ABSTRACT
Circular RNAs (circRNAs) are regulatory molecules involved in the modulation of gene expression. Although originally assumed as non-coding RNAs, recent studies have evidenced that animal circRNAs can act as translatable transcripts. The study of plant-circRNAs is incipient, and no autonomous coding plant-circRNA has been described yet. Viroids are the smallest plant-pathogenic circRNAs known to date. Since their discovery 50 years ago, viroids have been considered valuable systems for the study of the structure-function relationships in RNA, essentially because they have not been shown to have coding capacity. We used two pathogenic circRNAs (Hop stunt viroid and Eggplant latent viroid) as experimental tools to explore the coding potential of plant-circRNAs. Our work supports that the analysed viroids contain putative ORFs able to encode peptides carrying subcellular localization signals coincident with the corresponding replication-specific organelle. Bioassays in well-established hosts revealed that mutations in these ORFs diminish their biological efficiency. Interestingly, circular forms of HSVd and ELVd were found to co-sediment with polysomes, revealing their physical interaction with the translational machinery of the plant cell. Based on this evidence we hypothesize about the possibility that plant circRNAs in general, and viroids in particular, can act, under certain cellular conditions, as non-canonical translatable transcripts.
KEYWORDS: Circular RNAs, plant coding circRNAs, non canonical transcripts, plant pathogenic RNAs, viroids, viroid-derived peptides
Introduction
Circular RNAs (circRNAs) are covalently closed transcripts found in diverse organisms that play important regulatory roles[1]. According to its origin, circRNAs can be classified as endogenous (en-circRNAs), originated from the transcription of the own genome or exogenous (ex-circRNAs), associated with external agents [2]. Viroids (a class of ex-circRNAs) were discovered as replicating, low molecular weight infectious RNAs [3] with circular nature [4]. Viroids, thus, can be considered as the first circRNAs found in any organism. A few years later, the presence of virus-derived circRNAs in the cytoplasm of human cells was reported [5]. Endogenous circRNAs were first found in humans associated with the non-canonical splicing of cancer-related transcripts [6]. Diverse experimental evidence has revealed that multiple types of en-circRNAs are expressed in animals [7]. The first global identification of en-circRNAs in plants was done in rice and arabidopsis [8]. More recent studies have described the existence of en-circRNAs in diverse plant species [9–15]. The majority of human en-circRNAs are a few hundred nucleotides in length [2], while soybean en-circRNAs are mainly between 150–600 bp [16]. A common characteristic of both animal and plant en-circRNAs is that they are dependent on RNA polymerase II-mediated transcription and non-canonical RNA processing [17].
Under a functional viewpoint, animal en-circRNAs act predominantly as miRNA sponges [18]. Moreover, it has been proposed that they may also be involved in modulating the transcriptional activity [19], and in the memory of the transcriptional history of the cell [2]. The potential role of plant circRNA remains largely unknown as only two en-circRNAs have been functionally characterized [20,21]. Plant en-circRNAs possess features that differ from those described in animals, like a lower prediction to be potential miRNA sponges [8]. However, their functional characterization could leverage animal circRNA studies [22].
In general, circRNAs have been considered as non-coding RNAs (ncRNAs) [23]. However, recent evidence supports that certain animal en-circRNAs are translated in vivo [2] suggesting that the coding potential of these assumed ncRNAs has been underestimated and that may exert more biological functions than previously predicted. To date, no plant en-circRNAs has been reported to generate proteins and only non-autonomous coding activity has been reported for a virus-satellite circRNA in rice [24]. Nonetheless, the coding potential of plant circRNAs has not been deeply investigated. Elucidating the functional roles of circRNAs, in translation, might constitute a primary research topic in plant circRNAs [17].
Viroids are thus, naked ex-circRNAs, without apparent coding capacity, that infect host-plants causing phenotypic effects ranging from severe symptoms to latent infections [25]. In coincidence with en-circRNAs, viroid replication is dependent on plant-endogenous RNA polymerases, and their genome (≅ 300 nt) is comparable to the predominant size of en-circRNAs. Additionally, these autonomous pathogenic RNAs also exhibit differential subcellular compartmentalization; as members of the Pospiviroidae family replicate and accumulate in the nucleus, while the Avsunviroidae develop their life cycle in chloroplasts [26]. Due to their particular biological characteristics, viroids have emerged as a valuable tool to explore the regulatory pathways mediated by RNA in plants [27,28]. Prompted by the unexpected discovery of the coding activity of circRNAs in animals, here we use these pathogenic ex-circRNAs as an experimental model to explore the potential of plant circRNAs to encode functional peptides. In this opinion article we hypothesize about the possibility that under specific (and today unknown yet) cellular conditions, ex-circRNAS could act as protein-coding transcripts. We analysed representative members of the two viroid families replicating in either nucleus or chloroplasts and showed that HSVd and ELVd circRNAs contain conserved ORFs able to encode functional peptides carrying specific subcellular localization signals that direct fluorescent-fused proteins to the corresponding organelle where replication/accumulation takes place. Through bioassays in well-established hosts, we show that mutations in these ORFs diminish the biological efficiency of these pathogenic ex-circRNAs. Finally, circular forms of both, HSVd and ELVd RNA were found to co-sediment with plant polysomes in vivo, revealing their physical interaction with the translational machinery of the plant cell.
I. HSVd and ELVd contains putative ORFs able to encode peptides
In order to analyse their potential coding capability, we computationally inferred all the possible peptides derived from the plus strand of infectious cDNA clones [29] of Hop stunt viroid (HSVd) and Eggplant latent viroid (ELVd), members of families Pospiviroidae and Avsunviroidae, respectively. In viroids, the mature RNA form predominantly accumulated in the host is referred to as the plus strand. HSVd has been found inducing symptomatic infection in a wide range of hosts and has been frequently used to explore diverse aspects of viroid-host interactions [30–33]. ELVd induces asymptomatic infections and is assumed as a valuable model to study molecular aspects of the family Avsunviroidae [34].
