ABSTRACT
The ribonucleoprotein RNase MRP is responsible for the processing of ribosomal RNA precursors. It is found in virtually all eukaryotes that have been examined. In the Euglenozoa, including the genera Euglena, Diplonema and kinetoplastids, MRP RNA and protein subunits have so far escaped detection using bioinformatic methods. However, we now demonstrate that the RNA component is widespread among the Euglenozoa and that these RNAs have secondary structures that conform to the structure of all other phylogenetic groups. In Euglena, we identified the same set of P/MRP protein subunits as in many other protists. However, we failed to identify any of these proteins in the kinetoplastids. This finding poses interesting questions regarding the structure and function of RNase MRP in these species.
KEYWORDS: MRP, RNA, ribosomal RNA processing, bioinformatics, Euglenozoa, kinetoplastids
Introduction
RNase P and RNase MRP are ubiquitous ribonucleoprotein particles (RNPs) that process tRNA and ribosomal RNA precursors, respectively. MRP RNA contains the secondary structure elements as shown in Figure 1 with the Babesia bovis RNA as an example [1]. This figure also shows the presence of a more conserved catalytic (C) domain and a more variable specificity (S) domain. Protein subunits that are known to be part of MRP are Pop1, Rpp38, Rpp29, Pop5, Rpp25, Rpp20, Rpp14 and Rpp30 [2]. The three-dimensional structure of Saccharomyces cerevisiae MRP was recently elucidated using cryo-EM [3,4].
Figure 1.
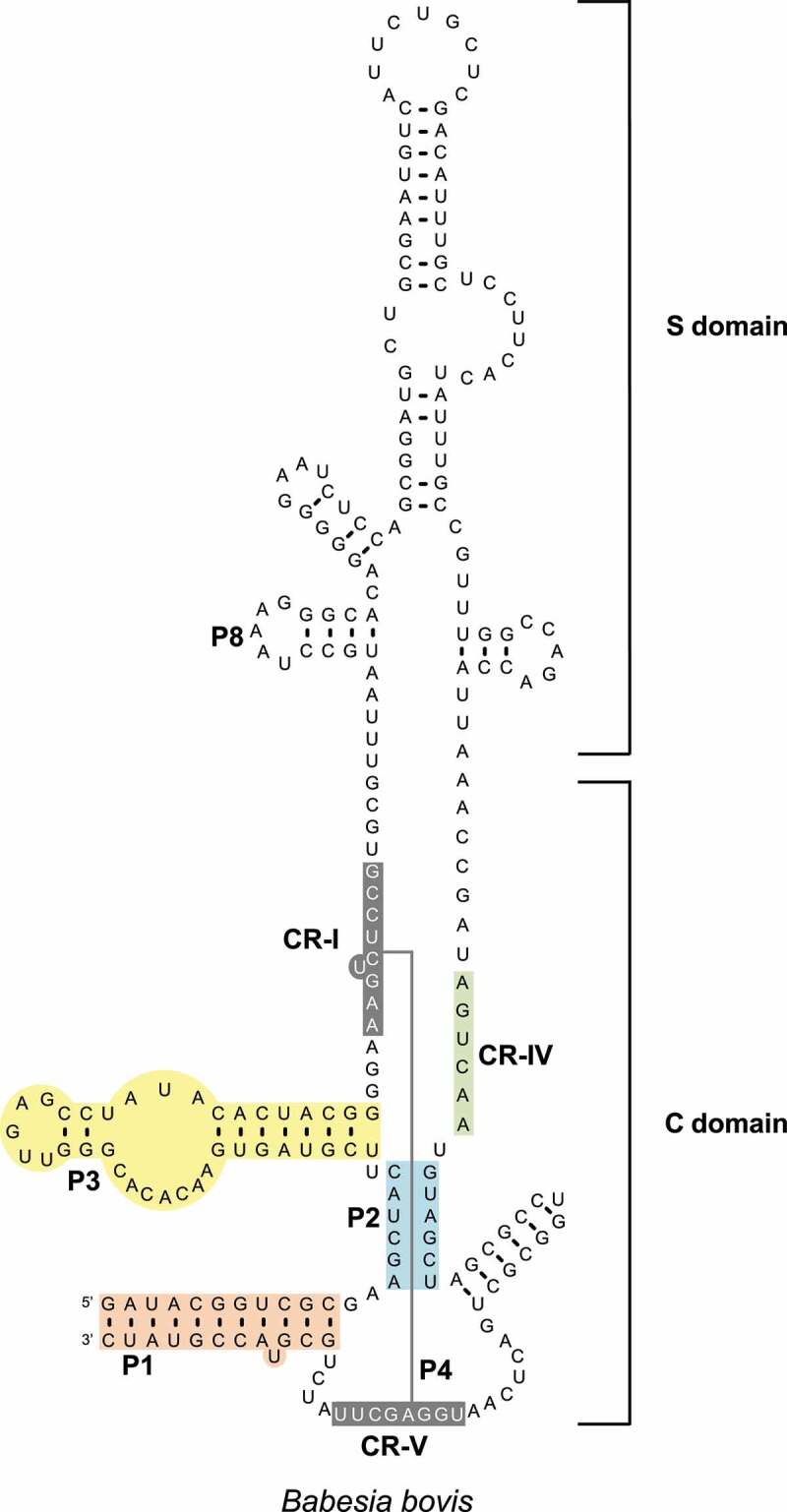
Secondary structure model of Babesia bovis MRP RNA. The conserved helices P1, P2, P3 and P4 (CR-I and CR-V) are highlighted with coloured backgrounds, as well as the conserved region CR-IV. The catalytic properties are within the highly conserved C domain and some of the specificity is within the less conserved S domain
The RNases P and MRP are structurally and evolutionary related, and they share a set of protein subunits. A pseudoknot structure, referred to as the P4 helix, of the P and MRP RNAs is critical for catalytic activity. In spite of these similarities of RNases P and MRP, they differ as to their phylogenetic distribution. RNase P is found in all kingdoms of life, whereas RNase MRP is restricted to eukaryotes.
