ABSTRACT
In Drosophila melanogaster, PD isoform of the double-stranded RNA binding protein (dsRBP) Loquacious (Loqs-PD) facilitates dsRNA cleavage to siRNA by Dicer-2. StaufenC (StauC) was discovered as a coleopteran-specific dsRBP required for dsRNA processing in coleopteran insects. Here, we show that StauC is essential for the high RNAi efficiency observed in coleopterans. Knockdown of StauC but not the homologs of Loqs-PD and R2D2 evoked a long-lasting insensitivity to RNAi in the coleopteran cell line, Ledp-SL1. The dsRNA insensitivity induced by StauC knockdown could not be overcome merely by an increase in dose or time of exposure to dsRNA or expression of Loquacious or R2D2. Furthermore, StauC but not Loqs and R2D2 are required for processing of dsRNA into siRNA. StauC overexpression also partly restored the impaired RNAi caused by the knockdown of Loqs-PD in D. melanogaster Kc cells. However, StauC was unable to compensate for the loss-of-the function of Dcr-2 or R2D2. Overall, these data suggest that StauC functions like Lops-PD in processing dsRNA to siRNA.
KEYWORDS: StaufenC, Loqs-PD, Dicer-2, dsRNA, siRNA, RNAi
Introduction
RNA interference (RNAi) is widely used as a powerful tool in functional genomics and is also being explored for developing pest management methods[1]. Indeed, transgenic plants expressing dsRNA targeting lethal genes in insect pests acquire effective resistance against the herbivorous insect [2]. RNAi works efficiently in many beetle species (belonging to order Coleoptera), including the Colorado potato beetle (Leptinotarsa decemlineata) [3–8], red flour beetle (Tribolium castaneum) [9,10], Western corn rootworm (Diabrotica virgifera virgifera) [2,11–14] and Asian longhorn beetle [15]. Therefore, many coleopteran insects are considered promising targets for RNAi-mediated insect pest management.
Coleopteran-specific StaufenC (StauC), a dsRNA binding protein (dsRBP), is identified as a key component in the processing of dsRNA to siRNA in coleopteran insects [15]. StauC expression decreased in the RNAi-resistant Lepd-SL1 cells selected by prolonged exposure to dsRNA targeting the inhibitor of apoptosis 1 (dsIAP) for multiple generations [16]. These observations suggest that the knockdown of StauC is a potential molecular mechanism of cellular resistance to RNAi in coleopteran insects. StauC knockdown decreases RNAi efficiency in cells, thereby conferring a resistance phenotype for dsRNA insecticide. However, the mechanisms employed by StauC to modulate siRNA production are not known. Also, further research is required to 1. understand the biological significance of StauC in potential resistance to RNAi and 2. to ascertain if knockdown of StauC is sufficient to develop a meaningful level of RNAi resistance in coleopteran insects, and if this resistance mechanism can be overcome by higher dose or longer exposure to dsRNA.
In Drosophila melanogaster, two Dicer enzymes (Dcr-1 and Dcr-2) are responsible for miRNA biogenesis and dsRNA into siRNA processing, respectively [17]. dsRBP Loquacious (Loqs) is a dicer partner protein and functions in both the siRNA and microRNA pathways [18]. The gene loqs encodes four isoforms in D. melanogaster, and the Loqs-PD isoform is demonstrated essential for facilitating exogenous siRNA production by Dcr-2 in vivo [19] as well as endogenous siRNA accumulation in D. melanogaster S2 cells [20]. Consistent with these findings, Trettin et al. recently showed that Loqs-PD is capable of enhancing the enzyme activity of Dcr-2 towards dsRNA substrates [21]. R2D2, a paralog of Loqs, was shown to associate with Dcr-2 and functions in loading the guide siRNA strand into the Argonaute 2 (Ago-2) protein complex [22,23]. In contrast to D. melanogaster, only one Loqs protein has been identified in L. decemlineata. The RNAi-mediated knockdown of Loqs and R2D2 homologs in L. decemlineata moderately affected RNAi efficiency in Lepd-SL1 cells [24], suggesting that they may contribute to RNAi efficiency in these cells. However, it is unknown if the Loqs and R2D2 homologs have functional redundancy with StauC in coleopterans and if StauC can substitute for Loqs or R2D2 in other insects.
The main goals of studies described here are to understand the interplay between StauC and Loqs/R2D2 homologs in coleopteran insects. We tested how the RNAi susceptibility of Lepd-SL1 cells is affected by the knockdown of StauC, Loqs, or R2D2. In a gain of function study to improve the RNAi efficiency, StauC was also heterologously expressed in Sf9 cells known to be refractory to RNAi. We also investigated to check if StauC can functionally replace Loqs-PD or R2D2 in D. melanogaster cells. Moreover, the protein crosslinking experiments were carried out to determine if StauC is physically associated with Dcr-2. The results from these studies advance our understanding of the biological significance of StauC, Loqs, and R2D2 in RNAi and the potential molecular mechanisms of resistance to dsRNA insecticides.
Materials and methods
Plasmid construction
The full-length coding sequences of L. decemlineata StauC were amplified by PCR and cloned into the pIEx4 vector at Nco I and Hind III restriction sites. The full-length coding sequence of L. decemlineata Dicer-2a was cloned into the pIZT-V5-His vector at Kpn I and Not I restriction sites. The primers used in gene cloning are shown in Table S1.
Cell culture and transfection
Cell lines developed from insects belonging to three orders Coleoptera (L. decemlineata, Lepd-SL1 cells where RNAi works efficiently), Diptera (Drosophila melanogaster Kc cells where RNAi works well) and Lepidoptera (Spodoptera frugiperda Sf9 cells where RNAi is inefficient) were used. Lepd-SL1 cells obtained from the Biological Control of Insects Research Laboratory, USDA-ARS, Columbia, MO. The cells were cultured in EX-CELL 420 (Sigma–Aldrich) containing 10% fetal bovine serum (FBS) (Life Technologies) Penicillin-Streptomycin (Life Technologies). Sf9 cells were maintained in Sf-900 III SFM medium (Thermo Fisher Scientific, Waltham, MA). Kc cells were cultured in Schneider’s insect medium (MilliporeSigma, Burlington, MA) supplemented with 5% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA) at 28°C. The day before transfection, 1.8 × 106 cells were seeded into a T-25 flask or 4 × 106 cells were seeded into each well of a 6-well plate. After reaching 80–90% confluency, the cells were then transfected using the TransIT-PRO Transfection Kit (Mirus Bio LLC, Madison, WI).
dsRNA synthesis
dsRNA targeting a specific gene was synthesized using the MEGAscript RNAi kit (Life Technologies) as described previously [25,16]. Briefly, primers containing a canonical T7 promoter sequence at the 5ʹend of both the forward primer and the reverse primer were utilized to amplify the 350–500bp amplicons (Table S1). The PCR products were used as templates for dsRNA synthesis using the Ambion MEGAscript transcription kit (Ambion, Austin, TX). The input template DNA was digested with Turbo DNase (Ambion, USA) at 37°C for 1 h. The dsRNA was purified by phenol/chloroform extraction, followed by ethanol precipitation. The quality of dsRNA was checked by running on an agarose gel, and the concentration was measured using NanoDrop2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). Primers used for dsRNA synthesis are shown in Table S1.