Viroid sequences were translated into the three possible frames using any codon as start. Only peptides longer than 45 amino acids (according to the criteria explained in experimental procedures) derived from each ORF were selected for further analysis (Figure 1 and Figure S1A and B). The putative HSVd-ORFs 1 and 2 (H-ORF1 and H-ORF2) were 48 and 98 amino acids in length, respectively. The third ORF (H-ORF3) constitutes a unique reading frame without any stop codon (Figure 1A). ELVd-ORFs were 110 (E-ORF1), 87 (E-ORF2), and 59 (E-ORF-3) amino acids in length (Figure 1B).
Figure 1.
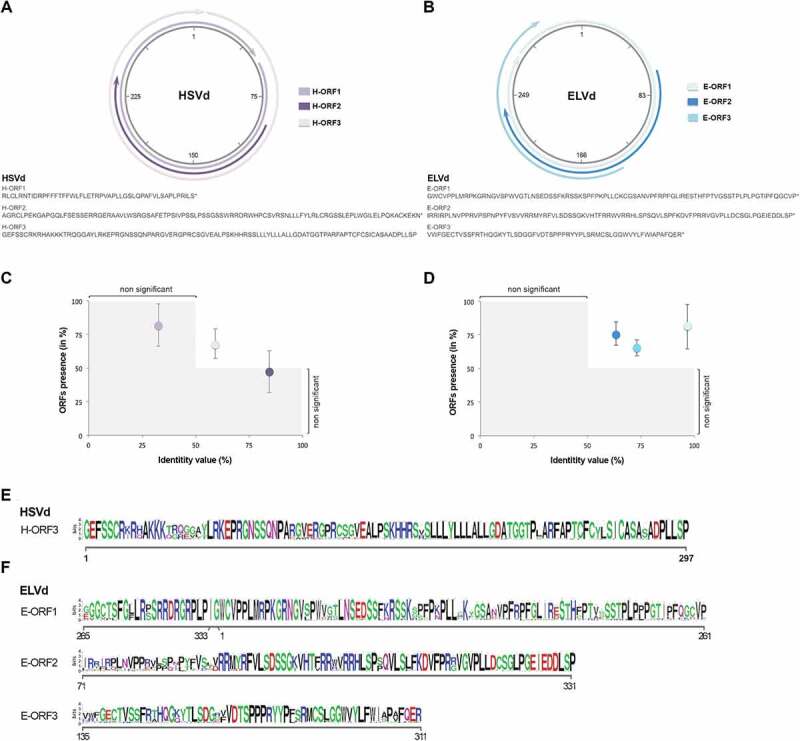
Computational prediction of putative ORFs in ex-circRNAs. Detailed location (upper panel) and peptide sequences (lower) of the ORFs identified in HSVd (A) and ELVd (B) infectious clones. Graphic representation of the conservation rates estimated for the ORFs inferred in HSVd (C) and ELVd (D) accessions recovered from public repository. Consensus sequence of the conserved peptides predicted to be encoded by HSVd (E) and ELVd (F). Error bars represent the standard error values
To determine if these ORFs are conserved in HSVd and ELVd, the predicted ORFs were aligned against the ORFs identified in the complete sequences publicly available of HSVd (779) and ELVd (104). The conservation rate was estimated considering the presence (in percentage) of the predicted ORFs in all analysed accessions with identity value (between potential peptides) higher than 50%. Conservation rates lower than 50% were considered as not significant. As it is shown in Figure 1C, H-ORF3 was detected in 59% of the analysed sequences with an identity value mean of 68.12%. The highest conservation value observed in ELVd was for the E-ORF1, which presents an identity mean of 81.17% in 96.6% of the accessions. The other predicted ORFs (E-ORF2 and E-ORF-3) present identity means of 75.93% and 65.31% in the 63.46% and 73.07% of the analysed accessions, respectively (Figure 1D). The consensus sequences of the most conserved ORFs detected in HSVd and ELVd circular forms are shown in the Figure 1E and respectively.
Peptides derived from conserved ORFs (H-ORF3, E-ORF1 and E-ORF2) were blasted against the non-redundant protein database of higher plants (taxid: 3193) at the NCBI. Significant similarities (>50%) were not found for any of the putative peptides. Only a region (51 amino acids) of the E-ORF2 showed a slight identity (31%) with Choline monooxygenase (Figure S1C), a nucleus-encoded ferredoxin-dependent enzyme located in the chloroplast stroma [35].
II. Putative viroid-derived peptides show specific cellular compartmentalization
To analyse their potential activity, we developed reporter constructs containing the ORFs derived from HSVd and ELVd fused in frame to the amino end of the green (for H-ORFs) and yellow (for E-ORFs) fluorescent proteins cDNAs (Figure S2). The chimeric cDNAs (H-ORFs/GFP and E-ORFs/YFP) were cloned into a binary vector and transiently expressed in N. benthamiana plants.
Observation of infiltrated leaves by confocal microscopy revealed that H-ORF1/GFP and H-ORF2/GFP show a cellular localization pattern similar to GFP (Figure 2A). Conversely, and in coincidence with a previous observation [36], H-ORF3/GFP was predominantly accumulated in the nucleus of the analysed cells (Figure 2A). Specifically, the fluorescence signal co-localized with fibrillarin, a well-established marker of the nucleolus [37]. Searching for potential domains responsible for this specific subcellular localization revealed the presence, in the peptide H-ORF3, of a region enriched in basic amino acids conserved in a high proportion of the HSVd sequences described to date (Figure S3A – left panel). This type of R/K motifs are involved in the specific targeting of peptides to the plant cell nucleolus, acting as Nucleolar Localization Signals (NoLS) [38]. To evaluate if this putative R/K motif is responsible for the specific subcellular localization of the H-ORF3 peptide, we generated a reporter (H-RK/GFP) carrying this potential domain fused to GFP. Transient expression assays revealed that the protein derived from the H-RK/GFP construct also accumulates in the nucleolus supporting that this H-ORF3 domain acts as a functional NoLS (Figure S3B). A similar domain enriched in basic amino acids was identified in Potato spindle tuber viroid (PSTVd), the type species of the family Pospiviroidae (Figure S3A – right panel). The functionality of this potential NoLS was validated by transient expression (Figure S3B).
Figure 2.