Another difference between RNases P and MRP is that certain RNases P have been replaced by protein-only enzymes that are not dependent on an RNA for catalytic activity. The proteinaceous RNase P has been termed ‘protein-only RNase P’ (PRORP). The presence of PRORP is the explanation that in some species such as many plants and algae, we fail to identify an RNase P RNA [1,5]. The PRORP protein occurs only in eukaryotes. A number of metazoa have a mitochondrial PRORP while retaining a nuclear RNA-based RNase P. Notably, in many plants and in Trypanosoma brucei, an RNA-based RNase P is missing entirely and instead PRORPs have replaced the RNA-based enzyme both in the nucleus and in the mitochondrion [5,6]. PRORP is also known to be present in the plastids of some plants and algae.
In contrast to RNase P, there are apparently no instances where an RNase MRP is replaced by a protein-only enzyme. Consistently, an MRP RNA is found in virtually all eukaryotes. We have previously reported on the phylogenetic distribution of this RNA, [1,2] and only in a few groups, we could not identify the RNA. In particular, in the kinetoplastids, we failed to observe the MRP RNA as well as the protein subunits characteristic of this RNP. Another group where we initially failed to identify an MRP RNA is Giardia, but an MRP RNA of Giardia intestinalis was identified by Chen et al. [7].
A question that subsequently arises is whether our failure to identify MRP in Euglenozoa is due to highly divergent homologs that cannot be identified by the currently available bioinformatic methods. Or is the RNase MRP made of yet unidentified components, be they proteins or RNAs, resembling the situation for the protein-only RNase P?
The Euglenozoa are flagellated unicellular protists that are considered monophyletic, and they have no close relatives. The major groups of Euglenozoa are Euglenida (euglenids), Diplonemea and Kinetoplastea (kinetoplastids). The kinetoplastids have a granule named ‘kinetoplast’; this organelle is contained within the single mitochondrion of kinetoplastids and contains multiple copies of the mitochondrial genome. There is one kinetoplastid species, Perkinsela amoebae, that seems to be phylogenetically basal to other kinetoplastids [8–10].
One gene located in a snoRNA cluster in T. brucei has been identified as a plausible candidate for an RNase MRP RNA [11]. However, the published secondary structure did not conform to the standard model of MRP RNA, and no homologs in genomes of related species could be identified, casting doubts as to the authenticity of this RNA. So far, no homologs have been identified in other Euglenozoa. Furthermore, no protein components have been identified.
For the kinetoplastids, there are now more than a hundred genome assemblies available for analysis. Genome and transcriptome information is also available for euglenids. We have now further examined the presence of MRP RNA and protein subunits in all of these Euglenozoa using bioinformatic methods and report on a number of novel MRP RNA homologs in these species.
Materials and methods
Sequence resources were from NCBI [12], including the NCBI assembly database [13], the NCBI transcriptome database (https://www.ncbi.nlm.nih.gov/genbank/tsa/), the NCBI SRA data (https://www.ncbi.nlm.nih.gov/sra) and protein sequences of GenBank (see supplement for a list of genome assembly identifiers (leftmost column of supplementary table S_Table1.xls)).
Profile HMM searches made use of the Pfam database, Pfam A versions 28.0 and 33.1 [14], and data for RNA analysis used data from Rfam version 14.3 [15].
