ABSTRACT
Colorectal cancer (CRC) is one of the leading causes of cancer-related deaths worldwide. Long non-coding RNAs (lncRNAs) have been increasingly reported to serve vital parts in malignancies including CRC. Although cancer susceptibility 21 (CASC21) has been uncovered to play a part in CRC, its mechanism still needs further explanation. Thus, our study aimed to further explore the influence and mechanism of CASC21 in CRC progression. Quantitative real-time RT-PCR and western blot were performed to detect gene expression; a series of functional assays were performed to investigate the effect of CASC21 on CRC cells; in vivo tumour growth was evaluated via the nude mice xenograft model. The results revealed that CASC21 facilitated CRC cell proliferation, migration, epithelial-mesenchymal transition (EMT) and stemness. In addition, CASC21 was co-expressed with and bound to transcription factor POU5F1B (POU class 5 homeobox 1B). CASC21 recruited POU5F1B to HGH1 promoter to activate the transcription of HGH1 homolog. Also, CASC21 served as a competitive endogenous RNA (ceRNA) to up-regulate HGH1 via endogenously sponging miR-485-5p. Moreover, HGH1 overexpression counteracted the suppression of CASC21 deficiency on CRC tumour growth. In summary, our study indicated that CASC21 enhanced the expression of HGH1 to promote the malignancy of CRC by recruiting POU5F1B and sponging miR-485-5p, suggesting a key role of CASC21 in CRC progression.
KEYWORDS: Colorectal cancer, CASC21, POU5F1B, HGH1, miR-485-5p
Graphical Abstract
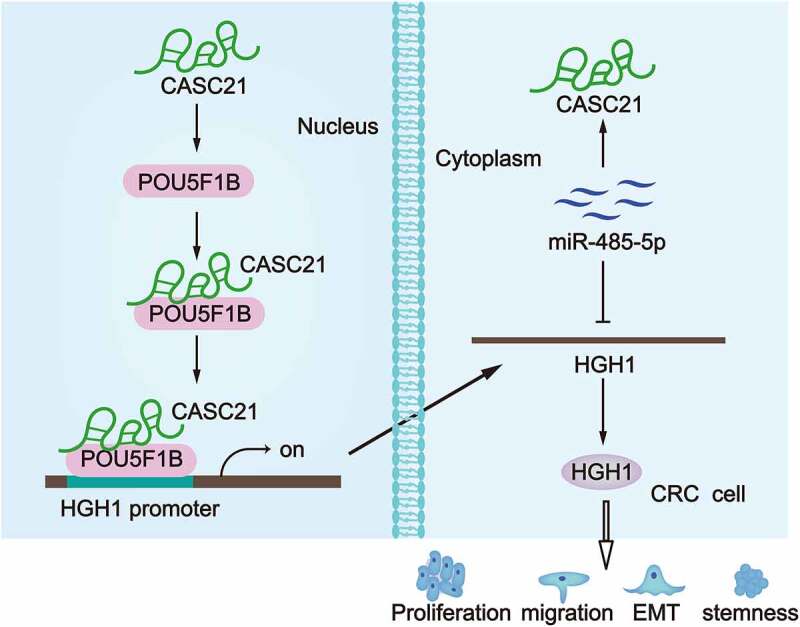
Introduction
Colorectal cancer (CRC) is one of the most prevalent cancers worldwide and takes up 8% of cancer-related deaths in 2018 [1]. The 5-year survival rates of CRC patients with primary tumours are about 90%; for patients with lymph metastasis or distal metastasis, the 5-year survival rates are respectively as low as 70.4% or even 12.5% [2]. Despite the improvement of traditional therapeutic methods for CRC, the efficacy of current therapies remains disappointing in CRC patients at advanced stage [3]. Thus, it is urgently in need to strengthen the researches on CRC to promote the treatment for CRC.
LncRNAs refer to a group of non-coding RNAs with the length more than 200 nucleotides and have been verified to exert crucial roles in tumour progression. Importantly, lncRNAs exert their functions by means of various mechanisms in cancers, CRC included. For instance, lncRNA RPPH1 interacts with TUBB3 and promotes the polarization of macrophage M2 to facilitate CRC metastasis [4]. LncRNA GAS5 is negatively modulated by m6A reader YTHDF3 and hinders CRC progression through activating YAP phosphorylation and degradation [5]. LncRNA ITIH4-AS1 accelerates CRC tumorigenesis by triggering JAK/STAT3 pathway at a FUS-dependent way [6]. LncRNA SATB2-AS1 inhibits CRC tumour metastasis by directly binding to WDR5 and GADD45A, cis-activating SATB2 transcription via mediating H3K4me3 deposition and DNA demethylation of SATB2 promoter [7].
Recently, a growing number of reports have verified the existence and function of ceRNA pattern in diverse cancers. CeRNA pattern is a typical post-transcription mechanism in which lncRNAs competitively bind to the complementary seed regions of microRNAs (miRNAs) to free message RNAs (mRNAs) from miRNA-caused inhibition. The ceRNA pattern is commonly reported in CRC development. For instance, lncRNA SLCO4A1-AS1 enhances autophagy through acting as an endogenous sponge of miR-508-3p to elevate PARD3 expression, thus facilitating CRC cell progression [8]. SP1-activated up-regulation of lncRNA TINCR contributes to CRC progression by sponging miR-7-5p to induce PI3K/AKT/mTOR pathway [9]. MALAT1 accelerates the invasion and metastasis of CRC via sponging miR-106b-5p to enhance SLAIN2-mediated microtubules mobility [10]. The up-regulation of lncRNA XIST is induced by CXCL12/CXCR4 and functions as a ceRNA to sponge miR-133a-3p, therefore alleviating the inhibitory effect of miR-133a-3p on RhoA in CRC progression [11].