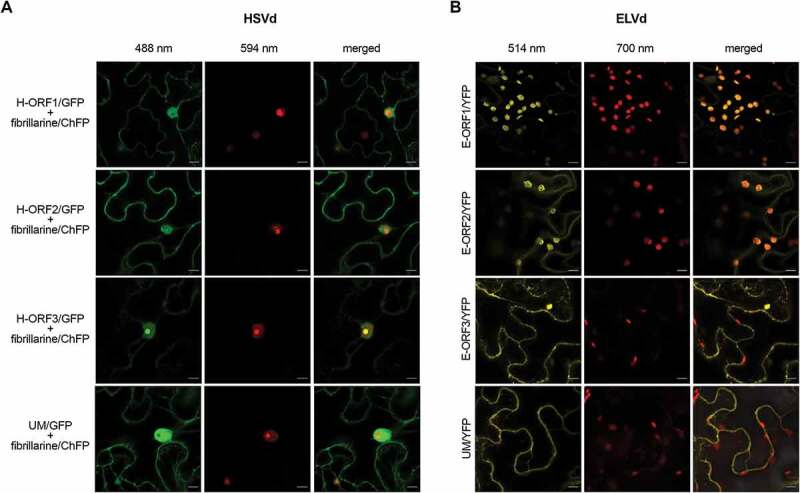
Subcellular localization of the putative ex-circRNAs-derived peptides. Confocal microscopy images of N. benthamiana leaves agro-infiltrated with constructs expressing H-ORFs fused to GFP (A) and E-ORFs in frame with YFP (B). Unmodified GFP/YFP (UM/GFP and UM/YFP, respectively) were used as control. The fusion fibrillarin-ChFP (central panels in A) was used as a nucleolus marker. Chloroplasts were determined by the chlorophyll auto-fluorescence recovered above 700 nm (central panels in B). Scale bars are 10 μm
Regarding the putative ELVd-derived peptides, we observed that the chimeric E-ORF1/YFP was predominantly localized in chloroplasts (Figure 2B), in agreement with the in silico prediction as a chloroplast transit peptide (ChTP) (Figure S4A). Remarkably, the second ORF identified in the ELVd (E-ORF2) was also predicted to act as a ChTP (Figure S4A). Transient expression assays revealed that E-ORF2/YFP peptide was also transported to chloroplasts (Figure 2B). Finally, E-ORF3/YFP showed a localization pattern comparable to YFP. The vast majority of the nucleus-encoded proteins destined to plastids are targeted into the chloroplast as a precursor whose transport is facilitated by a cleavable N-terminal ChTP [39]. To evaluate if the peptides E-ORF1 and E-ORF2 are transported to chloroplast through this canonical model, chimeric E-ORF1/YFP and E-ORF2/YFP proteins transiently expressed in N. benthamiana leaves were analysed by western blot assays. Our results revealed that both peptides derived from this ex-circRNA are cleaved during the transport process to chloroplasts (Figure S4B). To gain additional functional evidence, we transiently expressed two constructs containing E-ORF1 and E-ORF2 partial peptides lacking the predicted ChTP region fused to YPF. As expected the YFP fussed to both depleted peptides showed a localization pattern comparable to free-YFP (Figure S4C).
Furthermore, we also analysed the subcellular compartmentalization of H-ORF3/GFP and E-ORF1/YFP in N. benthamiana and eggplant leaves inoculated with HSVd and ELVd, respectively. The obtained data revealed that the specific accumulation (in nucleolus for H-ORF3/GFP and chloroplasts for E-ORF1/YFP) was also observed in viroid-infected tissues (Figure S5).
III. Inactivation of putative ORFs affects biological efficiency of ex-circRNAs
To evaluate if the potential peptides derived from ex-circRNAs can exert an effect on the viroid infection cycle, we analysed the viroid accumulation in cucumber and eggplant leaves infected with mutant HSVd and ELVd variants, respectively. Sequence modifications were designed to truncate the inferred H-ORF3 and E-ORF1 while maintaining (without relevant changes in the free energy values) the predicted RNA secondary structure of the viroids (Figure 3A and B – Figure S6 and S7). Furthermore, we specifically modified positions in which a stop codon could be introduced with a single nucleotide change, and designed control mutants that do not alter the putative ORF but are in the same structural domain.
Figure 3.
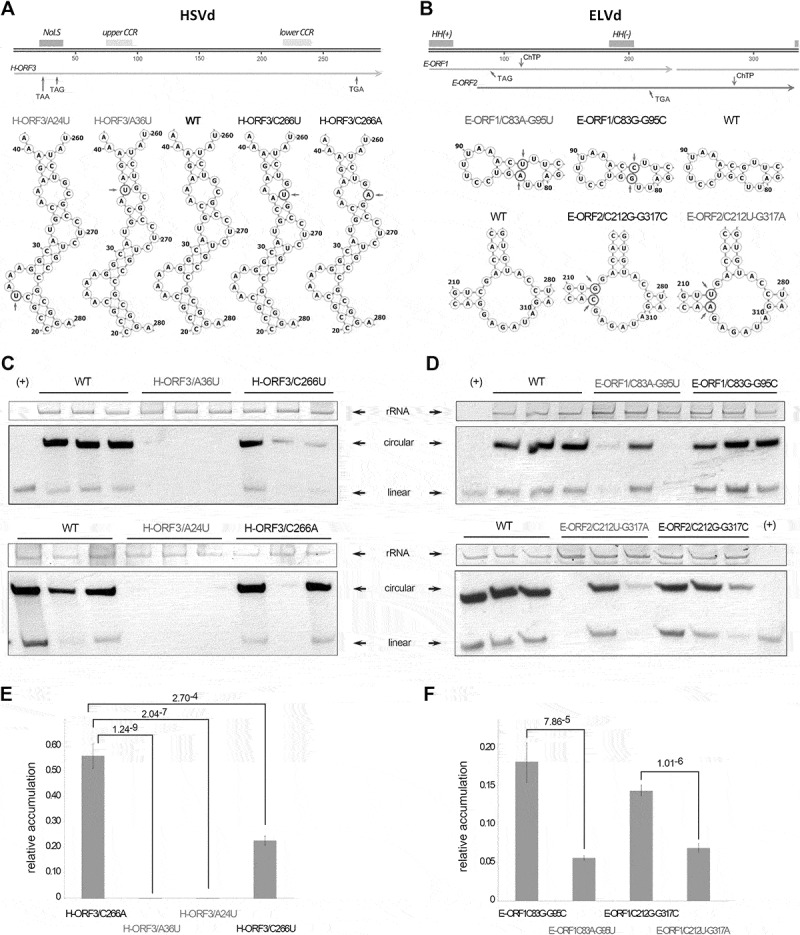
Truncated ORFs affect viroid biological efficiency. Representation of the HSVd (A – upper panel) and ELVd (B – upper panel) linear monomers (plus polarity). The Central Conserved Region (CCR) and the potential Nucleolar Localization Signal (NoLS) (in HSVd) and Hammerhead Ribozyme (HH) (in ELVd) are highlighted (boxes). The position of the nucleotides changed to introduce the stop codons in the conserved ORFs are marked with arrows. Predicted secondary structure of the regions of the HSVd (A – lower panel) and ELVd (B – lower panel) in which were performed the nucleotide substitutions (marked with circles and arrows). WT represents the structure of the unmodified HSVd and ELVd sequences. Representative northern blot hybridization of RNA extracted from cucumber an eggplant inoculated with the HSVd (C) and ELVd (D) mutant-variants. Ribosomal RNA (rRNA) was used as load control. HSVd and ELVd linear transcripts (+) were used as hybridization controls. Relative accumulation of HSVd (E) and ELVd (F) mutant-variants in inoculated plants, estimated by qRT-PCR. The statistical significance was estimated by paired T-tests and the obtained p-values are shown. Error bars represent the standard error values
Two HSVd-derived mutants (H-ORF3/A24U and H-ORF3/A36U) were designed harbouring the substitution A/U in the positions 24 and 36, respectively. These modifications introduce two stop codons UAA and UAG in the R/K region responsible for the nucleolar targeting. A third mutant (H-ORF3/C266A) contains a C/A substitution that introduces the stop codon UGA out of the functional NoLS but in the same region of the secondary structure of the viroid. As an additional control we modified the position 266 with a C/U silent substitution that maintains unaltered the H-ORF3 (H-ORF3/C266U) (Figure 3A).