Detection of apoptosis
The terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labelling (TUNEL) assay was performed using the In Situ Cell Death Detection Kit, Fluorescein (Roche Applied Science, Mannheim, Germany). Briefly, dsRNA-treated Lepd-SL1 cells were fixed in 4% paraformaldehyde for 15 min at room temperature and washed with PBS. The cells were then permeabilized with freshly prepared 0.2% Triton X-100 and 0.1% sodium citrate for 3 min on ice and washed with PBS. The slides were then incubated with a TUNEL reaction mixture for 1 h at 37°C and washed again with PBS and photographed under a fluorescence microscope.
Quantitative analysis (qRT-PCR)
The total RNA was extracted with TRIzol reagent (Life Technologies) following the manufacturer’s instructions. Briefly. 2 µg of total RNA was applied to synthesize cDNAs using SuperScript II reverse transcriptase (Life Technologies). qRT-PCR reactions were run in triplicate or quadruplicate on the iCycler iQ system using iQ SYBR Green Supermix (Bio-Rad). The relative mRNA levels were calculated using the ribosomal protein 49 (RP49) gene as a reference. The data were analysed by the Student’s t-test. Primers used for qRT-PCR are listed in Table S1.
Measuring cell viability
Cells were pre-incubated with dsRNA for 3 days, then seeded into a 96-well culture plate (5 x 103 cells per well). The cells were treated with different concentrations of dsIAP for 24 and 48 h. The cells were then counted using Cell Counting Kit-8 assay (Dojindo Molecular Technologies, Rockville, MD).
Luciferase assay
When Sf9 cells reached 80–90% confluency, they were co-transfected with 3 µg of the luciferase reporter vector and 9 µg of pIEx4/StauC or/and pIZT-V5-His/Dcr-2a constructs. At 4 h after transfection, the cells were re-seeded into a 48-well culture plates (1 x 105 cells per well) and exposed to 4 µg/ml dsLuc or dsGFP for 60 h. The cells were harvested and the luciferase assay was performed as described previously [26]. The luciferase activity was normalized to total protein concentration.
Protein-protein crosslinking and affinity chromatography
For protein crosslinking between adjacent proteins in cells, the cells transiently expressing StauC-S-tag and Dcr-2a-V5-6xHis were exposed to dithiobis succinimidyl propionate (DSP), a cell-permeable and reversible crosslinking agent (at a final concentration of 2.5 mM) at room temperature for 30 minutes. To stop the reaction, Tris-HCl solution was added at a final concentration of 20 mM and incubated for 5 minutes. The protein complexes containing the 6xHis-tagged Dcr-2a were isolated by metal affinity chromatography under denaturing conditions. Briefly, Ni-charged MagBeads from GenScript (Piscataway, NJ) were used to isolate the recombinant protein complexes. The cells were suspended in an extraction buffer (50 mM Tris, pH 7.4, 150 mM NaCl, and 6 M guanidine HCl) at 10 mg/ml of concentration followed by incubation for 30 min with shaking at 4°C. The suspension was then centrifuged at 100,000 x g for 30 min. The supernatant was loaded onto Ni-charged MagBeads (GenScript, Piscataway, NJ), which has been pre-equilibrated with washing buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 6 M guanidine HCl, and 10 mM imidazole). After extensive washing with the washing buffer, bound His-tagged proteins and protein complexes were eluted by stepwise gradient elution with imidazole in the presence of 6 M guanidine HCl and 150 mM NaCl. To remove salts in the eluents, methanol/chloroform precipitation was carried out. The purified proteins were analysed by SDS-PAGE under the reducing and non-reducing conditions.
Western blots
Western blot analysis was performed using rabbit anti-S-tag IgG obtained from Novus Biologicals (Centennial, CO), anti-V5 tag from Cell Signalling Technology (Danvers, MA) and polyclonal StauC antibody [16]. HRP-conjugated goat anti-rabbit IgG (Cell Signalling Technology, Danvers, MA) was used at 1:2000 as a secondary antibody. The protein bands were detected using the SuperSignal West Dura Extended Duration Substrate from Pierce Biotechnology (Waltham, MA).
Processing of 32P-labelled dsRNA
32P-labelled dsRNA processing assay was performed as described previously [16,27]. Briefly, Lepd-SL1 cells were incubated with the desired dsRNA for 3 days, followed by exposure to 1.6 × 106 CPM (count per million) 32P-labelled dsGFP for 24 h. Total RNA was isolated, and 2000 CPM RNA was loaded onto 8 M urea-16% polyacrylamide gels. After drying, the gels were exposed to a phosphorImager screen. The screen was scanned in a phosphorImager (Typhoon 9500, G.E. Healthcare Life Sciences, Pittsburgh, PA).
Statistical analysis
The GraphPad Prism 5.01 software (San Diego, CA) was used for performing statistical analyses. Comparisons of multiple factors were examined using the Student’s t-test analysis. p < 0.05 was considered statistically significant.
Results
StauC is required for RNAi in coleopteran Lepd-SL1 cells
Previous studies showed that RNAi-mediated knockdown of StauC inhibited RNAi response in coleopteran insects. We first determined if the protein levels of StauC are reduced in Lepd-SL1 cells exposed to dsRNA targeting StauC (dsStauC). Lepd-SL1 cells were exposed to 300 ng/ml dsStauC for 72 h, and the proteins harvested from these cells were analysed by western blots using StauC-specific antibody. Continuous exposure of these cells to dsStauC for 72 h is sufficient to reduce StauC protein levels (Fig. 1A). Interestingly, the decrease in StauC protein levels did not recover even after the subsequent incubation of the cells in the dsRNA-free medium for 48 h (Fig. 1B). The Lepd-SL1 cells exposed to dsIAP exhibited cell death as indicated by the presence of disintegrating cells and apoptotic bodies (Fig. 1C). Lepd-SL1 cells pre-treated with dsStauC prevented cell death induced by dsIAP. Consistent with these observations, in the Lepd-SL1 cells exposed to dsIAP, the TUNEL assay detected an increase in the 3ʹ-OH ends of the DNA fragments that are formed during apoptosis (Fig. 1D & Figs. S1& S2). These data confirm our previous report that StauC is required for RNAi in Lepd-SL1 cells [16].
Figure 1.
