ABSTRACT
The human genome contains 481 ultraconserved regions (UCRs), which are genomic stretches of over 200 base pairs conserved among human, rat, and mouse. The majority of these regions are transcriptionally active (T-UCRs), and several have been found to be differentially expressed in tumours. Some T-UCRs have been functionally characterized, but of those few have been associated to breast cancer (BC). Using TCGA data, we found 302 T-UCRs related to clinical features in BC: 43% were associated with molecular subtypes, 36% with oestrogen-receptor positivity, 17% with HER2 expression, 12% with stage, and 10% with overall survival. The expression levels of 12 T-UCRs were further analysed in a cohort of 82 Brazilian BC patients using RT-qPCR. We found that uc.147 is high expressed in luminal A and B patients. For luminal A, a subtype usually associated with better prognosis, high uc.147 expression was associated with a poor prognosis and suggested as an independent prognostic factor. The lncRNA from uc.147 (lnc-uc.147) is located in the nucleus. Northern blotting results show that uc.147 is a 2,8 kb monoexonic trancript, and its sequence was confirmed by RACE. The silencing of uc.147 increases apoptosis, arrests cell cycle, and reduces cell viability and colony formation in BC cell lines. Additionally, we identifed 19 proteins that interact with lnc-uc.147 through mass spectrometry and demonstrated a high correlation of lnc-uc.147 with the neighbour gene expression and miR-18 and miR-190b. This is the first study to analyse the expression of all T-UCRs in BC and to functionally assess the lnc-uc.147.
KEYWORDS: T-UCRs, uc.147, breast cancer, tumorigenesis, ultraconserved region, lncRNAs
Introduction
Breast Cancer (BC) is the most commonly diagnosed type of cancer and the leading cause of cancer death among women [1]. BC is a heterogeneous disease with a range of clinical and molecular characteristics [2]. Based on immunohistochemical analysis, BC is subdivided into four main groups: luminal A, luminal B, HER2 (Human Epidermal Growth Factor Receptor 2), and triple-negative [3,4]. This classification is essential for prognosis and treatment decisions. While luminal A and luminal B are sensitive to hormone therapy, HER2 positive patients can benefit from monoclonal antibody therapy [5]. Triple-negative remains the most challenging group to treat, but new targeted therapies are becoming available, as, for example, PARP inhibitors in BRCA mutated patients [6]. Besides that, BC can also be classified according to their gene expression profile. Here, BC can be stratified in five groups: luminal A, luminal B, HER2 enriched, basal, and normal-like [7,8]. More recently, two more groups have been added to the molecular classification: claudin-low [9] and molecular apocrine [10,11]. Despite the BC general classification system and the existing guidelines for the stratification of patients inside each group, the wide range of diversity observed in each subtype demands additional molecular markers, in order to offer personalized therapy and to improve BC survival rates [12,13].
Potential novel candidates to be used for molecular characterization of BC are the ultraconserved regions (UCRs). In 2004, Bejerano and colleagues described UCRs as genome regions larger than 200 nucleotides that are fully conserved among human, mouse, and rat and highly conserved across other disparate taxa [14]. Most UCRs are known to be transcribed ultraconserved regions (T-UCRs), in both normal and malignant tissues [15], but most of their transcripts remain uncharacterized [16]. Recently, these regions have been demonstrated to participate in several cellular processes, such as differentiation [17–19], metabolism [20,21], response to hypoxia [22], drug resistance [15,23], and cancer development and progression [15,16,24,25]. In addition, the differential expression of T-UCRs was associated with different types of cancer [26–28](Fabris and Calin, 2017; Rossi et al., 2008; Habic et al., 2019), including cervical cancer [29], colorectal cancer [15,30], hepatocellular carcinoma [31], neuroblastoma [32], and others [15,25,33,34]. In BC luminal A patients, overexpression of uc.63 is associated with poor prognosis [35], and uc.38 is downregulated in BC patients [36]. Thus, these studies highlights the importance of T-UCRs in the cellular environment and, consequently, in cancer biology. Moreover, the above mentioned associations suggest that T-UCRs-expression profiles can be a useful tool to differentiate human cancer types and improve its diagnosis and prognosis.
In this study, we expand the knowledge of T-UCRs in breast cancer by screening the expression of all 481 T-UCRs in BC patients. We found that 63% of transcripts correlated with some clinical and/or molecular parameter of BC. In addition, we further investigated 12 T-UCRs in a cohort of Brazilian BC patients and confirmed that two of those transcripts are differentially regulated in BC patients. Here, we provide a comprehensive characterization of the biology and the role of lnc-uc.147 in BC development.
Material and methods
Bioinformatic analysis
The statistical analyses for the TCGA data were performed in R (version 3.4.1) (http:///www.r-project.org/), and the statistical significance was defined as a p-value less than 0.05. The sequences of 481 ultraconserved elements (ucr) in the human genome, their genomic coordinates in GRCh34/hg16, as well their type (exonic, nonexonic), and the closest gene were acquired from https://users.soe.ucsc.edu/~jill/ultra.html (Bejerano et al., 2004). We used UCSC Genome Browser LiftOver to lift the coordinatesfrom GRCh34/hg16 to GRCh37/hg19 assembly. The expression of the ultraconserved regions, quantified as RPKM (reads per kilobase of transcript per million mapped reads) in primary tumours, were retrieved from TANRIC (https://ibl.mdanderson.org/tanric/_design/basic/main.html) [37]. We downloaded clinical information for the TCGA patients with invasive breast carcinoma (BRCA) from cbioPortal (http://www.cbioportal.org/) (cohort: TCGA provisional). The PAM50 subtype of the samples was acquired from [38]. We ended up with 827 cases with uc expression and clinical information. From Genomic Data Commons Data Portal, we downloaded FPKM (fragments per kilobase of transcript per million mapped reads) files in order to retrieve gene expression quantification data for the host genes of the ultraconserved regions in same tumours. The log2-transformation was applied to the mRNA data.
The relationship between overall survival and covariates (mRNA expression levels and clinical parameters age and stage) was examined using a Cox proportional hazard model. A multivariate Cox proportional hazard model was fitted, including the clinical parameters and mRNA expression significance in the univariate analysis. These analyses were conducted for all the breast cancer cohort and also separately on each subtype as defined by PAM50 or immunohistochemistry. To visualize the association between genes that identified significant prognostic factors and survival, we first used the log-rank test to find the point (cut-off) with the most significant (lowest p-value) split in high and low mRNA expression groups. Then, we used the Kaplan–Meier method to generate plots for this cut-off.
The t-test or analysis of variance (depending on the number of groups considered) was applied to normally distributed data. Otherwise, the Mann–Whitney–Wilcoxon test or Kruskal–Wallis test was applied to assess the relationship between mRNA expression and PAM50 status. The normality of the data was prior tested with a Shapiro–Wilk test. The data are presented as box-and-whisker plots (box plot represents first: lower bound; and third, upper bound quartiles, whiskers represent 1.5 times the interquartile range). The Pearson correlation test was applied to measure the strength of the association between the expressions of T-UCR, host genes, and other genes of interest. TANRIC platform (https://ibl.mdanderson.org/tanric/_design/basic/main.html) [37] also was used to analyse the correlation between the somatic copy number and microRNA expression with lnc-uc.147 expression. MiRDB was used for miRNA target prediction (http://mirdb.org/).
Patients samples
16 luminal A, 36 luminal B, 10 HER2-enriched, and 40 triple-negative Brazilian BC samples were obtained from Hospital Nossa Senhora da Graças, Curitiba, Paraná, Brazil. Tissue samples were obtained from fresh surgical specimens frozen in RNA later and stored at −80°C. All the samples were obtained with the patients’ informed consent, and the subtypes were immunohistochemically confirmed. The protocol for studying biological markers associated with disease outcome was approved by the medical ethics committee of the Universidade Federal do Paraná (19,870,319.3.0000.0102-CONEP).
Cell cultures and growth conditions
MDA-MB-231, MDA-MB-436, SK-BR-3, BT-474, MCF-7, T-47D, ZR-75-1, and CAMA-1 cells were cultured in Dulbecco’s Modified Eagle’s Medium DMEM (LONZA) supplemented with 10% FBS (Gibco) and Penicillin-Streptomycin (Gibco), 100 U/mL and 100 μg/mL respectively. ZR-75-1 were cultured in RPMI medium supplemented with 10% FBS and 1% Penicillin-Streptomycin. The MCF10A were cultured in DMEM/F12 (LONZA, USA) supplemented with 2.5 mM of L-glutamine, 20 ng/ml of epidermal factor growth (EFG), 0.01 mg/ml of insulin, 500 ng/ml of hydrocortisone, 0.5% of Penicillin-Streptomycin and 5% of horse serum.
RNA isolation, cDNA synthesis, and RT-qPCR
RNA from cells and tissues were extracted using the Quick RNA Miniprep Kit (ZymoResearch, USA). All RNAs were digested by DNase (Ambion, USA). The quality and concentration of the RNA were assessed using the nanodrop ND-1000 instrument (NanoDrop Technologies, Thermo Scientific, USA) and Bioanalyser (Agilent, USA). cDNA was synthesized using the SuperScript III cDNA kit (Invitrogen), and diluted cDNA was used for RT-PCR analysis using iQ SYBR Green Supermix (Bio-Rad) with the appropriate primers (Supplemental Table S4). The 2−ΔΔCt method was used to calculate the relative abundance of RNA genes compared with two of the following genes GAPDH, actin-beta, TBP, and u6 expression.