Bioinformatic methods used were BLAST (version 2.2.29+) [16], FASTA (version 36.3.6 f) [17], Clustal Omega [18], HMMER version 3.3 (http://hmmer.org) and the Infernal software for RNA analysis version 1.1.3 [19]. UNAfold version 3.8 [20] and Mfold version 3.6 [21] were used for the prediction of RNA secondary structure. In some cases, constraints to UNAfold and Mfold were used to avoid pairing of the CR-I and CR-V pseudoknot regions and to force the folding of specific helices. Sequence logos were created with software from Weblogo [22].
For the prediction of RNase P RNA and MRP RNA, we used, in addition to other methods as described under ‘Results’, the Infernal software with the Rfam covariance models RF00009 and RF00030, respectively. A covariance model specific to kinetoplastid MRP RNA was constructed as follows. An MRP RNA candidate sequence was previously identified in T. brucei [11]. As we believed the structure shown in that paper was not correct, we assigned the P1 helix, 5ʹ and 3ʹ ends differently and deduced a secondary structure consistent with already available MRP RNAs. Using the revised T. brucei sequence, we searched all available Euglenozoa genomes as well as transcriptome data for the presence of MRP RNA. With similarity searches, it was possible to identify MRP RNA homologs in many kinetoplastids. All candidates that we found were examined with respect to their secondary structure. Ultimately, we used a collection of all kinetoplastid MRP RNA sequences that we identified to create a secondary structural alignment that includes the P1, P2, P3, P4 and P8 helices and the CR-IV motif. The alignment was used to produce a covariance model (supplemental files S_kinetoplastid.stk.txt and S_kinetoplastid.cm.txt).
The reason the standard Rfam MRP RNA model is not effective in the identification of Euglenozoa homologs is that the primary sequences of Euglenozoa RNAs have diverged strongly from sequences being members of the RF00030 seed alignment. This is illustrated in a simplified structural alignment without the S domain (supplemental figure S_alignment_RF00030_euglenozoa.pdf).
The great majority of MRP RNA sequences were identified from analysis of genome assemblies, but the sequences of the diplonemids Diplonema ambulator, Rhynchopus euleeides, Rhynchopus humris, Lacrimia lanifica, the euglenid Eutreptiella gymnastica as well as Trypanosoma theileri were identified from SRA data. These MRP RNA sequences are shown in the supplemental file S_MRP_RNA.fa.txt.
For Pfam A searches, all genomes were translated into all six reading frames, and the resulting protein sequences were searched using hmmscan of the hmmer software with Pfam A as a database. Thus, all the domains present in this database were considered in this search. We also searched all available Euglenozoa proteins in GenBank.
Results
MRP RNA
We used a variety of methods to identify MRP RNA in Euglenozoa. First, for closely related species, it was possible to use standard similarity searches such as FASTA and BLAST. Second, we noted that in Euglenozoa the CR-I and CR-V regions are highly conserved, making possible a method similar to that we previously used [1] where we construct hmmer profiles of these regions and search instances where they are within a specific distance (100–1000 nt) from each other. Third, we exploited covariance models for MRP RNA in the context of the Infernal software ([19], see ‘Materials and Methods’). However, as we discovered the standard Rfam model of MRP RNA (RF00030) was not adequate for a large majority of Euglenozoa species, we constructed a novel covariance model based on selected kinetoplastid MRP RNAs as described under ‘Materials and Methods’. This model turned out highly effective for prediction of the RNA in kinetoplastids.
An overview of our results with respect to the prediction of MRP RNA in Euglenozoa is shown in the context of a schematic taxonomic tree in Figure 2 (open and filled green boxes). More detailed results of Infernal cmsearch are shown in supplemental file S_Table1.xls. Among the kinetoplastids, we failed to identify the RNA in Angomonas, Strigomonas, Bodo saltans and Hemistasia. In the case of Trypanoplasma, a full-length RNA was not identified, but only partial matches that covered the conserved 5ʹ and 3ʹ parts of the catalytic domain. In the case of Hemistasia, we analysed only raw short RNA sequence reads as no assemblies are available. Therefore, our failure to identify the RNA in Hemistasia is not conclusive.
Figure 2.
