ABSTRACT
The blood-brain barrier (BBB), which controls permeability into and out of the nervous system, is a tightly connected, structural, and functional separation between the central nervous system (CNS) and circulating blood. CNS diseases, such as Alzheimer’s disease, multiple sclerosis, traumatic brain injury, stroke, meningitis, and brain cancers, often develop with the increased BBB permeability and further leads to irreversible CNS injury. Non-coding RNAs (ncRNAs) are functional RNA molecules that generally lack the coding abilities but can actively regulate the mRNA expression and function through different mechanisms. Various types of ncRNAs, including microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs), are highly expressed in brain microvascular endothelial cells and are potential mediators of BBB permeability. Here, we summarized the recent research progress on miRNA, lncRNA, and circRNA roles regulating the BBB permeability in different CNS diseases. Understanding how these ncRNAs affect the BBB permeability shall provide important therapeutic insights into the prevention and control of the BBB dysfunction.
KEYWORDS: Blood-brain barrier, permeability, microRNAs, long non-coding RNAs, circular RNAs
Introduction
The blood-brain barrier (BBB) is composed of brain microvascular endothelial cells (BMECs), pericytes, and astrocyte endfeet [1]. It maintains brain homoeostasis and protects the brain from xenobiotics and neurotoxic metabolites circulating in the bloodstream [2]. BMECs are indispensable and distinct structural components of the BBB. They are characterized by the presence of tight junction proteins (TJs) that lead to high transendothelial electrical resistance and low paracellular permeability [3]. TJs are composed of several integral membrane proteins, including Occludin, Claudins, and junctional adhesion molecules, all of which are linked through cytoplasmic zonula occludens (ZO) family members (e.g. ZO-1,-2,-3) to the intracellular actin cytoskeleton [4]. A decrease or distribution changes of these TJs in BMECs could increase the BBB permeability, an important indicator of BBB dysfunction [5]. Increased BBB permeability is a key event in the pathogenesis of several central nervous system (CNS) diseases with inflammatory characteristics, such as Alzheimer’s disease, multiple sclerosis, traumatic brain injury, stroke, and meningitis [6–10].
Recent genome-wide studies using deep sequencing technology have revealed that eukaryotic genomes are extensively transcribed to produce thousands of non-coding RNAs (ncRNAs) [11]. The available data indicate that most of the mammalian genome produces RNA transcripts, despite only approximately 1.2% of the genomic DNA sequences code proteins [12]. Many ncRNAs, such as transfer RNA (tRNAs) and ribosomal RNA (rRNAs), are directly involved in translation [13]. Except for tRNAs and rRNAs, the ncRNAs can be broadly classified into small ncRNAs (<200 nucleotides [nt], including microRNA [miRNA], soRNA, snoRNA, siRNA, and piRNA) [14], long non-coding RNA (lncRNAs) >200 nt in length [15], and circular RNA (circRNAs), which have a particular covalent loop structure without a 5ʹ cap or 3ʹ poly-A tail [16]. Non-coding transcripts were once considered to have no biological function [17]. However, with the progress of research, ncRNAs have been increasingly identified as the critical regulators of gene expression and participate in multiple major biological and physiological processes that involve development, differentiation, and metabolism, as well as pathologies in a variety of human diseases [18–20]. In particular, the function of miRNAs has been most extensively studied compared to lncRNAs and circRNAs in CNS diseases.
Here, we provide a functional overview of the miRNAs, lncRNAs, and circRNAs in the regulation of BBB permeability. Understanding the molecular mechanism of the increased BBB permeability regulated by ncRNAs shall contribute to the development of safe and effective therapeutic approaches to maintain BBB integrity in many CNS diseases.
miRNAs and BBB Permeability
miRNAs are small, endogenous, single-stranded ncRNA molecules that are typically 19–25 nt in length and are involved in transcriptional and post-transcriptional gene expression regulation by affecting the translation as well as stability of the messenger RNAs (mRNAs) (Fig. 1) [21]. miRNAs are present and stable in many mammalian cell types and appear to target more than 50% of genes in humans [22]. They control various normal biological processes, including cell proliferation and differentiation, apoptosis, immune responses, angiogenesis, and inflammation [23,24]. Recently, many studies have demonstrated that miRNAs play central roles in the BBB dysfunction in CNS diseases (Table 1 and Fig. 2) [25].
Figure 1.

The structure of miRNAs, lncRNAs and circRNAs
Table 1.
miRNAs as therapeutic targets of pharmacological agents for CNS injuries
| CNS injuries | Cell/Animal species | miRNAs | Target mRNAs | References |
|---|---|---|---|---|
| Ischaemic stroke | Mice | miR-132 | MMP-9 | Zuo et al[27]. |
| bEnd.3/Rats | miR-539 | MMP-9 | Fan et al[28]. | |
| Rats | miR-21 | MAP2K3 | Yao et al[29]. | |
| BMEC/Rats | miR-34a | Not investigated | Ren et al[30]. | |
| bEnd.3 | miR-34a | Cytochrome c | Bukeirat et al[31]. | |
| Rats | miR-210 | Occludin/β-catenin | Ma et al[33]. | |
| Human BMEC | miR-155 | claudin-1 | Philippides et al[34]. | |
| BMEC/Mice | miR-155 | MFSD2A | Awad et al[35]. | |
| bEnd.3/ Mice | miR-182 | mTOR/FOXO1 | Zhang et al[36]. | |
| BMEC/Mice | miR-130a | HOXA5 | Wang et al[37]. | |
| Rat BMEC | miR-150 | Tie-2 | Fang et al[38]. | |
| Mice | miR-15a | claudin-5 | Ma et al[39]. | |
| Haemorrhagic stroke | Rat BMEC/Rats | miR-126-3p | PIK3R2 | Xi et al[40]. |
| Rat BMEC/Rats | miR-27a-3p | AQP11 | Xi et al[41]. | |
| Traumatic brain injury | Mouse BMEC | miR-212/132 | claudin-1/JAM3/TJP4 | Burek et al[42]. |
| Rat BMEC | miR-21 | Angiopoietin-1/Tie-2 | Ge et al[43]. | |
| Multiple sclerosis | hCMEC/Mice | miR-155 | annexin-2/claudin-1 | Ramirez et al[45]. |
| hCMEC | miR-125a-5p | VE-cadherin/ZO-1 | Reijerkerk et al[25]. | |
| Meningitis | Mouse BMEC | let-7/miR-98 | CCL2/CCL5 | Rom et al[46]. |
| HUVEC/Monkeys | miR-1303 | MMP-9 | Song et al[48]. | |
| Human BMEC | miR-101 | VE-cadherin | Mishra et al[49]. | |
| Mice | miR-155 | Not investigated | Barker et al[50]. | |
| Alzheimer′s Disease | hCMEC | miR-107 | Endophilin-1 | Liu et al[51]. |
| hCMEC | miR-424-5p | Endophilin-1 | Lin et al[52]. | |
| bEnd.3 | miR-501-3p | ZO-1 | Toyama et al[53]. | |
| Diabetes | Mouse BMEC | let-7a | Not investigated | Song et al[54]. |
| Rat BMEC | miR-200b | VEGFA | Yang et al[55]. | |
| Hyperhomocysteinemia | bEnd.3/Mice | miR-29b | DNMT3B, MMP-9 | Kalani et al[56]. |
| Methamphetamine | Human BMEC | miR-143 | PUMA | Bai et al[57]. |
| Cancer | BMEC | miR-181 c | PDPK1 | Tominaga et al[58]. |
Figure 2.