Strand-specific RT was made in 20 μL, where 100 ng of RNA was transcribed to strand-specific cDNA. Two reverse transcription reaction mix was made for strand-specific cDNA synthesis: one to detect uc.147 sense-oriented transcript (using a forward primer for uc.147 and reverse primer for actin beta) and other for uc.147 antisense-oriented transcript (using reverse primers both for uc.147 and actin beta). Of note, we used only actin beta reverse primer in both reaction mix because it can recognize actin beta’s mRNA. Strand-specific cDNAs were diluted 5-times with nuclease-free water to be used in qPCR analysis (Supplemental Table S4).
RACE cloning and northern blotting
To identify the 5′- and 3′-end of the uc.147 transcript, BT474 cell total RNAs were treated with DNase I (RNase-free) (Invitrogen), and the SMARTer RACE cDNA Amplification Kit (Clontech) was used, according to the manufacturer’s instructions. The cDNA ends were amplified with the Platinum Taq DNA Polymerase High Fidelity (Invitrogen), and gene-specific primers (listed in Supplemental) were used. We performed a nested PCR with the nested universal primer provided with the kit and the nested gene-specific primers listed in Supplemental Table S4. The PCR fragments were then run on a 1.5% agarose gel, and DNA was extracted with the QIAquick Gel Extraction Kit (Qiagen), according to the manufacturer’s instructions. The RACE products were cloned into a TOPO® TA pCR®2.1 cloning vector (Invitrogen) according to the manufacturer’s instructions, and the inserts were sequenced using the M13 primers. For northern blotting, a total RNA was electrophoresed on 15% PAA-urea gels (Calin et al., 2002). RNA source was BT474 cell line with no treatment, and cells treated with uc.147 siRNA.
siRNA treatment
CAMA-1 and BT474 cells were seeded 1 day before onto six-well plates, reaching a total of 200,000 for 50–70% confluence. The cells were transfected with 100 nM siRNA scramble, siRNA uc.147.1, and siRNA uc.147.2, and 20 nM siRNA LRBA.1, and siRNA LRBA.2. (Supplemental Table S4). For the transfection, we used lipofectamine 2000 reagent (Invitrogen) for 24, 48, and 72 h. We obtained the best results within 24 h. All siRNAs were provided by Sigma Aldrich.
Subcellular fractionation
The separation of nuclear and cytosolic fractions was performed using the PARIS Kit (Life Technologies) according to the manufacturer’s instructions in two different cell lines CAMA-1 and BT474. The detection level of U6 and GAPDH was used as a nuclear and cytosolic marker, respectively [39].
Cell viability
For cell viability assay, CAMA-1 and BT474 cells treated with siSCR, siuc.147, or siLRBA were plated in 96 well plates. A total of 5000 cells were seemed in each well. The cell viability was read after 24, 48, 72, and 96 hours after transfection. At each day the media were changed to 100ul of media + MTS, incubated for 4 hours, and read at 580 nm.
Apoptosis
The apoptosis ratio was analysed using the Annexin V-FITC Apoptosis Detection Kit. At 48 h after transfection, cells were harvested and resuspended in binding buffer containing Annexin V-FITC and PI according to the manufacturer’s instructions. The samples were analysed by flow cytometry (BD Biosciences, USA). Cells were discriminated into viable cells, necrotic cells, and apoptotic cells using BD FACSVantage™ cytofluorimeter (BD Biosciences, USA), and then the percentages of apoptotic cells from each group were compared.
Cell cycle
For cell-cycle analysis, the cells were harvested 24 h after treatment with siSCR, siuc.147 or siLRBA, fixed in 70% ethanol, and stained in a solution containing 10 μg ml−1 of propidium iodide (Sigma-Aldrich), 10,000 U ml−1 of RNase (Sigma-Aldrich) and 0.01% of NP40 (Sigma-Aldrich). After 30–60 min, the samples were analysed by flow cytometry using a BD FACSVantage™ cytofluorimeter (BD Biosciences). Analysis of samples were carried out acquiring 10,000–15,000 events/sample on FACS Calibur (BD). ModFit software was used to analyse cell cycle phases.
Colony-forming assay
The cells were seeded in 6-well plate (500 cells per well) and allowed to grow for 20 days at 37 C in a 5% CO2 humidified incubator after treatment with siRNA uc.147, siRNA LRBA, and siRNA scramble. The cells were fixed with 100% methanol at room temperature for 20 min, stained with 1% crystal violet at room temperature for 5 min, and washed with water until excess dye is removed. The number of colonies in each well was counted. Because of the low ability of BT474 to form colonies, we only have the results of CAMA-1. The experiment was performed in triplicate.
Pull-down assay
Lnc-uc.147 was first cloned into the TOPO TA vector following manufactory´s instructions (TAKARA, USA). The pull-down protocol was adapted from [40]. The TOPO TA with lnc-uc.147 was cloned inside the pMS2 vector. The digestion of TOPO TA was done by enzymes Xba1 (1ul- 20,000 U/ul) and Spe1 (2ul – 10,000 U/ul). Also, Xba1 (1ul- 20,000 U/ul) was used to digest the pMS2 vector and generate complementary extremities. The pMS2 vector with lnc-uc.147 and an empty control vector were co-transfected with the pMS2-GST vector in HEK293 cells. After 48 hours, the supernatant was collected, and the protein concentration was determinate by the Bradford method.
The GST fusion proteins were immobilized in the GSH beads (GE healthcare; cat 17–0756-01). The beads were washed (3 times) with NT2 buffer. Reduced glutathione was used to elute the complexes. The fractions were analysed in silver staining SDS-PAGE gel (Pierce™ Silver Stain for Mass Spectrometry). The obtained complexes were sent to the Proteomics and Metabolomics Facility core of MD Anderson Cancer Center to mass spectrometry analysis.
Western blotting
For immunoblotting analysis, proteins were quantified using Bradford assay (Bio-Rad, USA), with absorbance read at 595 nm on an ELISA reader (BioTek, USA), and the concentrations calculated using a standard curve. The proteins were not denaturated for the anti-LRBA. Overall, 40 μg of proteins were loaded on 8% gel and transferred on a nitrocellulose membrane using a semi-dry Transfer System, according to the manufacturer’s instructions. The membrane was stained with Ponceau S to make sure that equal amounts of proteins were loaded in each lane. The membranes were then incubated for 2 h at room temperature with TBST containing 5% non-fat dry milk. The membrane was probed overnight at 4 C with the primary antibody, then horseradish peroxidase-conjugated secondary antibody (Cell Signalling) was added at a dilution of 1:3000. The following primary antibodies were used: anti-LRBA rabbit monoclonal antibody (Bethyl Cat.# A304-478A) diluted in 1:1000; and anti-beta-actin rabbit monoclonal antibody (BioVision Cat.#3917-30 T) diluted in 1:3000. The used secondary antibody was the anti-rabbit HRP linked (Cell Signalling Cat.#: 7074S) diluted in 1:5000. The bound antibodies were detected by enhanced chemiluminescence and using the SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
Statistical analysis
Data are presented as mean ± SD. Statistical analysis of the data was performed by Student’s t-test or ANOVA. P-values of ≤0.05 were considered statistically significant. All experiments were performed in triplicate.
Results
T-UCRs expression in breast cancer correlates with patients´ clinical parameters and overall survival
In order to investigate the expression level of T-UCRs and their association with clinical parameters, we evaluated the expression of the 481 T-UCRs using TCGA (The Cancer Genome Atlas) data available from TANRIC website [37]. The expression levels were given as reads per kilobase of transcript per million mapped reads (RPKM). The TANRIC website provides p-values for each comparison of the expression level of the UCRs with PR and ER status, HER2 status, stage, therapy, overall survival, and PAM50 status.
A total of 302 (62.78%) T-UCRs were significantly associated with at least one BC clinical parameter. From those, 148 (49%) were overexpressed and 154 (51%) were downregulated when comparing expression levels between tumour and normal adjacent tissue. Each T-UCR can be associated with more than one clinical parameter. Therefore, we found that 209 (43.45%) T-UCRs were associated with PAM50, 172 (35.76%) with ER+, 158 (32.85%) with PR+, 80 (16.63%) with HER2+, 60 (12.47%) with stage, 176 (36.59%) with therapy outcome, and 46 (9,56%) were associated with overall survival (Supplemental Table S1). To enrich for novel lncRNA T-UCRs, we excluded all T-UCR which overlaped with mature mRNA sequences of protein coding genes. This left us with 125 intronic and 84 intergenic T-UCRs. T-UCRs identified in the majority of the breast cancer samples (>80%) were selected in order to avoid rare transcripts, generating a list of 33 T-UCRs. Analysing the subgroups of PAM50 gene signature from these 33 T-UCRs, we observed that 12 T-UCRs (uc.84, uc.138, uc.147, uc.193, uc.268, uc.271, uc.311, uc.376, uc.378, uc.427, uc.456, uc.475) had greater difference (p < 0.01) in expression levels among all subtypes. Finally, the expression level of these 12 T-UCRs were evaluated in a Brazilian cohort of BCs (Supplemental Figure S1 and Supplemental Table S2). Uc.147 and uc. 193 expression were confirmed associated with subtype classification (Figure 1).
Figure 1.