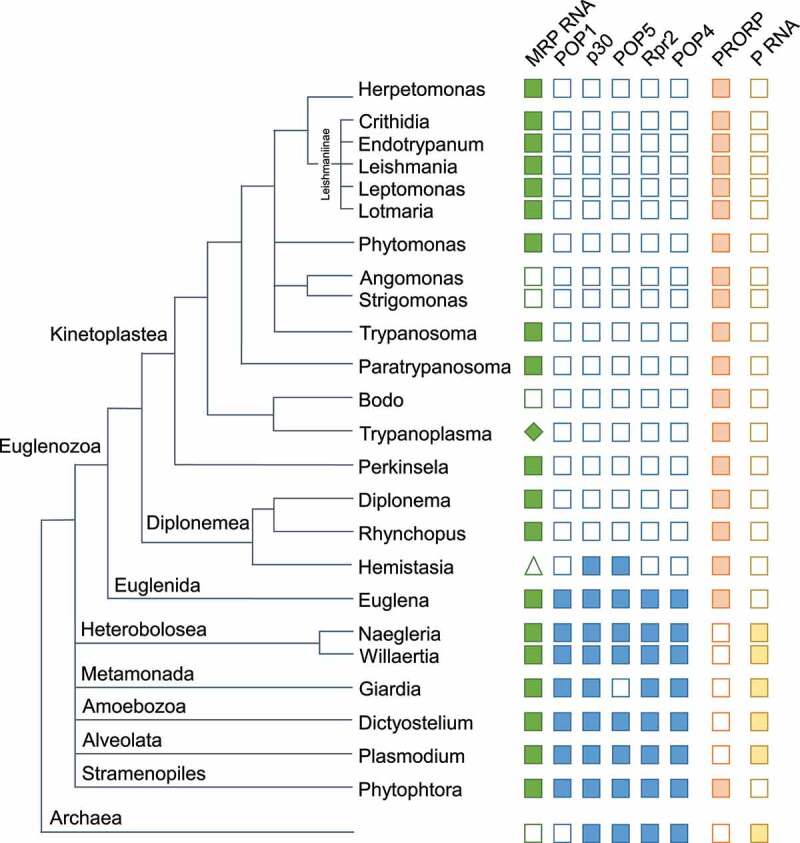
Schematic phylogenetic tree of Euglenozoa and other protists. Filled and open green squares indicate instances where an MRP RNA was identified/not identified, respectively. A triangle in the case of Hemistasia indicates that a genome assembly is not available and the apparent absence of MRP RNA is not conclusive. A filled diamond for Trypanoplasma indicates that partial matches to MRP RNA were identified. Filled and open blue squares indicate P/MRP proteins found/not found. Five such proteins were analysed and appear in the order POP1, p30, Pop5, Rpr2 and POP4. The occurrence of PRORP and RNase P RNA is indicated with light orange and light yellow squares, respectively
We also identified MRP RNA in the Euglenida and Diplonemea groups. In contrast to the kinetoplastids, Euglena gracilis and Euglena longa MRP RNA was identified with the standard Rfam model (RF00030). Neither this model nor our novel kinetoplastid model was adequate for Perkinsela and some species of Diplonemae, where we instead employed a method with hmmer profiles for the CR-I and CR-V regions [1] or fasta and blastn searches to identify the MRP RNA primary sequence.
Secondary structure models of selected Euglenozoa MRP RNAs are shown in Figure 3 and in supplemental file S_SS.pdf. (Predictions of structure were with mfold or UNAfold as described under ‘Materials and Methods’; actual sequences in FASTA format are in supplemental files S_cmsearch.fa.txt and S_MRP_RNA.fa.txt). All structures shown are tentative and based on what we previously knew about MRP RNA structure. In particular, the S domain has a highly variable