Moreover, lncRNAs can bind to specific proteins and pose an indirect regulatory effect on target genes. For instance, lncRNA RMST binds to HuR protein to enhance DNMT3 expression [12]. LncRNA 00324 binds to HuR to stabilize FAM83B, thereby promoting gastric cancer cell proliferation [13]. LncRNA FALEC recruits EZH2 at the promoter regions of ECM1 to epigenetically suppress ECM1, thus inhibiting cell proliferation in tongue squamous cell carcinoma [14]. SATB2-AS1 suppresses CRC development via serving as a scaffold to recruit p300 at the SATB2 promoter regions to elevate SATB2 expression [15]. LncRNA AFAP1-AS1 recruits EZH2 to the promoter of p21 and epigenetically represses p21 expression, thus facilitating cell proliferation in non-small cell lung cancer [16].
CASC21 has been reported to be integrated by HPV in cervical cancer tissues [17], while its mechanism on CRC has been scarcely reported. For example, CASC21 exerts an oncogenic role in colon cancer through regulating miR-7-5p/YAP1 axis [18], and it promotes colorectal cancer growth by regulating CDK6 [19]. What aroused our interest was that CASC21 is significantly up-regulated in colon adenocarcinoma based on GEPIA database. Besides, although CASC21 has been discovered to function in CRC, its mechanism was not substantially explored. Thus, present study was concentrated on CASC21, trying to probe into its function and underlying mechanisms in CRC.
Materials and methods
Patient samples
The paired CRC tumour tissue and adjacent non-tumour tissue samples were collected from 80 CRC patients after surgery, with the approval from the Ethical Committee of Changhai Hospital. All patents had signed the written informed consent. Tissues were preserved instantly in liquid nitrogen at −80°C.
Cell lines and culture
Human CRC cell lines (HCT116, SW620, RKO, SW116) and normal colorectal mucosal cell line (FHC), which were obtained from ATCC (Rockville, Maryland), were all maintained in DMEM (Invitrogen, Carlsbad, CA) under 5% CO2 at 37°C. 10% FBS and 1% pen/strep solution were required for cell culture.
Quantitative real-time RT-PCR (RT-qPCR)
Total RNAs from tissue samples and cell samples were extracted with Invitrogen TRIzol reagent for cDNA synthesis as required by supplier (Thermo Fisher, Waltham, MA). To measure gene expression, SYBR green Supermix (Thermo Fisher) was used for qPCR, following the 2−ΔΔCt method. GAPDH or U6 served as normalization. Related sequences of primers are provided in Supplementary Table 1.
Subcellular fractionation
1 × 106 SW620 and RKO cells were rinsed in PBS and centrifuged for 5 min, and the supernatant was collected. The cell nucleus and cell cytoplasm were finally separated using PARIS™ Kit (Invitrogen). RT-qPCR was performed for quantifying CASC21, with GADPH and U6 as cytoplasmic and nuclear controls, respectively.
Fluorescence in situ hybridization (FISH) assay
The CASC21-FISH probe (Ribobio, Guangzhou, China) was used as per direction and FISH assay was performed using Fluorescent in Situ Hybridization Kits according to the manufacturer’s requirements. SW620 and RKO cells were fixed by 4% formaldehyde, dehydrated, and then cultured with in hybridization buffer. DAPI was used for nuclear counterstaining. Finally, the images were photographed by using a fluorescence microscope.
Cell transfection
The designed shRNAs and NC-shRNAs were available from Genepharma (Shanghai, China) to silence CASC21, POU5F1B and HGH1 in SW620 and RKO cells employing the Lipofectamine 2000 (Invitrogen). In addition, the pcDNA3.1-HGH1, pcDNA3.1-CASC21, pcDNA3.1-POU5F1B and NC-pcDNA3.1 vectors, as well as miR-485-5p-mimics and NC mimics were all designed at Genepharma. Plasmid transfection was performed for 48 h. Sequences of the transfection plasmids are provided in Supplementary Table 2.
Colony formation assay
800 SW620 and RKO cells were cultured for 14 days in the 6-well plate. The culture medium was discarded and the cells were washed with PBS for two times. Cells were fixed in 4% formaldehyde and stained with 0.1% crystal violet for observation. Finally, the number of colonies was measured manually.
5-ethynyl-20-deoxyuridine (EdU) incorporation assay
After the 48-hour plasmid transfection, transfected SW620 and RKO cells were incubated in 96-well plate for EdU staining assay as guided by the supplier (Ribobio). Cell nuclei were detected by DAPI staining. Samples were observed by fluorescent microscope and cell proliferation was then measured by calculating the EdU-positive cells.
Transwell migration assay
Cells suspended in serum-free DMEM were seeded to the upper chamber of 8-mm pore size Transwell inserts (Corning Inc., Corning, NY), with lower chamber added with complete DMEM. Twenty-four hours later, cells in the upper layer were removed with caution by a cotton swab and then fixed in methanol solution for 15 min. 0.1% crystal violet was adopted to stain the membranes for 10 min, and then the migrated cells were observed and counted under a microscope (10 × 10).
Western blot assay
Cell protein samples were extracted for separation on the 12% SDS-PAGE, then transferred to PVDF membranes and sealed with 5% non-fat milk. The membranes were cultivated with primary antibodies over night at 4°C, followed by being cultivated with secondary antibody for 1 h. After washing in TBST, the secondary antibodies were added. Primary antibodies against POU5F1B, HGH1 and the loading control GAPDH, as well as HRP-tagged secondary antibodies were produced by Abcam (Cambridge, MA) and utilized after dilution. Blots were finally analysed by enhanced chemiluminescence (ECL) detection system.
Immunofluorescence (IF) assay
Cells were grown on coverslips for IF assay, fixed by 4% paraformaldehyde and blocked by 3% BSA. The coverslips were then probed with primary antibodies (Abcam) against E-cadherin and N-cadherin overnight at 4°C. Fluorescence-conjugated secondary antibodies were added for 2 h at room temperature, and cell nucleus was processed with DAPI for 5 min.
Sphere formation assay
Cells were seeded in the 96-well ultralow attachment plates adding sphere medium for 1 week, following the user manual (Corning Inc.). Cell clusters with diameter >50 mm were counted.