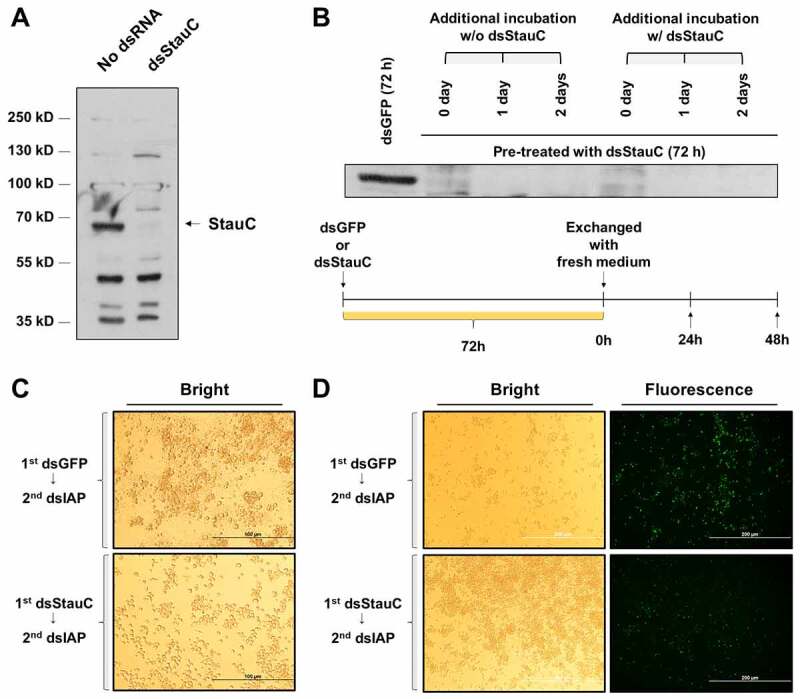
Exposure to dsStauC significantly knockdowns StauC protein in Lepd-SL1 cells. (A) Lepd-SL1 cells were exposed to 300 ng/ml dsStauC for 72 h before western blot analysis using StauC antibody. (B) Lepd-SL1 cells pre-exposed to dsStauC for 72 h were incubated for addition 24 and 48 h in the presence or absence of dsStauC before subjected to western blot analysis using StauC antibody. 50 µg of total cell lysate was utilized to measure endogenous StauC level. (C) Apoptosis phenotype was observed after exposing Lepd-SL1 cells to dsIAP, but not dsGFP. Lepd-SL1 cells pre-exposed to dsStauC or dsGFP were incubated with dsIAP (a final concentration of 30 ng/ml) and photographed 24 after treatment. (Scale bar = 100 µm). (D) TUNEL staining of dsRNA-treated Lepd-SL1 cells to examine the apoptotic cell death. (Scale bar = 200 µm) Images are representative of three replicates shown in
To study the biological significance of StauC in RNAi, we examined how StauC depletion affects the RNAi efficiency in Lepd-SL1 cells. Lepd-SL1 cells were exposed to dsStauC or dsGFP for 72 h prior to incubation with different concentrations of dsIAP (0.3 to 1,075 ng/ml) for 24 or 48 h. After dsIAP treatment for 24 h, the IC50 value of dsIAP in the Lepd-SL1 cells exposed to dsGFP was 11.40 ng/ml (Fig. 2A). After 48 h of continuous exposure, the dsIAP IC50 value decreased to 2.555 ng/ml. However, Lepd-SL1 cells exposed to dsStauC exhibited complete insensitivity to dsIAP (up to 1075 ng/ml) at least for 24 h. Although longer exposure (48 h) to dsIAP led to a significant reduction in the viability of the StauC-knockdown cells, with an estimated IC50 value of > 2000 ng/ml. This is 400-fold higher IC50 value than that detected in control cells exposed to dsGFP. These data suggest that the knockdown of StauC increases the resistance of Lepd-SL1 cells to apoptosis induced by knockdown of IAP gene.
Figure 2.
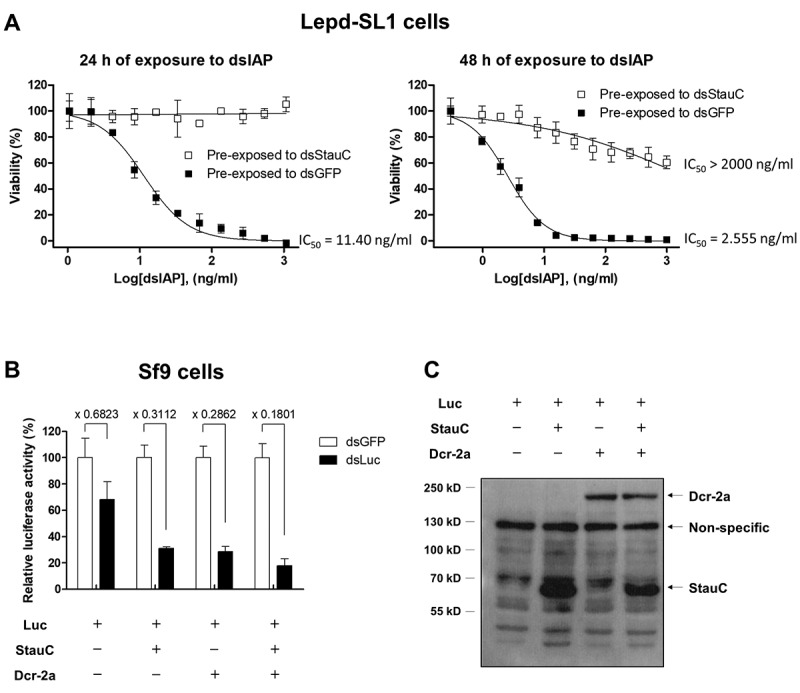
Effect of StauC expression on the RNAi efficiency in Lepd-SL1 and Sf9 cells. (A) StauC depletion induces high resistance to RNAi in Lepd-SL1 cells. Lepd-SL1 cells acquire high resistance to apoptosis induced by dsIAP. Lepd-SL1 cells pre-exposed to dsStauC were incubated with different concentrations of dsIAP for 24 h (1 to 1075 ng/ml)) and 48 h (0.3 to 1000 ng/ml). (B) StauC facilitates dsRNA-mediated gene silencing in the Spodoptera Sf9 system. Expression of genes coding for StauC and Dcr-2a decreased the luciferase gene expression in Sf9 cells exposed to dsLuc compared to the dsGFP treatment control. The measured luciferase activity was normalized to total protein concentration. Each dot refers to the average of triplicates, and its values were expressed as the mean ± standard deviation. Sf9 cells were transiently co-transfected with a luciferase reporter vector and gene expression construct encoding L. decemlineata StauC and Dcr-2a protein. The cells were then re-seeded into a 48-well culture plate (1 x 105 cells per well) at 4 h post-transfection, followed by incubation with 4 µg/ml dsLuc or dsGFP for an additional 60 h. The measured luciferase activity was normalized to total protein concentration. (C) Heterologous expression of StauC and Dcr-2 in Sf9 cells. These recombinant proteins were detected by western blot analysis using the antibodies specific to S tag and V5 tag at 60 h post-transfection.
Since longer exposure to dsIAP decreases the viability of Lepd-SL1 cells exposed to dsStauC, we wanted to know if the prolonged exposure of a high concentration of dsIAP induces higher levels of cell death in StauC-knockdown cells. Lepd-SL1 cells exposed to dsStauC were seeded into each well of a 48-well plate and treated with dsIAP or dsLuc at a final concentration of 1 µg/ml for different lengths of time. Continuous exposure of StauC-knockdown Lepd-SL1 cells to dsLuc for 72 to 96 h slightly decreased their viability (Fig. S3A). However, membrane-bound extracellular vesicles, apoptotic bodies, were not detected up to 96 h (Fig. S3C). In contrast, Lepd-SL1 cells exposed to dsGFP and treated with dsIAP began to show apoptosis by two days after dsIAP treatment (Fig. S3C). In line with these findings, the cell viability also decreased significantly in dsIAP-treated cells compared to the control cells treated with dsLuc (Fig. S3A). However, the overall results suggest that the Lepd-SL1 cells exposed to dsStauC retained strong resistance to cell death induced by dsIAP for up to 96 h (Figs. S3A & C). The StauC protein levels did not recover in Lepd-SL1 cells treated with dsStauC for three days, followed by culture in the dsRNA-free medium for up to 5 days (Fig. S3B). These observations suggest that even higher doses or prolonged exposure to dsIAP fail to induce robust cell death in StauC-knockdown Lepd-SL1 cells.
