ABSTRACT
MicroRNAs (miRNAs) can serve as activation signals for membrane receptors, a recently discovered function that is independent of the miRNAs’ conventional role in post-transcriptional gene regulation. Here, we introduce a machine learning approach, BrainDead, to identify oligonucleotides that act as ligands for single-stranded RNA-detecting Toll-like receptors (TLR)7/8, thereby triggering an immune response. BrainDead was trained on activation data obtained from in vitro experiments on murine microglia, incorporating sequence and intra-molecular structure, as well as inter-molecular homo-dimerization potential of candidate RNAs. The method was applied to analyse all known human miRNAs regarding their potential to induce TLR7/8 signalling and microglia activation. We validated the predicted functional activity of subsets of high- and low-scoring miRNAs experimentally, of which a selection has been linked to Alzheimer’s disease. High agreement between predictions and experiments confirms the robustness and power of BrainDead. The results provide new insight into the mechanisms of how miRNAs act as TLR ligands. Eventually, BrainDead implements a generic machine learning methodology for learning and predicting the functions of short RNAs in any context.
KEYWORDS: miRNA, TLR, ligand, machine learning, RNA structure
Introduction
MicroRNAs (miRNAs) are very short non-coding RNAs (~22 nt) that predominantly bind to the 3´ untranslated regions of mRNAs to regulate their expression post-transcriptionally. To date, more than 2,000 miRNAs have been discovered in humans, and it is believed that they collectively regulate about one-third of the genes in the human genome [1]. miRNAs play important roles in development and physiology, and have been linked to various human diseases. These days these small RNAs are increasingly being pursued as both clinical diagnostics and therapeutic targets relevant in many medical fields, ranging from cancer to neurodegenerative disease. In particular, miRNAs are considered as potential biomarkers for diseases and treatment responses [2,3]. Under certain conditions such as cellular stress and malignancy, miRNAs are released from cells, thereby potentially acting as extracellular signalling molecules enabling intercellular communication [4,5]. In line with this, it has been recently discovered that some extracellular miRNAs directly activate membrane receptors such as Toll-like receptors (TLRs) [4,6,7], thereby expanding the function of miRNAs beyond their conventional role in gene regulation.
TLRs are pattern recognition receptors detecting both pathogen-associated molecules and damage-associated factors, such as those derived from dying cells and tumour tissue. Upon activation, TLRs signal through a complex array of effector proteins, resulting in an inflammatory response [8,9]. Among the different TLR family members, TLR7 and TLR8 (TLR7/8) primarily recognize single-stranded RNA (ssRNA) 40 derived from human immunodeficiency virus-1 (HIV-1). The RNA’s GU-rich motifs are essential for species-specific TLR7/8 recognition [10–12], and a specific activating consensus sequence composed of GUUGUGU repeats (G, guanosine; U, uridine) was linked to the degree of receptor activation [13]. Forsbach et al. systematically narrowed down GU-rich and AU-rich nt tri- and tetramers to be crucial for activation of human TLR7/8. Furthermore, diverse motifs exhibit specific receptor preferences, thereby triggering the release of cytokines, such as TNF-α and IFN-α [14]. TLR7 was recently found to detect host-derived RNA, including miRNAs [4,6,15]. let-7 miRNA, when extracellularly present in the brain, activates TLR7 in microglia, the resident immune cells in the central nervous system (CNS). Consequently, microglia release inflammatory molecules and cause neurodegeneration in the cerebral cortex [4,16]. Moreover, cerebrospinal fluid of patients with Alzheimer’s disease (AD), the most common neurodegenerative disease in humans, exhibits elevated levels of let-7 copies [3,4,17]. Overall, these findings suggest a mechanistic contribution of the interaction between miRNAs and TLR7 to neurodegenerative processes.
Not only pre-miRNA, but also mature miRNAs can form stable secondary structures that potentially are not only important for their extracellular stability, but may also affect the presentation of specific sequence motifs to TLR7/8 [18]. To reduce time- and cost-intensive experiments on the mechanism of the interaction between miRNAs and TLRs in a given disease context effectively, reliable in silico prediction methods are needed for the identification of oligonucleotides serving as receptor ligands and activating an immune cell response. Previous studies on in silico classification of nt sequences have been mainly focussed on genomic DNA and RNA within the genomic context. Lee et al. introduced a method to predict putative enhancers in the mouse and human genomes based on DNA sequence [19]. kmer-SVM is a Support Vector Machine (SVM) that uses a string kernel operating on subsequences of length k the so-called k-mers [20]. Most of the follow-up algorithms focused on the identification of DNA genomic elements, for instance, from large Chip-seq datasets (e.g. gkmSVM [21]) or using DNA-specific structure properties (e.g. PseKNC [22]). Zhang et al. proposed a solution for the identification of piwi-interacting RNAs using k-mer features from the genome sequence without considering structure [23], while iMcRNA uses sequence- and structure-based features to identify precursor miRNAs via a pseudo amino acid composition approach [24]. The vectorization server repRNA [25] generates k-mer and pseudo-structure features of RNAs based on reduced representation of their minimum-free-energy (MFE) structures to enable machine learning tasks. However, to the authors’ knowledge, no accessible solution for the classification of short RNAs potentially serving as receptor ligands exists so far. It is also often desired to integrate previous knowledge of the applied features to better interpret and link the prediction process with knowledge from the literature and other experimental sources that cannot easily be incorporated without an interpretable methodology.
The let-7 miRNAs’ UUGU motif represents the required minimum motif to induce cytokine release from microglia through TLR7 [16]. Whether the structural features of a given oligonucleotide, e.g. a miRNA, are beneficial for TLR7/8 activation/binding or potentially mask/inhibit the association to its binding sites remains unexplored to date. Still, the secondary structure should be considered as an essential feature for predicting an oligonucleotide’s potential to activate TLR7/8. As mature miRNA is very short, transient hairpin structures can be formed. Thus, bioinformatics solutions designed to classify highly structured RNA molecules [26] are not suitable to predict oligonucleotides as receptor ligands. Instead, a fine-tuned flexible definition of structuredness accompanied by sequence information is required. In particular, as homo-dimerization likely occurs when miRNAs are released in larger quantities [18], base-pairing potential during homo-duplex formation should be taken into account by a model aiming to predict miRNAs as extracellular signalling molecules.