RNA immunoprecipitation (RIP) assay
Using Magna RIP™ RNA Binding Protein Immunoprecipitation Kit, RIP assay in SW620 and RKO cells was conducted as per manual (Millipore, Bedford, MA). Cell lysates from RIP lysis buffer were incubated with the magnetic beads conjugated with the AGO2 antibody or IgG antibody (negative control). Antibodies against human AGO2 (ab186733, Abcam) and negative control IgG (#3900, Cell Signalling) were procured from Millipore. Precipitated RNAs were analysed via RT-qPCR.
RNA pull down assay
For RNA pull down assay, protein extracts from SW620 and RKO cells were mixed with streptavidin magnetic beads bound to the biotinylated probes for CASC21 or miR-485-5p and their relative control probes. For DNA pull down, the biotin-labelled HGH1 promoter was bound to streptavidin magnetic beads and mixed with protein extracts. No-biotin HGH1 promoter served as control.
Chromatin immunoprecipitation (ChIP) assay
Formaldehyde cross-linked chromatin samples were acquired from CRC cells and fragmented for immunoprecipitation with POU5F1B antibody (ab230429, Abcam) or negative control IgG antibody (#3900, Cell Signalling). DNA enrichment was monitored by RT-qPCR.
Luciferase reporter assay
For promoter assay, SW620 and RKO cells were co-transfected in 24-well plate with the pGL3-HGH1 promoter, the pRL-TK-Renilla and indicated transfection plasmids for 48 h. In addition, cells were co-transfected with the constructed pmirGLO reporter vectors including CASC21-WT/MUT and HGH1-WT/MUT, and miR-485-5p mimics or NC mimics. Luciferase Reporter Assay System (Promega, Madison, WI) was applied for all luciferase activities.
In vivo tumour growth assay
Animal study was approved by the Institutional Animal Care and Use Committee (IACUC) of Changhai Hospital. 6-week old BALB/c nude mice were randomly applied for subcutaneous injection with 2 × 106 transfected SW620 cells. For each group, 3 mice were used and a total of 27 mice were used for in vivo assay. Mice were sacrificed on day 28 by cervical dislocation, and tumours were carefully excised and weighed. Tumour volume was calculated as length × width2/2.
Immunohistochemistry (IHC) assay
The 4-μm-thick sections were acquired after cutting the paraffin-embedded tumours from in vivo assay. After deparaffinization and rehydration, sections were probed with primary antibodies against PCNA and Ki-67 overnight, then with secondary antibody for 1 h.
Statistical analysis
All experimental results of independent bio-triplicates were displayed with the mean ± standard deviation (S.D.), and were processed by Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA). Kaplan-Meier analysis and log-rank test were used to figure out correlation between gene expression and overall survival of CRC patients. Pearson’s correlation analysis was used to calculate correlation between gene expressions in CRC samples. Statistical analyses were implemented in form of Student’s t-test or one-way ANOVA, with the threshold set as p-value < 0.05.
Results
CASC21 expression was up-regulated in CRC tissues and cells
To verify whether CASC21 played a role in CRC, we detected CASC21 expression in 80 pairs of CRC tumour tissues and adjacent non-tumour tissues. The results revealed that CASC21 was remarkably up-regulated in CRC samples in comparison to the adjacent non-tumour ones (Fig. 1(a)). Also, CRC samples were divided into low and high CASC21 expression group. Based on Kaplan-Meier analysis, we found the overall survival rate of CRC patients was low in CRC samples of high CASC21 expression group (Figure S1A). Next, CASC21 expression in CRC cell lines (HCT116, SW620, RKO, SW116) and normal colorectal mucosal FHC cell line was examined by RT-qPCR. Consequently, CASC21 expression level was much higher in CRC cells compared with normal FHC cells (Fig. 1(b)). As the expression of CASC21 in SW620 and RKO cells were the highest, we used these two cell lines in follow-up assays. Additionally, due to the fact that the subcellular location of lncRNAs largely determines their regulatory roles in cancer cells, we subsequently explored the subcellular location of CASC21. The prediction of LncLocator database [20] demonstrated that CASC21 was distributed in both nucleus and cytoplasm (Fig. 1(c)). According to the results of nucleus/cytoplasm fraction and FISH assays, we also proved the location of CASC21 in both the nucleus and cytoplasm of CRC cells (Fig. 1(d7#x002D;e)). Based on these data, we concluded that CASC21 was up-regulated in CRC tissues and cells, and it might have different functions in cytoplasm and nucleus in CRC.
Figure 1.

CASC21 expression was up-regulated in CRC tissues and cells
a. CASC21 expression in 80 pairs of CRC tumour tissues and adjacent non-tumour tissues was evaluated via RT-qPCR. b. CASC21 expression in CRC cell lines and normal FHC cells was detected by RT-qPCR. c. LncLocator database was applied to predict the subcellular location of CASC21. d-e. Nucleus/cytoplasm fraction and FISH assays (scar bar: 20 μm) were carried out to detect the subcellular location of CASC21 in SW620 and RKO cells. *P < 0.05, **P < 0.01.
CASC21 inhibition suppressed CRC cell proliferation, migration, EMT and stemness
Functional assays were subsequently conducted to explore the function of CASC21 in CRC cells. CASC21 was first knocked down by shRNAs and it was verified by RT-qPCR that CASC21 was successfully downregulated in SW620 and RKO cells by sh-CASC21#1/2 (Figure S1B). Through colony formation and EdU assays, we discovered that CASC21 depletion caused a remarkable decrease of CRC cell proliferation (Fig. 2(a-b)). Then, transwell assay was conducted to examine cell migration after CASC21 was knocked down in CRC cells. Result revealed that the migratory ability of SW620 and RKO cells was restrained under CASC21 depletion (Fig. 2(c)). Furthermore, we detected the impact of CASC21 on EMT, a hallmark of metastasis. Hence, the positivity of two EMT-related proteins, E-cadherin and N-cadherin, was estimated by IF assay. It was revealed that E-cadherin positivity was enhanced while N-cadherin positivity was weakened in SW620 and RKO cells after the transfection of sh-CASC21#1/2, indicating that CASC21 silencing suppressed the EMT process in CRC (Fig. 2(d), Figure S1C). Moreover, we explored the influence of CASC21 on cell stemness. Experimental result revealed that the sphere formation efficiency of CRC cells was evidently reduced in response to CASC21 knockdown, indicating that CASC21 knockdown repressed the stemness of CRC cells (Fig. 2(e)). Thus, we concluded that CASC21 accelerated CRC cell proliferation, migration, EMT and stemness.