Heterologous expression of StauC increases RNAi efficiency in Sf9 cells
To test if StauC expression increases RNAi efficiency in the lepidopteran cells that are recalcitrant to RNAi, StauC was overexpressed in Spodoptera frugiperda (belongs to order Lepidoptera) Sf9 cells where StauC homolog does not exist. The luciferase expression construct was co-transfected into Sf9 cells with the gene expression construct containing L. decemlineata StauC or Dicer-2a (Dcr-2a) genes. At 4 h after transfection, the cells were incubated with 4 µg/ml of dsLuc for an additional 60 h. The cells were harvested, and the luciferase levels were quantified. Exposure of Sf9 cells to dsLuc resulted in a significant reduction in the luciferase activity compared to that in the cells treated with dsGFP control (Fig. 2B). Notably, the overexpression of StauC or Dcr-2a increased RNAi in Sf9 cells, and the increased gene silencing effect was observed at 64 h post-transfection when StauC or Dcr-2a are expressed well in these cells (Fig. 2B & C). These data suggest that the heterologous expression of StauC enhances RNAi efficiency in Sf9 cells.
StauC expression partly compensates for the loss-of-function of Loqs-PD in Kc cells
In D. melanogaster, the enzyme Dcr-2 processes dsRNA to siRNA, which is integrated into RISC. The D. melanogaster Dcr-2 works with dsRBP partners, including Loquacious and R2D2, to facilitate this process. Loqs-PD is identified as the unique Loquacious isoform capable of promoting the Dcr-2-mediated processing of dsRNA into siRNA [18–20,28,29] while R2D2 mediates the loading of the resulting siRNA into the Ago2 of RISC [30]. This encouraged us to investigate if StauC mimics the function of Loqs-PD, Dcr-2, or R2D2 in the RNAi of D. melanogaster. D. melanogaster Kc cells were exposed to Loqs-PD, Dcr-2 or R2D2 dsRNA for 3 days to allow for efficient knockdown of the target protein. The cells were then co-transfected with a luciferase expression construct and the gene expression construct encoding L. decemlineata StauC or empty vector. Four hours after transfection, the cells were exposed to different concentrations of dsLuc for an additional 48 h. The luciferase assays showed that Kc cells pre-exposed to dsGFP were highly susceptible to dsRNA-mediated gene silencing, such that dsLuc in the ng/ml range induced a significant reduction in the luciferase activity. However, the RNAi efficiency was not improved further by overexpression of StauC (Fig. 3A).
Figure 3.
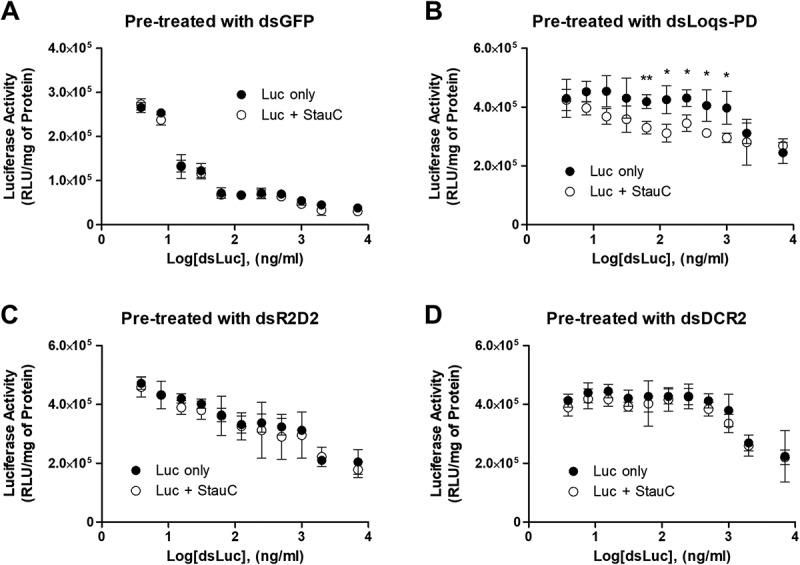
Heterologous expression of StauC in part functionally compensates for the loss-of-function of Loqs-PD in Kc cells. Kc cells were pre-incubated with dsRNA corresponding to (A) GFP or D. melanogaster (B) Loqs-PD, (C) R2D2, or (D) DCR2 for 3 days to allow for a significant reduction in the target protein. A luciferase expression construct was then transfected into the cells along with pIEx4 or pIEx4/StauC. 4 h after transfection; the cells were exposed to different concentrations of dsLuc (3.9 ng/ml – 7000 ng/ml) for 48 h. Incubation with 3.9 ng/ml dsLuc induced no significant reduction in luciferase activity in all the tested groups, compared to no dsRNA treatment (Fig. S7). The measured luciferase activity was normalized to total protein concentration. Mean ± standard deviation (n=3) are shown. Asterisks, significant differences of the mean values between two groups. p values were determined using the Student’s t test: *, p < 0.05; **, p < 0.01
In contrast, exposure of Kc cells to dsLoqs-PD, dsR2D2, or dsDCR-2 prior to treatment with dsLuc caused a reduction in the luciferase knockdown (Fig. 3(B,C,D)). Even 48 h of continuous exposure to 1 µg/ml dsLuc failed to knockdown the luciferase gene in Kc cells pre-treated with dsLoqs-PD or dsDcr-2. Interestingly, StauC overexpression increased the RNAi efficiency in Kc cells pre-treated with dsLoqs-PD (Fig. 3B). At the same time, no significant restoration of RNAi susceptibility was achieved by StauC overexpression in Kc cells pre-exposed to dsR2D2 or dsDCR-2 (Fig. 3(C,D)). Considering our previous observations [16] that showed StauC is required for efficient processing of dsRNA to siRNA in coleopteran insects, these findings support the hypothesis that the function of StauC is similar to that of Loqs-PD in the RNAi pathway.
Physical association of StauC with Dcr-2
To check if StauC is physically associated with Dcr-2, Sf9 cells were co-transfected with the two constructs expressing L. decemlineata Dicer-2a fused to V5 and 8xHis tags and StauC fused to S tag (Fig. 4A). The transfected cells were then incubated with or without dsGFP for 4 h before harvesting them at 48 h post-transfection (Fig. 4B). The formation of intermolecular crosslinks between the Dcr-2a protein and its neighbouring proteins was induced by exposing the cells to a crosslinking agent, dithiobis (succinimidyl propionate) (DSP). The proteins were isolated from these cells and purified by immobilized metal ion affinity chromatography (IMAC). The purified proteins were separated by SDS-PAGE, transferred to a membrane and probed with S-tag and V5 tag-specific antibodies. To reduce proteins and break crosslinking, a portion of the lysate or purified proteins were treated with β-mercaptoethanol (β-ME) before running them on gels. In Sf9 cells, more than half of the S-tagged StauC protein formed high molecular protein complexes that were almost completely resolved to the monomeric size of StauC upon treatment with a reducing agent, β-ME (Fig. 4C). These results suggest that the StauC forms complexes with other proteins in these cells.
Figure 4.