The main aim of this work was to identify miRNAs that act as TLR7/8 ligands in humans and mice. Since the experimental validation of a vast number of miRNA candidates able to activate TLR7/8 within a reasonable time frame is cumbersome and costly, we applied BrainDead, a novel machine learning (ML) approach for the identification of TLR7/8-activating miRNAs. The methodology assesses an RNA’s accessibility via its ensemble of most stable structures and combines this information with k-mer feature generation for a user-defined set of motifs. BrainDead was trained on a smaller set of previously validated miRNAs that in their extracellular form activated microglia, and used on all known human miRNAs. The predicted functional activity of a subset of in total 20 high- and low-scoring miRNAs, which in part have been previously linked to AD, were tested for their capacity to activate murine TLR7, as well as human TLR7, and human TLR8 expressed in HEK TLR reporter cells. We found that oligonucleotide-induced activation of TLR7/8 operated sequence-specifically and preferred binding of unpaired bases. The experimental validation results well support the in silico classification of BrainDead, highlighting its power to drive and support experimental design and studies.
Materials and methods
BrainDead pipeline
BrainDead is a machine learning (ML) approach to classify short RNA sequences/oligonucleotides such as miRNAs based on sequence and secondary structural features. The workflow is depicted in (Figure.1). First, BrainDead analyzes the occurrence of k-mers within different structural contexts. The respective feature sets of each RNA are subsequently used to train a machine learning model based on the available pre-classification. Four sets/types of features are supported by the BrainDead pipeline. Sequence features are defined as the presence or absence of short subsequences or their count. These so-called k-mers, of which k defines the length of the subsequence, are problem-specific. Their selection is discussed in a subsequent section. The collected data define the feature set ‘k-mer in any context’.
Figure 1.
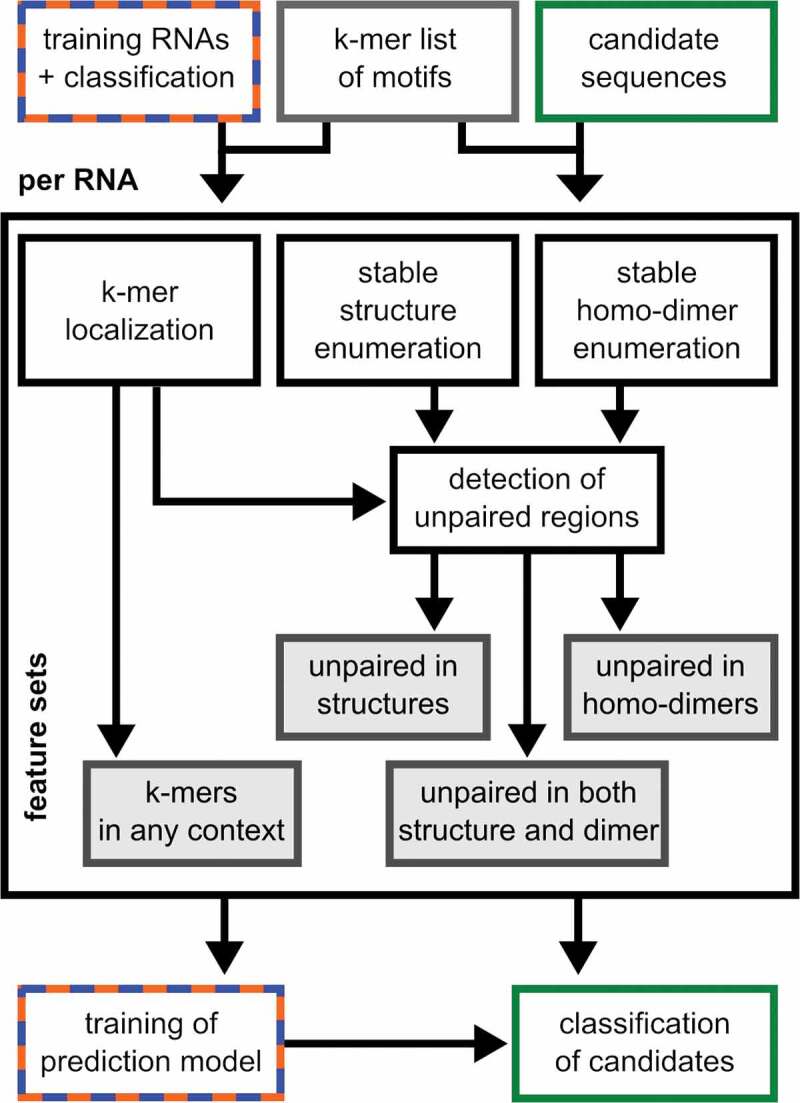
Depiction of BrainDead’s workflow of feature generation (centre), model trainingand candidate classification (bottom)
The considered k-mers are assumed to be important for the RNAs’ function, which typically involves direct interaction with target molecules. Thus, the structural context of each k-mer is important, i.e. whether it occurs within an unstructured/single-stranded region, or is involved in intra-molecular structure formation. To this end, the pipeline predicts all stable putative secondary structures via RNAsubopt [27]. A structure is considered stable if its predicted free energy is below a user-defined absolute threshold (default −3 kcal/mol). If a k-mer is not involved in base-pairing in any stable structure, it is considered ‘unpaired in intra-molecular context’. This defines a second set of features that encodes k-mers in unstructured regions.
We integrated a novel approach to consider intermolecular interactions under the assumption that oligonucleotides are present in high concentrations, which can occur in cells or extracellularly. When large amounts of mature miRNAs are released, it is likely that intermolecular homo–duplex interactions are formed [18]. The homo-duplex features are computed by predicting suboptimal homo-duplex RNA–RNA interactions using IntaRNA [28], with a subsequent ‘unpaired in homo-duplex’ feature generation analogue to the primary single-stranded features. The procedure is illustrated in (Figure 2), and an example of a mature miRNA sequence from the training dataset is provided in (Figure S2).
Figure 2.