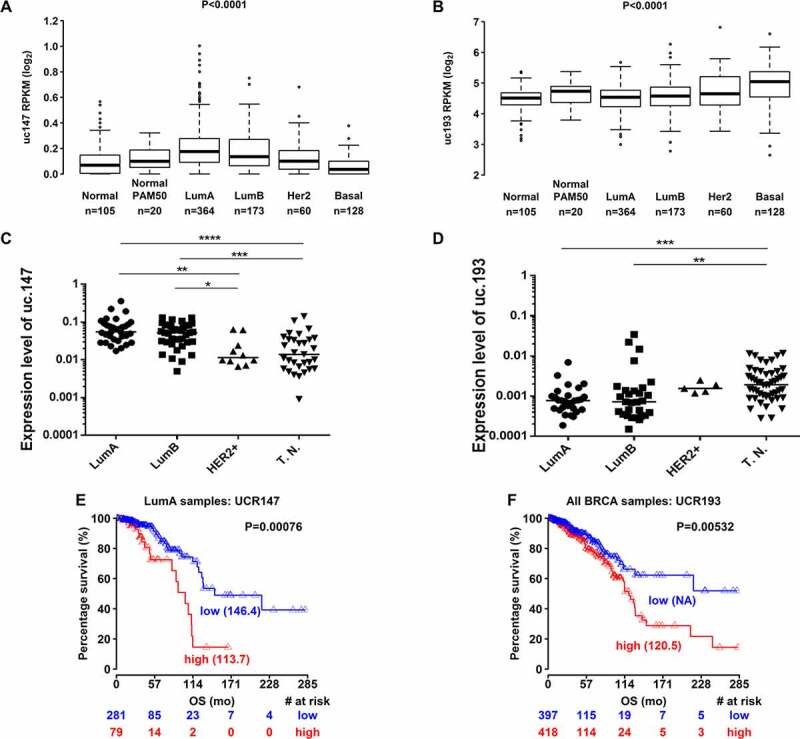
Expression analysis of uc.147 and uc.193. (a) and (b) Expression levels of T-UCRs (uc.147 in a and uc.193 in b) in all breast cancer subtypes from TCGA patients: Normal, Normal PAM50, Luminal a (LumA), Luminal b (LumB), HER2 overexpression (HER2+), and Basal. (c) and (d) Expression levels of T-UCRs (uc.147 in c and uc.193 in d) in the Brazilian BC cohort subtypes: Luminal A (LumA,n = 12), luminal b (LumB, = 15), HER2+(HER2+, n = 5), and triple negative (T.N.,n = 20). E. Overall survival for LumA BC patients according to uc.147 expression. f. Overall survival for BC patients according to uc.193 expression. The Kaplan–Meier method was used to generate the percentage of disease-free curves. *p < 0.05, **p < 0.01, ***p < 0.001
Our analysis based on TCGA data demonstrate that uc.147 has higher expression in luminal A subtype, while uc.193 has higher expression in basal subtype (Figure 1(a,b)). The differential expression of uc.147 and uc.193 was further confirmed in a Brazilian BC cohort (Figure 1(c,d)). In that cohort, we found that uc.147 is expressed at a significantly higher level in luminal A and B patients, when compared to HER2+ and triple-negative subtypes (Figure 1(c)). Moreover, we confirmed that uc.193 is highly expressed in triple-negative tumours versus luminal subtypes (Figure 1(d)).
We used univariate and multivariate Cox regression analysis on the TGCA cohort to predict the effect of T-UCRs on the patients’ overall survival. For luminal A, a subtype usually associated with better prognosis, high uc.147 expression was associated with a poor prognosis (Figure 1(e)) with the highest hazard ratio (HR = 7.19, IC 95%: 1.23–42.09, P < 0.05)(Figure 1(e)). Therefore, we suggest that uc.147 could be an independent prognostic factor. Moreover, we also show that uc.193 expression could be an independent prognostic marker of poor survival for all BC (HR = 1.75, IC 95%: 1.21–2.52, P < 0.01) (Table 1 and Figure 1(f)).
Table 1.
Summary of univariate and multivariate Cox regression analysis of overall survival and luminal A patients survival
| Univariate analysis |
Multivariate analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | HR | lower .95 | upper .95 | p-value | HR | lower .95 | upper .95 | p-value | |
| BRCA | Age (continuous) | 1.03 | 1.02 | 1.05 | 1.92E-05 | 2.34 | 1.55 | 3.52 | 5.30E-05 |
| Stage (III/IV vs. I/II) | 2.34 | 1.55 | 3.52 | 5.30E-05 | 1.04 | 1.02 | 1.05 | 5.9E-07 | |
| uc.147 | 2.64 | 0.75 | 9.25 | 0.12971 | |||||
| LRBA | 1.01 | 0.99 | 1.04 | 0.25445 | |||||
| uc.193 | 1.57 | 1.12 | 2.21 | 0.00962 | 1.75 | 1.21 | 2.52 | 0.0026 | |
| SYNCRIP | 1.01 | 0.99 | 1.03 | 0.22748 | |||||
| Luminal A | Age (continuous) | 1.03 | 1.01 | 1.05 | 0.01059 | 1.03 | 1.005 | 1.05 | 0.01752 |
| Stage (III/IV vs. I/II) | 1.75 | 0.89 | 3.45 | 0.10494 | |||||
| uc.147 | 8.25 | 1.57 | 43.47 | 0.01276 | 7.19 | 1.23 | 42.09 | 0.028 | |
| LRBA | 1.04 | 1.00 | 1.07 | 0.04808 | 1.03 | 0.99 | 1.07 | 0.123 | |
| uc.193 | 2.42 | 1.17 | 4.99 | 0.01692 | 2.31 | 1.1 | 4.85 | 0.02612 | |
| SYNCRIP | 1.04 | 0.99 | 1.10 | 0.10606 | |||||
uc.147 is located in an intronic region of the LRBA gene (ENSG00000198589). Even though in our first analysis the uc.193 appears to be intronic, a more detailed analysis demonstrated that part of its sequence overlap the 3´UTR of the SYNCRIP gene (ENSG00000135316). Interestingly, both uc.147 and uc.193 expression are correlated with their host genes (r = 0.6 and 0.9, respectly with p < 0.001) (Supplemental Figure S2). Despite of that, LRBA and SYNCRIP expression are not correlated with survival (Table 1 and Supplemental Figure S2), which indicates that the T-UCRs could be directly involved with BC.
Both uc.147 and uc.193 demonstrated promising prognostic value. As uc.193 have associated with the TNBC that we know to have worse prognosis, it was expected that higher levels of uc.193 was also associated with worse prognosis in BC patients. But when we looked specifically to the TNBC subtype, the expression levels of uc.193 is not associated with a better or poor prognosis (Supplemental Figure S3). Although, uc.147 expression level is not associated with overal survival in BC patients, it is is associate with overall survival inside the luminal A group (Supplemental figure S3 and Figure 1(e)). This suggest uc.147 expression levels as a potential additional prognostic marker for luminal A patients, which may be important for therapeutic decisions.
A novel long non-coding RNA resides within the 51st intron of LRBA
Taking into consideration that uc.147 expresses the highest levels in the luminal A sub-group among all PAM50 groups, and it also has the highest HR among the prognosis markers for the luminal A sub-group, we hypothesized that this transcript should have an important oncogenic role in luminal A subtype. Thus, we focused our further analysis on uc.147.
uc.147 is an ultraconserved region of 308 bp located inside the 51st intron of the LRBA gene (Figure 2(a)). To initiate genomic characterization of lnc-uc.147, we first aimed to determine whether this T-UCR is transcribed in the same orientation as its host gene in BC cell lines. Total RNA was isolated and DNAse treated from CAMA-1 (luminal A) and BT474 (luminal B) cell lines before we carried out two strand-specific RT-qPCR reactions, one with primer binding to the plus strand and one with primer binding to minus strand. We found that that the predominant forms of lnc-uc.147 expressed in CAMA-1 and BT474 lineages is an antisense transcript in regard to LRBA, and the sense transcript was only poorly detectable (Figure 2(b)). Finally, we separated the cytosolic and nuclear cell fractions and performed RT-qPCRs from each fraction to determine in which cell compartment the transcript is most abundant. The expression of lnc-uc.147 was only detectable at the nucleus (Figure 2(c)). The GAPDH was used as a cytosolic control and the U6 as nucleus control (Figure 2(d)).
Figure 2.