To analyse the functionality of the putative peptides derived from ELVd, we constructed mutants carrying a double substitution C/A-G/U in the positions 83 and 95, respectively, which truncates the E-ORF1 with the stop codon UAG (E-ORF1/C83A-G95U) (Figure 3B). The mutant E-ORF2/C212U-G317A contains a double substitution that introduces the stop codon UGA in the E-ORF2. Finally, two control mutants (E-ORF1/C83G-G95C and E-ORF2/C212G-G317C) containing substitutions in the same ELVd-sequence positions but preserving the secondary structure and not disrupting any ORF, were also obtained. Analysis of the cleavage efficiency of transcripts derived from the ELVd mutants in vitro revealed that the modifications do not affect the autocatalytic activity of the hammerhead ribozymes (Figure S8).
Dimeric constructs of HSVd and ELVd variants were used to inoculate cucumber and eggplant plants, respectively. At 21 dpi total RNAs were extracted to estimate viroid accumulation. Mature forms of H-ORF3/A24U and H-ORF3/A36U mutants (carrying stop codons in the R/K motif) were not detected by northern blot assays (Figure 3C – upper panel). In contrast, circular forms of H-ORF3/C266A and control mutants (although with variable accumulation levels) were identified in all the analysed samples (Figure 3C – lower panel). These results were reinforced by the observation that the symptom intensity in H-ORF3/A24U and H-ORF3/A36U inoculated plants was significantly lower than that observed in cucumber plants inoculated with the unmodified and/or control (H-ORF3/C266U) variants (Figure S9). Regarding ELVd, circular forms of all the analysed viroid-sequences were detected by northern blot (Figure 3D). However, it was evident that the accumulation of both E-ORF1/C83A-G95U and E-ORF2/C212U-G317A variants with truncated ORFs was decreased in relation to control mutants (Figure 3D). The sequence stability of the mutant viroid variants in the analysed plants was confirmed by sequencing 10 clones of each (Figure S10), observing that only in the case of the C83A-G95U mutant one out of the ten clones reverted to the wild type sequence revealing a high stability of the introduced mutations.
To perform a more precise determination of HSVd and ELVd levels, total RNAs from inoculated plants were also analysed by qRT-PCR (Figure 3E). As expected, all HSVd mutants presented lower accumulation levels than the wild type, being the control sequence H-ORF3/C266U the less affected variant. In contrast, mutants H-ORF3/A24U and H-ORF3/A36U showed accumulation values near zero (Figure 3E). The HSVd mutant H-ORF3/C266A, carrying a stop codon out of the NoLS presented accumulation values intermediate between both NoLS-disrupted mutants and H-ORF3/C266U control. Results obtained from the HSVd qRT-PCR were similar to those of northern blot analysis (Figure 3F). These results revealed that the estimated levels of ELVd variants carrying stop codons in their sequence are significantly lower than the observed for their respective controls with unaltered ORFs.
IV. Circular forms of HSVd and ELVd interact with translational machinery in vivo and HSVd contains m6A modification
It is generally assumed that the functional status of a translatable RNA is associated with the specific presence of such RNA in polysomes fractions. Consequently, we performed polysome profiling of viroid-infected plants to study the possibility that ex-circRNAs might be incorporated into plant translational complexes. Ribosome fractions from cucumber and eggplant leaves infected with HSVd and ELVd, respectively, were separated by ultracentrifugation on a sucrose density gradient, analysed by absorbance at 260 nm and collected for posterior analysis (Figure 4A and B). In order to monitor the presence of ex-circRNAs, total RNA was extracted from cytosolic (2 and 3), and polysome-associated (10 and 11) fractions, which were discriminated by the position of the 80S peak that corresponds to monosomes. As positive control we analysed endogenous mRNAs constitutively expressed in both analysed plants. The negative control consisted of a linear transcript of another ex-circRNA (PSTVd) added to the extracts before being loaded into the sucrose gradient. RT-PCR analysis revealed the presence of internal endogenous controls and HSVd and ELVd RNA in all the analysed fractions, whereas linear PSTVd transcripts were only detected in cytosolic fractions (Figure 4C and D). To confirm that circular forms of HSVd and ELVd were associated with polysome fractions, total RNAs extracted from fractions 10 and 11 were analysed by northern blot assays. Circular forms of plus-strand HSVd and ELVd (marked with arrows in the Figure 4E and F) were clearly detected in the polysome fractions in both analysed plants. These results confirm that both ex-circRNAs analysed here, are associated in vivo with plant polysomes. Regarding to the detection of linear forms of HSVd and ELVd, at this point, we are incapable to determine if these detected transcripts are derived from circular RNAs broken during the purification process or constitute evidence that some linear monomers of both analysed viroids might also be associated with polysomes.