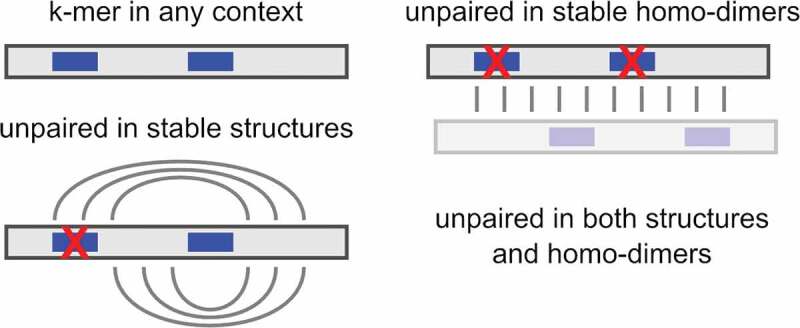
Illustration of context-sensitive k-mer counting for feature generation. One of the two occurrences of a fictive k-mer (blue bar) within an RNA (grey bar) is masked by intra-molecular structure formation while both locations are involved in homo-dimerization. See Supplementary Material for a miRNA example
Finally, both intra- and intermolecular structure information is combined into a fourth feature set encoding ‘k-mer unpaired in any context’. The feature sets (and the positions of each k-mer) are generated by the first module of the pipeline and provide the database for training and application of BrainDead’s ML models.
To train a model, a set of RNAs has to be provided that is accompanied by the reference labels or values for the biological function under study (e.g. whether the RNA can trigger some effect or not). In addition, the set of k-mers has to be given. One can provide the whole set of k-mers of specific lengths (e.g. all 3-mers or 4-mers). Alternatively, users can provide a set of kmers that are known to be important in the target problem either from previous studies or by following a feature-selection strategy (see Results). The latter approach can boost the classification performance through pruning the feature space. The motifs are used to generate the parameter space of the model and to integrate biological knowledge. Based on this, per default, a support vector machine (SVM) is trained, but other models such as logistic regression from the scikit package [29] can be selected. The SVM and its parameterization were chosen based on a comparative evaluation of four logistic and SVM models with and without hyperparameter optimization. Further details are discussed within the Supplementary Material.
Finally, the trained BrainDead model is used for an automated classification of RNAs with unknown activity. For each such candidate RNA, the feature sets are generated and the ML model is applied for its classification (i.e. its putative functional impact). The source code is freely available at https://github.com/BackofenLab/BrainDead.
BrainDead web server
To simplify BrainDead’s application for experimentalists, a web server is freely available as part of the Freiburg RNA tools [30] at http://rna.informatik.uni-freiburg.de/BrainDead/.
As input for training the ML model, the server only needs a set of sequences in FASTA format and a list of k-mers. Each sequence header from the training set must have a label from a binary pre-classification (+-1). This data is used to automatically train an ML prediction model. The generated feature tables, as well as training statistics are available for download and inspection. This model is applied on a user-provided set of candidate sequences with unknown classification to predict their outcome. Their classification is visualized in the result page.
The web server is supplemented with the data obtained from our analysis of immune cell activation, which is discussed in the following.
Microglial activation training data
Immune response data were obtained from the exposure of primary microglia derived from C57BL/6 mice to synthetic oligoribonucleotides. As activated microglia release inflammatory molecules, also in response to oligoribonucleotides that induce TLR7 signalling [4,15], we determined TNF-α amounts in the microglial supernatant after oligoribonucleotide treatment via ELISA, thereby assessing the degree of microglia activation. We included 50 oligoribonucleotide sequences with a large fraction of mature miRNA origin, of which we analysed concentrations of TNF-α released from microglia after 24 h exposure to the individual oligoribonucleotide. Setting a cut-off of Fold Change >12 compared to unstimulated control conditions relying on at least two biological repetitions, we defined 22 of the tested oligoribonucleotides as microglia-activating and the remaining 28 as non-activating miRNAs as reference classification for training BrainDead’s ML models (Table S1). These activation data are based on previous in-house experiments [4,15,31] and (Wallach et al., unpublished).
K-mers for microglial activation training data
We generated an exhaustive feature set covering all possible k-mers of lengths 1–4 for the analysed miRNAs of the murine microglia training set, since it is unknown what sequence k-mers and which structural features are important for classifying microglia activation. The range of lengths was chosen based on previous findings concerning sequence motives activating TLR7/8, considering both structural [32] and sequential aspects [14], to limit the search range, and to avoid long k-mers that might be too specific and not represent a general pattern. Given the reference classification of the training data, the resulting feature set was subsequently analysed to identify k-mer subsets associated with the biologically validated reference classification of the training set. We scored the features based on their importance for a robust classification. To this end, we applied the ReliefF algorithm [33] as implemented in the ReBATE package [34] and extracted the top-ranked features according to ReliefF scores as detailed in the Supplementary Material.
miRNA candidate selection for verification
We applied the BrainDead pipeline on all known human miRNAs to evaluate BrainDead predictions for the case of microglial activation as experimentally assessed and described above. To this end, BrainDead predictions for 2,656 human miRNAs from mirBase v22.1 [35] were ranked by BrainDead’s prediction score. The highest- and lowest-scored five miRNAs from that list were selected as candidate list 1 for verification. Noteworthy, the sequences from the candidate list do not overlap with the training data. We furthermore extracted the five highest-/lowest-scored candidates from the subset of human miRNAs that are linked to AD, serving as an example for a common disease affecting the human brain, as the second set of candidates. This selection was in particular motivated by our previous findings on let-7b-5p, which is (i) able to extracellularly induce mTLR7 signalling, thereby triggering inflammation and neurodegeneration in the CNS and (ii) specifically elevated in cerebrospinal fluid of AD patients [4,17]. Therefore, the overall list was pruned to miRNAs with the tag ‘Alzheimer’s’ and ‘increased expression’ in the disease annotation database PhenomiR v2.0 that includes expression profiles of the stored disease-associated miRNAs [36]. Table S4 provides details for both candidate lists that cover in total 20 miRNAs.
Validation experiments
Oligoribonucleotides
To validate the predicted miRNAs’ activation of immune cells and to test their potential to induce mTLR7 and/or hTLR7/8 signalling, we used miRNA mimics. Oligoribonucleotides were modified with 5´ phosphorylation and phosphorothioate bonds in every base (Integrated DNA Technologies, Coralville, IA, USA). Sequence information for experimentally tested miRNAs is provided in (Table S4). A non-activating oligoribonucleotide containing a mutated let-7b sequence, referred to as control in (Table S1), served as negative control for sequence-specific microglial activation and HEK TLR7/8 reporter cell induction [4].