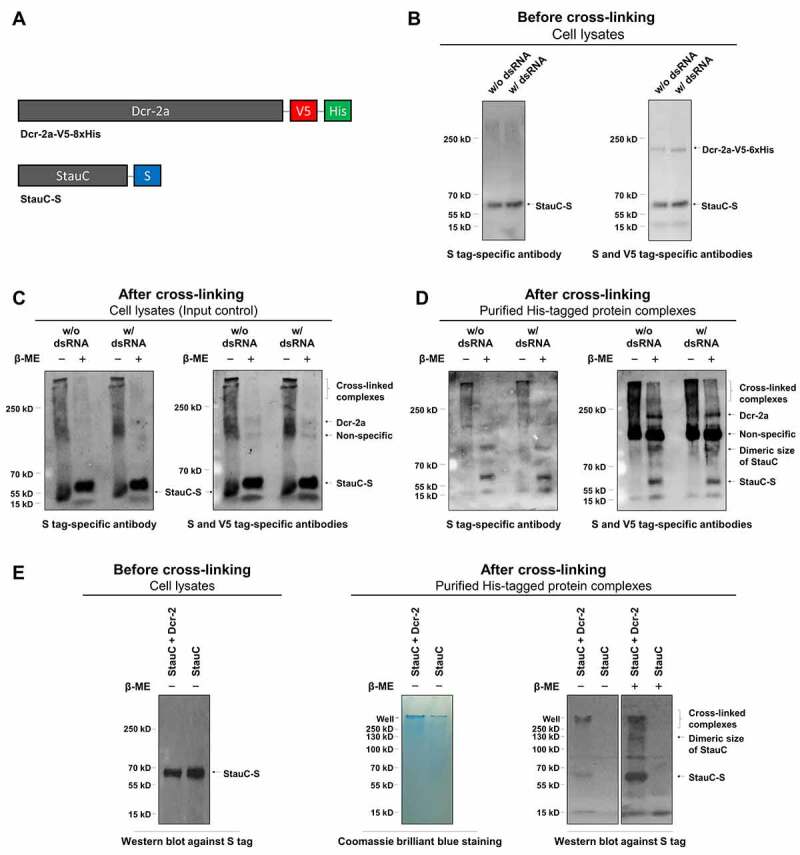
StauC forms a protein complex with Dcr-2a in cells. (A) Schematic representation for the recombinant StauC-S and Dcr-2a-V5-8xHis. (B) Sf9 cells were co-transfected with the gene expression constructs encoding L. decemlineata StauC-S and Dcr-2a-V5-8xHis. The transfected cells were incubated with or without dsGFP for 4 h before harvesting them at 48 h post-transfection. The proteins were etracted and used to perform western blot analysis using S and V5 tag-specific antibodies. 50 µg of total cell lysate was utilized to measure the levels of target proteins. (C) The same cells but exposed to DSP, a cell-permeable and reversible crosslinking agent, were analysed by western blot against S and V5 tags. 50 µg of total cell lysate was used for analysis. (D) The His-tagged protein and protein complexes extracted from the DSP-treated cells were subjected to immunoblotting using S and V5 tag-specific antibodies. (E) L. decemlineata StauC-S was expressed in Sf9 cells in the presence or absence of Dcr-2a. The protein complexes isolated from the DSP-treated cells by immortalized metal affinity chromatography were subjected to SDS-PAGE and western blot analysis
The his-tag protein-containing complexes extracted from the DSP-treated cell lysates were purified by immobilized metal ion affinity chromatography (IMAC) under strong denaturing conditions. The purified protein complexes were subjected to immunoblotting using S tag antibodies. The proteins with a molecular weight higher than the size of StauC (~60 kDa) were recognized by S tag antibodies in the absence of β-ME treatment (Fig. 4D). After treatment with β-ME, the high molecular weight complexes dissociated and the S-tag antibodies recognized StauC monomer and dimer in these proteins suggesting that the His-tagged protein complexes isolated by IMAC chromatography contained S-tagged StauC protein (Fig. 4D). The immunoblotting results using V5-tag antibodies showed that these antibodies recognized the His and V5-tagged Dcr-2a in the purified protein complexes. In a control experiment, the S-tagged StauC protein was isolated by IMAC chromatography only when co-expressed with the V5 and His tagged Dcr-2a protein (Fig. 4E). Also, the presence of dsRNA did not have a significant effect on the formation of StauC and Dcr-2a complexes (Fig. 4). Overall, these observations suggest that StauC and Dcr-2a are nearby and crosslinked into a complex in Sf9 cells.
Effect of knockdown of StauC, Loquacious, or R2D2 on RNAi efficiency in L. decemlineata cells
We also evaluated the contributions of StauC, Loquacious, and R2D2 to RNAi efficiency in Lepd-SL1 cells to determine which one of them is essential for RNAi in L. decemlineata. BLAST searches in the GenBank database identified XP_023022366.1 and XP_023028942.1 as the orthologs of D. melanogaster Loquacious and R2D2 in L. decemlineata, respectively. The Loquacious ortholog from L. decemlineata has the two dsRNA-binding domains (dsRBDs) sharing high protein sequence homology with the first two dsRBDs of the D. melanogaster Loquacious isoforms (Fig. S4A). However, relatively less sequence similarity was observed between R2D2 orthologs from these two insect species (Fig. S4B). Lepd-SL1 cells were pre-incubated with the dsRNA targeting StauC, Loqs, R2D2 or GFP (control) for 3 days. The cells were then treated with different concentrations of dsIAP for 24 h. As shown in Fig. 5(A,B,C,D), at 24 h after dsIAP treatment, the IC50 values of dsIAP in Lepd-SL1 cells pre-exposed to either dsLoqs or dsR2D2 were 823.1 and 138.7 ng/ml, respectively, which are considerably higher than that in Lepd-SL1 cells pre-exposed to dsGFP (9.552 ng/ml). However, treatment with up to 4 µg/ml dsIAP did not induce any significant reduction in the cell viability in Lepd-SL1 cells pre-exposed to dsStauC. Although 48 h of continuous exposure to dsIAP induced some reduction in the viability of Lepd-SL1 cells pre-exposed to dsStauC, the cells still exhibited a considerable level of resistance to dsIAP (Fig. 5D). At the same time, the IC50 values for dsIAP in Lepd-SL1 cells pre-exposed to dsLoqs and dsR2D2 were drastically decreased to 2.305 ng/ml and 4.106 ng/ml, respectively (Fig. 5(A,B) & Table 1). The dsIAP IC50 value in the dsGFP pre-treatment control was also changed from 9.552 ng/ml to 4.243 ng/ml (Fig. 5C & Table 1) by increasing incubation time with dsIAP. These results clearly show that although Loquacious and R2D2 make contributions to RNAi in Lepd-SL1 cells, StauC is the primary key component for the high susceptibility of L. decemlineata to dsRNA-mediated RNAi.
Figure 5.