structure, and in this domain, only the P8 helix can be reliably predicted. For the previously published T. brucei MRP RNA [11,23,24] (see ‘Materials and Methods’), we now identified the RNA in the same genomic region, but our sequence is shorter at its 5ʹ and 3ʹ ends as compared to the previously published sequences (see also under ‘Discussion’), and we inferred a structure which conforms to the consensus structure of MRP RNA (Figure 3(a)).
Figure 3.
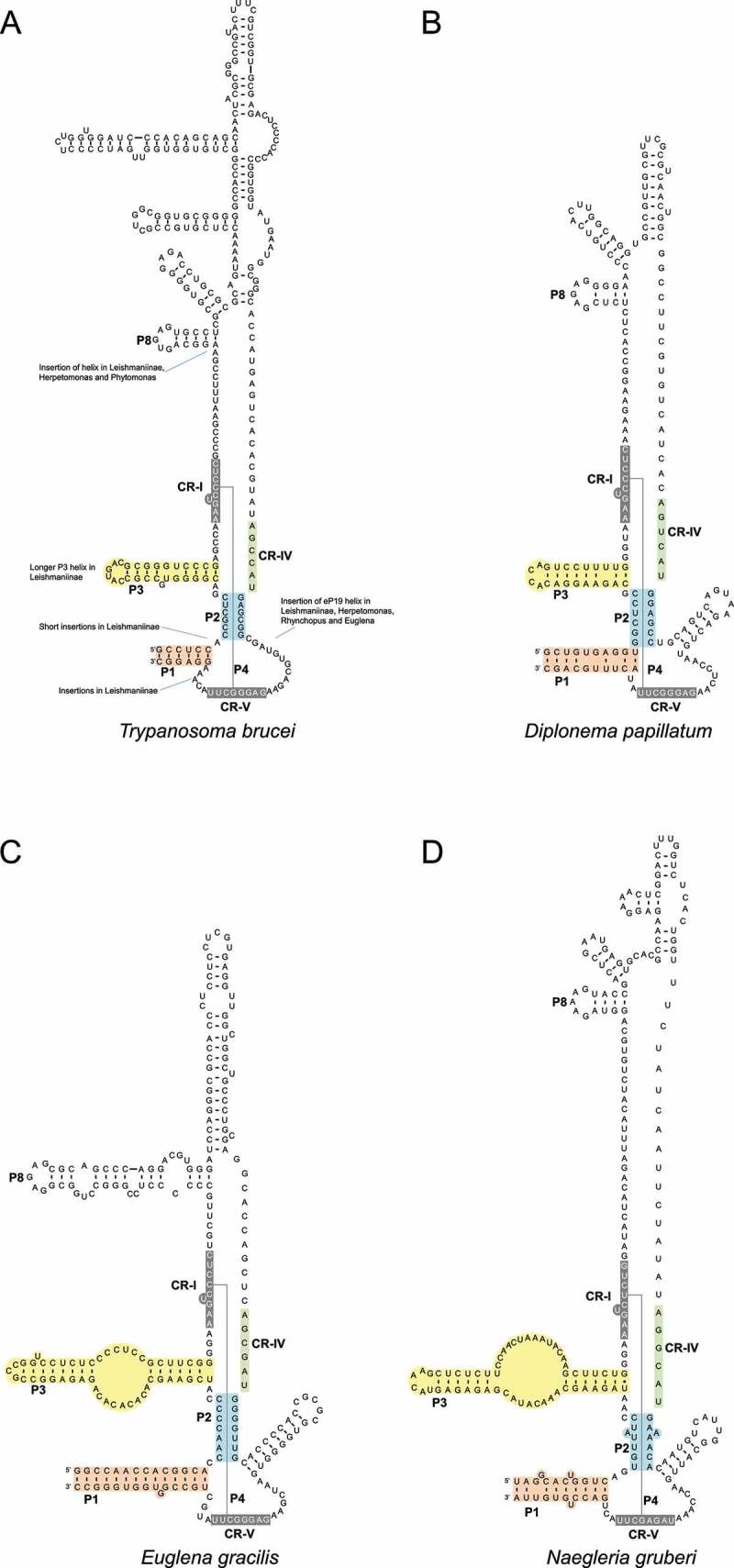
Secondary structure models of MRP RNAs. Conserved elements are highlighted as in Figure 1. (a). T. brucei. Positions where sequences are inserted in other kinetoplastids are indicated. (b). D. papillatum. (c). E. gracilis. (d). N. gruberi.
The predicted structures of the Diplonema papillatum and E. gracilis RNAs are shown in Figures 3(b,c). For reference, we also systematically searched all non-Euglenozoa protist genome assemblies (for some example species, see Figure 2) and derived secondary structures of the predicted MRP RNAs. In Figure 3(d), it is shown a predicted structure of Naegleria gruberi, a species of the phylum Heterolobosea which is believed to be one of the closest relatives to the Euglenozoa [25].
As with the MRP RNA in many other phylogenetic groups, the catalytic domain in Euglenozoa is conserved, with the exception of helices or single-stranded regions that have been inserted or deleted (Figure 3(a)). The 8–9 nucleotides in the P4 helix (CR-I and CR-V regions) are extremely well conserved in kinetoplastids as highlighted with the sequence logos in Figure 4 and supplemental Figure S_alignment_RF00030_euglenozoa.pdf.
Figure 4.