Mice and cell lines
C57BL/6 mice were bred at the FEM, Charité – Universitätsmedizin Berlin, Germany. All animals were maintained according to the guidelines of the committee for animal care. All animal procedures were approved by the Landesamt für Gesundheit und Soziales (LAGeSo) Berlin, Germany. HEK-BlueTM cells expressing mouse TLR7, human TLR7, or human TLR8, as well as the respective control cell lines HEK-BlueTM Null2-k, Null1-k and Null-1 (Invivogen, San Diego, CA, USA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen #41,965,062, Carlsbad, CA, USA). The DMEM was supplemented with 10% heat-inactivated foetal calf serum (FCS, Gibco #10,082-147, Thermo Fisher Scientific, Waltham, MA, USA) and penicillin (100 U/ml)/streptomycin (100 μg/ml; Gibco #15,140-122, Thermo Fisher Scientific, Waltham, MA, USA). Cells were cultured at 37°C in humidified air with 5% (v/v) CO2.
Primary cultures of microglia
Primary cell cultures of microglia were generated as previously described [37]. Briefly, microglia were isolated from mouse brains on postnatal day 1–4. Meninges, superficial blood vessels and cerebellum were removed from the cortices. The cortices were then homogenized with 3 ml Trypsin (2.5%; Gibco #15,090-046, Thermo Fisher Scientific, Waltham, MA, USA) for 25 min at 37°C. The trypsin reaction was stopped with FCS (Gibco #10,082-147, Thermo Fisher Scientific, Waltham, MA, USA). 100 µl DNase (Roche #ROD 1,284,932, Basel, Switzerland) were added. The cell suspension was centrifuged at 1200 rpm at 4° C for 5 min. Pellets were resuspended in DMEM (Invitrogen #41,965,062, Carlsbad, CA, USA) supplemented with 10% FCS (Gibco #10,082-147, Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin/streptomycin (Gibco #15,140-122, Thermo Fisher Scientific, Waltham, MA, USA), mechanically disassociated and passed through a 70-µm-cell strainer. Microglia were grown in T75 flasks for 10–14 d in 12 ml of DMEM (Invitrogen #41,965,062, Carlsbad, CA, USA) at 37°C in humidified air with 5% (v/v) CO2. The cells were seeded in 96-well plates. On the following day cells were transfected with the synthetic oligonucleotides (10 µg/ml) or control oligonucleotide (10 µg/ml) complexed to the transfection agent LyoVec (InvivoGen #LYEC-RNA, San Diego, CA, USA) according to the manufacturer’s instructions.
HEK-Blue TLR activation assays
Human Embryonic Kidney 293 Blue (HEK-Blue) SEAP reporter cells overexpressing murine TLR7, human TLR7, or human TLR8 were used in activation assays. The parental control cell lines HEK-Blue Null2-k, Null1-k and Null1 were used as control. All cell lines were purchased from InvivoGen (San Diego, CA, USA). Cells were seeded into 96-well plates (5 × 104/well). After 24 h, cells were transfected with the synthetic oligonucleotides (5 µg/ml) or control oligonucleotide complexed to the transfection agent LyoVec (InvivoGen #LYEC-RNA, San Diego, CA, USA) according to the manufacturer’s instructions. Cells were stimulated with indicated agents dissolved in HEK-Blue detection reagent (InvivoGen #hb-det2, San Diego, CA, USA). Each condition was performed in duplicate. The reporter protein SEAP was detected using the Varioskan Flash device (Thermo Fisher Scientific, Waltham, MA, USA) at a wavelength of OD 655 nm.
Results
Sequence-structure features associated with microglial activation
Using feature selection techniques, we identified a specific set of k-mers that are important for the classification of microglial activation, which was considered to represent an immune cell response. The identified k-mers were AA, AGA, AGGU, AGU, AGUU, CU, GAA, GAGG, GG, GGG, GU, GUU, UGA, UGU, UU, UUG, UUGU and UUU. For most top-ranked k-mers, occurrence in a structure-free context, i.e. unpaired/accessible within the folded structure, was important (see Figure S3(a)), indicating the impact of structure on activation. However, homo-dimerization (inter-molecular pairing of the same miRNA species) was found to be less important. k-mers that correlate with microglial activation aligned around central motifs (G)UU(G) and AGU, while k-mers that correlate with non-activation aligned around (U)GG(A) and AGAA. Further details on the k-mer selection and their properties are provided in the Supplementary Material.
Training of the BrainDead model on microglial activation
We trained an ML classifier to learn a model of microglial activation and to predict oligonucleotides as TLR7/8 ligands given their extracellular mode of function and sequence. The model uses the k-mers identified in the previous step in each structural context (any single-stranded, unpaired in homo-dimer, unpaired in both structure and dimer). As applied for the training model, we evaluated several ML classifiers with a stratified 3-fold strategy on the training data to identify the suitable algorithm. Among the scikit models, support vector machines (SVM) and logistic regression (logit) kernels had the best classification score measured by F-score as the harmonic mean of precision and recall (see Supplementary Material Section 3). Both SVM and logit achieved high F-scores. However, since it was crucial for our experimental validation studies to have a low false-positive rate, the model with the highest precision, i.e. SVM-rbf, was selected as the final model for the prediction of microglial activation.
BrainDead predictions and candidate selection
(Figure 3) summarizes the distribution of predicted scores with respective activation potential classification of all human miRNAs identified so far. The major portion of human miRNAs has exhibited a low BrainDead score (<0.3). This was expected, since only a limited subset of human miRNAs are anticipated to function as microglia-activating receptor ligands. The learned model has set a score of 0.54 as the ligand classification threshold. While scores higher than the threshold are predicted to be activating, we would expect candidates scored in the boundary region as unlikely to activate despite being predicted to be positive. The bottom plot in (Figure 3) shows the score distribution of the 93 miRNAs that are listed with the tags ‘increased expression’, and ‘Alzheimer’ in the PhenomiR database. Their scores are distributed over the whole BrainDead scoring range.
Figure 3.