Figure 2.

CASC21 inhibition suppressed CRC cell proliferation, migration, EMT and stemness
a-b. Colony formation and EdU assays (scar bar: 100 μm) were conducted to evaluate CRC cell proliferation ability after the depletion of CASC21. c.Transwell assay (scar bar: 100 μm) was carried out to examine the number of migrated CRC cells with CASC21 inhibition. d. IF staining (scar bar: 10 μm) was applied to analyse E-cadherin and E-cadherin positivity in CRC cells upon CASC21 silencing. e.Sphere formation assay (scar bar: 50 μm) was carried out to examine the sphere formation efficiency of CRC cells after silencing CASC21. **P < 0.01.
CASC21 recruited POU5F1B to synergistically up-regulate HGH1
According to UCSC genome browser (http://genome.ucsc.edu/), POU5F1B is found to be the nearby gene of CASC21. Also, based on data processing in GEPIA database (http://gepia.cancer-pku.cn/), POU5F1B was positively co-expressed with CASC21 in CRC samples (Fig. 3(a)). In addition, we unveiled that POU5F1B expression was significantly increased in CRC tissues compared with that in the adjacent non-tumour cells (Fig. 3(b)). Also, it was verified through RT-qPCR that POU5F1B was with higher expression in CRC cells than in normal colorectal mucosal FHC cell line (Fig. 3(c)). Of note, the data from RPISeq (RNA-Protein Interaction Prediction) (http://pridb.gdcb.iastate.edu/RPISeq/) prediction displayed a high possibility of the interaction between CASC21 and POU5F1B protein (Prediction using RF classifier: 0.85; Prediction using SVM classifier: 0.81) (Fig. 3(d)). Next, we conducted RNA pull down assay and found that POU5F1B could be pulled down by biotin-labelled CASC21. Moreover, it was found that biotin-labelled PCAT2 (a lncRNA with the similar length of CASC21) could not pull down POU5F1B (Fig. 3(e)). Furthermore, the results of RIP assay revealed that CASC21 was abundantly enriched by anti-POU5F1B (Fig. 3(f)). For the following assays, POU5F1B expression was silenced in CRC cells and the interference efficiency of POU5F1B was examined by RT-qPCR (Figure S1D). Then, RT-qPCR was conducted to examine the expression of POU5F1B upon CASC21 knockdown. We figured out that CASC21 knockdown could not impact POU5F1B at neither mRNA nor protein level, and POU5F1B also had no influence on CASC21 expression level (Fig. 3(g-h), Figure S1E).
Figure 3.

CASC21 bound to POU5F1B to synergistically up-regulate HGH1
a.CASC21 was co-expressed with POU5F1B based on GEPIA database.b-c.POU5F1B expression in CRC tissues versus adjacent normal tissue and in CRC cell lines versus normal FHC cells was evaluated through RT-qPCR.d.A high possibility of the interaction between CASC21 and POU5F1B protein was predicted via RPISeq based on the Random Forest (RF) and Support Vector Machine (SVM) classifiers. e.RNA pull down and western blot was conducted to verify the binding ability between POU5F1B and CASC21, antisense CASC21 and bio-PCAT2 serving as negative control. f.RIP assay was carried out to examine CASC21 enrichment in anti-IgG and anti-POU5F1B groups. g. RT-qPCR and western blot were conducted to detect the influence of CASC21 depletion on POU5F1B expression. h. RT-qPCR was used to detect the influence of POU5F1B depletion on CASC21 expression. i. Six genes were predicted to be co-expressed with both CASC21 and POU5F1B. j-k. Influence of respective or combined knockdown of CASC21 and POU5F1B depletion on the expression and protein levels of HGH1 was tested by RT-qPCR and western blot. *P < 0.05, **P < 0.01. The symbol ‘n.s’. means no significance.
After that, we decided to identify the downstream target which was regulated by both POU5F1B and CASC21. As was predicted from GEPIA database, six genes (CSE1L, TOP1MT, PSMA7, DCAF13, BOP1 and HGH1) were found to be co-expressed with both POU5F1B and CASC21 (Fig. 3(i)). RT-qPCR was then conducted to examine the expression of the above genes in CRC cells, and it was revealed that only HGH1 was remarkably up-regulated in both SW620 and RKO cells compared to normal FHC cell line (Figure S1F). In addition, HGH1 was also found to be up-regulated in colon adenocarcinoma and rectum adenocarcinoma according to GEPIA database (Figure S1G). Therefore, we speculated that HGH1 was the target for POU5F1B and CASC21 in CRC. Then, we found that CASC21 knockdown reduced HGH1 level in CRC cells (Fig. 3(j)). Transfection of sh-POU5F1B#1 also decreased HGH1 level in CRC cells, however, co-transfection of sh-CASC21#1/2 failed to cause a more significant decrease in HGH1 level in CRC cells (Fig. 3(k)). We deduced that this may be because sh-POU5F1B#1/2 has already brought HGH1 level down to a large extent, co-transfection of sh-CASC21#1/2 may also decrease HGH1 level but the effect was not significant due to the already low HGH1 expression caused by sh-POU3F1B#1/2 in CRC.
After that, the interference efficiency of HGH1 in CRC cells was confirmed via RT-qPCR (Figure S2A), and then a series of functional assays were implemented to examine the role of HGH1 in CRC cells. As a result, we uncovered that HGH1 knockdown inhibited CRC cell proliferation, migration, EMT and stemness (Figure S2B-F). Also, HGH1 expression was up-regulated in CRC cells by pcDNA3.1-HGH1 (Figure S3A). And the effects of HGH1 overexpression on the biological behaviours of CRC cells were explored through related functional assays. Results showed that HGH1 overexpression accelerated CRC cell proliferation and EMT process in vitro (Figure S3B-E). Also, CRC cells with HGH overexpression generate tumours with greater volume and weight at a faster speed in vivo (Figure S3F-H), and CRC tumours in mice with HGH1 overexpression were confirmed to contain higher HGH1 level (Figure S3I). Besides, HGH1 overexpression increased E-cadherin and decreased N-cadherin in CRC cells in vitro (Figure S3J-K). According to these findings, we drew the conclusion that CASC21 could bind to POU5F1B to synergistically up-regulate HGH1 in CRC cells.