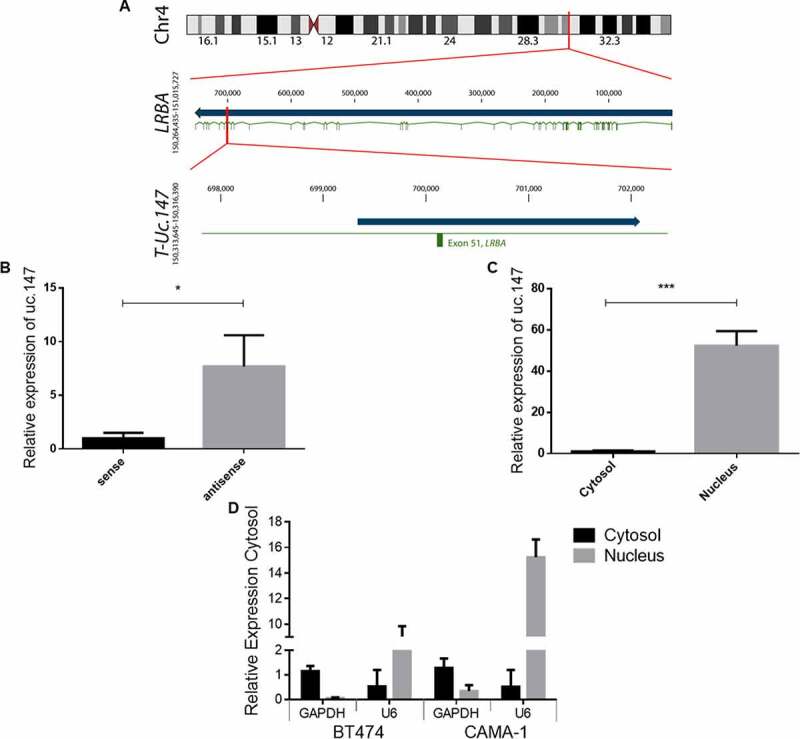
Characterization of lnc-uc.147. (a) Scheme of lnc-uc.147 transcript and locus in chromosome 4 (grey: LRBA gene/orange: uc.147 ultraconserved sequence and lnc-uc.147 transcript/ blue: LRBA exons). (b) Strand-specific RT-qPCR for lnc-uc.147. (c) RT-qPCR assays for detection of lnc-uc.147 performed on RNA extracted from nuclear and cytosolic fractions obtained from two different cell lines, CAMA-1 and BT474. (d) Detection level of GAPDH and U6 as cytosolic and nuclear markers, respectevely. *p < 0.05. ***p < 0.001
Northern blotting analysis were performed in BC cell lines (BT474 and CAMA-1) to determine the precise size and relative amount of lnc-uc.147, and it revealed that the transcript is approximately 3 kb long. To further define the exact size of lnc-uc.147, 3′ and 5´ RACE analysis were performed. For the cloning by RACE analysis, we first did a poly A+ tailing in both RNA ends. From the 3´ end we uncovered 1800 nt from the lnc-uc.147 (Supplemental Figure S4). The sequence was colinear with the LRBA antisense sequence available at NCBI (National Center for Biotechnology Information). To completely uncover the 5´ region of the lnc-uc.147 transcript, we designed PCR primers based on the LRBA gene sequence corresponding to the region obtained from 3ʹ RACE results. We performed a primer walking using the cDNA from lnc-uc.147 as a reference. We then sequenced each amplified fragment and uncovered the final and complete sequence of the lnc-uc.147 transcript – a mono-exonic RNA with 2746 nucleotides.
Knock-down of uc.147 decrease cell viability and induces apoptosis in luminal breast cancer cells
Due to the high conservation of T-UCRs, we hypothesize that those transcripts have important cellular function in humans, as it is known that evolutionary conservation is a hallmark of many biological mechanisms. Using silico analysis, we were able to identify that the lnc-uc.147 has 95% homology with Zebrafish sequences and 99% homology with sequences of other mammals. Further analysis on the sequence of this genomic region suggested that the evolution of this locus started within the first vertebrates (Supplemental Figure S5).
RT-qPCR was performed in nine BC cell lines to identify lnc-uc.147 high-expressing cell lines (Figure 3(a)). To highlight the lnc-uc.147 as a proper transcript and not only a passenger intronic expression, we performed an RT-qPCR in other intron of LRBA gene and compared the Ct values of both and with expressed exon of LRBA (Figure 3(b)). We could observe a major expression of the lnc-uc.147 in comparasion with a non-specific intron (Figure 3(b)). In addition, using TCGA data, we compared the expression levels of the lnc.uc.147 with the expression levels of all introns from LRBA and all exons, and also observed a greater expression of lnc.uc.147 in comparison with the all LRBA introns expression (Figure 3(c)).
Figure 3.
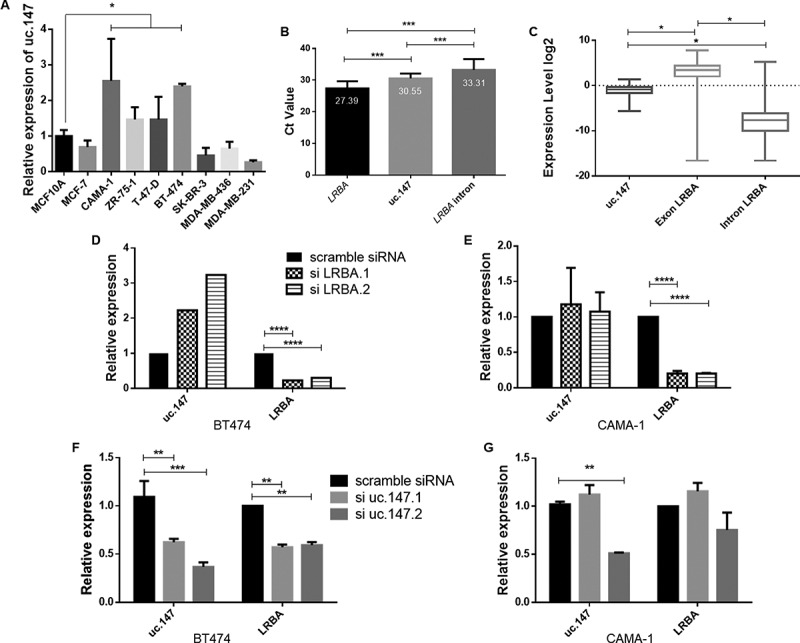
Lnc-uc.147 knockdown reduces cell viability, colony formation and increases apoptosis. (a) Expression levels of lnc-uc.147 in different BC cell lines. (b) Expression levels of LRBA, uc.147 and a intron of LRBA in CAMA-1. The representative CT values are 27.39, 30.55 and 33.31 respectively. (c) Expression levels of uc.147, all LRBA exons and all LRBA introns retrieve from TANRIC website. (d) Relative expression of lnc-uc.147 and LRBA after siRNA uc.147.1 and siRNA uc.147.2 (100 nM) treatment in BT474. (e) Relative expression of lnc-uc.147 and LRBA after siRNA uc.147.1 and siRNA uc.147.2 (100 nM) treatment in CAMA-1. (f) Relative expression of LRBA and lnc-uc.147 after siRNA LRBA.1 and siLRBA.2 (20 nM) treatment in BT474. (g) Relative expression of LRBA and lnc-uc.147 after siRNA LRBA.1 and siLRBA.2 (20 nM) treatment in CAMA-1
To investigate the biological role of lnc-uc.147 in BC, two luminal cell lines with high expressing levels of lnc-uc.147 were chosen (CAMA and BT474). A siRNA-based approach, using two specific siRNAs targeting the uc.147 ultraconserved sequence, was applied to investigate the phenotypic effects of lnc-uc.147 knockdown (KD) (Figures 3 and 4). In BT474 cells, the siuc.147#1 reduces expression by 40%, and the siuc.147#2 reduces 60% (Figure 3(d)). On the other hand, in CAMA-1 cells, siRNA uc.147.1 does not have any effect, but siRNA uc.147.2 has the potential to inhibit 50% of lnc-uc.147 expression (Figure 3(e)).
Figure 4.
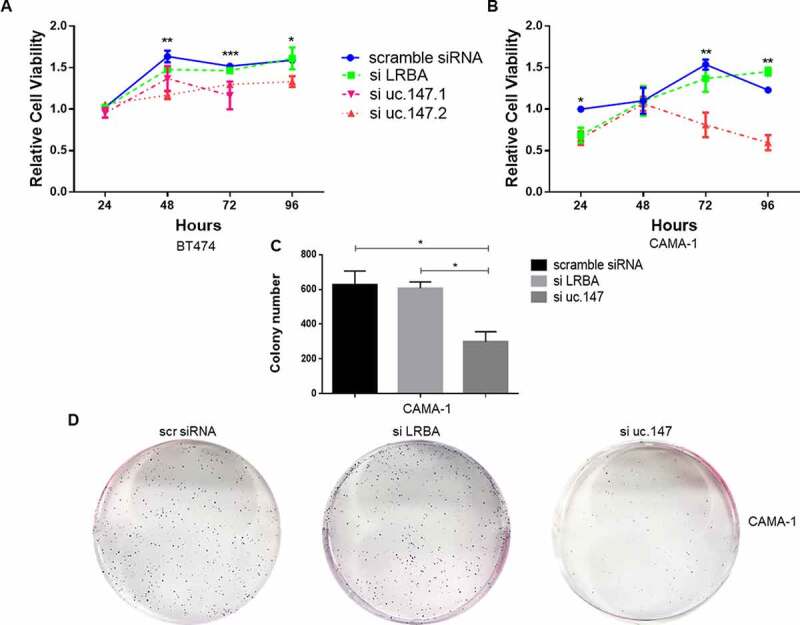
Cell viability and colony formation assays of lnc-uc.147. (a) and (b) Cell viability in BT474 (a) and CAMA-1 (b) after treatment with siRNA uc.147.1 (100 nM), uc.147.2 (100 nM), and siRNA LRBA (20 nM), at 24, 48, 72, and 96 hours.(c) Number of CAMA-1 colonies after siRNA treatment with uc.147.2 (100 nM) and siRNA LRBA (20 nM). (d) Representative plates of the colony formation
In BT474 cells, we saw a decrease in LRBA expression after siRNA treatment (Figure 3(d)). In addition, in CAMA-1 cells, the siRNA against uc.147 has a weak effect on the LRBA expression (Figure 3(e)). Assays using a specific siRNA against the LRBA gene was performed and here only LRBA expression level was reduced (Figure 3(f,g)). Furthermore, silencing the LRBA gene leads to the complete knockdown of its protein, but the LRBA protein is still detectable when the siRNA against uc.147 is used (Supplemental Figure S6).
After lnc-uc.147 downregulation, BT474 and CAMA-1 cell viability decreased, but this effect was not observed after LRBA downregulation (Figure 4(a,b)). On BT474, both siRNAs against lnc-uc.147 led to decreased cell viability (Figure 4(a)). Additionally, the knockdown of lnc-uc.147 in CAMA-1 cells has an inhibitory effect in colony formation (Figure 4(c,d)). Colony formation was not assessed on BT474 due to its low ability to form colonies. In addition, we also performed the cell viability after the silence of lnc-uc.147 in the MDA-MB-231, a cell line that did not express the lnc-uc.147 and no effect was observed (Supplemental Figure S7).