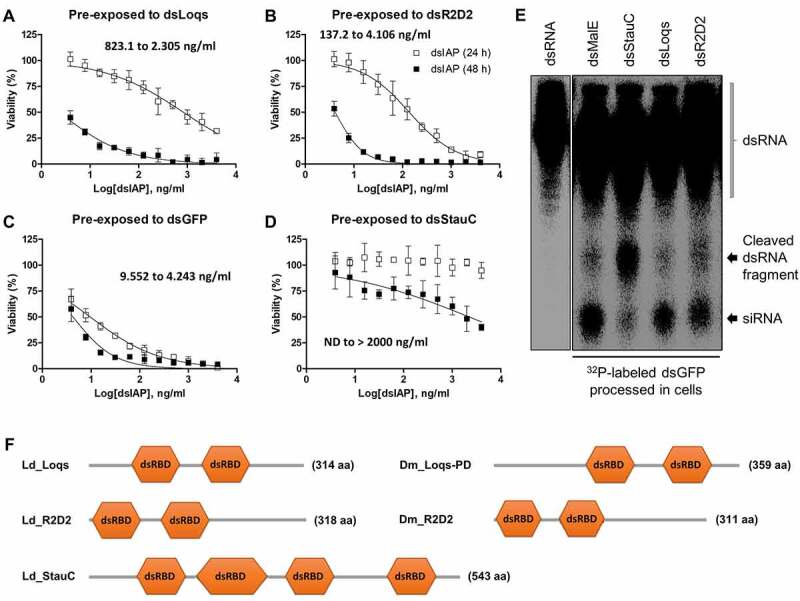
Effect of the knockdown of StauC, Loquacious, and R2D2 on the RNAi susceptibility of Lepd-SL1 cells. (A-D) The Lepd-SL1 cells were pre-treated with dsRNA targeting L. decemlineata Loquacious (A), R2D2 (B), GFP (C, control) StauC, (D) for 3 days. The cells were exposed to different concentrations of dsIAP (3.9 ng/ml – 4000 ng/ml) for 48 h. The cell viability was determined by Cell Counting Kit 8. Mean ± standard deviation (n = 3) are shown. (E) Processing of dsRNA to siRNA in Lepd-SL1 cells treated with dsLoqs, dsR2D2, dsStauC, or dsmalE (control). The Lepd-SL1 cells were seeded in six-well plates and exposed to dsLoqs, dsR2D2, dsStauC, or dsmalE for 3 days. The cells were then exposed to 1.6 million CPM []32P-labelled dsGFP. At 48 h after treatment with the second dsRNA, the total RNA was isolated from the cells and resolved on 16% acrylamide-urea gel. The dried gel was then analysed using the phosphorImager. The first lane shows dsGFP used as a marker. (F) Schematic representation of protein domain organization for StauC, Loquacious and R2D2 orthologs in L. decemlineata (Ld) and D. melanogaster (Dm)
Table 1.
IC50 values (ng/ml) of dsIAP after 24 and 48 h of exposure (n = 3, mean ± SD)
| 24 h of exposure to dsIAP | 48 h of exposure to dsIAP | |||
|---|---|---|---|---|
| Lepd-SL1 | IC50 value | R2 | IC50 value | R2 |
| pre-exposed to dsGFP | 9.552 ± 1.548 | 0.957 | 4.243 ± 0.901 | 0.8519 |
| pre-exposed to dsStauC | N/D | >2000 | 0.6399 | |
| pre-exposed to dsLoqs | 823.1 ± 187.5 | 0.92 | 2.305 ± 0.689 | 0.8893 |
| pre-exposed to dsR2D2 | 137.2 ± 25.3 | 0.9505 | 4.106 ± 0.343 | 0.9615 |
Consistent with these findings, the dsRNA-processing assay results also showed that a significant amount of the conversion of 32P-labelled dsGFP to siRNA occurred in Lepd-SL1 cells pre-exposed to either dsLoqs or dsR2D2 within 24 h of incubation with the radioactive dsGFP. The detected siRNA levels seem to be similar to or somewhat lower than that of Lepd-SL1 cells pre-treated with non-specific dsmalE. In contrast, Lepd-SL1 cells pre-exposed to dsStauC showed only minimal siRNA production under the same assay condition. Instead, another length of RNA fragment longer than siRNA was detected in these cells (Fig. 5E). These data suggest that knockdown of Loquacious or R2D2 cannot substantially block the conversion of dsRNA into functional siRNAs. The endogenous Loquacious and R2D2 are also unable to functionally compensate for the loss of StauC, especially in dsRNA to siRNA processing.
The results of quantitative real-time PCR analysis showed that pre-exposure of Lepd-SL1 cells to dsRNA significantly decreased its target gene mRNA levels when compared to those in dsGFP or no dsRNA treated control cells (Fig. S5). Interestingly, the RNAi-mediated depletion of R2D2 also led to significant induction in the expression of Loqs-PD and StauC genes (Fig. S5) in Lepd-SL1 cells. Treatment with dsStauC also upregulated Loquacious, but not R2D2 (Fig. S5). However, silencing of Loquacious did not affect the expression of R2D2 and StauC (Fig. S5).
StauC is highly expressed in L. decemlineata larvae and adults
Previous studies showed that feeding dsRNA induces knockdown of the target gene and mortality in L. Decemlineata [4,5]. It is possible that higher expression of StauC in the alimentary canal than in other tissues may facilitate this effect. To test this hypothesis, StauC mRNA levels were determined in different tissues dissected from L. Decemlineata larvae and adults. Significantly higher StauC mRNA levels were detected in the alimentary canal of larvae and adults than in any other tissues tested (Fig. 6(A,B)). These data suggest that the cells in the alimentary canal of L. Decemlineata may be more susceptible to dsRNA-mediated gene silencing.
Figure 6.
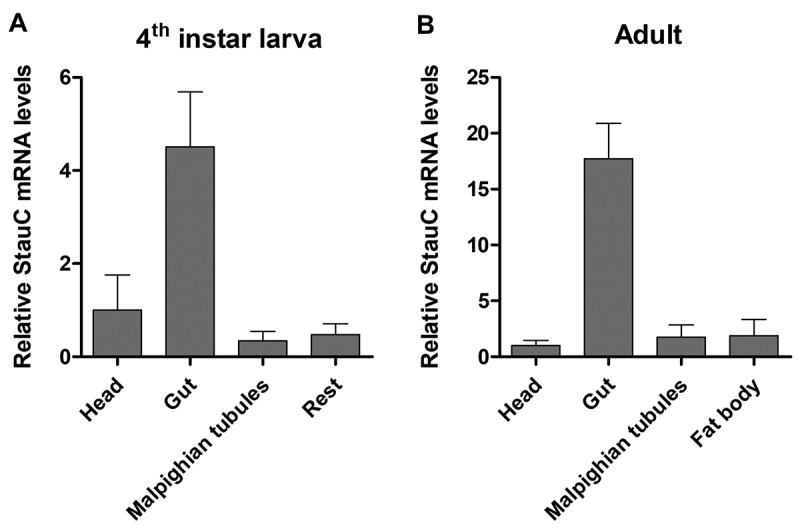
Tissue-specific expression of StauC in L. decemlineata. (A) qRT-PCR analysis of StauC mRNA levels in different tissues of L. decemlineata 4th instar larvae. (B) qRT-PCR analysis of StauC mRNA levels in various tissues of L. decemlineata adults
Discussion
In the current studies, we determined whether Lepd-SL1 cells can develop RNAi resistance by reducing StauC levels. Lepd-SL1 cells exposed to dsStauC show an extremely high level of resistance to dsIAP (Figs. 2A & Figs. 5D). Also, the RNAi insensitivity induced by StauC knockdown is not overcome by higher doses or longer exposure to dsIAP (Fig. 2A & Fig. S3), suggesting that StauC expression is essential for dsRNA-mediated RNAi in Lepd-SL1 cells. Intriguingly, although some induction in Loquacious expression, but not R2D2, was detected in Lepd-SL1 cells exposed to dsStauC, it is unlikely that the endogenous levels of Loquacious and R2D2 in these cells are sufficient to compensate for the loss-of-function of StauC. These data suggest that Loquacious and R2D2 probably do not have functional redundancy with StauC in coleopteran insects.