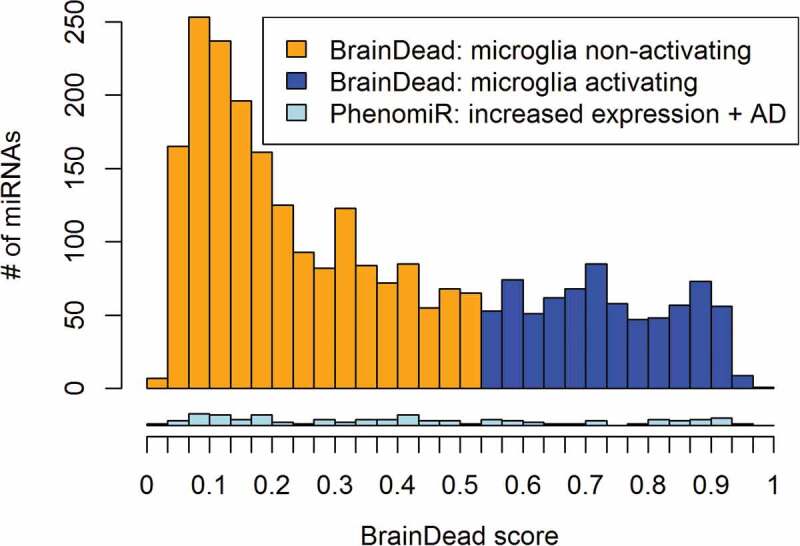
Distribution of BrainDead scores and predicted activation potential (orange and blue) for all 2,656 human miRNAs annotated in mirBase. The bottom histogram (light blue) provides the distribution of 93 BrainDead scores of Alzheimer’s disease (AD)-associated miRNAs according to the PhenomiR database
The ‘high-5’ miRNAs with highest activation score among all human miRNAs were: hsa-miR-6888-3p, hsa-miR-374b-3p, hsa-miR-130b-5p, hsa-miR-4288, hsa-miR-5701; the ‘low-5’ were: hsa-miR-4727-3p, hsa-miR-3198, hsa-miR-361-5p, hsa-miR-422a, and hsa-miR-541-3p (list 1, Table S4). The ‘high-5’-scored human miRNAs associated with AD were: hsa-miR-30a-3p, hsa-miR-9-5p, hsa-miR-30e-3p, hsa-miR-375-3p, hsa-miR-381-5p; the ‘low-5’ were: hsa-miR-191-5p, hsa-miR-216a-3p, hsa-miR-501-3p, hsa-miR-204-3p, and hsa-miR-422a (list 2, Table S4). Noteworthy, both high- as well as low-scored miRNAs from list 2 are AD-associated. Both lists were used for the downstream experimental verification. Further details are provided in the Supplementary Material.
Experimental candidate verification
For validation, we tested all miRNA candidates from list 1 and list 2 (in total 20 miRNAs) using primary mouse microglia, i.e. the same cellular system that the microglial activation training data is based on. To do so, microglia isolated from C57BL/6 (wild-type, WT) mice were exposed to miRNA mimics for 24 h. Subsequently, supernatants were collected, and TNF-α concentration was measured via ELISA (Figure 4, Table S5). Four out of the five top-scored candidates predicted by the BrainDead pipeline from list 1 significantly induced TNF-α release from microglia (Figure 4(a), blue bars), whereas all low-5 candidates did not induce significant TNF-α release (Figure 4(a), orange bars). In addition, all tested high-5 AD-associated miRNAs from list 2, but none of the corresponding low-5 candidates significantly induced TNF-α release from microglia (Figure 4(b)). In both experimental approaches testing miRNA candidates of list 1 and list 2, the non-activating mutant control (ctrl) oligonucleotide did not induce TNF-α production in microglia.
Figure 4.
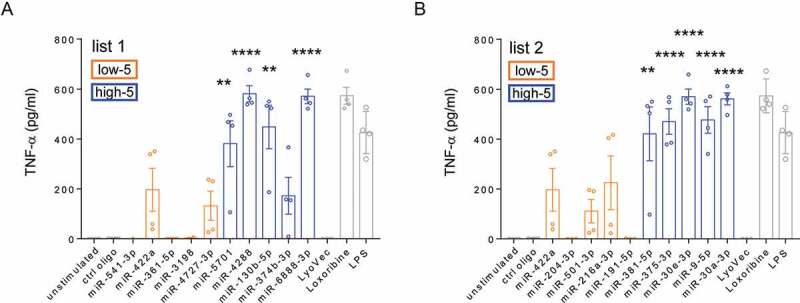
Experimentally assessed TNF-α release from microglia. (a) list 1 – miRNA candidates that were selected based on BrainDead score only and (b) list 2 – AD-associated miRNAs. miRNAs are arranged by ascending BrainDead prediction score. Blue and orange colouring refers to BrainDead prediction, i.e. activating (high-5) and non-activating (low-5), respectively. Control conditions are indicated by grey colour. Microglia were exposed to 10 µg/ml of the indicated miRNA mimic for 24 h. The established TLR7 agonist loxoribine (1 mM) and the TLR4 agonist lipopolysaccharide (LPS, 100 ng/ml) served as positive control for microglial activation. Control mutant oligonucleotide (10 µg/ml), unstimulated cells, and the transfection agent LyoVec were used as negative control. Bars represent mean values ± SEM (n = 4) of depicted measurements (dots). **P < 0.01; ****P < 0.0001 compared to unstimulated condition, two-tailed Student’s t-test
To further validate the oligonucleotide-induced effects observed in microglia and to analyse the miRNA candidates’ capacity to activate mTLR7, we made use of HEK-Blue reporter cells overexpressing mTLR7. In these cells, the Secreted Embryonic Alkaline Phosphatase (SEAP) reporter gene was inserted directly after the NF-B/AP-1-promoter, a well-established output of TLR7/8 signalling [8]. SEAP activity was determined via colorimetric change of the SEAP-substrate reporter media. Four out of high-5 miRNAs of list 1, miR-6888-3p, miR-130b-5p, miR-4288, and miR-5701, significantly activated mTLR7 (Figure S6(a)). Exposure of mTLR7 HEK reporter cells to the high-5 list 1 candidate miR-374b-3p led to NF-kB induction compared to control, although not reaching statistical significance (Figure S6(a)). Exposure of mTLR7 HEK reporter cells to the low-5 candidates of list 1 did not induce any response (Figure S6(a)). The high-5 AD-associated miRNAs of list 2, miR-30e-3p, miR-375-3p, and miR-381-5p significantly induced mTLR7 reporter activation (Figure S6(b)). The high-5 AD-associated candidates miR-9-5p and miR-30a-3p induced NF-kB responses compared to control, although not reaching significance. Out of the low-5 AD-associated candidates of list 2, only miR-216a-3p significantly induced mTLR7 activation, while all other tested miRNAs of the low-5 AD-associated candidate list 2, miR-422a, miR-204-3p, miR-501-3p, and miR-191-5p did not induce receptor activation (Figure S6(b)). Results on activation of mTLR7 expressed in HEK TLR reporter cells (see Figure S6, Figure 5) were in line with the experiments on microglial activation described above (see Figure 4, Figure 5). For instance, miR-4288 (classified as activating miRNA) consistently induced strong responses in both cell systems compared to control conditions, while only a weak response in terms of microglial activation and mTLR7 induction was assessed in the case of miR-374b-3p (also classified as activating miRNA). A consistent trend is observed in (Figure S7), which shows in total 38 miRNAs that were experimentally tested for receptor activation within our study. This includes both the 20 candidates classified by BrainDead (see above) as well as 18 additional miRNAs from the ML training data set that were also analysed in the HEK mTLR7 reporter cell system. Similar and consistent results obtained from the experiments analysing activation of mouse microglia and HEK TLR reporter cells overexpressing mTLR7 indicate that mouse microglial activation is likely mediated through mTLR7 signalling.