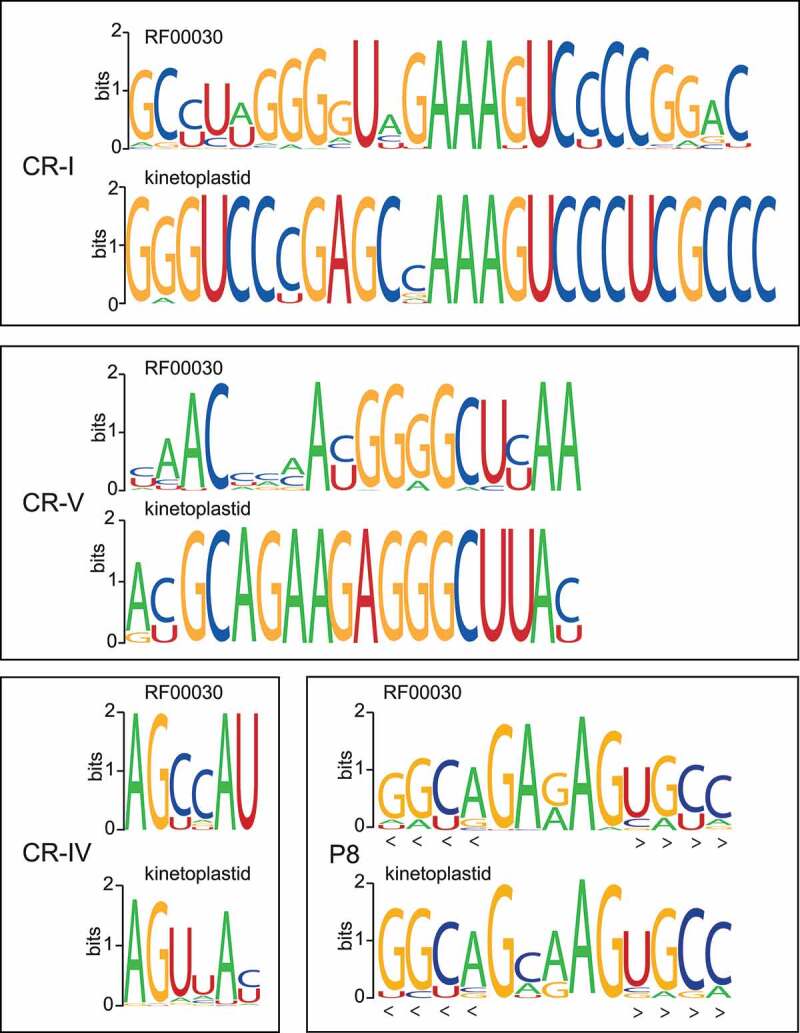
Sequence logos of CR-I, CR-IV, CR-V and P8 regions of MRP RNA. For each region are shown logos based on the Rfam RF00030 seed alignment and kinetoplastid sequences being the basis of our novel covariance model, respectively
There is a large size variation within the group of kinetoplastid MRP RNAs (supplemental table S_Table1.xls). Thus, the RNAs of Trypanosoma are in a small size range (about 300 nt, Figure 3(a)), while the Crithidia RNAs represent the longest members with up to 992 nt. As compared to Trypanosoma, there are multiple insertions for instance in the RNAs of Leishmaniinae (Crithidia, Leptomonas, Leishmania, Lotmaria, and Endotrypanum) as indicated in Figure 3(a) (see also supplemental Figure S_SS.pdf). Examples are, in Leishmaniinae and trypanosomatids, a CA-rich insertion of varying length between CR-V and the 3ʹ part of the P1 helix, an insertion not observed in non-kinetoplastid MRP RNA. A shorter CA-rich region is also inserted between helices P1 and P2 in the MRP RNAs of Leishmaniinae (Figure 3(a)). Finally, the P8 helix, with the ‘GARAR’ loop, is located at a varying distance from the P4 5ʹ region (compare, for instance, T. brucei, Figure 3(a), and Rhynchopus euleeides, Supplemental Figure S_SS.pdf).
MRP protein subunits
To obtain a better understanding of MRP in Euglenozoa, we searched all available genomes for the presence of proteins known to be part of MRP of other species. As described under ‘Materials and Methods’, Euglenozoa protein sequences were searched using hmmscan of the hmmer software (http://hmmer.org) with Pfam A [14] as a database. Remarkably, only in Euglena gracilis and Euglena longa, we identified a number of Pfam families related to MRP proteins, namely Rpp1/Rpp30, Pop1, Pop4/Rpp29, Pop5 and Rpr2/Rpp21 (Figure 2, supplemental table S_Table2.xls). The inferred protein sequences (not full length) are shown in supplemental file S_proteins.fa.txt.
For comparison, we also analysed many other non-Euglenozoa protists in the same way. The same proteins as in Euglena are typically identified (Figure 2, supplemental Table S_Table2.xls).
In contrast, we could not identify a single protein characteristic of RNA-based P/MRP in any of the kinetoplastids or in Diplonema (Figure 2). However, in Hemistasia, another member of the Diplonemea group, p30 and Pop5 homologs are identified. Therefore, Hemistasia seems to represent an intermediate between Euglena and the kinetoplastids with regard to protein subunits.
We also searched proteins with the Pfam model for PRORP and identified proteins with this domain in a large majority of Euglenozoa genomes. Two major groups of PRORP proteins are found. It is possible that these groups correspond to the nuclear and organellar proteins, respectively, but they cannot be assigned an intracellular location based on the available sequences only.
RNase P RNA has previously not been identified in Euglenozoa. Because of its relationship to PRORP, we now searched all available genomes of Euglenozoa and other protists using the Infernal software. The results suggest that RNase P RNA is indeed missing in Euglenozoa, while it is present in the many of the other protist groups (Figure 2). We encounter a situation where in most species either PRORP or RNase P RNA is present.