The dsRNA-mediated depletion of Loquacious or R2D2 also affords dsIAP insensitivity in Lepd-SL1 cells at least for 24 h (IC50 values of dsIAP after 24 h of exposure are 823.1 and 137.2 ng/ml, respectively when compared to 9.552 ng/ml the dsGFP treated control cells) (Fig. 5(A,B,C)). However, in contrast to the case of StauC-silenced Lepd-SL1 cells, their dsIAP susceptibility was restored by prolonged incubation with dsIAP (Fig. 5(A,B)). Consistent with these observations, the dsRNA-processing assay also revealed that the pre-exposure to dsLoqs or R2D2 does not block dsRNA conversion into siRNA in Lepd-SL1 cells (Fig. 5E). However, dsRNA-mediated depletion of StauC makes Lepd-SL1 cells produce RNA fragments longer than siRNA from dsGFP (Fig. 5E), which implies that StauC is essential for dsRNA processing into siRNA or the enhanced enzyme activity of Dcr proteins. This is an interesting issue that has to be further investigated in future studies using purified StauC and Dcr proteins. Overall, these observations indicate that dsRNA processing to siRNA in Lepd-SL1 cells requires StauC, but not Loquacious and R2D2. The data also suggest that RNAi response can be triggered in Lepd-SL1 cells without the strong activity of Loquacious and R2D2.
The dsRNA-mediated depletion of Loquacious induced no substantial increase in StauC and R2D2 levels (Fig. S5) and the measured IC50 value for dsIAP in Lepd-SL1 cells exposed to dsLoqs is 86-fold higher than in the cells treated with dsGFP (Table 1). These data suggest that the endogenous levels of StauC and R2D2 are insufficient to compensate for the loss-of-the function of Loquacious. In contrast to dsLoqs treatment, incubation with dsR2D2 activates both StauC and Loquacious genes in Lepd-SL1 cells (Fig. S5). However, the cells exposed to dsR2D2 still exhibited significant resistance to dsIAP (14.3-fold higher than the dsGFP pre-treatment control), implying that R2D2 also contributes to RNAi functioning in L. decemlineata probably in a way distinct from StauC and Loquacious. It is still unclear if StauC and Loquacious can mimic the function of R2D2 in the RNAi of Lepd-SL1 cells, but the gene/genetic compensation for R2D2 occurring in Lepd-SL1 cells seems not that efficient.
Our previous studies showed that StauC mRNA levels were reduced by ~80% in the RNAi-resistant Lepd-SL1 cells selected by the prolonged exposure to increasing concentration of dsIAP for multiple generations compared to its levels in naïve Lepd-SL1 cells [16]. In the present study, we demonstrate that the Lepd-SL1 cells pre-exposed to dsGFP quickly die within 24 to 48 h after dsIAP treatment (Fig. S2). However, prolonged exposure to a high concentration of dsIAP still fails to induce strong apoptosis in the Lepd-SL1 cells pre-exposed to dsStauC (Figs. S3A and C). These results suggest that RNAi-resistant Lepd-SL1 cells’ appearance after treatment with dsIAP is mediated by reducing StauC expression and not selecting the cells with low StauC production. These observations may implicate that Lepd-SL1 cells possibly develop RNAi resistance at molecular levels via inducing changes in StauC protein turnover after continuous exposure to dsRNA insecticide.
Another important finding of this paper is that the heterologous expression of StauC increases RNAi efficiency in RNAi refractive Sf9 cells. Many reports have demonstrated that lepidopteran insects have several biological barriers for dsRNA delivery [31], including the endosomal entrapment of dsRNA [32] and high levels of endogenous dsRNases [33]. Besides, our present (Fig. 2B) and previous studies [32] have shown that the heterologous expression of the L. decemlineata gene related to RNAi increases the RNAi efficiency in Sf9 cells, suggesting that the insufficient expression of RNAi genes is probably another reason for lower RNAi efficiency in Lepidopteran insects. Intriguingly, StauC overexpression can also facilitate RNAi in Sf9 cells where no StauC homologs are expressed (Fig. 2B). In contrast, overexpression of StauC does not further improve RNAi efficiency in Drosophila Kc cells except if their high efficiency to RNAi declines by the knockdown of Loqs-PD (Fig. 3). These data suggest that StauC can enhance RNAi efficiency in RNAi refractive insect cells by offering Loqs-PD-like activity or functionally compensating for their insufficient Loqs-PD activity.
Loqs-PD is a unique Loquacious isoform capable of enhancing the rate of dsRNA cleavage by Dcr-2 in D. melanogaster [18,21]. Our cell-based assay using D. melanogaster Kc cells showed that Kc cells pre-exposed to dsGFP are highly susceptible to dsRNA in the ng/ml range (Fig. 3A). However, 48 h of continuous exposure to 1 µg/ml dsRNA still failed to induce any significant gene silencing in Kc cells pre-exposed to dsLoqs-PD or dsDcr-2, or both (Fig. 3B and D). Considering the unique function of Loqs-PD in RNAi in Drosophila, these findings may suggest that the intracellular levels of dsRNA generally achieved in Kc cells are probably not high enough to be recognized in the cells by Dcr-2 enzyme as a substrate without the help of Loqs-PD. Similar results were observed in StauC-silenced Ledp-SL1 cells but not in the cells with Loquacious or R2D2 knockdown. The depletion of StauC strongly blocks the conversion of dsRNA into siRNA in Lepd-SL1 cells (Fig. 5A), conferring resistance to RNAi (Figs. 2A & Figs. 5B). Furthermore, the heterologous expression of StauC partly recovered the impaired RNAi in Loqs-PD knockdown Kc cells, but not in the cells pre-exposed to dsDcr-2 or dsR2D2 (Fig. 3). Although StauC and Loqs-PD do not share a marked homology in their sequence (Fig. 5F & Fig. S6), these results suggest that, like Loqs-PD in D. melanogaster, StauC is required to activate Dcr-2 in Lepd-SL1 cells.
The helicase domain of Dcr-2 consists of two RecA-like helicase subdomains, Hel1 and Hel2. Trettin et al. [21] showed that D. melanogaster Loqs-PD interacts with the Hel2 subdomain of the Dcr-2’s through an FDF-like motif in the C-terminus and this interaction is required to activate dsRNA cleavage by Dcr-2. Loqs-PD bound to Dcr-2 helps to recruit dsRNA substrates into the helicase domain of Dcr-2 [21]. In the present study, our protein-protein crosslinking experiment demonstrated that StauC forms a protein complex with Dcr-2a (Fig. 4). However, since our protein crosslinking studies are not quantitative and we could not find any putative FDF-like motif on StauC sequence, it is not clear if StauC directly interacts with Dcr-2 via its other protein-interacting motifs. Also, Dcr-2 could indirectly associate with StauC through its binding partners. However, considering the importance of StauC in the processing of dsRNA to siRNA in Lepd-SL1 cells (Fig. 5E), these observations support the hypothesis that StauC increases the rate of Dcr-2 cleavage via its physical association with Dcr-2. In future studies, a series of biochemical analyses using purified proteins are required to understand the mechanism by which StauC modulates Dcr-2 cleavage.
Coleopteran insects are well-known for their high vulnerability to dsRNA-mediated RNAi. However, how long the dsRNA-mediated gene silencing effect lasts in coleopteran insects is not well understood. Our cell-based assays using Lepd-SL1 cells revealed that the duration of gene silencing induced by dsRNA was sufficiently long enough to cause a prolonged knockdown of a target protein in these cells. For instance, exposure to dsStauC resulted in a significant knockdown of this protein in Lepd-SL1 cells within 48 to 72 h of the treatment (Fig. 1A). The protein levels were not restored completely even after the subsequent incubation of the cells in dsRNA-free medium for 5 days (Fig. S3B), indicating that the gene silencing complexes formed after dsRNA treatment are functional for a lengthy period in Lepd-SL1 cells.