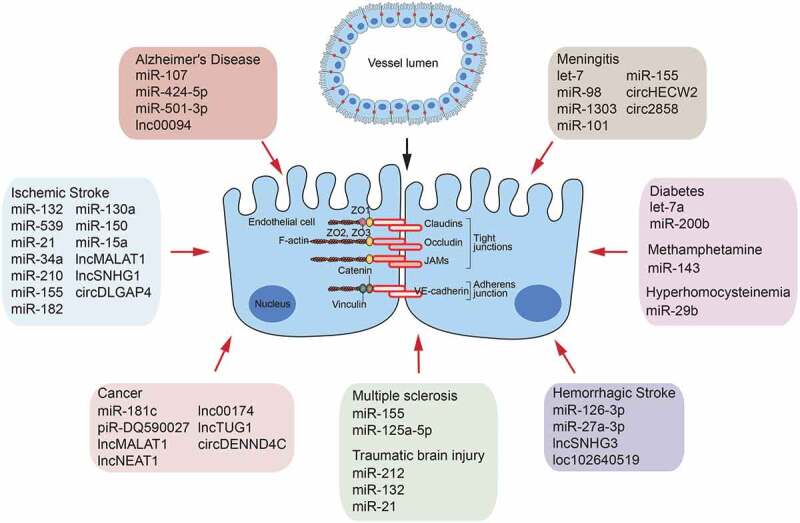
A brief summary of miRNAs, lncRNAs and circRNAs as a factor controling BBB permeability
In ischaemic stroke, matrix metallopeptidase (MMP)-9 showed an affinity for BBB permeability by degrading TJs expression [26]. miR-132 and miR-539 were reported to regulate BBB permeability by directly targeting MMP-9 [27,28], and miR-21 indirectly regulated MMP-9 by activating the mitogen-activated protein kinase signalling pathway [29]. Studies have noted the increase of miR-34a expression in the brains of mice suffering experimental stroke [30], and another study found that miR-34a triggered the breakdown of BBB by targeting cytochrome c in the BMECs monolayer in vitro [31,32]. In neonatal rat hypoxic-ischaemic brain injury, overexpression of miR-210 negatively regulated BBB integrity and preserved the expression of Occludin and β-catenin [33]. In a cerebral ischaemia BMECs model, a decrease in the miR-155 level enhanced the BMECs transepithelial electrical resistance and decreased the monolayer permeability by directly targeting Claudin-1 [34]. Another study demonstrated that high levels of miR-155 targeted MFSD2A in BMECs to increase BBB permeability, and in the miR-155-deleted mouse model, ischaemia-induced brain injury was effectively alleviated [35]. And miR-182 was reported to exacerbate BBB disruption by downregulating the mTOR/FOXO1 pathway [36]. In other studies, miR-130a could downregulate Occludin and enhance the BBB permeability by inhibiting HOXA5 expression [37], and miR-150 could regulate Claudin-5 expression by targeting Tie-2 to affect the permeability of BBB [38]. In addition, evidence has suggested that BMECs-selective deletion of the miR-15a/16-1 clusters attenuates BBB pathology after ischaemic stroke [39]. As for haemorrhagic stroke, exogenous miR-126-3p can alleviate the BBB disruption by targeting PIK3R2 to suppress angiopoietin (ANGPT)-1 and vascular endothelial growth factor A (VEGFA)-induced activation of Akt [40]. miR-27a-3p may protect against BBB disruption and brain injury by targeting endothelial Aqp11 [41]. In an in vitro model of traumatic brain injury, it was reported that overexpression of miR-212/132 led to the decrease in both mRNA and protein expression of Claudin-1, JAM3, and TJP4 [42], and the up-regulated miR-21 level could alleviate traumatic brain injury-induced secondary BBB damage and loss of TJs by targeting ANGPT-1 and Tie-2 [43].
Multiple sclerosis is a prototypical inflammatory disease of the CNS [44]. MiR-155 was reported to modulate monolayer BMECs function by targeting Annexin-2 and Claudin-1 in multiple sclerosis [45]. Similarly, miR-125a-5p was a key regulator of BMECs tightness by affecting the expression of vascular endothelial (VE)-cadherin and ZO-1 in the TNF-α/IFNγ-treated-BMECs multiple sclerosis model in vitro[25]. In the study of aseptic meningitis, the low expression levels of let-7 and miR-98 resulted in BBB dysfunction by targeting CCL2 and CCL5 [46,47]. Besides, evidences have suggested that miRNAs played essential roles in BBB damage during pathogenic infections. It was shown that Coxsackievirus penetrated the BBB by downregulating miR-1303, which disrupted Claudin-5, Claudin-4, VE-cadherin, and ZO-1 by directly regulating MMP-9 [48]. HIV-1 Tat C increased the expression of miR-101, which led to the down-regulation of VE-cadherin to enhance BBB permeability [49]. Moreover, the increase of BBB permeability induced by malaria infection was significantly reduced in miR-155 knocking-down mice compared to control mice [50].