Silencing of lnc-uc.147 also leads to cell death in CAMA-1 (Figure 5(a)), but not in BT474 (Figure 5(b)). The cell death in CAMA-1 could be associated with an increase in G0/G1 cells and a reduction in G2/M events (Figure 5(a)). Although we observe a higher percentage of the G0/G1 phase in BT474, this difference was not significant (Figure 5(b)). These effects in the cell cycle, however, are only seen when lnc-uc.147 is knocked down, but not when the LRBA host gene is knocked down (Figures 4 and 5). This suggests that lnc-uc.147 and LRBA are two independent transcripts with different roles. Taken together, these findings show that lnc-uc.147 plays an important role in the survival of BC cells.
Figure 5.
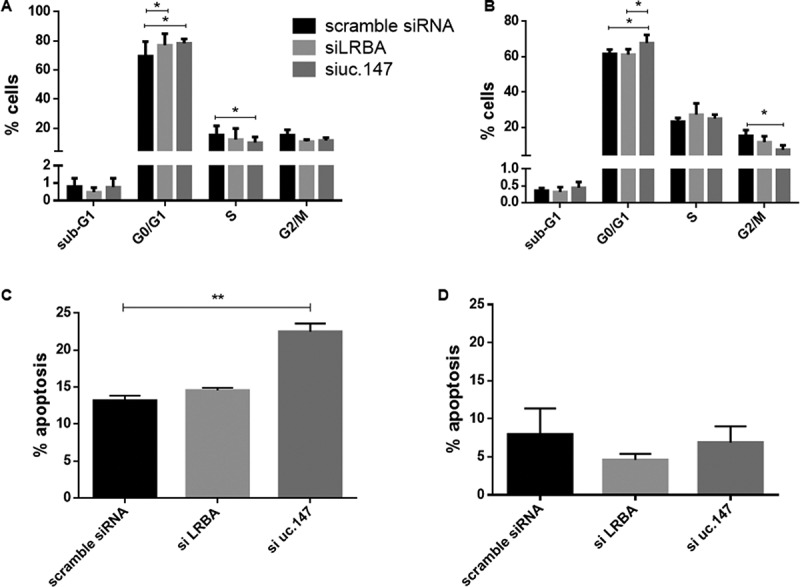
Cell cycle and apoptosis after silence of lnc-uc.147 (a) and (b) Cell cycle analysis in CAMA-1 (a) and BT474 (b) after siRNA uc.147.2 (100 nM) and siLRBA (20 nM). (c) and (d) Apoptosis ratio after CAMA-1 (c) and BT474 (d) treatment with siRNA uc.147.2 (100 nM) and siLRBA (20 nM). *p < 0.05. **p < 0.01. ***p < 0.001
Protein binding sites in lnc-uc.147
To understand the interactions of lnc-uc.147 and to uncover its function we performed a pull-down assay [40]. We constructed two pMS2 vectors, one with the lnc-uc.147 sequence and the other without, and then we co-transfected the constructed pMS2-GST vectors in HEK293 cells. The supernatant of each was collected separately and precipitated using GSH beads. After the precipitation, the proteins were added in SDS-PAGE gel and stained with silver. Two experiments were done independently. The gel was cut, and the proteins were identified using mass spectrometry.
The proteins identified in each gel were compared. First, we compared the two gels of the vector with the lnc-uc.147 transcript and obtained a list of proteins. Second, we compared the proteins extracted from the gels of the empty vector. The protein lists were compared within each other, between gels, and we excluded the proteins that were common in both to finally obtain the proteins bound only to the lnc-uc.147 (Table 2).
Table 2.
Results of pull-down assay indicating the coverage of proteins binding to the lnc-uc.147
| Protein | Protein ID | Official Symbol (local) |
Coverage | Function |
|---|---|---|---|---|
| Nascent Polypeptide Associated Complex Subunit Alpha | ENSG00000196531 | NACA (N) | 40.85% | Epithelial–Mesenchymal Transition |
| Keratin 17 | ENSG00000128422 | KRT17 (+C) | 26.16% | Cytoskeletal System |
| Dyskerin Pseudouridine Synthase 1 | ENSG00000130826 | DKC1 (+N) | 21.4% | rRNA processing and TERC stabilization |
| ATP synthase subunit alpha | ENSG00000152234 | ATP1 (+C) | 19.48% | ATP production |
| Zinc finger antiviral 1 | ENSG00000105939 | ZC3HAV1 (N/C) | 17.6% | mRNAs Stability |
| Calmodulin Like 5 | ENSG00000178372 | CALML5 (N/C) | 16.44% | Epithelial–Mesenchymal Transition |
| Pescadillo Ribosomal Biogenesis factor 1 | ENSG00000100029 | PES1 (N/C) | 14.25% | Contain a BRCA1 Domain |
| Scaffolding protein | ENSG00000083544 | TDRD3 (+N) | 13.85% | Transcription Activation |
| Spermatid Perinuclear RNA Binding Protein | ENSG00000165209 | STRBP (+N) | 11.25% | RNA binding |
| POM121 Transmembrane Nucleoporin Pseudogene | ENSG00000196313 | LOC102725072 (+N) | 8.66% | Transmembrane Nucleoporin |
| WD Repeat Domain 18 | ENSG00000065268 | WDR18 (N) | 7.84% | DNA replication |
| Ras Homolog Family Member A | ENSG00000067560 | RHOA (N/C) | 7.77% | Cytoskeletal System |
| Glioma Tumour Suppressor Candidate Region Gene 2 Protein | ENSG00000105373 | NOP53 (N/C) | 4.60% | Chromosome Stability |
| Leucine Rich Repeat Containing 59 | ENSG00000108829 | LRRC59 (N) | 3.91% | ER binding |
| Centrosomal Protein 68kDa | ENSG00000011523 | CEP68 (C/N) | 3.39% | Chromosome Stability |
| Chaperonin Containing TCP1 Subunit 8 | ENSG00000156261 | CCT8 (+C) | 2.11% | Cytoskeletal System |
| Centrosomal protein 131 | ENSG00000141577 | CEP131 (+C) | 2.1% | Cytoskeletal System |
| Testis Expressed 10 | ENSG00000122966 | TEX10 (N/C) | 1.97% | Epithelial–Mesenchymal Transition |
| Citron Rho-Interacting Serine/Threonine Kinase | ENSG00000122966 | CIT (N/C) | 0.45% | Cytoskeletal System |
The coverage indicates the quantite of bounding proteins. Local = subcellular localization. N = nucleus; C = cytosol; +N = most nuclear; +C = most cytosolic.
After the pull-down results, we did a literature search on the function of all proteins. We observed that most of them are related to cancerous cell processes, with emphasis in the cytoskeletal structure, centrossomal organization, and epithelial–mesenchymal transition (EMT). Some of these proteins have already been associated with BC: high expression of KRT17 was associated with poor survival and aggressiveness of several cancers including BC [41,42]; and CCT8, TEX10, TDRD3, and DKC1 overexpression were associate with BC progression [43–47]. CALML5 was found as a biomarker of BC [48].
PES1 contains a BC–associated gene 1 (BRCA1) C-terminal (BRCT) protein domain, and it is oestrogen inducible. The sensibility to oestrogen is interesting because lnc-uc.147 was associated with oestrogen receptor-presence, and its upregulation was associated with survival only in Luminal A patients. In addition, PES1 expression gradually increases during BC development and progression [49,50]. Another protein that interact with the oestrogen receptor and could potentially have a correlation with lnc-uc.147 is LRRC9, and its overexpreesion is associated with poor prognosis in BC [51,52].
Despite the fact that some of these proteins had already been associated with BC, no studies have reported any complexes among them. In addition, we analysed the overall survival ratio in BC of this proteins through the Oncolnc website (http://www.oncolnc.org/). Of 19 proteins, only two (KRT17 and LRRC59) are associate with overal survival in BC (data not shown).
To better understand the interaction among these proteins and lnc-uc.147, we analysed protein interactions in tools such as String (https://string-db.org/) and Interactome (http://www.interactome-atlas.org/) (Supplemental Figure S8 and S9). In String, we searched for the interactions that had already been confirmed in other studies. We observed that some of these proteins interact with each other, forming networks such as the one including RHOA and CIT, and WDR18, TEX10, PES1, NOP53 (GLTSCR2), DKC1 and ZC3HAV1 (Supplemental Figure S8).
Of the 19 proteins found in the pull-down followed by mass spectrometry, 17 of the genes encoding these proteins demonstrated a negative or positive correlation with the lnc-uc.147 (Supplemental Table S3), reinforcing a potential association among these molecules. However, more studies will be necessary to fully uncover the interactions of lnc-uc.147 and the network of proteins that this transcript interacts.
Correlation with neighbours lnc-uc.147 genes, somatic copy number and microRNAs
Aiming to identify potential cis-acting mechanisms, we analysed the correlation expression of lnc-uc.147 with the neighbour genes MAB21L2, PITX2, TACR3, CXXC4, HNRPD, HNRPDL, CDKL2, G3BP2, and DCLK2. Eight from nine genes analysed showed a significant correlation with lnc-uc.147 (Supplemental Table S3). A positive correlation was also found between lnc-uc.147 and two downstream UCRs uc.148 and uc.149 (Supplemental Table S3). It was analysing a correlation between the somatic copy number aberration in the lnc-uc.147 locus and neighbours gene that we found a positive correlation of this transcript with the host gene LRBA and its downstream gene MAB21L2 .