Figure 4.
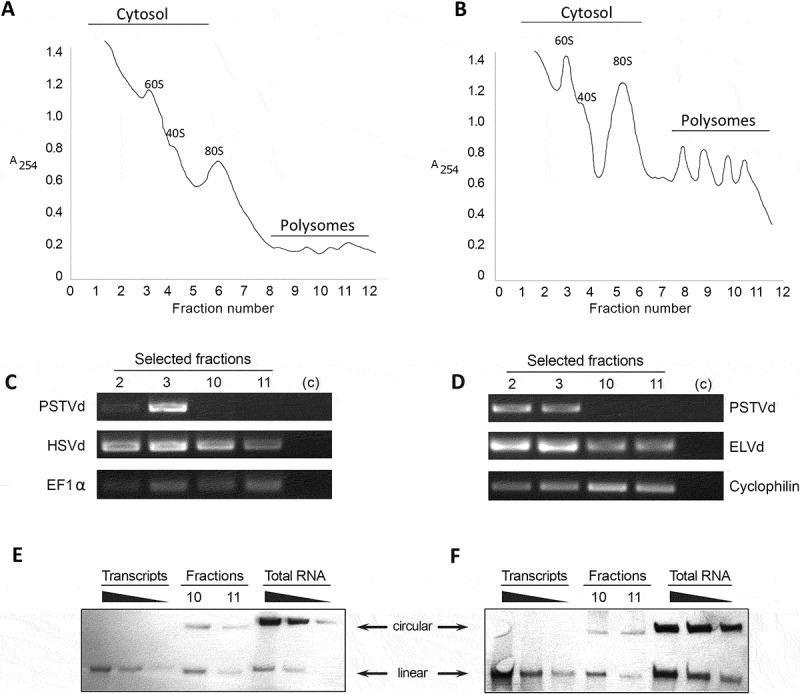
Ex-circRNAs are associated with translational machinery. Sucrose density gradient analysis of fractions from leaves infected with HSVd (A) or ELVd (B). The position of the 80S monosomes is indicated to separate the polysome fractions from the cytosolic ones. Total RNAs extracted from fractions 2 and 3 (cytosol), and 10 and 11 (polysomes) of HSV- and ELVd-infected plants (C and D, respectively) were analysed by RT-PCR. Detection of EF1α (in cucumber) and Cyclophilin (eggplant) transcripts was used as endogenous control. An in vitro generated transcript of PSTVd (added to the plant extract before being loaded into the sucrose gradient) was used as negative control. HSVd and ELVd RNA and endogenous controls were detected in all analysed fractions. In contrast, exogenous control (PSTVd) expected to be not associated with polysomes, was only present in cytosolic fractions. Northern blot of RNA isolated from polysome fractions derived from leaves infected with HSVd (E) or ELVd (F). Serial dilutions of monomeric transcripts and total RNAs extracted from infected plants were used as hybridization controls. Analyses were performed in duplicate and only representative results are shown
One issue to be solved is how the translation of these viroid-derived peptides might start in the absence of an initial methionine. Nevertheless, there are a growing number of reports on alternative mechanisms of translation initiation [40]. Among them, N6-Methyladenosine (m6A) in RRACH sites (being R [A or G] and H [A or C or U]) can promote internal initiation of polysome-associated endogenous circular RNAs (circRNA) in cells [41]. In this regard, canonical RRACH sites were computationally identified in HSVd and ELVd (Figure S11, upper and lower panel respectively). Moreover, immunoprecipitation of total RNA extracted from HSVd-infected cucumber plants revealed the presence of the plus polarity of HSVd in the RNAs immunoprecipitated with m6A antibody but not in those RNAs recovered after immunoprecipitation with GFP antibody, which was used as negative control (Figure S11B). Besides, circular forms of HSVd were isolated, digested into nucleosides and analysed by UPLC-PDA-Tof-MS. A putative m6A peak (recovered at 6,48 s) was identified in comparison with the pattern obtained with a commercial N6-methyladenosine marker (Figure S11C).
Discussion and future perspectives
CircRNAs were originally regarded as a novel class of ncRNAs. However, recent studies have provided evidence that circRNAs can also act as non-canonical translatable transcripts [42]. Different functional peptides derived from circRNAs have been described in animal cells [41,43–48], supporting the emerging notion that the true coding-potential of circRNAs may have been underestimated. Although circRNAs were described initially in plants [4], research on this topic is still in its infancy as no autonomous coding plant-circRNA has been described yet [17]. Given this new scenario, we used HSVd and ELVd, two well-known pathogenic circRNAs representatives of the two viroid families characterized by replicating and accumulating in nucleus and chloroplasts, respectively, as experimental tools to explore the coding potential of circRNAs in plants.
Computational analysis revealed the existence of conserved ORFs in both analysed ex-circRNAs. H-ORF3 and E-ORF1 were the predicted peptides showing the highest conservation rates in HSVd and ELVd accessions, respectively. It is important to consider that ELVd is a more recently described viroid and that the reported variants come from a unique host (eggplant). In contrast, HSVd is a viroid largely studied with multiple sequence-variants recovered from diverse host-plants and (in some cases) submitted to sequence repositories without exhaustive previous quality controls (for example, 73 sequences were excluded from this analysis because they contained some nucleotide indeterminacy and/or incomplete sequence). Consequently, it cannot be excluded the possibility that the conservation rate inferred for H-ORF3 (lower than that observed for E-ORF1 and E-ORF2) can be underestimated.