CASC21 recruited POU5F1B to activate the transcription of HGH1
We went on to explore the regulatory mechanism underlying the regulation of CASC21/POU5F1B on HGH1. HGH1 was shown to be up-regulated in CRC tissues relative to adjacent non-tumour tissues (Fig. 4(a)). Moreover, it was confirmed through Pearson’s correlation analysis that there was a positive correlation between CASC21/POU5F1B expression and HGH1 expression (Fig. 4(b)). Then, we upregulated the expression of CASC21 and POU5F1B in CRC cells and used RT-qPCR to examine their overexpression efficiency respectively (Figure S4A). After that, it was examined through RT-qPCR that up-regulation of CASC21 or POU5F1B induced a remarkable increase on HGH1 expression (Fig. 4(c)). Since POU5F1B is a transcription factor, we then speculated that CASC21-POU5F1B complex affected HGH1 transcription in CRC. Using JASPAR (http://jaspar.genereg.net/), we compared the DNA motif of POU5F1B and HGH1 promoter sequence, finding two potential binding sites of POU5F1B on HGH1 promoter (Fig. 4(d)). Next, the results of DNA pull down assay identified that POU5F1B could be significantly pulled down by biotin-labelled HGH1 promoter. Additionally, as PCAT2 is a lncRNA similar to CASC21 in length, we also conducted pull down assay using biotin PCAT2 promoter, and it was found that POU5F1B could not be pulled down by biotin-labelled PCAT2 promoter (Fig. 4(e)). The following ChIP assay results further verified the binding between POU5F1B and HGH1 promoter (Fig. 4(f)). Furthermore, we verified HGH1 promoter was less enriched in ChIP products for anti-POU5F1B in CRC cells after the transfection with sh-CASC21#1 and sh-CASC21#2, which showed that CASC21 knockdown reduced the binding of POU5F1B to HGH1 promoter (Figure S4B). Moreover, as shown in Fig. 4(g), luciferase reporter assay revealed that CASC21 or POU5F1B knockdown reduced the luciferase activity of HGH1 promoter whereas the up-regulation of CASC21 or POU5F1B exerted the opposite effects. Importantly, double knockdown of CASC21 and POU5F1B failed to exert more significant inhibitory effect on HGH1 transcription activity compared with single knockdown of CASC21 or POU5F1B. However, co-overexpression of CASC21 and POU5F1B up-regulation could exert more significant promoting effect on HGH1 transcription than compared with single overexpression of CASC21 or POU5F1B. Afterwards, we mutated the binding sites of POU5F1B on HGH1 promoter. The follow-up luciferase reporter assay results depicted that the luciferase activity of wild-type HGH1 promoter was enhanced by POU5F1B overexpression; the luciferase activity of Site 1-mutated HGH1 promoter was slightly enhanced by POU5F1B overexpression, but that of HGH1 promoter with mutant Site 1/2 was not altered under POU5F1B up-regulation (Fig. 4(h)), which suggested that POU5F1B bound to HGH1 promoter at both the two predicted sites. Based on the abovementioned results, we concluded that CASC21 cooperated with POU5F1B to activate HGH1 transcription in CRC.
Figure 4.

CASC21 cooperated with POU5F1B to activate the transcription of HGH1
a. RT-PCR was used to examine HGH1 expression in 80 pairs of CRC tissues and adjacent non-tumour tissues. b. Pearson’s correlation analysis was carried out to examine the correlation between HGH1 expression and CASC21/POU5F1B expression in CRC tumours. c. RT-qPCR was used to examine the influence of CASC21/POU5F1B overexpression on HGH1 expression. d. DNA motif of POU5F1B and binding sites of POU5F1B on HGH1 promoter were predicted by JASPAR. e. Pull down assay followed by western blot was carried out to verify the interaction between POU5F1B and HGH1 promoter, biotin PCAT2 promoter acting as negative control. f. ChIP assay was conducted to verify the binding between POU5F1B and HGH1 promoter. g-h. Luciferase reporter assay was conducted to assess the luciferase activity of indicated HGH1 promoter under different transfections. *P < 0.05, **P < 0.01.
CASC21 sponged miR-485-5p to elevate HGH1
Since CASC21 was also distributed in the cytoplasm of CRC cells, we sought to explore whether CASC21 regulated HGH1 expression at post-transcriptional level. As ceRNA is a typical post transcriptional pattern, we aimed at verifying whether CASC21 served as a ceRNA to induce HGH1 level in CRC. As revealed by RIP assay, CASC21 and HGH1 were both highly enriched by AGO2 in RISC (Figure S4C), indicating the potential ceRNA pattern involving CASC21 and HGH1 in CRC cells. Based on the prediction of both ENCORI [21] and miRDB [22], miR-485-5p was identified as the specific miRNA that bound to both CASC21 and HGH1 (Fig. 5(a)). Then, miR-485-5p-mimics were transfected into CRC cells and its overexpression efficiency was verified by RT-qPCR (Figure S4D). We figured out that miR-485-5p overexpression caused a decrease in HGH1 expression in both SW620 and RKO cells (Fig. 5(b)). However, miR-485-5p mimics failed to alter CASC21 level and sh-CASC21#1/2 failed to alter mR-485-5p level (Figure S4E-F), further implying that CASC21 and miR-485-5p formed a ceRNA network instead of a regulatory relation of each other’s expression in CRC cells. Also, miR-485-5p was unveiled to be significantly down-regulated in CRC tissues and cells (Fig. 5(c-d)). Pearson’s correlation analysis depicted the negative correlation between miR-485-5p expression and CASC21/HGH1 expression (Fig. 5(e)). Next, the binding sequences in CASC21/HGH1 for miR-485-5p were predicted in ENCORI database (Fig. 5(f)). We mutated the binding sequences to explore whether these sites were responsible for the interactions. As unveiled by luciferase reporter assay, the luciferase activities of CASC21-WT and HGH1-WT were decreased by miR-485-5p overexpression, whereas that of CASC21-MUT and HGH1-MUT were not apparently altered (Fig. 5(g)). Moreover, the results of RNA pull down assay revealed that both CASC21 and HGH1 could be pulled down by biotin-labelled miR-485-5p (Fig. 5(h)). Furthermore, CASC21, miR-485-5p and HGH1 were all found to be abundantly enriched in AGO2 RIP precipitates (Fig. 5(i)) and the corresponding copy numbers were shown in Figure S4G. Thus, we came to the conclusion that CASC21 served as a ceRNA to up-regulate HGH1 via sponging miR-485-5p in CRC cells.
