protein [104–107], and its function in mitochondria is secondary to that of mtEXO. Also, in spite of superficial similarities, the mitochondrial genomes of S. cerevisiae and C. albicans show significant differences, consistent with their evolutionary distance (discussed in detail in [15]), that are particularly manifested in the greater length and number of intergenic noncoding regions in the former. Our results are the first indication of pervasive mitochondrial transcription among yeasts, and further studies are necessary to determine to what extent the phenomenon is prevalent in this very diverse group of Fungi.
The phenomenon of pervasive transcription of noncoding regions was observed in many different genomic systems. It is still not clear whether it is just a ‘wasteful’ process that is not detrimental enough to be eliminated by evolution, or perhaps a mechanism with a biological function that is yet to be identified. In the nucleus it could be involved in maintaining genome stability through transcription-coupled DNA repair [108]. There is, however, no clear evidence of such a repair mechanism operating in mitochondria. Reductive evolution of the endosymbiont genome that gave rise to modern organellar genomes entailed not only a reduction in coding capacity, but also a significant simplification of the regulatory mechanisms, particularly those related to transcriptional control [38]. Relying on a very simple, two-component RNase complex for the shaping of transcriptome could be another indication of this evolutionary trend.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Maciej Kotliński for his advice on MS data analysis.
Funding Statement
This work was supported by the National Science Centre of Poland [UMO-2015/19/B/NZ2/00201, SYMFONIA 00463].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI Sequence Read Archive (SRA) repository at http://www.ncbi.nlm.nih.gov/bioproject/662600, and in ProteomeXchange at https://www.doi.org/10.6019/PXD024934.
Author contributions
Karolina Łabędzka-Dmoch, Magdalena Karpińska, Sonia Dębek, and Thi Hoang Diu Bui performed genetic and Northern blot experiments. Adam Kolondra performed RNA-seq experiments. Jakub Piątkowski performed proteomic analysis. Joanna Jabłońska, Maciej Grochowski, Karolina Łabędzka-Dmoch, and Thi Hoang Diu Bui performed physiological measurements. Paweł Golik supervised and designed the study, participated in data analysis, and wrote the initial draft. All authors read and approved the final manuscript.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Kapranov P, Willingham AT, Gingeras TR.. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. [DOI] [PubMed] [Google Scholar]
- [2].Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. [DOI] [PubMed] [Google Scholar]
- [3].Tudek A, Candelli T, Libri D. Non-coding transcription by RNA polymerase II in yeast: hasard or nécessité. Biochimie. 2015;117:28–36. [DOI] [PubMed] [Google Scholar]
- [4].Wade JT, Grainger DC. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol. 2014;12:647–653. [DOI] [PubMed] [Google Scholar]
- [5].Gudipati RK, Xu Z, Lebreton A, et al. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell. 2012;48:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Graur D, Zheng Y, Price N, et al. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5:578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. [DOI] [PubMed] [Google Scholar]
- [10].Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. [DOI] [PubMed] [Google Scholar]
- [11].Burger G, Gray MW, Forget L, et al. Strikingly Bacteria-Like and Gene-Rich Mitochondrial Genomes throughout Jakobid Protists. Genome Biol Evol. 2013;5:418–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. [DOI] [PubMed] [Google Scholar]
- [13].Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. [DOI] [PubMed] [Google Scholar]
- [14].Turk EM, Das V, Seibert RD, et al. The mitochondrial RNA landscape of Saccharomyces cerevisiae. PLoS One. 2013;8:e78105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kolondra A, Labedzka-Dmoch K, Wenda JM, et al. The transcriptome of Candida albicans mitochondria and the evolution of organellar transcription units in yeasts. BMC Genomics. 2015;16:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shang J, Yang Y, Wu L, et al. The S. pombe mitochondrial transcriptome. RNA. 2018;24:1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mercer TR, Neph S, Dinger ME, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Houseley J, Tollervey D. The Many Pathways of RNA Degradation. Cell. 2009;136:763–776. [DOI] [PubMed] [Google Scholar]
- [19].Szczesny RJ, Borowski LS, Malecki M, et al. RNA degradation in yeast and human mitochondria. Biochim Biophys Acta. 2012;1819:1027–1034. [DOI] [PubMed] [Google Scholar]