Emerging pieces of evidence suggested that miRNAs played significant roles in the disruption of BBB integrity in Alzheimer’s disease, and miR-107 and miR-424-5p were reported to affect the expression of ZO-1 and Occludin in an in vitro model with Aβ-incubated BMECs by targeting Endophilin-1 [51,52]. TNF-α induced-miR-501-3p was considered the upstream regulator of ZO-1 in vascular dementia [53]. In a diabetes study, a high level of miR-Let7a was suggested to attenuate BMECs damage by controlling TJ Claudin-5 and ZO-1 expression [54], and a high level of miR-200b was also reported to attenuate BBB dysfunction by inhibiting VEGFA [55]. Moreover, miR-29b was suggested to be a novel nucleic acid target for maintaining BBB permeability by inhibiting DNMT3B and MMP-9 up-regulation during hyperhomocysteinemia [56]. Another study found that silencing miR-143 increased TJs’ expression and protected BBB integrity against the effects of methamphetamine treatment in a monolayer BMECs model [57]. Interestingly, a recent study found that miRNAs in extracellular vesicles can also mediate the BBB destruction. Brain metastatic cancer cell-derived miR-181 c can increase the BBB permeability through its target gene PDPK1 [58]. And in the study of reversible cerebral vasoconstriction syndrome, the circulating miRNAs were functionally linked to the headache, BBB integrity, and vasomotor function [59].
miRNAs are important components involving in epigenetic regulation. Current studies have confirmed that many miRNAs can negatively regulate the expression of TJs by directly targeting TJs or indirectly regulating upstream molecules of TJs, thereby affecting the integrity of BBB. However, because of the diversity of mechanisms and functions of miRNAs, more in-depth studies are still needed to reveal their specific contributions.
LncRNAs regulation of vascular endothelial permeability via TJs
Transcripts longer than 200 nt without coding capacity are generally defined as lncRNAs (Fig. 1) [60]. lncRNAs are further subdivided into long intergenic ncRNAs, long intronic ncRNAs, telomeric ncRNAs, pseudogene transcripts, enhancer RNAs, and promoter-associated long RNAs [61]. It has been predicted that the human genome encodes more than 50,000 lncRNAs; however, most of them are uncharacterized [62]. lncRNAs were reported to regulate gene expression through various mechanisms, including modulation of transcription factor activity and splicing machinery, transcription enhancers, increasing mRNA stability, acting as architectural components in the assembly of protein complexes, as well as played as molecular decoys for miRNAs [63–65]. Although only a small proportion of the identified lncRNAs have been studied in-depth, they have pivotal roles in various physiological and pathological processes, such as cell differentiation, tumorigenesis, metastasis, immune response, ageing, and others [66].
Accumulating evidence has shown that lncRNAs acted as key regulators of vascular function, including senescence, angiogenesis, vascular repair, and inflammatory signalling cascades (Table 2 and Fig. 2) [67]. For example, lnc-tie-1AS was universally transcribed in zebrafish, mice, and humans and controlled the transcription of its parental gene tie-1 by directly binding to tie-1 mRNA, thus leading to specific defects in vascular endothelial TJs [68]. The blood-tumour barrier (BTB) in brain, similar to BBB, is characterized by the presence of TJs between brain capillary endothelial cells and forms a major obstacle in brain tumour therapy by preventing the delivery of sufficient therapeutic drugs [69]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved lncRNA in mammals, and its function has been shown in numerous cancers [70]. In glioma endothelial cells, knockdown of MALAT1 induced the down-regulation of ZO-1, Occludin, and Claudin-5 by sponging miR-140 [71]. Nuclear enriched abundant transcript 1 (NEAT1), also known as nuclear paraspeckle assembly transcript 1, is an essential lncRNA for the integrity of the nuclear paraspeckle substructure [72]. A recent study found that knockdown of NEAT1 increased BTB permeability by binding to miR-181d-5p and reduced TJs expression by targeting SOX5 [73]. Knockdown of lnc00174 increased BTB permeability by activating the miR-138-5p (miR-150-5p)/FOSL2 feedback loop [74]. Moreover, lnc-taurine upregulated gene 1 (lncTUG1) is another highly expressed lncRNA in glioma vascular endothelial cells from glioma tissues [75]. Knockdown of lncTUG1 increased the BTB permeability by binding to miR-144 and reduced glioma endothelial cell TJs expression by targeting HSF2 [76]. Together, these findings largely indicated that aberrant expression of lncRNAs could act as competing endogenous RNAs (ceRNA) to affect the permeability of BTB.
Table 2.