Figure 5.

CASC21 sponged miR-485-5p to elevate HGH1
a. ENCORI and miRDB databases were applied to identify potential micRNA binding to both CASC21 and HGH1. b. RT-qPCR was used to explore the influence of miR-485-5p overexpression on HGH1 expression. c. RT-qPCR was used to evaluate miR-485-5p expression in 80 pairs of tumour samples. d. The expression of miR-485-5p in CRC cells and control cells was examined via RT-qPCR. e. Pearson’s correlation analysis examined the correlation between miR-485-5p expression and CASC21/HGH1 expression in CRC samples. f. Binding sequences between CASC21/HGH1 and miR-485-5p, as well as the mutant CASC21/HGH1 sequences were presented. g. Luciferase reporter assay was conducted to examine the luciferase activity of wild-type or mutant CASC21/HGH1 in CRC cells transfected with miR-485-5p-mimics. h. RNA pull down assay was used to detect the relative enrichment of CASC21 and HGH1 pulled down by biotin-labelled miR-485-5p. i. RIP assay was carried out to measure the enrichment of CASC21, miR-485-5p and HGH1 in anti-AGO2 RIP precipitates. *P < 0.05, **P < 0.01. The symbol ‘n.s’. means no significance.
HGH1 mediated the function of CASC21 in regulating CRC cell proliferation, migration, EMT and stemness
Subsequently, we designed a series of rescue assays to explore whether HGH1 participated in the modulatory effect of CASC21 on CRC progression. Experimental groups were divided into sh-NC, sh-CASC21#1, sh-CASC21#2, sh-CASC21#1+ pcDNA3.1-HGH1 and sh-CASC21#2+ pcDNA3.1-HGH1. At first, colony formation and EdU assays revealed that HGH1 up-regulation could reverse the suppressive effect of CASC21 depletion on CRC cell proliferation (Fig. 6(a-b)). Transwell assay result illustrated that the migratory ability of SW620 and RKO cells was declined by CASC21 depletion, while this effect was reversed by the co-transfection of pcDNA3.1-HGH1 (Fig. 6(c)). Furthermore, it was observed that the rising E-cadherin level and declining N-cadherin level caused by CASC21 knockdown was then reversed by HGH1 up-regulation (Fig. 6(d)). Moreover, the sphere formation efficiency lowered by CASC21 depletion was restored by the up-regulation of HGH1 (Fig. 6(e)). Based on these data, we concluded that HGH1 mediated the function of CASC21 in regulating CRC cell proliferation, migration, EMT and stemness.
Figure 6.

HGH1 mediated the function of CASC21 in regulating CRC cell proliferation, migration, EMT and stemness
a-b. Colony formation and EdU assays (scar bar: 100 μm) were carried out to evaluate CRC cell proliferation ability in sh-NC, sh-CASC21#1/2 and sh-CASC21#1/2+ pcDNA3.1-HGH1 groups. c. Transwell assay (scar bar: 100 μm) was performed to detect the number of migrated CRC cells in different groups. d. IF staining (scar bar: 10 μm) was applied to examine E-cadherin (red) and N-cadherin (greed) expression in different groups. e. Sphere formation assay (scar bar: 50 μm) was applied to examine the sphere formation efficiency of indicated CRC cells. **P < 0.01.
CASC21 and HGH1 promoted CRC tumour growth in vivo
As we have verified the effect of HGH1 on CRC progression by the in vitro assays above, a series of rescue assays were conducted to explore the function of CASC21/HGH1 axis on CRC tumour growth. CRC cells were divided into 7 transfection groups: sh-NC, sh-CASC21#1, sh-CASC21#1+ pcDNA3.1, sh-CASC21#1+ pcDNA3.1-HGH1, sh-CASC21#2, sh-CASC21#2+ pcDNA3.1, and sh-CASC21#2+ pcDNA3.1-HGH1. As shown in Fig. 7(a-c), CASC21 knockdown hampered the tumour growth in CRC and reduced tumour volume and weight, and such effect was restored by the overexpression of HGH1. Next, we validated that CASC21 and HGH1 expression levels were indeed decreased in tumour cells with sh-CASC21#1/2 transfection, while the co-transfection of pcDNA3.1/HGH1 could reverse the decline of HGH1 but not CASC21 (Fig. 7(d-e)). Also, the expression of HGH1 was increased in CRC cells transfected with pcDNA3.1-CASC21 and decreased by the co-transfection with sh-POU5F1B#1/2 or miR-485-5p mimics. Moreover, the co-transfection with pcDNA3.1-CASC21+ sh-POU5F1B#1+ miR-485-5p mimic could lead to a more significant decrease in HGH1 expression in CRC cells compared with the co-transfection of only miR-485-5p mimics or sh-POU5F1B#1 (Figure S4H). Likewise, the positivity of two proliferative markers (Ki67 and PCNA) was reduced in tumours with CASC21 depletion but recovered in tumours with CASC21 inhibition plus HGH1 elevation (Fig. 7(f)). Moreover, we detected the changes in EMT process of CRC cells under the above transfection groups (Fig. 7(g) and Figure S4I). As shown by the result, the increase in E-cadherin level and decline in N-cadherin level was caused by the transfection of sh-CASC21#1/2, but these changes were reversed by the transfection with sh-CASC21 + pcDNA3.1/HGH1. According to these data, we concluded that CASC21 promoted CRC tumour growth through HGH1 in vivo.