- [20].Stepien PP, Margossian SP, Landsman D, et al. The yeast nuclear gene suv3 affecting mitochondrial post-transcriptional processes encodes a putative ATP-dependent RNA helicase. Proc Natl Acad Sci USA. 1992;89:6813–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hoffmann B, Nickel J, Speer F, et al. The 3ʹ ends of mature transcripts are generated by a processosome complex in fission yeast mitochondria. J Mol Biol. 2008;377:1024–1037. [DOI] [PubMed] [Google Scholar]
- [22].Gagliardi D, Kuhn J, Spadinger U, et al. An RNA helicase (AtSUV3) is present in Arabidopsis thaliana mitochondria. FEBS Lett. 1999;458:337–342. [DOI] [PubMed] [Google Scholar]
- [23].Mattiacio JL, Read LK. Evidence for a degradosome-like complex in the mitochondria of Trypanosoma brucei. FEBS Lett. 2009;583:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Minczuk M, Piwowarski J, Papworth MA, et al. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Borowski LS, Dziembowski A, Hejnowicz MS, et al. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013;41:1223–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Holec S, Lange H, Kühn K, et al. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol Cell Biol. 2006;26:2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dziembowski A, Piwowarski J, Hoser R, et al. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem. 2003;278:1603–1611. [DOI] [PubMed] [Google Scholar]
- [28].Mattiacio JL, Read LK. Roles for TbDSS-1 in RNA surveillance and decay of maturation by-products from the 12S rRNA locus. Nucleic Acids Res. 2008;36:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Szczesny RJ, Borowski LS, Brzezniak LK, et al. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pietras Z, Wojcik MA, Borowski LS, et al. Controlling the mitochondrial antisense - role of the SUV3-PNPase complex and its co-factor GRSF1 in mitochondrial RNA surveillance. Mol Cell Oncol. 2018;5:e1516452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pietras Z, Wojcik MA, Borowski LS, et al. Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria. Nat Commun. 2018;9:2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dhir A, Dhir S, Borowski LS, et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018;560:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Malecki M, Jedrzejczak R, Stepien PP, et al. In vitro reconstitution and characterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J Mol Biol. 2007;372:23–36. [DOI] [PubMed] [Google Scholar]
- [34].Razew M, Warkocki Z, Taube M, et al. Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components. Nat Commun. 2018;9:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rogowska AT, Puchta O, Czarnecka AM, et al. Balance between transcription and RNA degradation is vital for Saccharomyces cerevisiae mitochondria: reduced transcription rescues the phenotype of deficient RNA degradation. Mol Biol Cell. 2006;17:1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Golik P, Szczepanek T, Bartnik E, et al. The S. cerevisiae nuclear gene SUV3 encoding a putative RNA helicase is necessary for the stability of mitochondrial transcripts containing multiple introns. Curr Genet. 1995;28:217–224. [DOI] [PubMed] [Google Scholar]
- [37].Dmochowska A, Golik P, Stepien PP. The novel nuclear gene DSS-1 of Saccharomyces cerevisiae is necessary for mitochondrial biogenesis. Curr Genet. 1995;28:108–112. [DOI] [PubMed] [Google Scholar]
- [38].Lipinski KA, Kaniak-Golik A, Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome - from genetics to evolution and systems biology. Biochim Biophys Acta. 2010;1797:1086–1098. [DOI] [PubMed] [Google Scholar]
- [39].Dziembowski A, Malewicz M, Minczuk M, et al. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol Gen Genet. 1998;260:108–114. [DOI] [PubMed] [Google Scholar]
- [40].Szczesny RJ, Wojcik MA, Borowski LS, et al. Yeast and human mitochondrial helicases. Biochim Biophys Acta. 2013;1829:842–853. [DOI] [PubMed] [Google Scholar]
- [41].Chen XJ, Clark-Walker GD. The petite mutation in yeasts: 50 years on. Int Rev Cytol. 2000;194:197–238. [DOI] [PubMed] [Google Scholar]
- [42].Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Noble SM, Johnson AD. Genetics of Candida albicans, a diploid human fungal pathogen. Annu Rev Genet. 2007;41:193–211. [DOI] [PubMed] [Google Scholar]
- [44].Hernday AD, Noble SM, Mitrovich QM, et al. Genetics and molecular biology in Candida albicans. Methods Enzymol. 2010;470:737–758. Elsevier. [DOI] [PubMed] [Google Scholar]