The study of specific cellular localization has been frequently assumed as key knowledge for elucidating biological activity and protein functions in the cell [49,50]. Consequently, the observation that peptides encoded by ORFs detected in ex-circRNAs analysed here show predominant localization in the nucleolus (H-ORF3) and chloroplasts (E-ORF1) provides the primary observation suggesting a potential functional role. Interestingly, the specific subcellular compartmentalization of these putative peptides is coincident with the organelles where both plant-pathogenic RNAs predominantly accumulate, nucleolus for a member of the family Pospiviroidae (HSVd) and chloroplasts for a member of the family Avsunviroidae (ELVd). Previously, it has been suggested that the specific compartmentalization of ELVd-derived constructs may be regulated by the viroid-RNA sequence [51]. However, the coding potential of those sequences could have been undervalued because of the primary consideration as plant-pathogenic ncRNAs. Remarkably, ELVd contains an additional ORF (E-ORF2), potentially encoding a peptide with redundant biological activity (localization in chloroplasts). It has been demonstrated that viroids in the family Avsunviroidae exhibit the highest mutation rate reported for any biological entity [52], whereas the mutation rates of pospiviroids are significantly lower [53]. Under this strict evolutionary constriction, it may be expected that, according to the role of genetic redundancy in robustness [54], the functionality played by a determined component (E-ORF1 encoded peptide, in this case) may be guaranteed by another (E-ORF2) with a functional overlap. Finally, the additional identification of ORFs potentially encoding peptides carrying functional NoLS in a representative member of the family Pospiviroidae (PSTVd), provides additional support to the notion that potential circRNA-derived peptides inferred here, might possess biological activity.
The protein-coding capability of a small sub-set of endogenous circRNAs in animals has been validated by the identification of circRNA-encoded peptides by mass spectrometry (MS) [2]. Unfortunately, our attempts to detect the predicted peptides by Liquid Chromatography MS (LC-MS) in total protein-extracts recovered from infected tissues were unsuccessful. However, it could not be excluded the possibility that the analysed tissues could represent a mix of infected and non-infected cells [55], thus prompting that peptide accumulation might be below the LC-MS detection limits in crude extracts [56]. Therefore, additional peptide-enrichment steps [43] and/or single-cell proteomic analysis [57] could be needed in order to unequivocally confirm, in the future, the presence in infected-plants of the putative circRNA-derived peptides described here. Searching for biological evidence supporting the involvement of circRNA-derived peptides in the infection cycle of viroids, we constructed diverse mutant-variants that truncate the identified ORFs. Infection bioassays revealed that variants carrying a stop codon in their sequence exhibit a significant lower accumulation in comparison with control mutants. The most evident effect was observed in cucumber plants inoculated with HSVd, where mature forms of H-ORF3/A24U and H-ORF3/A36U were practically undetectable. Regarding ELVd mutants, our results revealed that although ORF-truncated variants showed lower accumulation levels than their corresponding controls, the biological effect of the introduction of stop codons in both mutants was less evident than the observed in HSVd. At this point, it is important to remark that according to our prediction, ELVd possesses two putative ORFs with hypothetical functional activity, consequently in any of the two analysed mutants we were not able to generate a true potentially non-functional variant.
Diverse studies have shown that translatome technologies are the most reliable method to find coding circRNAs [45,46]. The demonstration that circular forms of HSVd and ELVd are physically associated with translating polysomes, reveals their involvement in the plant translational machinery as recently described for the circular forms of the Citrus exocortis viroid (another member in the family Pospiviroidae) [58]. In this latter work authors suggested that the interaction viroid-ribosome is related to the pathogenesis process in infected tomato plants as a consequence of ribosomal stress. However, the demonstration that besides HSVd (a viroid causing strong phenotypic alterations), circular forms of the ELVd also interact with the polysomes in non-symptomatic infected eggplant, suggests that this in vivo association between ex-circRNAs and the plant translational machinery may possess additional biological functions beyond the physiological alteration manifested as symptoms expression in infected plants.
The coding potential of circRNAs raises the interesting question of which mechanisms regulate the translation initiation of those covalently closed transcripts lacking 5´cap structure [42]. Although the predominant mode of translation initiation is cap-dependent, translation does not always start at the first AUG encountered by the ribosome and diverse alternative initiation mechanisms (re-initiation, initiation at non-AUG codons, internal ribosomal entry, N6-Methyladenosine (m6A), etc.) have also been described in plants [59] and/or mammalians [60]. There are also cases in which the mechanisms regulating translation in plants remain unknown [61,62]. Interestingly, these non-canonical translation initiation processes are generally activated under stress [42], like that induced in the plant cell as a consequence of viroid infection. The prediction of canonical RRACH domains (the sequence context in which m6A modifications predominantly occur in mRNAs) [63,64] in HSVd and ELVd RNAs (Figure S11A), and the detection of potential m6A residues in HSVd circular RNAs (Figure S11B and C), allow us to speculate about the existence of non-canonical initiation mechanisms of translation in viroid RNAs, perhaps dependent on m6A modification as it has been reported for circRNAs of mammals [41,42,65]66-79. However, further studies are needed to decipher the mechanistic aspects of how the potential translation of ex-circRNAs would be regulated in plants.
In our opinion, these results generated through computational approaches, assays of subcellular compartmentalization, analysis of functional activity and polysome profiling, provide novel insights about the possible translation of exogenous plant-circRNAs (using viroids as experimental models) and contributed to expand the coding landscape of the cell transcriptome, suggesting the existence of an unexplored layer of gene activity in plants related to the potential protein-coding circRNAs, whose biological functions are largely yet to be revealed [17].
We are aware that the non-coding nature of circRNAs in general and viroids in particular constitutes a dogma that is difficult to refute, but progress showing that some animal circRNAs can code for small peptides [2,42] and the recent demonstration that CEVd causes ribosomal stress in tomato plants [58] are emerging insights that prompted us to reconsider this notion. Here, we provide experimental observations favouring that the coding capacity of viroids cannot be ruled out, although the definitive evidence (detection of the circRNA-encoded peptides) is a technological challenge to be addressed in future research lines focused on deciphering the molecular biology of the circular RNAs in plants.