In addition, looking for microRNAs and lnc-uc.147 expression, we found a negative correlation with mir-18a (r = −0.31, P = 1.18 e-12) and a positive correlation with mir-190b (r = 0.30, P = 3.57 e-12). We also predict the binding between lnc-uc.147 and miRNAs through the miRDB website [53]. A total of 125 miRNAs were predicted to bind to lnc-uc.147 (data not shown), we claimed attention for the mir-190 with a target score of 100% and correlated with this transcript.
Discussion
We analysed all 481 T-UCRs in TCGA data and found that most of them (~63%) are associate with some clinical parameters. In addition, uc.147 and uc.193 differential expression were evidenced in a Brazilian cohort. Also, the overexpression of uc.147 is associate with poor prognosis in luminal A patients and overexpression of uc.193 with both luminal A and BC poor survival. Previously, only two T-UCRS were investigated in BC, the overexpression of uc.63 is associated with poor prognosis in Luminal A patients [35], and downregulation of uc.38 is associated with the worst outcome of BC patients [36].
Uc.147 is a non-characterized T-UCRs, and our study helps to better understand these transcripts lnc-uc.147 is antisense and independent from its host gene LRBA. This transcript has a 2746 nucleotides, and the sequence includes an intron and one exon of the LRBA gene. The independent transcription indicates that this T-UCRs is a long coding RNA and could participate in some cellular processes of cancer development.
As lnc-uc.147 was highly expressed in ER-positive subtypes and associated with poor survival in luminal A patients, we investigated its role in luminal cell lines. Lnc-uc.147 silencing decreased cell viability in both cell lines (CAMA-1 and BT474). Lnc-uc.147 silencing also reduced colony formation, number of cells in G2/M during the cell cycle, and increased apoptosis in CAMA-1. These experiments suggests an oncogenic effect of the lnc-uc.147, mainly in luminal A. Moreover, the silencing of the host gene LRBA did not lead to the same cell phenotyping of the lnc-uc.147 silencing.
Understanding lncRNA functions can be accomplished by the identification of lncRNA-bound proteomes [54]. Thinking about that, we performed a pull-down to define the proteins that could interact with the lnc-uc.147 and gain some insight into which molecular pathway and functions. We identify 19 proteins that are precipited with the lnc-uc.147. Among these protein, 15 (79%) are present also in nucleus, the same subcellular localization that lnc-uc.147. Seventeen (90%) of the genes encoding these co-precipited proteins demonstrated negative or positive correlation with the lnc-uc.147.
There are lncRNAs that specifically bind with one protein and others that bind with multiple proteins [55]. For example, many proteins interact with HOTAIR lncRNA [56–59]. However, the lncRNA lnc-DC seems to interact only with the STAT 3 protein [60]. The lnc-uc.147 apparently have multiple interactions, either direct or indirectly, with several proteins. Overall lnc-uc.147 interact with proteins related to the cellular process involved in cancer development, such as KRT17 [61–63], NACA [64], CALML5 [65], CEP131 [66,67], POM121 [68], and others (Table 2).
Also, it is already known that the lncRNAs may interact with protein complexes [69] The complexes are mostly formed by RNA binding proteins (RBP) [54]. The ribonucleoproteins in those complexes may control the availability and function of lncRNAs in various steps from RNA biogenesis to RNA metabolism. The cells take advantage of these interactions in forming subcellular structures and regulating the activity of various proteins with functions unrelated to RNA processing. Other interest of these cell is to help the formation of subcellular structures and membrane organelles [70].
Through the STRING website, we could see that proteins binding to lnc-uc.147 form two networks: one between RHOA and CIT, and another among six proteins (WDR18, TEX10, PES1, NOP53, DKC1, and ZC3HAV1). We hypothesize that the lnc-uc.147 could be a scaffold for these two complexes.
The interaction between RHOA and CIT was the only interaction already described in the literature. CIT regulates cytokinesis at a step after RHOA in the contractile process. CIT maintains RHOA localization at the cleavage site, which is necessary for proper RhoA activity and contractile ring dynamics at the end of cytokinesis [71,72]. The lnc-uc.147 could be acting as a guide for these two proteins during cell division.
Another network is formed by the interaction of WDR18, TEX10, PES1, NOP53, DKC1, and ZC3HAV1 proteins and the lnc-uc.147. These proteins participate of RNA processing and regulation [73–79]. Disruptions in RNA regulation have consequences for the cell as it is seen in developmental defects and diseases including cancers in humans where the underlying cause involve RBPs or their target sites within RNAs [80–86].
LncRNAs may act as multiple function in human cells [87], and as lnc-uc.147 has been described for the first time at the present study, the exact mechanisms of action must be deeply investigated in future studies. But, trying to hypothesize other possible mechanisms for this lnc-uc.147, we also analysed the potential cis-acting regulation.
Lnc-uc.147 expression was found correlated with the neighbour genes, including MAB21L2, TACR3, CXXC4, CDKL2 and G3BP2 with a positive correlation, while HNRPD, HNRPDL and DCLK2 demonstrated a negative correlation. A positive correlation was also found between lnc-uc.147 and the two downstream UCRs uc.148 and uc.149.
The downstream gene MAB21L2 is conserved in most species and has a role in eye development [88]. In cancer, this gene was downregulated in APC-mutants adenomas [89]. TACR3 expression in the nervous system is essential for the proper function of the human reproductive axis and is highly expressed in the female genital tract [90,91]. The TAC receptor 3 is strongly regulated by oestrogens, and its activation leads to nuclear translocation affecting chromatin structure and gene expression [90]. Alterations in TACR3 expression are associated with vasomotor symptoms of menopause and leiomyomas [90,91]. CXXC4 is another gene related to oestrogen receptor, and lower expression of CXXC4 is associate with tamoxifen resistance [92]. In addition, CXXC4 knockdown accelerates cell proliferation in vitro and in vivo and renders breast cancer cells insensitive to tamoxifen, whereas CXXC4 overexpression inhibits cancer cell growth and increases tamoxifen sensitivity of resistant cells [92]. The correlation of lnc-uc.147 with TACR3 and CXXC4 could be related to the oestrogen receptor. One hypothesis is for the lnc-uc.147 and CXXC4 is the lnc-uc.147 bind to the CXXC4 and inhibit its expression, which increases the tumour aggressiveness. The CDKL2 is a well-described epidermal growth factor signalling, which is more expressed in mesenchymal breast cancer cells [93]. In addition, higher methylation of CDKL2 is associate with HER2+ tumours and poor response to neoadjuvant chemotherapy [94,95]. Increased expression of CDKL2 and its correlation with lnc-uc.147 is not clear at the moment. The G3BP2 gene is well associated with stress granule, but it also associate with cancer initiation, EMT, tamoxifen resistance, and target therapy in breast cancer [96–100]. The correlation between GEBP and lnc-uc.147 could also be related to the oestrogen receptor due to G3BP role in tamoxifen resistance. The HNRPD and HNRPDL code for RNA binding proteins [101]. The HNRPD, also known as AUF-1, is responsible for the stability of the cyclin-dependent kinase inhibitor p16 (INK4a) and p21 (WAF1). Both p16 and p21 are involved in various anti-cancer processes, including regulating the critical G1 to S phase transition of the cell cycle, senescence and apoptosis [102–104]. The negative correlation of HNRPD with lnc-uc.147 seems to be correlated with a more aggressive phenotype of breast cancer tumours. For last, we have a negative correlation of DCKL2 with lnc-uc.147. The DCLK2 is a well-described gene in neuronal activity, and in only one recent study, its higher expression was associated with reduced patient survival n chronic lymphocytic leukaemia [105–107]. So, it is not clear the correlation expression of this gene and the lnc-uc.147.
Lnc-uc.147 expression was also negatively correlated with mir-18a and a positive correlation with mir-190. In addition, mir-190 bind to the lnc-uc.147 according to a prediction analysis on miRDB website (http://mirdb.org/) [53]. Mir-18 was already demonstrated in EMT in BC and is associated with poor survival in BC [108]. The overexpression of mir-190 suppresses metastasis, inhibits migration and invasion, and enhances endocrine therapy sensitivity in BC [109–111]. The lnc-uc.147 could be acting as a decoy sequestering this miRNA and increasing the BC aggressiveness. This analysis indicates another possible regulatory mechanism of lnc-uc.147 in BC.
Despite all informations, the pull-down results and correlation analysis gave us some clues for lnc-uc.147 mechanisms and reinforce the relevant lnc-uc.147 role in BC cells. However, more studies of how this lncRNA influence tumorigenesis process is necessary.
We show here in several evidence that neoplastic BC cells exhibit a unique expression profile of T-UCRs, specific uc.147, and uc.193, suggesting a significant role of T-UCRs in the malignant process. In this study, we provide more evidence for the role of T-UCRs in BC. We also characterized the lnc-uc.147, T-UCR that have never been described before, which elucidates one more fraction about these complex regions. Indeed, lnc-uc.147 has an oncogenic effect in the luminal BC cell line and can interact with proteins. Furthermore, uc.147 and uc.193 expression have a potential as a BC prognosis marker in luminal patients.
Supplementary Material
Acknowledgments
G.A.C and MD Anderson Cancer Center for receiving E.P.Z to develop part of the research. Hospital Nossa Senhora das Graças and patients to disponibility the samples. CAPES, Fundação Araucária, and CNPq to funding. The MD Anderson Cancer Center Proteomics and Metabolomics Facility core for the mass spectrometry analysis.
Funding Statement
This work was supported by the CNPq and CAPES [001 and CAPES/PRINT grant number 88887.194815/2018-00].
Disclosure statement
Dr. Calin is the scientific founder of Ithax Pharmaceuticals.