In conclusion, MRP RNA is found in a majority of Euglenozoa, and the Euglena and Hemistasia RNase MRPs seem to be similar with respect to their protein components to other non-Euglenozoa MRPs. However, Diplonema, Rhynchopus and all the kinetoplastids are apparently lacking the protein subunits characteristic of MRP. As to the situation with RNase P, PRORP is present in Euglenozoa, while RNase P RNA is missing.
Discussion
Using bioinformatic methods, we have here provided strong evidence of an MRP RNA in the vast majority of all Euglenozoa examined. These results indicate that in every phylogenetic group examined, the MRP RNA has now been identified, demonstrating that the RNA is truly ubiquitous in eukaryotes.
The only group for which we have access to several assemblies and still did not find an MRP RNA is Strigomonadinae (Angomonas and Strigomonas). Even in a recently published full chromosome assembly of Angomonas deanei (GCA_903995115), we failed to identify the RNA. Thus, it is possible that this group is lacking the MRP RNA.
All the Euglenozoa MRP RNAs that we now identified conform to the secondary structure characteristic of MRP RNA in general. However, there are distinct features, which is why previously used bioinformatic methods failed to identify these RNAs. Mainly, the primary sequences of kinetoplastids have diverged strongly from other phyla. Furthermore, the helix P3 in kinetoplastids is missing the symmetric loop characteristic of other metazoan MRP RNAs. In addition, some of the kinetoplastid species have very unusual insertions close to the P1 helix. Our assignment of P8 may be regarded as tentative in some species, but if this assignment is correct, we notice that the distance between CR-I of P4 and helix P8 is highly variable when comparing Euglena, Diplomena and the kinetoplastids. This extent of variation is unusual in other phyla, and only in certain fungi, this distance is somewhat longer than for the typical MRP RNA.
For the previously published T. brucei MRP RNA, we now inferred a structure which conforms to the standard structure of MRP RNA (Figure 3(a)). The main difference is that our sequence is truncated at its 5ʹ and 3ʹ ends as compared to the previously published sequences [11,23,26]. Although the published sequence was shown to be transcribed [11,23], it is not clear whether it may be processed to yield a shorter mature sequence. It is also possible that Trypanosoma MRP RNA has additional structural elements not present in any other species.
There is earlier evidence that Trypanosoma is missing an RNA-based RNase P. Instead, both the nuclear and mitochondrial RNase P activities have been replaced by a protein-only enzyme (PRORP) [5]. It would seem that this is the case throughout Euglenozoa as we have not been able to identify an RNase P RNA in any of these genomes, and we have noted that PRORP proteins are present in virtually all Euglenozoa genomes examined. This means that in case we do observe a P/MRP-related protein, it is not likely to be a partner of RNase P RNA, but rather to MRP RNA, or possibly to some other hitherto unknown RNA.
We did identify P/MRP protein homologs in Euglena; Rpp1/Rpp30, Pop1, Pop4/Rpp29, Pop5 and Rpr2/Rpp21. It seems likely that these proteins are components of the Euglena MRP. These are the same protein subunits that are present in other non-Euglenozoa protists such as in N. gruberi, a species being one of the closest relatives to Euglenozoa. With the exception of Pop1, the subunits are also found in the archaeal RNase P ([2], Figure 2).
The presence of P/MRP protein subunits in Euglena suggests that its MRP has a structure similar to that of other metazoan MRPs. We know from the structure of the yeast MRP that Pop1 is a main structural brace of the MRP, interacting with regions that include P4 and P8. In the yeast MRP, Pop5, Pop8 (Rpp14) and two copies of Rpp1 (Rpp30) form a tetramer that interacts with the C domain [4].
Whereas P/MRP protein homologs are present in Euglena, a search for RNA-dependent RNase P/MRP protein homologs failed to identify any such proteins in the kinetoplasts and in Diplonema. Only two such proteins seem to be present in Hemistasia. It must be noted that there are at least two possible reasons for the apparent lack of P/MRP proteins. First, the proteins in kinetoplastids may be highly divergent and escape detection using the standard hmm profiles. Second, there might be entirely different proteins that have replaced the standard MRP proteins. While Euglena MRP might have an RNP structure similar to that in human and yeast MRP, the corresponding enzyme in kinetoplastids could be very different.
In case kinetoplastids are missing the standard P/MRP protein components, what are the implications? The sequences of kinetoplastid MRP RNA are highly conserved, indicating that these RNAs are physiologically significant. What is the function of kinetoplastid MRP RNA? In studies of yeast and human, the RNase MRP is active in processing of pre-ribosomal RNA, cutting transcripts at the A3 site close to the 5.8S rRNA. For T. brucei, it has been suggested that the RNase MRP cut at B1, a site close to the A3 site, which seems to be the first step in the pre-rRNA processing cascade in T. brucei [11]. However, other MRP functions have been demonstrated, such as a role in cell cycle regulation in S. cerevisiae [27–29]. In two recent publications, the 3D structures of RNase P and MRP have been elucidated [3,4]. In these investigations, it was noticed that the MRP could be flexible in its substrate recognition, further pointing to the possibility of multiple substrates. In addition, MRP RNA might have other partners than the proteins of RNase MRP. Indeed, it has been observed that an RNA-dependent RNA polymerase is formed through a complex of MRP RNA with the protein telomerase reverse transcriptase (hTERT) [30]. One would also need to elucidate the intracellular localization of kinetoplastid MRP RNA. Is it present in mitochondria (kinetoplastids), in the nucleolus or elsewhere?