Figure 5.
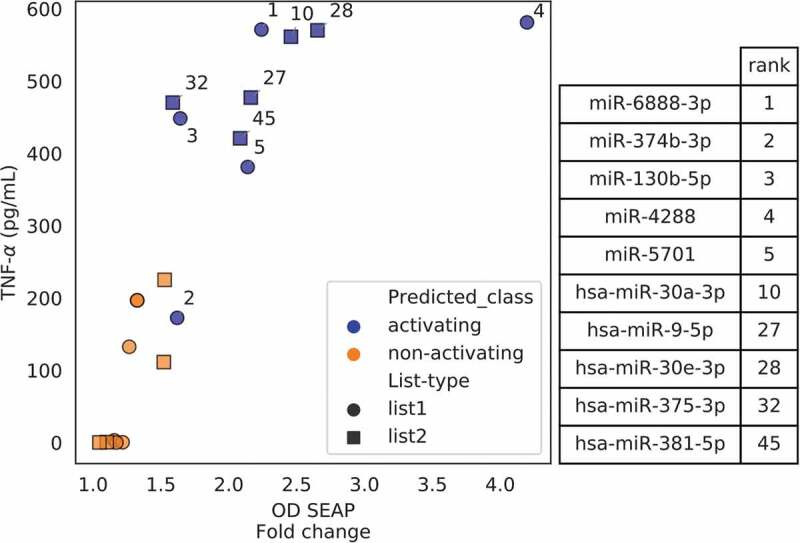
Relation of activity measurements from mouse microglia and mTLR7 reporter cells. Each point represents a miRNA from the respective candidate list, i.e. list 1 includes candidates that were selected based on BrainDead score only (circles), while list 2 includes AD-associated miRNAs classified by BrainDead (squares). TNF-α concentrations (mouse microglia, y-axis) and SEAP activity expressed as fold change (mTLR7 reporter activation, x-axis) averaged from four replicates are shown. The annotated numbers indicate the ranking predicted by BrainDead for the high-5 activating miRNAs of the two lists. See Figure S7 for an extended version of the plot
To transfer the results obtained from the ML approach described above to the human system, we analysed the miRNA candidates of list 1 and list 2 with respect to their potential to activate human TLR7 and/or human TLR8 using HEK reporter cells overexpressing hTLR7 or hTLR8. As TLRs are highly conserved among species, we expected the model trained on mouse microglia data as being able to predict miRNAs that activate human TLRs. Indeed, testing for hTLR7 activation we observed a similar response pattern (Figure S8) as observed for mTLR7 activation (see Figure S6) described above. From list 1, hTLR7 was significantly activated by the high-5 ranked miR-6888-3p, miR-4288, and miR-5701, while miR-374b-3p and miR-130b-5p incubation resulted in receptor activation by trend compared to control. In contrast, none of the tested low-5 miRNA candidates induced hTLR7 activation (Figure S8(a)). Among the high-5 AD-related miRNAs (list 2), miR-9-5p induced significant hTLR7 activation, while exposure to miR-30a-3p, miR-30e-3p, miR-375-3p and miR-381-5p led to NF-kB activation compared to control, although not reaching statistical significance (Figure S8(b)). miR-501-3p of list 2, categorized as low-5 candidate, significantly induced hTLR7, while miR-191-5p, miR-216a-3p, miR-204-3p, and miR-422a from this test group did not induce any response (Figure S8(b)).
Testing for hTLR8 activation revealed that four out of the five high-5 list 1 candidates, namely miR-6888-3p, miR-374b-3p, miR-130b-5p, and miR-5701, significantly induced hTLR8 reporter activation, while miR-4288, classified as activating candidate, and all tested miRNAs of the low-5 list 1 candidate group, miR-4727-3p, miR-3198, miR-361-5p, miR-422a and miR-541-3p did not induce such a response (Figure S9(a)). From list 2, the AD-related candidates according to the PhenomiR database [36], miR-30a-3p, miR-9-5p, miR-30e-3p, and miR-381-5p ranked as the high-5 candidate group, significantly induced hTLR8 activation, while only one of the high-5 candidates, namely miR-375-3p, did not induce such a response (Figure S9(b)). Out of the low-5 candidate group from list 2, miR-216a-3p significantly induced hTLR8 activation, while miR-191-5p, miR-501-3p, miR-204-3p, and miR-422a did not induce receptor activation (Figure S9(b)).
Discussion
BrainDead – generic and customizable RNA classification
BrainDead is a generic and customizable RNA classification pipeline that can be tailored to predict activity of any biological problem with a binary classification nature. This machine learning approach considers both sequence k-mers and their structural context, and requires a reference pre-classified dataset for training. Since tailored to short RNAs, it can take all (semi)-stable structures into account and is not restricted to a single putative structure per RNA, e.g. only the minimum-free-energy structure as considered by repRNA [25]. That way, stable structure alternatives are considered that are otherwise ignored. Furthermore, BrainDead has a simple but powerful definition of ‘stability’ via a customizable absolute energy threshold. This allows, in contrast to alternatives based on an unpaired probability [38], a fine-tuned classification of stability adjusted for the studied RNA. The indirect incorporation of structure via k-mer context allows to integrate a low evolutionary structure conservation and to investigate context – rather than localization-based structural similarities without requiring an overall or local similarity. This distinguishes BrainDead from the available solutions for structure-based classification and clustering that are designed to identify similar folds and homology analysis [26,39].
The customizable sequence feature generation based on a user-provided list of k-mers enables a fast and problem-specific feature generation. Thus, besides its application as an all-in-one classifier, BrainDead can be used as a feature generator, similar to the functionality of repRNA, which only provides exhaustive feature generation. BrainDead’s feature tables can be employed in any other pipeline if the BrainDead model has to be extended. The latter is also possible by direct modification of its open Python source code.