Author contributions
J.C.O, G.A.C., and E.P.Z. designed this study. E.P.Z, J.C.O, and G.A.C wrote the manuscript. E.P.Z, R.B, T.S.J, A.C.R performed most of the wet-lab experiments. C.I., E.K., and C.M. reviewed and performed most bioinformatical analysis. D.F.G and E.M.S.R.F assisted with interpretation. E.P.Z., J.C.O. and G.A.C analyzed the data. All authors read and approved the manuscript’s content.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. Nov;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers. 2019. Sep;5(1):66. [DOI] [PubMed] [Google Scholar]
- [3].Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011. Aug;22(8):1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Goldhirsch A. Personalized adjuvant therapies: lessons from the past: the opening address by the St. Gallen 2013 award recipient. Breast. 2013. Aug;22(Suppl 2):S3–7. [DOI] [PubMed] [Google Scholar]
- [5].Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012. Oct;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Beniey M, Haque T, Hassan S. Translating the role of PARP inhibitors in triple-negative breast cancer. Oncoscience. 2019. Jan;6(1–2):287–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000. Aug;406(6797):747–752. [DOI] [PubMed] [Google Scholar]
- [8].Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001. Sep;98(19):10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005. Jul;24(29):4660–4671. [DOI] [PubMed] [Google Scholar]
- [11].Sanga S, Broom BM, Cristini V, et al. Gene expression meta-analysis supports existence of molecular apocrine breast cancer with a role for androgen receptor and implies interactions with ErbB family. BMC Med Genomics. 2009;2(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Andre F, Ismaila N, Stearns V. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: ASCO clinical practice guideline update summary. J Oncol Pract. 2019. Sep;15(9):495–497. [Google Scholar]
- [13].Curigliano G, Burstein HJ, Winer EP, et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the primary therapy of early breast cancer 2017. Ann Oncol. 2017. August;28(8):1700–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bejerano G, Pheasant M, Makunin I, et al. Ultraconserved elements in the human genome. Science. 2004. May;304(5675):1321–1325. [DOI] [PubMed] [Google Scholar]
- [15].Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007. Sep;12(3):215–229. [DOI] [PubMed] [Google Scholar]
- [16].Pereira Zambalde E, Mathias C, Rodrigues AC, et al. Highlighting transcribed ultraconserved regions in human diseases. Wiley Interdiscip Rev RNA. 2019. Sep;11:e1567. [DOI] [PubMed] [Google Scholar]
- [17].Luo HL, Chen J, Luo T, et al. Downregulation of Macrophage-Derived T-UCR uc.306 associates with poor prognosis in hepatocellular carcinoma. Cell Physiol Biochem. 2017;42(4):1526–1539. [DOI] [PubMed] [Google Scholar]
- [18].Wang JY, Cui YH, Xiao L, et al. Regulation of Intestinal Epithelial barrier function by long noncoding RNA. Mol Cell Biol. 2018. Jul;38(13). doi: 10.1128/MCB.00010-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xiao L, Wu J, Wang JY, et al. Long noncoding RNA uc.173 promotes renewal of the intestinal Mucosa by inducing degradation of microRNA 195. Gastroenterology. 2018. February;154(3):599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cui X, You L, Li Y, et al. A transcribed ultraconserved noncoding RNA, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J. 2016. December;30(12):4301–4312. [DOI] [PubMed] [Google Scholar]
- [21].Guo J, Fang W, Sun L, et al. Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nat Commun. 2018. February;9(1):612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ferdin J, Nishida N, Wu X, et al. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ. 2013. Dec;20(12):1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sekino Y, Sakamoto N, Ishikawa A, et al. Transcribed ultraconserved region Uc.63+ promotes resistance to cisplatin through regulation of androgen receptor signaling in bladder cancer. Oncol Rep. 2019. May;41(5):3111–3118. [DOI] [PubMed] [Google Scholar]
- [24].Olivieri M, Ferro M, Terreri S, et al. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget. 2016. Apr;7(15):20636–20654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhou J, Wang C, Gong W, et al. uc.454 inhibited growth by targeting heat shock protein family a member 12B in Non-Small-Cell lung cancer. Mol Ther Nucleic Acids. 2018. Sep;12:174–183. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [26].Fabris L, Calin GA. Understanding the Genomic Ultraconservations: t-UCRs and cancer. Int Rev Cell Mol Biol. 2017;333:159–172. [DOI] [PubMed] [Google Scholar]
- [27].Rossi S, Sevignani C, Nnadi SC, et al. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm Genome. 2008. Aug;19(7–8):526–540. [DOI] [PubMed] [Google Scholar]
- [28].Habic A, Mattick JS, Calin GA, et al. Genetic variations of ultraconserved elements in the human genome. OMICS. 2019. November;23(11):549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Q, Shen F, Wang C. TUC338 promotes cell migration and invasion by targeting TIMP1 in cervical cancer. Oncol Lett. 2017. Jun;13(6):4526–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pichler M, Rodriguez-Aguayo C, Nam SY, et al. Therapeutic potential of FLANC, a novel primate-specific long non-coding RNA in colorectal cancer. Gut. 2020. Oct;69(10):1818–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bo C, Li N, Li X, et al. Long noncoding RNA uc.338 promotes cell proliferation through association with BMI1 in hepatocellular carcinoma. Hum Cell. 2016. Oct;29(4):141–147. [DOI] [PubMed] [Google Scholar]
- [32].Mestdagh P, Fredlund E, Pattyn F, et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010. Jun;29(24):3583–3592. [DOI] [PubMed] [Google Scholar]
- [33].Honma R, Goto K, Sakamoto N, et al. Expression and function of Uc.160+, a transcribed ultraconserved region, in gastric cancer. Gastric Cancer. 2017. Nov;20(6):960–969. [DOI] [PubMed] [Google Scholar]
- [34].Sekino Y, Sakamoto N, Goto K, et al. Transcribed ultraconserved region Uc.63+ promotes resistance to docetaxel through regulation of androgen receptor signaling in prostate cancer. Oncotarget. 2017. Nov;8(55):94259–94270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marini A, Lena AM, Panatta E, et al. Ultraconserved long non-coding RNA uc.63 in breast cancer. Oncotarget. 2017;8(22):35669–35680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang LX, Xu L, Zhang CH, et al. uc.38 induces breast cancer cell apoptosis via PBX1. Am J Cancer Res. 2017;7(12):2438–2451. [PMC free article] [PubMed] [Google Scholar]
- [37].Li J, Han L, Roebuck P, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015. Sep;75(18):3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive Lobular breast cancer. Cell. 2015. Oct;163(2):506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kong R, Zhang EB, Yin DD, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015. Apr;14(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yoon JH, Gorospe M. Identification of mRNA-Interacting factors by MS2-TRAP (MS2-Tagged RNA affinity purification). Methods Mol Biol. 2016;1421:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Escobar-Hoyos LF, Shah R, Roa-Peña L, et al. Keratin-17 promotes p27KIP1 nuclear export and degradation and offers potential prognostic utility. Cancer Res. 2015. Sep;75(17):3650–3662. [DOI] [PubMed] [Google Scholar]
- [42].Merkin RD, Vanner EA, Romeiser JL, et al. Keratin 17 is overexpressed and predicts poor survival in estrogen receptor-negative/human epidermal growth factor receptor-2-negative breast cancer. Hum Pathol. 2017. April;62:23–32. [DOI] [PubMed] [Google Scholar]
- [43].Alawi F, Lin P. Dyskerin is required for tumor cell growth through mechanisms that are independent of its role in telomerase and only partially related to its function in precursor rRNA processing. Mol Carcinog. 2011. May;50(5):334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Montanaro L, Calienni M, Ceccarelli C, et al. Relationship between dyskerin expression and telomerase activity in human breast cancer. Cell Oncol. 2008;30(6):483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shaw PG, Chaerkady R, Wang T, et al. Integrated proteomic and metabolic analysis of breast cancer progression. PLoS One. 2013;8(9):e76220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Huang X, Wang X, Cheng C, et al. Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS. 2014. Nov;122(11):1070–1079. [DOI] [PubMed] [Google Scholar]
- [47].Qiu X, He X, Huang Q, et al. Overexpression of CCT8 and its significance for tumor cell proliferation, migration and invasion in glioma. Pathol Res Pract. 2015. Oct;211(10):717–725. [DOI] [PubMed] [Google Scholar]
- [48].Debald M, Franken S, Heukamp LC, et al. Identification of specific nuclear structural protein alterations in human breast cancer. J Cell Biochem. 2011. Nov;112(11):3176–3184. [DOI] [PubMed] [Google Scholar]
- [49].Cheng L, Li J, Han Y, et al. PES1 promotes breast cancer by differentially regulating ERα and ERβ. J Clin Invest. 2012. Aug;122(8):2857–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li J, Yu L, Zhang H, et al. Down-regulation of pescadillo inhibits proliferation and tumorigenicity of breast cancer cells. Cancer Sci. 2009. Dec;100(12):2255–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhen Y, Sørensen V, Skjerpen CS, et al. Nuclear import of exogenous FGF1 requires the ER-protein LRRC59 and the importins Kpnα1 and Kpnβ1. Traffic. 2012. May;13(5):650–664. [DOI] [PubMed] [Google Scholar]
- [52].Toda H, Kurozumi S, Kijima Y, et al. Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: antitumor miR-204-5p targets AP1S3. J Hum Genet. 2018. Dec;63(12):1197–1210. [DOI] [PubMed] [Google Scholar]
- [53].Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015. Jan;43(Database issue):D146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yang Y, Wen L, Zhu H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015;5(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kazimierczyk M, Kasprowicz MK, Kasprzyk ME, et al. Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci. 2020. Feb;21(3):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016. September;41(9):761–772. [DOI] [PubMed] [Google Scholar]