Ingestion of dsRNA is regarded as the preferred administration for controlling pests, which can be achieved by feeding them with the dsRNA targeting an essential gene directly or through its expression in transgenic plants or microorganisms [34–37]. In the present study, we demonstrated that the alimentary canal where ingested dsRNA is absorbed has substantially higher StauC levels than in the other tissues of L. decemlineata (Fig. 6). This may contribute to increased RNAi-induced by ingested dsRNA in this insect [3–6]. These data suggest a possibility that StauC could be used as one of the crucial indicators for predicting the susceptibility to ingested dsRNA insecticide.
Supplementary Material
Funding Statement
The research is supported by grants from NIH (GM070559‐15 and 1R21AI131427‐01), the NSF (Industry/University Cooperative Research Centers, the Center for Arthropod Management Technologies under Grant IIP‐1821936), Agriculture and Food Research Initiative Competitive Grant No. 2019‐67013‐29351 and the National Institute of Food and Agriculture, U.S. Department of Agriculture (under HATCH Project 2353057000).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authorship Contributions:
Participated in research design: Kim & Palli
Conducted experiments: Kim & Koo
Contributed new reagents or analytical tools: Palli
Performed data analysis: Kim & Yoon
Wrote or contributed to the writing of the manuscript: Kim & Palli
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Zhu KY, Palli SR.. Mechanisms, applications, and challenges of insect RNA interference. Annu Rev Entomol. 2020;65:293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baum JA, Bogaert T, Clinton W, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. [DOI] [PubMed] [Google Scholar]
- [3].Palli SR. RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci. 2014;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhu F, Xu J, Palli R, et al. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci. 2011;67:175–182. [DOI] [PubMed] [Google Scholar]
- [5].Petek M, Coll A, Ferenc R, et al. Validating the potential of double-stranded RNA targeting Colorado Potato Beetle Mesh gene in laboratory and field trials. Front Plant Sci. 2020;11:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mehlhorn SG, Geibel S, Bucher G, et al. Profiling of RNAi sensitivity after foliar dsRNA exposure in different European populations of Colorado potato beetle reveals a robust response with minor variability. Pestic Biochem Physiol. 2020;166:104569. [DOI] [PubMed] [Google Scholar]
- [7].Maximo WPF, Howell JL, Mogilicherla K, et al. Inhibitor of apoptosis is an effective target gene for RNAi-mediated control of Colorado potato beetle, Leptinotarsa decemlineata. Arch Insect Biochem Physiol. 2020;104:e21685. [DOI] [PubMed] [Google Scholar]
- [8].Chereddy S, Gurusamy D, Howell JL, et al. Double-stranded RNAs targeting inhibitor of apoptosis gene show no significant cross-species activity. Arch Insect Biochem Physiol. 2020;104:e21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Knorr E, Fishilevich E, Tenbusch L, et al. Gene silencing in Tribolium castaneum as a tool for the targeted identification of candidate RNAi targets in crop pests. Sci Rep. 2018;8:2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cao M, Gatehouse JA, Fitches EC. A systematic study of RNAi effects and dsRNA stability in Tribolium castaneum and Acyrthosiphon pisum, following injection and ingestion of analogous dsRNAs. Int J Mol Sci. 2018;19:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Niu X, Kassa A, Hu X, et al. Control of Western Corn Rootworm (Diabrotica virgifera virgifera) Reproduction through plant-mediated RNA interference. Sci Rep. 2017;7:12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rangasamy M, Siegfried BD. Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera: chrysomelidae) adults. Pest Manag Sci. 2012;68:587–591. [DOI] [PubMed] [Google Scholar]
- [13].Khajuria C, Ivashuta S, Wiggins E, et al. Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. PLoS One. 2018;13:e0197059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hu X, Steimel JP, Kapka-Kitzman DM, et al. Molecular characterization of the insecticidal activity of double-stranded RNA targeting the smooth septate junction of western corn rootworm (Diabrotica virgifera virgifera). PLoS One. 2019;14:e0210491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dhandapani RK, Duan JJ, Palli SR. Orally delivered dsRNA induces knockdown of target genes and mortality in the Asian long-horned beetle, Anoplophora glabripennis. Arch Insect Biochem Physiol. 2020;104:e21679. [DOI] [PubMed] [Google Scholar]
- [16].Yoon JS, Mogilicherla K, Gurusamy D, et al. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proc Natl Acad Sci U S A. 2018;115:8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Obbard DJ, Gordon KH, Buck AH, et al. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hartig JV, Forstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fukunaga R, Han BW, Hung JH, et al. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hartig JV, Esslinger S, Bottcher R, et al. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Trettin KD, Sinha NK, Eckert DM, et al. Loquacious-PD facilitates Drosophila Dicer-2 cleavage through interactions with the helicase domain and dsRNA. Proc Natl Acad Sci U S A. 2017;114:E7939–E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pham JW, Pellino JL, Lee YS, et al. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. [DOI] [PubMed] [Google Scholar]
- [23].Tomari Y, Matranga C, Haley B, et al. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. [DOI] [PubMed] [Google Scholar]
- [24].Yoon JS, Shukla JN, Gong ZJ, et al. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: identification of key contributors. Insect Biochem Mol Biol. 2016;78:78–88. [DOI] [PubMed] [Google Scholar]
- [25].Gaddelapati SC, Kalsi M, Roy A, et al. Cap ‘n’ collar C regulates genes responsible for imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Biochem Mol Biol. 2018;99:54–62. [DOI] [PubMed] [Google Scholar]
- [26].Cui Y, Sui Y, Xu J, et al. Juvenile hormone regulates Aedes aegypti Kruppel homolog 1 through a conserved E box motif. Insect Biochem Mol Biol. 2014;52:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shukla JN, Kalsi M, Sethi A, et al. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016;13:656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou R, Czech B, Brennecke J, et al. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Miyoshi K, Miyoshi T, Hartig JV, et al. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010;16:506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Leuschner PJF, Obernosterer G, Martinez MJ. MicroRNAs: loquacious speaks out. Curr Biol. 2005;15(15):R603–5. [DOI] [PubMed] [Google Scholar]
- [31].Terenius O, Papanicolaou A, Garbutt JS, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. [DOI] [PubMed] [Google Scholar]
- [32].Yoon JS, Gurusamy D, Palli SR. Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem Mol Biol. 2017;90:53–60. [DOI] [PubMed] [Google Scholar]
- [33].Arimatsu Y, Kotani E, Sugimura Y, et al. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2007;37:176–183. [DOI] [PubMed] [Google Scholar]
- [34].Nandety RS, Kuo YW, Nouri S, et al. Emerging strategies for RNA interference (RNAi) applications in insects. Bioengineered. 2015;6:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Joga MR, Zotti MJ, Smagghe G, et al. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front Physiol. 2016;7:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang J, Khan SA, Heckel DG, et al. Next-generation insect-resistant plants: rNAi-mediated crop protection. Trends Biotechnol. 2017;35:871–882. [DOI] [PubMed] [Google Scholar]
- [37].Whitten MM. Novel RNAi delivery systems in the control of medical and veterinary pests. Curr Opin Insect Sci. 2019;34:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