BrainDead web server
To simplify applications and enable reproducibility, a web server interface of BrainDead is available. Given a pre-classified set of RNAs (FASTA format with binary class label in each header) and a problem-specific set of k-mers, the web server will generate the respective feature tables and train a classification model. Features, as well as the model and training statistics are available for download and inspection. For a provided set of candidate RNAs (FASTA format), classification results are visualized on the result page and available for download (CSV format). Thus, the BrainDead web server provides a simple yet powerful platform to develop and use a problem-specific RNA prediction model, thereby supporting the design of experimental studies.
BrainDead microglial activation model
Sequence motifs identified and used to train BrainDead for receptor-mediated microglial activation, i.e. activation of an immune response by extracellular host-derived RNA, fall into two classes based on their occurrence in the training data, i.e. whether they are mainly found in (i) activating or (ii) non-activating RNAs (see Figure S3). The latter class distinguishes the BrainDead model from classic approaches that focus on activation only [14]. Based on such studies, it is known that GG- and/or GU-rich motifs are important for TLR activation. This knowledge was independently revealed by our (uninformed) feature extraction performed to select important motifs (Table S2), thereby demonstrating the power of automated systems. Most activation-related k-mers are UG-/GU-rich and some, like UUGU, were also top-ranked in the study by Forsbach et al. [14].
Three-dimensional structural analysis of TLR7 revealed that this receptor harbours two different ligand-binding sites, which can act synergistically on receptor dimerization and consequent immune cell activation (Z. Zhang et al. 2016). The first binding site exhibits a preference for G over U, while the second binding interface co-crystallizes with G- and U-rich ssRNA fragments. The second site requires a trimer of bases with one U present in the central position. These receptor features regarding structure and sequence are well-matched by the k-mers identified in our current study. Forsbach and colleagues used a battery of 4-mer sequence motifs to generate TLR7/8 activation data based on TNF-α and IFN-α release from peripheral immune cells [14]. However, this study did not consider sequence information and thus the impact of a whole mature miRNA. Since different binding sites with different RNA base preferences are located within TLR7 (see above [32]) it is likely that bases within one miRNA bind to both receptor sites to achieve activation. Thus, miRNAs may be considered as TLR-activating chimeras. Consequently, we used the activation data generated from short single-stranded oligonucleotides of 21–26 nt length (Table S1), including a large fraction of mature miRNA sequences for our training paradigm. The U and GU content of miRNAs was previously described to correlate with the degree of TLR7/8 activation [40,41]. However, specific sequence and structural features that enable a miRNA to act as a functional ligand for TLR7/8 remained unexplored so far. In our current study, we not only raised the question of which sequential features of a given miRNA are required to activate/bind to TLR7/8 but also whether these motifs are (not) masked (i.e. free accessible) for TLR7/8 binding by intramolecular and homo-dimerization structure formation. Our results indicate that activating k-mers are likely structure-free (unpaired/accessible), whereas homo-dimerization was not important for TLR7/8 activation.
Experimental candidate verification
The finding that four out of five high-scored miRNA candidates (list 1) defined by BrainDead significantly activated primary mouse microglia was reproduced in experiments using HEK reporter cells overexpressing mTLR7. However, out of the high-5 AD-linked candidates (list 2), which all induced microglial activation, only three (miR-30e-3p, miR-375-3p and miR-381-5p) induced statistically significant mTLR7 activation, and out of the five candidates from the low-5 group, which did not induce a significant response in microglia, miR-216-3p significantly activated mTLR7 in the HEK TLR reporter cells. These different findings regarding the statistical significance obtained from the experiments testing microglial activation and mTLR7 reporter induction is likely due to a higher variation of the measured values derived from the mTLR7 reporter induction analysis. Still, in general, the activation of mTLR7 by low-scored miRNAs expressed as Fold change was much lower compared to the activation induced by the high-scored miRNA candidates. The validation experiments testing for human TLR7 and human TLR8 activation also supported the consistent prediction results of BrainDead. In addition to minor exceptions, only high-ranked candidate miRNAs activated the respective tested TLR. These findings point to the presence of specific miRNAs’ sequence motifs relevant for the interaction with both receptors, TLR7 and TLR8, in mouse and human. Thus, a model trained on data obtained from experiments on mouse immune cells such as BrainDead seems to be capable of supporting research on RNA acting as ligands of human TLRs, especially in a human disease context. Furthermore, the consistent scoring of AD-related list 2 candidates and the uniform distribution of AD association within the BrainDead scoring scheme (see Figure 3) suggests that candidate selection purely based on AD database annotation would provide a much lower rate of activating candidates compared to BrainDead-based filtering.
Conclusion
We present here a novel, customizable, and generic machine learning approach for the functional classification of small oligonucleotides. The method was applied for the prediction of human miRNAs serving as TLR7/8 ligands and activating immune cells. While our training dataset was based on mouse microglial activation, the results obtained from validation experiments on mTLR7 and hTLR7/8 activation demonstrated the ligand character of the tested candidate miRNAs. The experimentally assessed potential of 20 tested miRNAs regarding TLR7/8 activation was congruent with the classification predicted by our in silico machine learning pipeline. The BrainDead model takes the structural context of k-mers concerning unpairedness/accessibility in intramolecular, as well as homo-dimer structure formation into account. Future work will broaden the supported context types to e.g. motifs occurring in RNA helices, specific substructures like hairpin loops, or tertiary motifs. We plan to incorporate more generic k-mer motif definitions via sequence logos or regular expressions, and the integration of measured affinity information of specific k-mers into the model. Overall, our study shows that BrainDead is well suited to support experimental study design based on its comprehensible model definition, simple user interface, and predictive power. While miRNAs play important roles in human health and diseases, TLR7 and TLR8 are key regulators of immune responses, are involved in organ-specific processes, such as neurodegeneration in the CNS, and also play complex roles in human diseases, e.g. rare TLR7 variants can implicate COVID-19 severity [42]. The power of the presented and online-provided model trained on immune cell activation can be used for any short RNA molecule to be tested for ligand-mediated TLR activation, considering any cell type capable of functional TLR7/8 signalling.