Effects of lncRNAs and circRNAs on BBB dysfunction
| CNS injuries | Cell/Animal species | lncRNAs/circRNAs | Molecular mechanisms | References |
|---|---|---|---|---|
| Alzheimer’s disease | hCMEC/pericytes co-culture | lnc00094 | miR-224-5p (miR-497-5p)/Endophilin-1 | Zhu et al[78]. |
| Ischaemic stroke | mouse BMEC | lncMALAT1 | miR-145/VEGFA (ANGPT2) | Ren et al[82]. |
| mouse BMEC and Mice | lncSNHG1 | miR-18a (miR-199a)/HIF-1α/VEGF | Zhang et al[83]. | |
| Haemorrhagic Stroke | Rat BMEC/Rats | lncSNHG3 | TWEAK/Fn14/STAT3 | Zhang et al[80]. |
| BMEC/astrocytes co-culture and Mice | loc102640519 | HOXC13 | Wu et al[85]. | |
| Glioma | hCMEC | piR-DQ590027 | MIR17HG/miR-133 (miR-377)/FOXR2 | Leng et al[79]. |
| hCMEC/glioblastoma co-culture | lncMALAT1 | miR-140/NFYA | Ma et al[71]. | |
| hCMEC/glioblastoma co-culture | lncNEAT1 | miR-181d-5p/SOX5 | Guo et al[73]. | |
| hCMEC/glioblastoma co-culture | lnc00174 | miR-138-5p (miR-150-5p)/FOSL2 | Guo et al[74]. | |
| hCMEC/glioblastoma co-culture | lncTUG1 | miR-144/HSF2 | Cai et al[76]. | |
| hCMEC/glioblastoma co-culture | circDENND4C | miR-577 | Wu et al[103]. | |
| Neuroinflammatory | Human BMEC and Mice | circHECW2 | miR-30d/ATG5 | Yang et al[104]. |
| Ischaemic stroke | bEnd.3 and Mice | circDLGAP4 | miR-143/HECTD1 | Bai et al[105]. |
| Bacterial meningitis | Human BMEC | circ2858 | miR-93-5p/ VEGFA | Yang et al[106]. |
Recent studies have also recognized that lncRNAs are closely involved in controlling the BBB permeability [77]. In Alzheimer’s disease, lnc00094 was dramatically increased in Aβ-incubated BMECs of the BBB in an in vitro model. Reduction of lnc00094 inhibited Endophilin-1 expression by up-regulating miR-224-4p and miR-497-5p, thus promoting the expression of ZO-1, Occludin, and Claudin-5, and ultimately alleviating BBB permeability [78]. In another study on BMECs, piR-DQ590027 was reported to regulate the permeability and transendothelial electrical resistance by activating the MIR17HG/miR-133 (miR-377)/FOXR2 pathway [79]. In the intracerebral haemorrhage study, the upregulation of lncRNA small nucleolar RNA host gene 3 (Snhg3) was confirmed to increase BBB permeability by activating the TNFSF12 /Fn14/STAT3 signalling pathway [80]. After the stroke, inhibition of VEGF can reduce BBB permeability [81]. Accumulating evidences have shown that lncRNAs are novel endogenous nucleic acid regulatory molecules of VEGFA in stroke. The levels of lncMALAT1 were up-regulated in oxygen-glucose deprivation-induced BMECs, and lncMALAT1 enhanced the expression of VEGFA and ANGPT-2 by targeting miR-145 to affect BBB integrity [82]. Meanwhile, the expression of the small nuclear RNA host gene 1 (lncSNHG1) was remarkably increased in isolated cerebral microvessels of a middle cerebral artery occlusion mouse model and oxygen-glucose deprivation-cultured mice BMECs. Moreover, lncSNHG1 could function as a ceRNA for miR-18a and miR-199a to regulate the hypoxia-inducible factor-1α/VEGFA signalling pathway [83,84]. Interestingly, VEGFA significantly aggravated cerebral ischaemia injury by up-regulating LOC102540519 and HOXC13. LOC102640519 positively regulated the expression of HOXC13, thus negatively regulating the expression of ZO-1, Occludin, and Claudin-5 to enhance BBB permeability [85]. These findings collectively suggest that the abnormal expression of lncRNAs could be an approach to regulating BBB permeability.
The above researches indicate that lncRNAs are considerable endogenous regulators that are responsible for the BBB permeability regulation. However, the precise regulatory mechanisms involving lncRNAs in BBB damage still require more in vitro and in vivo experimental evidences.
Roles of CircRNAs in BBB Permeability
The current advances in circRNAs study have shown that circRNAs are another vital component of regulatory ncRNAs [86]. CircRNAs, consisting of a circular configuration through a typical 5′ to 3′-phosphodiester bond, are composed of exonic or intronic sequences (Fig. 1) [87]. Because of the absence of the defined 5′ caps and 3′ poly-A tails, circRNAs are extremely stable then linear forms of RNAs [88]. circRNAs are highly abundant, tissue-specific, and evolutionarily conserved RNAs in mammalian cells and tissues [89–91]. In humans, circRNAs are particularly abundant in the brain, peripheral blood, and exosomes [92,93]. Their ability to cross the BBB makes them perfect candidates as potential diagnostic markers for CNS disorders [94]. Interestingly, circRNAs were found not to be uniformly distributed throughout the brain, as some regions were more enriched than the others [95], and their molecular functions were also diverse, including acting as miRNA sponges, regulators of transcription and splicing, ribosomal RNA processing, and adaptors for protein-protein interaction [96]. CircRNAs have also been reported to play important roles in pathophysiological processes, including Alzheimer’s diseases, neuronal diseases, cardiovascular disease, and cancer progression [97]. Compared to the researches on miRNAs and lncRNAs regulation of BBB dysfunction, there is currently little work focusing on the roles of circRNAs in the development of CNS diseases, particularly in the regulation of BBB permeability.
CircRNAs are prominently involved in numerous CNS diseases, and their transcriptional profiles have been increasingly reported in different disease models, such as stroke, meningitis, Parkinson’s disease, and Alzheimer′s disease (Table 2 and Fig. 2) [98–102]. The circ-DENND4C was enriched in glioma endothelial cells and acted as a molecular sponge to bind miR-577 and inhibit its negative regulation of the target genes ZO-1, Occludin, and Claudin-1, thus positively regulate the BTB permeability [103]. In the endothelial-mesenchymal transition study, circRNAs were reported to regulate BBB permeability by acting as miRNA sponges. Treatment of BMECs with methamphetamine or lipopolysaccharide significantly increased circHECW2 expression, and circHECW2 competitively sponged miR-30d to up-regulate the expression of autophagy-related 5 (Atg5), which was involved in the endothelial-mesenchymal transition and resulted in the damage of BBB integrity by promoting the degradation of ZO-1, Occludin, and Claudin-5. Therefore, specific blockage of circHECW2 may be envisioned as a potential therapeutic target for the treatment of BBB integrity damage [104]. In a stroke model, overexpression of circDLGAP4 significantly attenuated neurological deficits, decreased infarct areas, and BBB damage in a mouse stroke model with transient middle cerebral artery occlusion by competitively binding miR-143 to regulate HECTD1, thus increasing TJs expression. This circDLGAP4 was thus suggested as a potential therapeutic candidate in acute ischaemic injury [105]. Currently, our group profiled the expression of 41,504 circRNAs in BMECs after meningitic Escherichia coli infection and found that 308 were significantly altered compared with the control group. A ceRNA analysis was further performed, and the potential regulatory network was preliminarily constructed and validated. For the first time, this work showed that circRNAs were responsive to meningitic bacterial challenges, and their alterations might be involved in the infection-enhanced BBB permeability [100]. In another work, we further reported meningitic E. coli-induced upregulation of circ_2858 in BMECs and demonstrated this circRNA could facilitate the VEGFA expression by competitively sponging miR-93-5p, thus causing the TJs disruption and BBB dysfunction [106]. These findings mentioned above suggested that circRNAs could be a potential nucleic acid approach to secure BBB dysfunction in CNS disorders.