- [57].Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010. Apr;464(7291):1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011. May;7(5):e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011. Oct;71(20):6320–6326. [DOI] [PubMed] [Google Scholar]
- [60].Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014. Apr;344(6181):310–313. [DOI] [PubMed] [Google Scholar]
- [61].Depianto D, Kerns ML, Dlugosz AA, et al. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet. 2010. Oct;42(10):910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang L, Zhang S, Wang G. Keratin 17 in disease pathogenesis: from cancer to dermatoses. J Pathol. 2019. February;247(2):158–165. [DOI] [PubMed] [Google Scholar]
- [63].Mikami T, Maruyama S, Abé T, et al. Keratin 17 is co-expressed with 14-3-3 sigma in oral carcinoma in situ and squamous cell carcinoma and modulates cell proliferation and size but not cell migration. Virchows Arch. 2015. May;466(5):559–569. [DOI] [PubMed] [Google Scholar]
- [64].Murayama E, Sarris M, Redd M, et al. NACA deficiency reveals the crucial role of somite-derived stromal cells in haematopoietic niche formation. Nat Commun. 2015. Sep;6(1):8375. [DOI] [PubMed] [Google Scholar]
- [65].Méhul B, Bernard D, Schmidt R. Calmodulin-like skin protein: a new marker of keratinocyte differentiation. J Invest Dermatol. 2001. Jun;116(6):905–909. [DOI] [PubMed] [Google Scholar]
- [66].Li X, Song N, Liu L, et al. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat Commun. 2017. March;8(1):14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Staples CJ, Myers KN, Beveridge RD, et al. The centriolar satellite protein Cep131 is important for genome stability. J Cell Sci. 2012. Oct;125(Pt 20):4770–4779. [DOI] [PubMed] [Google Scholar]
- [68].Ma H, Li L, Jia L, et al. POM121 is identified as a novel prognostic marker of oral squamous cell carcinoma. J Cancer. 2019;10(19):4473–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015. Jan;22(1):29–35. [DOI] [PubMed] [Google Scholar]
- [70].Kilchert C, Sträßer K, Kunetsky V, et al. From parts lists to functional significance-RNA-protein interactions in gene regulation. IWIREs RNA. 2019. Dec;e1582. [DOI] [PubMed] [Google Scholar]
- [71].Bassi ZI, Verbrugghe KJ, Capalbo L, et al. Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J Cell Biol. 2011. Nov;195(4):595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Madaule P, Eda M, Watanabe N, et al. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998. Jul;394(6692):491–494. [DOI] [PubMed] [Google Scholar]
- [73].Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007. Dec;28(5):773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Gaudet P, Livstone MS, Lewis SE, et al. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011. Sep;12(5):449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Heiss NS, Knight SW, Vulliamy TJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998. May;19(1):32–38. [DOI] [PubMed] [Google Scholar]
- [76].Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012. Jun;149(6):1393–1406. [DOI] [PubMed] [Google Scholar]
- [77].Grimm T, Hölzel M, Rohrmoser M, et al. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34(10):3030–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011. Mar;30(6):1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kim JY, Cho YE, Park JH. The nucleolar protein GLTSCR2 is an upstream negative regulator of the Oncogenic Nucleophosmin-MYC axis. Am J Pathol. 2015. Jul;185(7):2061–2068. [DOI] [PubMed] [Google Scholar]
- [80].Brinegar AE, Cooper TA. Roles for RNA-binding proteins in development and disease. Brain Res. 2016. September;1647:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009. Feb;136(4):777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Corbett AH. Post-transcriptional regulation of gene expression and human disease. Curr Opin Cell Biol. 2018. June;52:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Montes M, Sanford BL, Comiskey DF, et al. RNA splicing and disease: animal models to therapies. Trends Genet. 2019. January;35(1):68–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Pereira B, Billaud M, Almeida R. RNA-Binding proteins in cancer: old players and new actors. Trends Cancer. 2017. July;3(7):506–528. [DOI] [PubMed] [Google Scholar]
- [85].Ratnadiwakara M, Mohenska M, Änkö ML. Splicing factors as regulators of miRNA biogenesis - links to human disease. Semin Cell Dev Biol. 2018. July;79:113–122. [DOI] [PubMed] [Google Scholar]
- [86].Sterne-Weiler T, Sanford JR. Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol. 2014. Jan;15(1):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zhang X, Wang W, Zhu W, et al. Mechanisms and functions of long Non-Coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019. Nov;20(22):5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Huang ZX, Xiang JW, Zhou L, et al. The male abnormal gene family 21 (Mab21) members regulate eye development. Curr Mol Med. 2016;16(7):660–667. [DOI] [PubMed] [Google Scholar]
- [89].Segditsas S, Sieber O, Deheragoda M, et al. Putative direct and indirect Wnt targets identified through consistent gene expression changes in APC-mutant intestinal adenomas from humans and mice. Hum Mol Genet. 2008. Dec;17(24):3864–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Cañete H, Dorta I, Hernández M, et al. Differentially regulated expression of neurokinin B (NKB)/NK3 receptor system in uterine leiomyomata. Hum Reprod. 2013. Jul;28(7):1799–1808. [DOI] [PubMed] [Google Scholar]
- [91].Crandall CJ, Manson JE, Hohensee C, et al. Association of genetic variation in the tachykinin receptor 3 locus with hot flashes and night sweats in the Women’s Health Initiative Study. Menopause. 2017. March;24(3):252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Fu Y, Wang Z, Luo C, et al. Downregulation of CXXC finger protein 4 leads to a Tamoxifen-resistant phenotype in breast cancer cells through activation of the Wnt/β-catenin pathway. Transl Oncol. 2020. Feb;13(2):423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Li L, Liu C, Amato RJ, et al. CDKL2 promotes epithelial-mesenchymal transition and breast cancer progression. Oncotarget. 2014. Nov;5(21):10840–10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lindqvist BM, Wingren S, Motlagh PB, et al. Whole genome DNA methylation signature of HER2-positive breast cancer. Epigenetics. 2014. Aug;9(8):1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Pineda B, Diaz-Lagares A, Pérez-Fidalgo JA, et al. A two-gene epigenetic signature for the prediction of response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Clin Epigenetics. 2019. February;11(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].De Marchi T, Kuhn E, Dekker LJ, et al. Targeted MS assay predicting tamoxifen resistance in Estrogen-Receptor-Positive breast cancer tissues and Sera. J Proteome Res. 2016. Apr;15(4):1230–1242. [DOI] [PubMed] [Google Scholar]
- [97].French J, Stirling R, Walsh M, et al. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem J. 2002. May;34(5):223–231. [DOI] [PubMed] [Google Scholar]
- [98].Gupta N, Badeaux M, Liu Y, et al. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc Natl Acad Sci U S A. 2017. January;114(5):1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wei SC, Fattet L, Tsai JH, et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015. May;17(5):678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhang Y, Yue C, Krichevsky AM, et al. Repression of the stress granule protein G3BP2 inhibits immune checkpoint molecule PD-L1. Mol Oncol. 2021. Feb. DOI: 10.1002/1878-0261.12915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kim DH, Langlois MA, Lee KB, et al. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33(12):3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Al-Ansari MM, Hendrayani SF, Shehata AI, et al. p16(INK4A) represses the paracrine tumor-promoting effects of breast stromal fibroblasts. Oncogene. 2013. May;32(18):2356–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Al-Khalaf HH, Aboussekhra A. p16(INK4A) positively regulates p21(WAF1) expression by suppressing AUF1-dependent mRNA decay. PLoS One. 2013;8(7):e70133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Al-Khalaf HH, Colak D, Al-Saif M, et al. p16(INK4a) positively regulates cyclin D1 and E2F1 through negative control of AUF1. PLoS One. 2011;6(7):e21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Barrow TM, Wong Doo N, Milne RL, et al. Analysis of retrotransposon subfamily DNA methylation reveals novel early epigenetic changes in chronic lymphocytic leukemia. Haematologica. 2021. January;106(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Edelman AM, Kim WY, Higgins D, et al. Doublecortin kinase-2, a novel doublecortin-related protein kinase associated with terminal segments of axons and dendrites. J Biol Chem. 2005. Mar;280(9):8531–8543. [DOI] [PubMed] [Google Scholar]
- [107].Nawabi H, Belin S, Cartoni R, et al. Doublecortin-Like kinases promote neuronal survival and induce growth cone reformation via distinct mechanisms. Neuron. 2015. Nov;88(4):704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Luengo-Gil G, García-Martínez E, Chaves-Benito A, et al. Clinical and biological impact of miR-18a expression in breast cancer after neoadjuvant chemotherapy. Cell Oncol (Dordr). 2019. Oct;42(5):627–644. [DOI] [PubMed] [Google Scholar]
- [109].Sun G, Liu M, Han H. Overexpression of microRNA-190 inhibits migration, invasion, epithelial-mesenchymal transition, and angiogenesis through suppression of protein kinase B-extracellular signal-regulated kinase signaling pathway via binding to stanniocalicin 2 in breast cancer. J Cell Physiol. 2019. August;234(10):17824–17838. [DOI] [PubMed] [Google Scholar]
- [110].Yu Y, Luo W, Yang ZJ, et al. miR-190 suppresses breast cancer metastasis by regulation of TGF-β-induced epithelial-mesenchymal transition. Mol Cancer. 2018. March;17(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yu Y, Yin W, Yu ZH, et al. miR-190 enhances endocrine therapy sensitivity by regulating SOX9 expression in breast cancer. J Exp Clin Cancer Res. 2019. Jan;38(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