Figure 7.

CASC21 depended on HGH1 to promote CRC tumour growth in vivo.
a.mages and the growth curves of tumours generated in mice by the injection of CRC cells transfected with sh-NC, sh-CASC21#1/2, sh-CASC21#1/2+ pcDNA3.1 or sh-CASC21#1/2+ pcDNA3.1-HGH1 were presented. b-c. Volume and weight of xenograft tumours were examined in the abovementioned groups. d. RT-PCR was used to detect CASC21 and HGH1 expression in tumours from different groups. e. Western blot analysis of HGH1 protein in tumours from different groups. f. IHC assay was carried out to detect the positivity of Ki67 and PCNA in these tumours from different groups. G. EMT markers of CRC cells in different groups was detected by western blot. **P < 0.01. The symbol ‘n.s’. means no significance.
Discussion
CRC is a prevalent cancer in China and has claimed numerous lives. The molecular functions and mechanisms in CRC have drawn more and more attention. LncRNAs have been reported to be involved in many cancers, including in CRC. For example, lncRNA NR4A1AS enhances NR4A1 expression through inhibiting UPF1-mediated mRNA degradation of NR4A1, thus facilitating CRC cell proliferation, migration and invasion [23]. EBF1-mediated up-regulation of PNO1 facilitates CRC development through negative regulation on p53 pathway [24]. CASC21 has been reported to exert oncogenic roles in CRC development [18,19]. Herein, we found that CASC21 was up-regulated in colon adenocarcinoma according to GEPIA database. Considering that CASC21 mechanism in CRC has not been substantially explained yet, we tried to further explore its function and mechanism in CRC. Expectedly, CASC21 was verified to show high expression in CRC tissues and cells, and was confirmed to facilitate CRC cell proliferation, EMT and stemness. Thus, our data were concordant with previous reports, suggesting that CASC21 was an oncogenic lncRNA in CRC.
Next, by bioinformatics analysis, we identified that POU5F1B was positively co-expressed and potentially interacted with CASC21. POU5F1B is a processed pseudogene which is highly homologous to OCT4. Recently, POU5F1B was verified to be transcribed in cancer cells. For example, Hayashi H et al. have revealed that POU5F1B is a significant predictor of unfavourable prognosis in gastric cancer patients and promotes an aggressive phenotype in gastric cancer [25]. Barry KH et al. reveal that six correlated sites and two intergenic sites in POU5F1B have a connection with prostate cancer risk [26]. POU5F1B has a positive correlation with activated AKT and facilitates carcinoma proliferation through AKT activation [27]. CCAT2 positively modulates POU5F1B gene to induce cell as well as inhibit cell apoptosis and autophagy in BGC-823 cells [28]. Present study first verified that CASC21 bound to POU5F1B but had no regulatory effects on POU5F1B expression. Also, POU5F1B had no regulatory effects on CASC21. These data indicated that CASC21 possibly worked synergistically with POU5F1B in CRC cells.
Subsequently, through bioinformatics analysis, HGH1 was identified as the mRNA that was positively co-expressed with both CASC21 and POU5F1B. Both CASC21 and POU5F1B positively modulated HGH1 expression in CRC cells. Function of HGH1 has not been studied in other diseases before. Herein, we first identified that HGH1 was up-regulated in colon adenocarcinoma and rectum adenocarcinoma according to GEPIA database. We then identified POU5F1B functioned as the transcription factor to bind to HGH1 promoter and activate transcription of HGH1. In addition, CASC21 knockdown reduced the binding between POU5F1B and HGH1 promoter. In summary, our results indicated that CASC21 served as a scaffold to recruit POU5F1B to HGH1 promoter, thus activating transcription of HGH1. Previously, the fact that lncRNAs recruit proteins has been reported in CRC. For instance, lncRNA CCAL promotes chemo-resistance of CRC cells via directly interacting with HuR to increase mRNA and protein levels of β-catenin [29]. LncRNA CASC9 interacts with CPSF3 to stabilize TGF-β2 mRNA and to trigger TGF-β pathway in CRC [30]. LINC01354 interacts with RNA-binding protein hnRNP-D to stabilize β-catenin, thus activating Wnt/β-catenin signalling pathway and contributing to proliferation and metastasis in CRC [31]. However, the binding between CASC21 and POU5FB1 and their synergistic effect on HGH1 transcription were first discovered in this study.
Besides, CASC21 was distributed not only in nucleus, but also in cytoplasm of CRC cells. Also, CASC21 as well as HGH1 was discovered to be abundantly enriched in the RIP products of AGO2, a core protein of RNA-induced silencing complex (RISC). Thus, we wondered whether CASC21 regulated HGH1 in the ceRNA pattern. The experimental data of mechanism assays revealed that miR-485-5p bound to both CASC21 and HGH1 in CRC cells, and CASC21 could sponge miR-485-5p to modulate HGH1 expression. MiR-485-5p has been verified to play as a tumour suppressor in CRC by former works. For example, miR-485-5p regulates O-GlcNAcylation and stability of Bmi-1 through targeting OGT to repress CRC cell proliferation [32]. MiR-485-5p negatively modulates CD147 expression to promote CRC cell proliferation and migration [33]. Apart from present study, the ceRNA pattern in CRC was commonly studied. LINC02418 functions as a ceRNA to elevate MELK expression through absorbing miR-1273 g-3p and serves as a diagnostic biomarker for CRC [34]. LncRNA ZNFX1-AS1 accelerates CRC metastasis via acting as a ceRNA of miR-144 to up-regulate EZH2 expression [35]. However, the ceRNA network formed by CASC21/miR-485-5p/HGH1 was first demonstrated in CRC by our study.