Due to their extensive involvement in different physiological and pathological processes, circRNAs have attracted increasing attention in recent years. As an important type of ncRNAs, circRNAs are considered to have significant regulatory potential due to their high expression in brain tissue and have been experimentally shown to be involved in a variety of CNS-related diseases. However, the roles of circRNAs in regulating the BBB function have not been well documented.
Conclusion
The field of BBB physiology and pathology, particularly the study of TJ complexes, has rapidly advanced in recent years. It is now recognized that TJs of BMECs are redistributed and degraded in response to stroke, meningitis, traumatic brain injury, and other CNS diseases. These TJs changes lead to increased BBB permeability, which will cause irreversible damage to the CNS; thus, regulation of the TJs expression and distribution is an essential direction for future studies. Meanwhile, many other proteins play important roles in maintaining BBB permeability and functions, such as the MMPs, transforming growth factor β1, VEGFA, TNF-α, Tie-2, Angiopoietin-1, Endophilin-1, and Neuropilin-1 [107]. Emerging evidence also suggests important roles of astrocyte-derived factors in BBB dysfunction and recovery after brain injury. The astrocyte-derived vascular permeability enhance factors include Vascular endothelial growth factors, MMPs, Glutamate, Nitric oxide, and Endothelin-1. In contrast, the astrocyte-derived protective factors include Apolipoprotein E, Sonic hedgehog, Angiopoietin-1, Glial-derived neurotrophic factor, Retinoic acid, and Insulin-like growth factor-1 [108,109]. Meanwhile, pericytes are also recognized as key players in BBB genesis and vessel stabilization. Pericytes-derived PDGF-B, Angiopoietin-1, TGF-β, IL-8, MMP2, and MMP9 play critical roles in regulating the BBB permeability [110–113]. These findings have identified multiple potential targets that can be exploited for protection against BBB dysfunction. However, the upstream regulatory mechanisms of these targets remain unclear. Studying these regulatory mechanisms in terms of ncRNAs in BBB permeability will lead to a better understanding of ncRNAs’ roles, which may help prevent and therapy BBB dysfunction and reduce neurological sequelae of CNS diseases.
In the past decade, numerous studies have been published on investigating the roles of miRNAs in CNS diseases. There has been accumulating evidence regarding miRNAs’ crucial roles in BBB permeability as post-transcriptional regulators of gene expression. Elevation of certain miRNAs can cause the degradation of TJs-related mRNAs translation, such as ZO-1, Occludin, β-catenin, VE-cadherin, and Claudins. Meanwhile, suppression of miRNAs can increase several target mRNAs, such as MMP9, Tie-2, VEGFA, Endophilin-1, ANGPT-1, and Neuropilin-1 to affect junction formation and maintenance. Studying the dysregulation and specific function of miRNAs in regulating BBB permeability will help identify new nucleic acid targets for CNS disease control. Although miRNAs as valuable therapeutic targets have been explored extensively, at the time of writing, lncRNAs and circRNAs are emerging regulatory ncRNAs that have been in the spotlight of research so far and are still in their preliminary stages in terms of research. A few studies on lncRNAs and circRNAs regulation of increased BBB permeability still reported the indirect mechanisms mediating through miRNAs. The lncRNAs and circRNAs provide the very stable molecular targets that act as the magnet for the miRNAs, thus hampering the regulatory roles of the specific miRNAs that lncRNAs or circRNAs bind. In this way, more strategies for the protection of BBB permeability through lncRNAs and circRNAs need to be further explored.
So far, ncRNAs researches have significantly advanced our understanding of the mechanisms underlying the BBB dysfunction. Accumulating pieces of evidence have already supported the active roles of ncRNAs regulatory networks in BBB integrity; however, further researches are urgently needed to comprehensively illustrate the molecular details associated with enhancement of BBB permeability, as well as explore new possibilities for drug targets, which shall ultimately lead to better outcomes for patients and animals afflicted with CNS diseases.
Funding Statement
This work was funded by grants from the National Natural Science Foundation of China, China (NSFC) (31772736), the earmarked fund for China Agriculture Research System (CARS-35), the Hubei Postdoctoral Preferential Foundation, China (590320102); and the China Postdoctoral Science Foundation, China (2020M672379).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Abbott NJ, Patabendige AAK, Dolman DEM, et al. Structure and function of the blood–brain barrier. Neurobiol Dis. 2010;37(1):13–25. [DOI] [PubMed] [Google Scholar]
- [2].Burkhart A, Thomsen LB, Thomsen MS, et al. Transfection of brain capillary endothelial cells in primary culture with defined blood–brain barrier properties. Fluids and Barriers of the CNS. 2015;12(1):19. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ballabh P, Braun A, Nedergaard M.. The blood–brain barrier: an overview. Neurobiol Dis. 2004;16(1):1–13. [DOI] [PubMed] [Google Scholar]
- [4].Gonzalez-Mariscal L, Betanzos A, Nava P, et al. Tight junction proteins. Prog Biophys Mol Biol. 2003;81:1–44. [DOI] [PubMed] [Google Scholar]
- [5].Candelario-Jalil E, Yang Y, Rosenberg GA. Diverse roles of matrix metalloproteinases and tissue inhibitors of metalloproteinases in neuroinflammation and cerebral ischemia. Neuroscience. 2009;158:983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Greenwood J. Mechanisms of blood-brain barrier breakdown. Neuroradiology. 1991;33:95–100. [DOI] [PubMed] [Google Scholar]
- [7].Yang R, Liu W, Miao L, et al. Induction of VEGFA and Snail-1 by meningitic Escherichia coli mediates disruption of the blood-brain barrier. Oncotarget. 2016;7:63839–63855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang RC, Qu XY, Xiao SY, et al. Meningitic Escherichia coli-induced upregulation of PDGF-B and ICAM-1 aggravates blood-brain barrier disruption and neuroinflammatory response. J Neuroinflammation. 2019;16:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pan YB, Sun ZL, Feng DF. The role of microRNA in traumatic brain injury. Neuroscience. 2017;367:189–199. [DOI] [PubMed] [Google Scholar]