Kinetoplastid MRP RNA could thus have other functions than to process rRNA precursors. If proteins are missing, could the RNA be catalytic on its own? Has it recruited novel protein partners? To improve our understanding of the kinetoplastids, it will be vital to experimentally examine the kinetoplastid MRP and to identify its protein partners. A priority would be to investigate what proteins, if any, bind to the MRP RNA or to use knock-down of this gene to see whether the pre-rRNA processing is affected.
Supplementary Material
Acknowledgments
We are grateful for a grant from Wilhelm and Martina Lundgrens Vetenskapsfond to MAR and a grant from the Swedish Foundation for Strategic Research (RIF14–0081) to MDL.
Funding Statement
This work was supported by the Stiftelsen för Strategisk Forskning [RIF14–0081]; Stiftelserna Wilhelm och Martina Lundgrens.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Piccinelli P, Rosenblad MA, Samuelsson T.. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33(14):4485–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rosenblad MA, López MD, Piccinelli P, et al. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34(18):5145–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lan P, Zhou B, Tan M, et al. Structural insight into precursor ribosomal RNA processing by ribonuclease MRP. Science. 2020;369(6504):656–663. [DOI] [PubMed] [Google Scholar]
- [4].Perederina A, Li D, Lee H, et al. Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP. Nat Commun. 2020;11(1):3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pinker F, Bonnard G, Gobert A, et al. PPR proteins shed a new light on RNase P biology. RNA Biol. 2013;10(9):1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lechner M, Rossmanith W, Hartmann RK., et al. Distribution of ribonucleoprotein and protein-only RNase P in Eukarya. Mol Biol Evol. 2015;32(12):3186–3193. [DOI] [PubMed] [Google Scholar]
- [7].Chen XS, Penny D, Collins LJ. Characterization of RNase MRP RNA and novel snoRNAs from Giardia intestinalis and Trichomonas vaginalis. BMC Genomics. 2011;12:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gile GH, Faktorová D, Castlejohn CA, et al. Distribution and phylogeny of EFL and EF-1alpha in Euglenozoa suggest ancestral co-occurrence followed by differential loss. PLoS One. 2009;4(4):e5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22(4):168–174. [DOI] [PubMed] [Google Scholar]
- [10].Yazaki E, Ishikawa SA, Kume K, et al. Global Kinetoplastea phylogeny inferred from a large-scale multigene alignment including parasitic species for better understanding transitions from a free-living to a parasitic lifestyle. Genes Genet Syst. 2017;92(1):35–42. [DOI] [PubMed] [Google Scholar]
- [11].Barth S, Shalem B, Hury A, et al. Elucidating the role of C/D snoRNA in rRNA processing and modification in Trypanosoma brucei. Eukaryot Cell. 2008;7(1):86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Coordinators NR . Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44(D1):D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kitts PA, Church DM, Thibaud-Nissen F, et al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016;44(D1):D73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kalvari I, Argasinska J, Quinones-Olvera N, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46(D1):D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- [17].Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227(4693):1435–1441. [DOI] [PubMed] [Google Scholar]
- [18].Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. [DOI] [PubMed] [Google Scholar]
- [19].Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. [DOI] [PubMed] [Google Scholar]
- [21].Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244(4900):48–52. [DOI] [PubMed] [Google Scholar]
- [22].Crooks GE, Hon G, Chandonia, JM. et al. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liang XH, Uliel S, Hury A., et al. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA. 2005;11(5):619–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6(4):459–474. [DOI] [PubMed] [Google Scholar]
- [25].Rodriguez-Ezpeleta N, Brinkmann H, Burger G, et al. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17(16):1420–1425. [DOI] [PubMed] [Google Scholar]
- [26].Michaeli S, Doniger T, Gupta SK, et al. RNA-seq analysis of small RNPs in Trypanosoma brucei reveals a rich repertoire of non-coding RNAs. Nucleic Acids Res. 2012;40(3):1282–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cai T, Aulds J, Gill T, et al. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics. 2002;161(3):1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gill T, Aulds J, Schmitt ME. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J Cell Biol. 2006;173(1):35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gill T, Cai T, Aulds J, et al. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol Cell Biol. 2004;24(3):945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Maida Y, Yasukawa M, Furuuchi M, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461(7261):230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