- [45].Noble SM, French S, Kohn LA, et al. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Samaranayake DP, Hanes SD. Milestones in Candida albicans gene manipulation. Fungal Genet Biol. 2011;48:858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hewitt VL, Heinz E, Shingu-Vazquez M, et al. A model system for mitochondrial biogenesis reveals evolutionary rewiring of protein import and membrane assembly pathways. Proc Natl Acad Sci U S A. 2012;109:E3358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Berman J, Sudbery PE. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. [DOI] [PubMed] [Google Scholar]
- [49].Ohama T, Suzuki T, Mori M, et al. Non-universal decoding of the leucine codon CUG in several Candida species. Nucleic Acids Res. 1993;21:4039–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Butler G, Rasmussen MD, Lin MF, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nobile CJ, Bruno VM, Richard ML, et al. Genetic control of chlamydospore formation in Candida albicans. Microbiology. 2003;149:3629–3637. [DOI] [PubMed] [Google Scholar]
- [52].Richard ML, Nobile CJ, Bruno VM, et al. Candida albicans biofilm-defective mutants. Eukaryot Cell. 2005;4:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Walther A, Wendland J. PCR-based gene targeting in Candida albicans. Nat Protoc. 2008;3:1414–1421. [DOI] [PubMed] [Google Scholar]
- [55].Colot HV, Park G, Turner GE, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Reuss O, Vik A, Kolter R, et al. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. [DOI] [PubMed] [Google Scholar]
- [57].Vyas VK, Barrasa MI, Fink GR. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv. 2015;1:e1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jeong J-Y, Yim H-S, Ryu J-Y, et al. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol. 2012;78:5440–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sprouffske K, Wagner A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics. 2016;17. DOI: 10.1186/s12859-016-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Li D, Chen H, Florentino A, et al. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryot Cell. 2011;10:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wittig I, Karas M, Schägger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6:1215–1225. [DOI] [PubMed] [Google Scholar]
- [62].Spinazzi M, Casarin A, Pertegato V, et al. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat Protoc. 2012;7:1235–1246. [DOI] [PubMed] [Google Scholar]
- [63].Somlo M. Induction and repression of mitochondrial ATPase in yeast. Eur J Biochem. 1968;5:276–284. [DOI] [PubMed] [Google Scholar]
- [64].Malecki M, Jedrzejczak R, Puchta O, et al. In vivo and in vitro approaches for studying the yeast mitochondrial RNA degradosome complex. Methods Enzymol. 2008;447:463–488. [DOI] [PubMed] [Google Scholar]
- [65].Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Skrzypek MS, Binkley J, Binkley G, et al. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017;45:D592–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. [DOI] [PubMed] [Google Scholar]
- [68].Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ziemann M Accuracy, speed and error tolerance of short DNA sequence aligners. BioRxiv. 2016053686.
- [70].Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ramírez F, Ryan DP, Grüning B, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ramírez F, Bhardwaj V, Arrigoni L, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- [74].Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wiśniewski JR. Filter-aided sample preparation for proteome analysis. Methods Mol Biol. 2018;1841:3–10. [DOI] [PubMed] [Google Scholar]
- [76].Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. [DOI] [PubMed] [Google Scholar]
- [77].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. [DOI] [PubMed] [Google Scholar]
- [78].Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. [DOI] [PubMed] [Google Scholar]
- [79].Wiśniewski JR, Wegler C, Artursson P. Multiple-enzyme-digestion strategy improves accuracy and sensitivity of label- and standard-free absolute quantification to a level that is achievable by analysis with stable isotope-labeled standard spiking. J Proteome Res. 2019;18:217–224. [DOI] [PubMed] [Google Scholar]
- [80].Gola S, Martin R, Walther A, et al. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20:1339–1347. [DOI] [PubMed] [Google Scholar]
- [81].Helmerhorst EJ, Murphy MP, Troxler RF, et al. Characterization of the mitochondrial respiratory pathways in Candida albicans. Biochim Biophys Acta. 2002;1556:73–80. [DOI] [PubMed] [Google Scholar]
- [82].Yan L, Li M, Cao Y, et al. The alternative oxidase of Candida albicans causes reduced fluconazole susceptibility. J Antimicrob Chemother. 2009;64:764–773. [DOI] [PubMed] [Google Scholar]