Supplementary Material
Acknowledgments
This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft DFG) [BA2168/16-1, BA2168/21-1 and BA2168/3-3 to R.B.; LE2420/2-1, SFB-TRR167/B03 to S.L.], and by the Germany’s Excellence Strategy (CIBSS-EXC-2189-Project ID 390939984 to R.B.). We thank the Lehnardt lab for the helpful discussion. The article processing charge was funded by the Baden–Wuerttemberg Ministry of Science, Research and Art and the University of Freiburg in the funding programme Open Access Publishing.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft [BA2168/16-1]; Deutsche Forschungsgemeinschaft [BA2168/21-1]; Deutsche Forschungsgemeinschaft [SFB-TRR167/B03]; Deutsche Forschungsgemeinschaft [CIBSS-EXC-2189-Project ID 390939984]; Deutsche Forschungsgemeinschaft [LE2420/2-1]; Deutsche Forschungsgemeinschaft [BA2168/3-3].
conflicts of interest
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. [DOI] [PubMed] [Google Scholar]
- [3].Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis JAD’. 2008;14(1):27–41. [DOI] [PubMed] [Google Scholar]
- [4].Lehmann SM, Krüger C, Park B, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15(6):827–835. [DOI] [PubMed] [Google Scholar]
- [5].Zhang J, Li S, Li L, et al. Exosome and exosomal MicroRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].He X, Jing Z, Cheng G. MicroRNAs: new regulators of Toll-like receptor signalling pathways. BioMed Res Int. 2014;2014:945169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. [DOI] [PubMed] [Google Scholar]
- [9].Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8(4):279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Diebold SS, Kaisho T, Hemmi H, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. [DOI] [PubMed] [Google Scholar]
- [11].Jurk M, Heil F, Vollmer J, et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol. 2002;3(6):499–499. [DOI] [PubMed] [Google Scholar]
- [12].Lund JM, Alexopoulou L, Sato A, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Nat Acad Sci. 2004;101(15):5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. [DOI] [PubMed] [Google Scholar]
- [14].Forsbach A, Nemorin J-G, Montino C, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol (Baltimore, MD). 2008;1950(180):3729–3738. [DOI] [PubMed] [Google Scholar]
- [15].Dembny P, Newman AG, Singh M, et al. Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI Insight. 2020;5(5). DOI: 10.1172/jci.insight.131093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Buonfiglioli A, Efe IE, Guneykaya D, et al. let-7 MicroRNAs regulate microglial function and suppress glioma growth through Toll-Like receptor 7. Cell Reports. 2019;29(3460–3471.e7):3460–3471.e7. [DOI] [PubMed] [Google Scholar]
- [17].Derkow K, Rössling R, Schipke C, et al. Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer’s disease. PloS One. 2018;13(7):e0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Belter A, Gudanis D, Rolle K, et al. Mature MiRNAs form secondary structure, which suggests their function beyond RISC. PLoS ONE. 2014;9(11):e113848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee D, Karchin R, Beer MA. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 2011;21(12):2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fletez-Brant C, Lee D, McCallion AS, et al. kmer-SVM: a web server for identifying predictive regulatory sequence features in genomic data sets. Nucleic Acids Res. 2013;41(W1):W544–W556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ghandi M, Mohammad-Noori M, Ghareghani N, et al. gkmSVM: an R package for gapped-kmer SVM. Bioinformatics. 2016;32(14):2205–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen W, Zhang X, Brooker J, et al. PseKNC-general: a cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics. 2015;31(1):119–120. [DOI] [PubMed] [Google Scholar]
- [23].Zhang Y, Wang X, Kang L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics. 2011;27(6):771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu B, Fang L, Liu F, et al. Identification of real MicroRNA precursors with a pseudo structure status composition approach. PLOS ONE. 2015;10(3):e0121501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu B, Liu F, Fang L, et al. repRNA: a web server for generating various feature vectors of RNA sequences. Mol Genet Genomics. 2016;291(1):473–481. [DOI] [PubMed] [Google Scholar]
- [26].Miladi M, Sokhoyan E, Houwaart T, et al. GraphClust2: annotation and discovery of structured RNAs with scalable and accessible integrative clustering. GigaScience. 2019;8(12):giz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lorenz R, Bernhart SH, Höner zu Siederdissen C, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 2017;45(W1):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- [30].Raden M, Ali SM, Alkhnbashi OS, et al. Freiburg RNA tools: a central online resource for RNA-focused research and teaching. Nucleic Acids Res. 2018;46(W1):W25–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wallach T, Wetzel M, Dembny P, et al. Identification of CNS injury-related microRNAs as novel Toll-Like receptor 7/8 signaling activators by small RNA sequencing. Cells. 2020;9(9):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang Z, Ohto U, Shibata T, et al. Structural analysis reveals that Toll-like receptor 7 is a dual receptor for guanosine and single-stranded RNA. Immunity. 2016;45(4):737–748. [DOI] [PubMed] [Google Scholar]
- [33].Kononenko I, Šimec E, Robnik-Šikonja M. Overcoming the myopia of inductive learning algorithms with RELIEFF. Appl Intell. 1997;7(1):39–55. [Google Scholar]
- [34].Urbanowicz RJ, Olson RS, Schmitt P, et al. Benchmarking relief-based feature selection methods for bioinformatics data mining. J Biomed Inform. 2018;85:168–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ruepp A, Kowarsch A, Schmidl D, et al. PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes. Genome Biol. 2010;11(1):R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lehnardt S, Lachance C, Patrizi S, et al. The toll-like receptor TLR4 is necessary for lipopolysaccharide-induced oligodendrocyte injury in the CNS. J Neurosci Off J Soc Neurosci. 2002;22(7):2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mückstein U, Tafer H, Hackermüller J, et al. Thermodynamics of RNA-RNA binding. Bioinforma Oxf Engl. 2006;22(10):1177–1182. [DOI] [PubMed] [Google Scholar]
- [39].Will S, Joshi T, Hofacker IL, et al. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA NYN. 2012;18(5):900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Salvi V, Gianello V, Busatto S, et al. Exosome-delivered microRNAs promote IFN-α secretion by human plasmacytoid DCs via TLR7. JCI Insight. 2018;3(3). DOI: 10.1172/jci.insight.98204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Feng Y, Zou L, Yan D, et al. Extracellular MicroRNAs induce potent innate immune responses via TLR7/MyD88-dependent mechanisms. J Immunol. 2017;199(6):2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].van der Made CI, Simons A, Schuurs-Hoeijmakers J, et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324(7):663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