Supplementary Material
Acknowledgments
The authors thank Dr. J.A. Daròs (IBMCP) for kindly providing the plasmids containing PSTVd-RG1 cDNA and the original ELVd sequence; and also thank Dr. M. De la Peña for assistance with the prediction of RNA tertiary structure.
Funding Statement
This work was supported by the Spanish Ministry of Economy and Competitiveness (co-supported by FEDER) PID2020-115571RB-I00 (VP) and AGL2016-79825-R (GG), and by the Spanish Ministry of Science and Innovation (co-supported by FEDER) Grant PID2019-104126RB-I00 (GG). JMM was the recipient of a predoctoral contract supported by the Conselleria d´Educació, Investigació, Cultura i Esport Generalitat Valenciana - ACIF Programme (ACIF-2017-114). The funders had no role in the experiment design, data analysis, decision to publish, or preparation of the manuscript;Conselleria d’Educació, Investigació, Cultura i Esport [ACIF-2017-114];Ministerio de Economia, Industria y Competitividad, Gobierno de España [AGL2016-79825-R];Ministerio de Econom?a, Industria y Competitividad, Gobierno de España [PID2020-115571RB-I00];Ministerio de Ciencia, Innovación y Universidades [PID2019-104126RB-I00].
Author contributions
J.M.M. performed experiments. J.M.M., J.A.N., V.P. and G.G.: analyzed and discussed the results. L.C.S and J.M.M.: performed computational analysis. J.A.N., V.P. and G.G.: designed the experiments. G.G.: wrote the main manuscript text and prepared the figures. All authors read, revised and approved the final manuscript.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Disclosure statement
The author(s) declare no competing interests.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Ji P, Wu W, Chen S, et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26(12):3444–3460.e5. [DOI] [PubMed] [Google Scholar]
- [2].Patop IL, Wüst S, Kadener S.. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Diener TO. Potato spindle tuber “virus”: IV. A replicating, low molecular weight RNA. Virology. 1971;45(2):411–428. [DOI] [PubMed] [Google Scholar]
- [4].Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci. 1976;73(11):3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].M-t HSU, COCA-PRADOS M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. [DOI] [PubMed] [Google Scholar]
- [6].Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64(3):607–613. [DOI] [PubMed] [Google Scholar]
- [7].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- [8].Ye C-Y, Chen L, Liu C, et al. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208(1):88–95. [DOI] [PubMed] [Google Scholar]
- [9].Darbani B, Noeparvar S, Borg S. Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley [Internet]. Front Plant Sci. 2016;7:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tan J, Zhou Z, Niu Y, et al. Identification and functional characterization of tomato CircRNAs derived from genes involved in fruit pigment accumulation. Sci Rep. 2017;7(1):8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang Y, Yang M, Wei S, et al. Identification of circular RNAs and their targets in leaves of triticum aestivum l. under dehydration stress. Front Plant Sci. 2017;7:2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang Z, Liu Y, Li D, et al. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion [Internet]. Front Plant Sci. 2017;8:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen L, Zhang P, Fan Y, et al. RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol. 2018;217(3):1292–1306. [DOI] [PubMed] [Google Scholar]
- [14].Zhu Y-X, Jia J-H, Yang L, et al. Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol. 2019;19(1):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Han Y, Li X, Yan Y, et al. Identification, characterization, and functional prediction of circular RNAs in maize. Mol Genet Genomics. 2020;295(2):491–503. [DOI] [PubMed] [Google Scholar]
- [16].Zhao W, Cheng Y, Zhang C, et al. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci Rep. 2017;7(1):5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhao W, Chu S, Jiao Y. Present scenario of circular RNAs (circRNAs) in plants [Internet]. Front Plant Sci. 2019;10:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- [19].Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. [DOI] [PubMed] [Google Scholar]
- [20].Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3(5):17053. [DOI] [PubMed] [Google Scholar]
- [21].Fan J, Quan W, Li G-B, et al. circRNAs are involved in the rice-magnaporthe oryzae interaction. Plant Physiol. 2020;182(1):272–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lai X, Bazin J, Webb S, et al. CircRNAs in plants. In: Circular RNAs: biogenesis and functions. Singapore: Springer Singapore; 2018. p. 329–343. DOI: 10.1007/978-981-13-1426-1_26. [DOI] [Google Scholar]
- [23].Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].AbouHaidar MG, Venkataraman S, Golshani A, et al. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A. 2014;111(40):14542–14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Flores R, Minoia S, Carbonell A, et al. Viroids, the simplest RNA replicons: how they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015;209:136–145. [DOI] [PubMed] [Google Scholar]
- [26].Di Serio F, Flores R, Verhoeven JTJ, et al. Current status of viroid taxonomy. Arch Virol. 2014;159(12):3467–3478. [DOI] [PubMed] [Google Scholar]
- [27].Gómez G, Pallás V. Viroids: a light in the darkness of the lncRNA-directed regulatory networks in plants. New Phytol. 2013;198(1):10–15. [DOI] [PubMed] [Google Scholar]
- [28].Steger G, Riesner D. Viroid research and its significance for RNA technology and basic biochemistry. Nucleic Acids Res. 2018;46:10563–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marquez-Molins J, Navarro JA, Pallas V, et al. Highly efficient construction of infectious viroid-derived clones. Plant Methods. 2019;15(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marquez-Molins J, Gomez G, Pallas V. Hop stunt viroid: a polyphagous pathogenic RNA that has shed light on viroid–host interactions. Mol Plant Pathol. 2020:13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Martinez G, Castellano M, Tortosa M, et al. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 2014;42(3):1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Castellano M, Pallas V, Gomez G. A pathogenic long noncoding RNA redesigns the epigenetic landscape of the infected cells by subverting host histone deacetylase 6 activity. New Phytol. 2016;211:1311–22. [DOI] [PubMed] [Google Scholar]