- [83].Lahens NF, Ricciotti E, Smirnova O, et al. A comparison of Illumina and Ion Torrent sequencing platforms in the context of differential gene expression. BMC Genomics. 2017;18:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Loman NJ, Misra RV, Dallman TJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. [DOI] [PubMed] [Google Scholar]
- [85].Szczepińska T, Kalisiak K, Tomecki R, et al. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015;25:1622–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Soudet J, Stutz F. Regulation of Gene expression and replication initiation by non-coding transcription: a model based on reshaping nucleosome-depleted regions: influence of pervasive transcription on chromatin structure. BioEssays. 2019;41:e1900043. [DOI] [PubMed] [Google Scholar]
- [87].Porrua O, Boudvillain M, Libri D. Transcription Termination: variations on Common Themes. Trends Genet. 2016;32:508–522. [DOI] [PubMed] [Google Scholar]
- [88].Ju X, Li D, Liu S. Full-length RNA profiling reveals pervasive bidirectional transcription terminators in bacteria. Nat Microbiol. 2019;4:1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Georg J, Hess WR. Widespread Antisense Transcription in Prokaryotes. Microbiol Spectr. 2018;6. DOI: 10.1128/microbiolspec.RWR-0029-2018 [DOI] [PubMed] [Google Scholar]
- [90].Zhelyazkova P, Sharma CM, Förstner KU, et al. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012;24:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Dietrich A, Wallet C, Iqbal RK, et al. Organellar non-coding RNAs: emerging regulation mechanisms. Biochimie. 2015;117:48–62. [DOI] [PubMed] [Google Scholar]
- [92].Hotto AM, Germain A, Stern DB. Plastid non-coding RNAs: emerging candidates for gene regulation. Trends Plant Sci. 2012;17:737–744. [DOI] [PubMed] [Google Scholar]
- [93].Hotto AM, Schmitz RJ, Fei Z, et al. Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the Arabidopsis transcriptome. G3 (Bethesda). 2011;1:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Small ID, Rackham O, Filipovska A. Organelle transcriptomes: products of a deconstructed genome. Curr Opin Microbiol. 2013;16:652–658. [DOI] [PubMed] [Google Scholar]
- [95].Kucharczyk R, Ezkurdia N, Couplan E, et al. Consequences of the pathogenic T9176C mutation of human mitochondrial DNA on yeast mitochondrial ATP synthase. Biochim Biophys Acta. 2010;1797:1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Fontanesi F, Soto IC, Horn D, et al. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am J Physiol Cell Physiol. 2006;291:C1129–47. [DOI] [PubMed] [Google Scholar]
- [97].Schmidt U, Maue I, Lehmann K, et al. Mutant alleles of the MRS2 gene of yeast nuclear DNA suppress mutations in the catalytic core of a mitochondrial group II intron 1 1Edited by J. Karn. J Mol Biol. 1998;282:525–541. [DOI] [PubMed] [Google Scholar]
- [98].Rossignol R, Faustin B, Rocher C, et al. Mitochondrial threshold effects. Biochem J. 2003;370:751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Linder T, Park CB, Asin-Cayuela J, et al. A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet. 2005;48:265–269. [DOI] [PubMed] [Google Scholar]
- [100].Fekete Z, Ellis TP, Schonauer MS, et al. Pet127 Governs a 5′ → 3′-Exonuclease Important in Maturation of Apocytochrome b mRNA in Saccharomyces cerevisiae. J Biol Chem. 2008;283:3767–3772. [DOI] [PubMed] [Google Scholar]
- [101].Valach M, Farkas Z, Fricova D, et al. Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. 2011;39:4202–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Dujon B, Sherman D, Fischer G, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. [DOI] [PubMed] [Google Scholar]
- [103].Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11:512–524. [DOI] [PubMed] [Google Scholar]
- [104].Chlebowski A, Lubas M, Jensen TH, et al. RNA decay machines: the exosome. Biochim Biophys Acta. 2013;1829:552–560. [DOI] [PubMed] [Google Scholar]
- [105].Lubas M, Chlebowski A, Dziembowski A, et al. Biochemistry and Function of RNA Exosomes. Enzymes. 2012;31:1–30. [DOI] [PubMed] [Google Scholar]
- [106].Chlebowski A, Tomecki R, Gas López ME, et al. Catalytic properties of the eukaryotic exosome. Adv Exp Med Biol. 2011;702:63–78. [DOI] [PubMed] [Google Scholar]
- [107].Lorentzen E, Basquin J, Tomecki R, et al. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. [DOI] [PubMed] [Google Scholar]
- [108].Kamarthapu V, Nudler E. Rethinking transcription coupled DNA repair. Curr Opin Microbiol. 2015;24:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in NCBI Sequence Read Archive (SRA) repository at http://www.ncbi.nlm.nih.gov/bioproject/662600, and in ProteomeXchange at https://www.doi.org/10.6019/PXD024934.