On the whole, present study uncovered that CASC21 recruited POU5F1B to the promoter of HGH1 and activated the transcription of HGH1. Moreover, CASC21 served as a ceRNA to up-regulate HGH1 through endogenously sponging miR-485-5p. CASC21 accelerated malignant phenotypes of CRC cells through enhancing HGH1 expression. Consistent with the previous outcomes of CASC21 researches on CRC, our study also revealed the oncogenic role of CASC21 in the regulation of CRC cell proliferation, migration, EMT as well as tumour growth. Importantly, we uncovered the novel and detailed mechanism behind CASC21 regulating HGH1 in CRC. Hopefully, this study may provide new insight for the future identification of therapeutic options and treatment for CRC.
Supplementary Material
Acknowledgments
We sincerely appreciate all lab members.
Funding Statement
The authors have no funding to report.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A.. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- [3].Watson AJ, Collins PD. Colon cancer: a civilization disorder. Dig Dis. 2011;29(2):222–228. [DOI] [PubMed] [Google Scholar]
- [4].Liang ZX, Liu H-S, Wang F-W, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10(11):829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liang C, Zhao T, Li H, et al. Long Non-coding RNA ITIH4-AS1 accelerates the proliferation and metastasis of colorectal cancer by activating JAK/STAT3 signaling. Mol Ther Nucleic Acids. 2019;18:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xu M, Xu X, Pan B, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer. 2019;18(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang Z, Jin J. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging (Albany NY). 2019;11(14):4876–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yu S, Wang D, Shao Y, et al. SP1-induced lncRNA TINCR overexpression contributes to colorectal cancer progression by sponging miR-7-5p. Aging (Albany NY). 2019;11(5):1389–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhuang M, Zhao S, Jiang Z, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yu X, Wang D, Wang X, et al. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J Exp Clin Cancer Res. 2019;38(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peng WX, Pratirodh K, Zhang W, et al. lncRNA RMST enhances DNMT3 expression through interaction with HuR. Mol Ther. 2020;28(1):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [13].Zou Z, Ma T, He X, et al. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression. Cell Death Dis. 2018;9(7):717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jia B, Xie T, Qiu X, et al. Long noncoding RNA FALEC inhibits proliferation and metastasis of tongue squamous cell carcinoma by epigenetically silencing ECM1 through EZH2. Aging (Albany NY). 2019;11(14):4990–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang YQ, Jiang D-M, Hu -S-S, et al. SATB2-AS1 suppresses colorectal carcinoma aggressiveness by inhibiting SATB2-Dependent snail transcription and Epithelial–Mesenchymal transition. Cancer Res. 2019;79(14):3542–3556. [DOI] [PubMed] [Google Scholar]
- [16].Yin D, Lu X, Su J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li W, Qi Y, Cui X, et al. Characteristic of HPV integration in the genome and transcriptome of cervical cancer tissues. Biomed Res Int. 2018;2018:6242173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zheng Y, Nie P, Xu S. Long noncoding RNA CASC21 exerts an oncogenic role in colorectal cancer through regulating miR-7-5p/YAP1 axis. Biomed Pharmacother. 2020;121:109628. [DOI] [PubMed] [Google Scholar]
- [19].Gong T, Li Y, Feng L, et al. CASC21, a FOXP1 induced long non-coding RNA, promotes colorectal cancer growth by regulating CDK6. Aging (Albany NY). 2020;12(12):12086–12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cao Z, Pan X, Yang Y, et al. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34(13):2185–2194. [DOI] [PubMed] [Google Scholar]
- [21].Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xie X, Lin J, Liu J, et al. A novel lncRNA NR4A1AS up-regulates orphan nuclear receptor NR4A1 expression by blocking UPF1-mediated mRNA destabilization in colorectal cancer. Clin Sci (Lond). 2019;133(13):1457–1473. [DOI] [PubMed] [Google Scholar]
- [24].Shen A, Chen Y, Liu L, et al. EBF1-Mediated upregulation of ribosome assembly factor pno1 contributes to cancer progression by negatively regulating the p53 signaling pathway. Cancer Res. 2019;79(9):2257–2270. [DOI] [PubMed] [Google Scholar]
- [25].Hayashi H, Arao T, Togashi Y, et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2015;34(2):199–208. [DOI] [PubMed] [Google Scholar]
- [26].Barry KH, Moore LE, Sampson JN, et al. Prospective study of DNA methylation at chromosome 8q24 in peripheral blood and prostate cancer risk. Br J Cancer. 2017;116(11):1470–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pan Y, Zhan L, Chen L, et al. POU5F1B promotes hepatocellular carcinoma proliferation by activating AKT. Biomed Pharmacother. 2018;100:374–380. [DOI] [PubMed] [Google Scholar]
- [28].Yu ZY, Wang Z, Lee K, et al. Effect of silencing colon cancer-associated transcript 2 on the proliferation, apoptosis and autophagy of gastric cancer BGC-823 cells. Oncol Lett. 2018;15(3):3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Deng X, Ruan H, Zhang X, et al. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2020;146(6):1700–1716. . [DOI] [PubMed] [Google Scholar]
- [30].Luo K, Geng J, Zhang Q, et al. LncRNA CASC9 interacts with CPSF3 to regulate TGF-beta signaling in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li J, He M, Xu W, et al. LINC01354 interacting with hnRNP-D contributes to the proliferation and metastasis in colorectal cancer through activating Wnt/beta-catenin signaling pathway. J Exp Clin Cancer Res. 2019;38(1):161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chai Y, Du Y, Zhang S, et al. MicroRNA-485-5p reduces O-GlcNAcylation of Bmi-1 and inhibits colorectal cancer proliferation. Exp Cell Res. 2018;368(1):111–118. [DOI] [PubMed] [Google Scholar]
- [33].Hu XX, Xu X-N, He B-S, et al. microRNA-485-5p functions as a tumor suppressor in colorectal cancer cells by targeting CD147. J Cancer. 2018;9(15):2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao Y, Du T, Du L, et al. Long noncoding RNA LINC02418 regulates MELK expression by acting as a ceRNA and may serve as a diagnostic marker for colorectal cancer. Cell Death Dis. 2019;10(8):568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shi L, Hong X, Ba L, et al. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10(3):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