- [33].Wei S, Bian R, Andika IB, et al. Symptomatic plant viroid infections in phytopathogenic fungi. Proc Natl Acad Sci. 2019;116(26):13042–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Daròs JA. Eggplant latent viroid : a friendly experimental system in the family avsunviroidae. Mol Plant Pathol. 2016;17(8):1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rathinasabapathi B, Burnet M, Russell BL, et al. Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc Natl Acad Sci. 1997;94(7):3454–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gómez G, Pallás V. A peptide derived from a single-modified viroid-RNA can be used as an “in vivo” nucleolar marker. J Virol Methods. 2007;144(1–2):169–171. [DOI] [PubMed] [Google Scholar]
- [37].Kalinina NO, Makarova S, Makhotenko A, et al. The multiple functions of the nucleolus in plant development, disease and stress responses. Front Plant Sci. 2018;9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2(6):107–112. [DOI] [PubMed] [Google Scholar]
- [39].Bruce BD. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 2000;10(10):440–447. [DOI] [PubMed] [Google Scholar]
- [40].Kwan T, Thompson SR. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol. 2019;11(4):a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Diallo LH, Tatin F, David F, et al. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie. 2019;164:45–52. [DOI] [PubMed] [Google Scholar]
- [43].Liang W-C, Wong C-W, Liang -P-P, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. [DOI] [PubMed] [Google Scholar]
- [48].Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9(1):4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nair R, Rost B. Better prediction of sub-cellular localization by combining evolutionary and structural information. Proteins Struct Funct Bioinforma. 2003;53(4):917–930. [DOI] [PubMed] [Google Scholar]
- [50].Lin H-N, Chen C-T, Sung T-Y, et al. Protein subcellular localization prediction of eukaryotes using a knowledge-based approach. BMC Bioinformatics. 2009;10:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gómez G, Pallás V, Zhang B. Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS One. 2010;5(8):e12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gago S, Elena SF, Flores R, et al. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323(5919):1308. [DOI] [PubMed] [Google Scholar]
- [53].López-Carrasco A, Ballesteros C, Sentandreu V, et al. Different rates of spontaneous mutation of chloroplastic and nuclear viroids as determined by high-fidelity ultra-deep sequencing. PLoS Pathog. 2017;13(9):e1006547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Fares MA. The origins of mutational robustness. Trends Genet. 2015;31(7):373–381. [DOI] [PubMed] [Google Scholar]
- [55].Harders J, Lukács N, Robert-Nicoud M, et al. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 1989;8(13):3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Karpievitch YV, Polpitiya AD, Anderson GA, et al. Liquid chromatography mass spectrometry-based proteomics: biological and technological aspects. Ann Appl Stat. 2010;4(4):1797–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shi T, Gaffrey MJ, Fillmore TL, et al. Facile carrier-assisted targeted mass spectrometric approach for proteomic analysis of low numbers of mammalian cells. Commun Biol. 2018;1(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cottilli P, Belda-Palazón B, Adkar-Purushothama CR, et al. Citrus exocortis viroid causes ribosomal stress in tomato plants. Nucleic Acids Res. 2019;47(16):8649–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Merchante C, Stepanova AN, Alonso JM. Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J. 2017;90(4):628–653. [DOI] [PubMed] [Google Scholar]
- [60].Chen J, Brunner A-D, Cogan JZ, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367(6482):1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hua XJ, Van De Cotte B, Van Montagu M, et al. The 5′ untranslated region of the At-P5R gene is involved in both transcriptional and post-transcriptional regulation. Plant J. 2001;26(2):157–169. [DOI] [PubMed] [Google Scholar]
- [62].Hansen ER, Petracek ME, Dickey LF, et al. The 5ʹ end of the pea ferredoxin-1 mRNA mediates rapid and reversible light-directed changes in translation in tobacco. Plant Physiol. 2001;125(2):770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Arribas-Hernández L, Brodersen P. Occurrence and functions of m6 A and other covalent modifications in plant mRNA. Plant Physiol. 2020;182(1):79–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yang Y, Hsu PJ, Chen Y-S, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Prats AC, David F, Diallo LH, et al. Circular rna, the key for translation. Int J Mol Sci. 2020;21(22):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wernersson R. Virtual ribosome–a comprehensive DNA translation tool with support for integration of sequence feature annotation. Nucleic Acids Res. 2006;34(Web Server):W385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sanjuán R, Daròs J-A. One-step site-directed mutagenesis of viroid dimeric cDNA. J Virol Methods. 2007;145(1):71–75. [DOI] [PubMed] [Google Scholar]
- [68].Herranz MC, Sanchez-Navarro JA, Aparicio F, et al. Simultaneous detection of six stone fruit viruses by non-isotopic molecular hybridization using a unique riboprobe or polyprobe. J Virol Methods. 2005;124(1–2):49–55. [DOI] [PubMed] [Google Scholar]
- [69].Sanz-Carbonell A, Marques MC, Martinez G, et al. Dynamic architecture and regulatory implications of the miRNA network underlying the response to stress in melon. RNA Biol. 2020;17(2):292–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- [71].Lecampion C, Floris M, Fantino JR, et al. An easy method for plant polysome profiling. J Vis Exp. 2016;114:54231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hellens RP, Brown CM, Chisnall MAW, et al. The emerging world of small ORFs. Trends Plant Sci. 2016;21:317–328. [DOI] [PubMed] [Google Scholar]
- [73].Hsu PY, Benfey PN. Small but mighty: functional peptides encoded by small ORFs in plants. Proteomics. 2018;18(10):1700038. [DOI] [PubMed] [Google Scholar]
- [74].Fadda Z, Daròs JA, Fagoaga C, et al. Eggplant latent viroid, the candidate type species for a new genus within the family avsunviroidae (hammerhead viroids). J Virol. 2003;77(11):6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kofalvi SA, Marcos JF, Cañizares MC, et al. Hop stunt viroid (HSVd) sequence variants from Prunus species: evidence for recombination between HSVd isolates. J Gen Virol. 1997;78(12):3177–3186. [DOI] [PubMed] [Google Scholar]
- [76].Kerpedjiev P, Hammer S, Hofacker IL. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31(20):3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Gruber AR, Lorenz R, Bernhart SH, et al. The vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server):1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].López-Carrasco A, Gago-Zachert S, Mileti G, et al. The transcription initiation sites of eggplant latent viroid strands map within distinct motifs in their in vivo RNA conformations. RNA Biol. 2016;13(1):83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