- [10].Sargento-Freitas J, Aday S, Nunes C, et al. Endothelial progenitor cells enhance blood–brain barrier permeability in subacute stroke. Neurology. 2018;90(2):e127–e34. . [DOI] [PubMed] [Google Scholar]
- [11].Schonrock N, Harvey RP, Mattick JS. Long Noncoding RNAs in cardiac development and pathophysiology. Circ Res. 2012;111(10):1349–1362. [DOI] [PubMed] [Google Scholar]
- [12].Clark MB, Amaral PP, Schlesinger FJ, et al. The reality of pervasive transcription. PLoS Biol. 2011;9(7):e1000625. discussion e1102. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun P, Liu DZ, Jickling GC, et al. MicroRNA-based therapeutics in central nervous system injuries. J Cereb Blood Flow Metab. 2018;38(7):1125–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roberts TC, Morris KV, Wood MJ. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652):20130507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang L, Wang H. Long non-coding RNA in CNS injuries: a new target for therapeutic intervention. Mol Ther Nucleic Acids. 2019;17:754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. [DOI] [PubMed] [Google Scholar]
- [20].Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13(8):528–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Strub GM, Perkins JA. MicroRNAs for the pediatric otolaryngologist. Int J Pediatr Otorhinolaryngol. 2018;112:195–207. [DOI] [PubMed] [Google Scholar]
- [23].Igaz P, Igaz I, Nagy Z, et al. MicroRNAs in adrenal tumors: relevance for pathogenesis, diagnosis, and therapy. Cell Mol Life Sci. 2015;72(3):417–428. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wienholds E, Plasterk RHA. MicroRNA function in animal development. FEBS Letters. 2005;579(26):5911–5922. [DOI] [PubMed] [Google Scholar]
- [25].Reijerkerk A, Lopez-Ramirez MA, van Het Hof B, et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: implications for multiple sclerosis. J Neurosci. 2013;33(16):6857–6863. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang Y, Estrada EY, Thompson JF, et al. Matrix metalloproteinase-mediated disruption of tight junction proteins in cerebral vessels is reversed by synthetic matrix metalloproteinase inhibitor in focal ischemia in rat. J Cereb Blood Flow Metab. 2007;27(4):697–709. [DOI] [PubMed] [Google Scholar]
- [27].Zuo X, Lu J, Manaenko A, et al. MicroRNA-132 attenuates cerebral injury by protecting blood-brain-barrier in MCAO mice. Exp Neurol. 2019;316:12–19. [DOI] [PubMed] [Google Scholar]
- [28].Fan F, Yang J, Xu Y, et al. MiR-539 targets MMP-9 to regulate the permeability of blood–brain barrier in ischemia/reperfusion injury of brain. Neurochem Res. 2018;43(12):2260–2267. [DOI] [PubMed] [Google Scholar]
- [29].Yao X, Wang Y, Zhang D. microRNA-21 Confers Neuroprotection against cerebral ischemia-reperfusion injury and alleviates blood-brain barrier disruption in rats via the MAPK signaling pathway. J Mol Neurosci. 2018;65(1):43–53. [DOI] [PubMed] [Google Scholar]
- [30].Ren X, Engler-Chiurazzi EB, Russell AE, et al. MiR-34a and stroke: assessment of non-modifiable biological risk factors in cerebral ischemia. Neurochem Int. 2019;127:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bukeirat M, Sarkar SN, Hu H, et al. MiR-34a regulates blood-brain barrier permeability and mitochondrial function by targeting cytochrome c. J Cereb Blood Flow Metab. 2016;36:387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hu H, Hone EA, Provencher EAP, et al. MiR-34a interacts with cytochrome c and shapes stroke outcomes. Sci Rep. 2020;10:3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ma Q, Dasgupta C, Li Y, et al. MicroRNA-210 suppresses junction proteins and disrupts blood-brain barrier integrity in neonatal rat hypoxic-ischemic brain injury. Int J Mol Sci. 2017;18:1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pena-Philippides JC, Gardiner AS, Caballero-Garrido E, et al. Inhibition of microRNA-155 supports endothelial tight junction integrity following oxygen-glucose deprivation. J Am Heart Assoc. 2018;7:e009244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Awad H, Bratasz A, Nuovo G, et al. MiR-155 deletion reduces ischemia-induced paralysis in an aortic aneurysm repair mouse model: utility of immunohistochemistry and histopathology in understanding etiology of spinal cord paralysis. Ann Diagn Pathol. 2018;36:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang T, Tian C, Wu J, et al. MicroRNA-182 exacerbates blood-brain barrier (BBB) disruption by downregulating the mTOR/FOXO1 pathway in cerebral ischemia. FASEB J. 2020;34:13762–13775. [DOI] [PubMed] [Google Scholar]
- [37].Wang Y, Wang MD, Xia YP, et al. MicroRNA-130a regulates cerebral ischemia-induced blood-brain barrier permeability by targeting Homeobox A5. FASEB J. 2018;32:935–944. [DOI] [PubMed] [Google Scholar]
- [38].Fang Z, He QW, Li Q, et al. MicroRNA-150 regulates blood-brain barrier permeability via Tie-2 after permanent middle cerebral artery occlusion in rats. FASEB J. 2016;30:2097–2107. [DOI] [PubMed] [Google Scholar]
- [39].Ma F, Sun P, Zhang X, et al. Endothelium-targeted deletion of the miR-15a/16-1 cluster ameliorates blood-brain barrier dysfunction in ischemic stroke. Sci Signal. 2020;13:eaay5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xi T, Jin F, Zhu Y, et al. MicroRNA-126-3p attenuates blood-brain barrier disruption, cerebral edema and neuronal injury following intracerebral hemorrhage by regulating PIK3R2 and Akt. Biochem Biophys Res Commun. 2017;494:144–151. [DOI] [PubMed] [Google Scholar]
- [41].Xi T, Jin F, Zhu Y, et al. MiR-27a-3p protects against blood-brain barrier disruption and brain injury after intracerebral hemorrhage by targeting endothelial aquaporin-11. J Biol Chem. 2018;293:20041–20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Burek M, Konig A, Lang M, et al. Hypoxia-induced microRNA-212/132 alter blood-brain barrier integrity through inhibition of tight junction-associated proteins in human and mouse brain microvascular endothelial cells. Transl Stroke Res. 2019;10:672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ge X, Han Z, Chen F, et al. MiR-21 alleviates secondary blood-brain barrier damage after traumatic brain injury in rats. Brain Res. 2015;1603:150–157. [DOI] [PubMed] [Google Scholar]
- [44].Aung LL, Mouradian MM, Dhib-Jalbut S, et al. MMP-9 expression is increased in B lymphocytes during multiple sclerosis exacerbation and is regulated by microRNA-320a. J Neuroimmunol. 2015;278:185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lopez-Ramirez MA, Wu D, Pryce G, Simpson JE, Reijerkerk A, King-Robson J, et al . MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J. 2014;28:2551–2565. [DOI] [PubMed] [Google Scholar]
- [46].Rom S, Dykstra H, Zuluaga-Ramirez V, et al. MiR-98 and let-7g* protect the blood-brain barrier under neuroinflammatory conditions. J Cereb Blood Flow Metab. 2015;35:1957–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bernstein DL, Rom S. Let-7g* and miR-98 reduce stroke-induced production of proinflammatory cytokines in mouse brain. Front Cell Dev Biol. 2020;8:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Song J, Hu Y, Li H, et al. MiR-1303 regulates BBB permeability and promotes CNS lesions following CA16 infections by directly targeting MMP9. Emerg Microbes Infect. 2018;7:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mishra R, Singh SK. HIV-1 Tat C modulates expression of miRNA-101 to suppress VE-cadherin in human brain microvascular endothelial cells. J Neurosci. 2013;33:5992–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Barker KR, Lu Z, Kim H, et al. MiR-155 modifies inflammation, endothelial activation and blood-brain barrier dysfunction in cerebral malaria. Mol Med. 2017;23:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Liu W, Cai H, Lin M, et al. MicroRNA-107 prevents amyloid-beta induced blood-brain barrier disruption and endothelial cell dysfunction by targeting Endophilin-1. Exp Cell Res. 2016;343:248–257. [DOI] [PubMed] [Google Scholar]
- [52].Lin M, Zhu L, Wang J, et al. MiR-424-5p maybe regulate blood-brain barrier permeability in a model in vitro with Abeta incubated endothelial cells. Biochem Biophys Res Commun. 2019;517:525–531. [DOI] [PubMed] [Google Scholar]
- [53].Toyama K, Spin JM, Deng AC, et al. MicroRNA-mediated therapy modulating blood-brain barrier disruption improves vascular cognitive impairment. Arterioscler Thromb Vasc Biol. 2018;38:1392–1406. [DOI] [PubMed] [Google Scholar]
- [54].Song J, Yoon SR, Kim OY. MiR-Let7a controls the cell death and tight junction density of brain endothelial cells under high glucose condition. Oxid Med Cell Longev. 2017;2017:6051874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yang MC, You FL, Wang Z, et al. Salvianolic acid B improves the disruption of high glucose-mediated brain microvascular endothelial cells via the ROS/HIF-1alpha/VEGF and miR-200b/VEGF signaling pathways. Neurosci Lett. 2016;630:233–240. [DOI] [PubMed] [Google Scholar]
- [56].Kalani A, Kamat PK, Familtseva A, et al. Role of microRNA29b in blood-brain barrier dysfunction during hyperhomocysteinemia: an epigenetic mechanism. J Cereb Blood Flow Metab. 2014;34:1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bai Y, Zhang Y, Hua J, et al. Silencing microRNA-143 protects the integrity of the blood-brain barrier: implications for methamphetamine abuse. Sci Rep. 2016;6:35642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tominaga N, Kosaka N, Ono M, et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 2015;6:6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen SP, Chang YA, Chou CH, et al. Circulating microRNAs associated with reversible cerebral vasoconstriction syndrome. Ann Neurol. 2021;89:459–473. [DOI] [PubMed] [Google Scholar]
- [60].Yang R, Huang F, Fu J, et al. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci Rep. 2016;6:38903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Vemuganti R. All’s well that transcribes well: non-coding RNAs and post-stroke brain damage. Neurochem Int. 2013;63:438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Iyer MK, Niknafs YS, Malik R, et al. The landscape of long non-coding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yao RW, Wang Y, Chen LL. Cellular functions of long non-coding RNAs. Nat Cell Biol. 2019;21:542–551. [DOI] [PubMed] [Google Scholar]
- [64].Rinn JL, Chang HY. Genome regulation by long non-coding RNAs. Annu Rev Biochem. 2012;81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Anfossi S, Babayan A, Pantel K, et al. Clinical utility of circulating non-coding RNAs-an update. Nat Rev Clin Oncol. 2018;15:541–563. [DOI] [PubMed] [Google Scholar]
- [66].Peng H, Li H. The encouraging role of long non-coding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol Carcinog. 2019;58:1465–1480. [DOI] [PubMed] [Google Scholar]
- [67].Yin KJ, Hamblin M, Chen YE. Non-coding RNAs in cerebral endothelial pathophysiology: emerging roles in stroke. Neurochem Int. 2014;77:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li K, Blum Y, Verma A, et al. A non-coding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou W, Chen C, Shi Y, et al. Targeting glioma stem cell-derived pericytes disrupts the blood-tumor barrier and improves chemotherapeutic efficacy. Cell Stem Cell. 2017;21:591–603 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sun Y, Ma L. New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel). 2019;11:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ma J, Wang P, Yao Y, et al. Knockdown of long non-coding RNA MALAT1 increases the blood-tumor barrier permeability by up-regulating miR-140. Biochim Biophys Acta. 2016;1859:324–338. [DOI] [PubMed] [Google Scholar]
- [72].Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Guo J, Cai H, Zheng J, et al. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, Occludin, and claudin-5. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2240–2254. [DOI] [PubMed] [Google Scholar]
- [74].Guo J, Shen S, Liu X, et al. Role of linc00174/miR-138-5p (miR-150-5p)/FOSL2 feedback loop on regulating the blood-tumor barrier permeability. Mol Ther Nucleic Acids. 2019;18:1072–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Young TL, Matsuda T, Cepko CL. The non-coding RNA taurine up-regulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501–512. [DOI] [PubMed] [Google Scholar]
- [76].Cai H, Xue Y, Wang P, et al. The long non-coding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget. 2015;6:19759–19779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ma SC, Li Q, Peng JY, et al. CLDN5 affects lncRNAs acting as ceRNA dynamics contributing to regulating blood brain barrier permeability in tumor brain metastasis. Oncol Rep. 2018;39:1441–1453. [DOI] [PubMed] [Google Scholar]
- [78].Zhu L, Lin M, Ma J, et al. The role of LINC00094/miR-224-5p (miR-497-5p)/Endophilin-1 axis in memantine mediated protective effects on blood-brain barrier in AD microenvironment. J Cell Mol Med. 2019;23:3280–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Leng X, Ma J, Liu Y, et al. Mechanism of piR-DQ590027/MIR17HG regulating the permeability of glioma conditioned normal BBB. J Exp Clin Cancer Res. 2018;37:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhang J, Dong B, Hao J, et al. LncRNA Snhg3 contributes to dysfunction of cerebral microvascular cells in intracerebral hemorrhage rats by activating the TWEAK/Fn14/STAT3 pathway. Life Sci. 2019;237:116929. [DOI] [PubMed] [Google Scholar]
- [81].Zhang ZG, Zhang L, Jiang Q, et al. VEGF enhances angiogenesis and promotes blood-brain barrier leakage in the ischemic brain. J Clin Invest. 2000;106:829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ren L, Wei C, Li K, et al. LncRNA MALAT1 up-regulates VEGF-A and ANGPT2 to promote angiogenesis in brain microvascular endothelial cells against oxygen-glucose deprivation via targetting miR-145. Biosci Rep. 2019;39:BSR20180226. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [83].Zhang L, Luo X, Chen F, et al. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem. 2018;119:5460–5472. [DOI] [PubMed] [Google Scholar]
- [84].Wang Z, Wang R, Wang K, et al. Up-regulated long non-coding RNA Snhg1 promotes the angiogenesis of brain microvascular endothelial cells after oxygen-glucose deprivation treatment by targeting miR-199a. Can J Physiol Pharmacol. 2018;96:909–915. [DOI] [PubMed] [Google Scholar]
- [85].Wu L, Ye Z, Pan Y, et al. Vascular endothelial growth factor aggravates cerebral ischemia and reperfusion-induced blood-brain-barrier disruption through regulating LOC102640519/HOXC13/ZO-1 signaling. Exp Cell Res. 2018;369:275–283. [DOI] [PubMed] [Google Scholar]
- [86].Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. [DOI] [PubMed] [Google Scholar]
- [87].Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- [91].Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- [92].Filippenkov IB, Kalinichenko EO, Limborska SA, et al. Circular RNAs-one of the enigmas of the brain. Neurogenetics. 2017;18:1–6. [DOI] [PubMed] [Google Scholar]
- [93].Gao Y, Ma H, Lv C, et al. Exosomes and exosomal microRNA in non-targeted radiation bystander and abscopal effects in the central nervous system. Cancer Lett. 2021;499:73–84. [DOI] [PubMed] [Google Scholar]
- [94].Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. 2017;14:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of non-coding RNA with novel functions. Exp Biol Med (Maywood). 2017;242:1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhang S, Zhu D, Li H, et al. Characterization of circRNA-associated-ceRNA networks in a senescence-accelerated mouse prone 8 brain. Mol Ther. 2017;25:2053–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lin SP, Ye S, Long Y, et al. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. 2016;471:52–56. [DOI] [PubMed] [Google Scholar]
- [100].Yang R, Xu B, Yang B, et al. Circular RNA transcriptomic analysis of primary human brain microvascular endothelial cells infected with meningitic Escherichia coli. Mol Ther Nucleic Acids. 2018;13:651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kumar L, Shamsuzzama S, Jadiya P, et al. Functional characterization of novel circular RNA molecule, circzip-2 and its synthesizing gene zip-2 in C. elegans model of Parkinson’s disease. Mol Neurobiol. 2018;55(8):6914–6926. [DOI] [PubMed] [Google Scholar]
- [102].Liu W, Jia C, Luo L, et al. Novel circular RNAs expressed in brain microvascular endothelial cells after oxygen-glucose deprivation/recovery. Neural Regen Res. 2019;14(12):2104–2111. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wu P, Gao Y, Shen S, et al. KHDRBS3 regulates the permeability of blood–tumor barrier via cDENND4C/miR-577 axis. Cell Death Dis. 2019;10(7):536. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Yang L, Han B, Zhang Y, et al. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14(3):404–418. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bai Y, Zhang Y, Han B, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood–brain barrier integrity. J Neurosci. 2018;38(1):32–50. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yang R, Chen J, Xu B, et al. Circ_2858 helps blood-brain barrier disruption by increasing VEGFA via sponging miR-93-5p during Escherichia coli meningitis. Mol Ther Nucleic Acids. 2020;22:708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Mone P, Gambardella J, Wang X, et al. MiR-24 targets the transmembrane glycoprotein Neuropilin-1 in human brain microvascular endothelial cells. Noncoding RNA. 2021;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Morad G, Daisy CC, Otu HH, et al. Cdc42-dependent transfer of mir301 from breast cancer-derived extracellular vesicles regulates the matrix modulating ability of astrocytes at the blood–brain barrier. Int J Mol Sci. 2020;21(11):3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Michinaga S, Koyama Y. Dual roles of astrocyte-derived factors in regulation of blood-brain barrier function after brain damage. Int J Mol Sci. 2019;20(3):571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Liu Q, Yang Y, Fan X. Microvascular pericytes in brain-associated vascular disease. Biomed Pharmacother. 2020;121:109633. [DOI] [PubMed] [Google Scholar]
- [111].Hellstrom M, Gerhardt H, Kalen M, et al. Lack of pericytes leads to endothelial hyperplasia and abnormal vascular morphogenesis. J Cell Biol. 2001;153(3):543–554. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Underly RG, Levy M, Hartmann DA, et al. Pericytes as inducers of rapid, matrix metalloproteinase-9-dependent capillary damage during ischemia. J Neurosci. 2017;37(1):129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Winkler EA, Bell RD, Zlokovic BV. Central nervous system pericytes in health and disease. Nat Neurosci. 2011;14(11):1398–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]


