ABSTRACT
Considered to be a field that is continuously growing, epitranscriptomics analyzes the modifications that occur in RNA transcripts and their downstream effects. As epigenetic modifications found in DNA and histones exhibit specific roles on various biological processes, also epitranscriptomic marks control gene expression patterns that are crucial for proper cell proliferation, differentiation and tissue development. Thus, various epitranscriptomic signatures have been identified to play specific roles during stem cell differentiation towards the neuronal and glial lineages, axonal guidance, synaptic plasticity, thus leading to the development of the mature brain tissue. Here we describe in-depth molecular mechanism underlying the most important RNA modifications with emerging roles in the nervous system.
KEYWORDS: Epitranscriptomic marks, m6A. m5C, ψ, Nm, neural stem cell differentiation, neural development, neurodegenerative disease
1. Introduction
Gene transcription processes are highly regulated by RNA modifications that are included under the umbrella of epitranscriptomics. Based on different chemical methods such as two-dimensional thin-layer chromatography and mass spectrometry around 170 modifications have been identified so far, among them N6-methyladenosine (m6A), 5-methylcytosine (m5C), pseudouridine (ψ) and 2ʹ-O methylation (Nm). These modifications have been found to exist in different RNA species such as messenger RNA (mRNA), ribosomal RNA (rRNAs), transfer RNA (tRNA), but also in microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). RNA modifications have been recently revealed to exhibit a certain control over important biological processes via RNA metabolism: stability, maturation, transport and translation [1].
The continuously changing epitranscriptomic landscape has been correlated with the development of the nervous system and also with numerous neurodegenerative disorders, raising interest in advanced fundamental research. It is known that embryonic stem cells (ESCs) are rich in RNA modifications, which have been found to also be present when they undergo differentiation to neural stem cells (NSCs) [2]. To establish neural circuits, NSCs migrate and differentiate to mature neurons and glial cells, thus leading to the development of adult nervous tissue with a significantly increased number of RNA-modified sites [3]. Accordingly, epitranscriptomic modifications are responsible for the proper regulation of central and peripheral nervous systems (CNS, PNS respectively) development, maintaining cellular plasticity and correct function. Therefore, any dysfunction that may occur in the post-transcriptional programme can potentially lead to malformations of the CNS and PNS.
Known as one of the most abundant change of mRNA molecules, m6A sites in mRNAs regions are localized in long exons and near stop codons, in the 3ʹ-untranslated regions (UTR) [4,5], mostly overlap with one specific motif: a DRACH sequence (D = A/G/U, R = A/G, H = A/C/U) [6]. The molecular machinery behind the m6A modifications is very complex, comprising of several enzymatic complexes with well-established contributions: (a) enzymes that are responsible for producing the modification, also known as ‘writers’, (b) enzymes that bind the modified motif by recognition, the ‘readers’ and finally (c) ‘erasers’ enzymes which can remove the modification and bring the RNA to its initial structure [7,8]. m6A deposition (Fig. 1) is co-transcriptionally generated by the conserved writers complex (RNA methyltransferases), which consists of a catalytic heterodimer, methyltransferase-like 3 (METTL3), and a catalytically inactive component, methyltransferase-like 14 (METTL14) which serves as an enzymatic activator of METTL3 [9]. Lastly, Wilms’ tumour 1-associating protein (WTAP) is responsible for the direct interaction with METTL3, specifically for the methylation of the complex, ensuring nuclear localization [10]. It was recently reported that zinc finger CCHC-type containing 4 (ZCCHC4) [11,12] and methyltransferase-like 16 (METTL16) acts on 28S rRNA, respectively on U6 snRNA as m6A methyltransferases [13,14]. Methylated adenine state can be reversed through the action of the erasers (RNA demethylases) complex. This complex is formed of two demethylases belonging to the Fe2+/α-KG-dependent enzymes, namely the mass- and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5) [15]. The methylation site can act as an anchor for reader proteins, such as YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2 [16–20].
Figure 1.
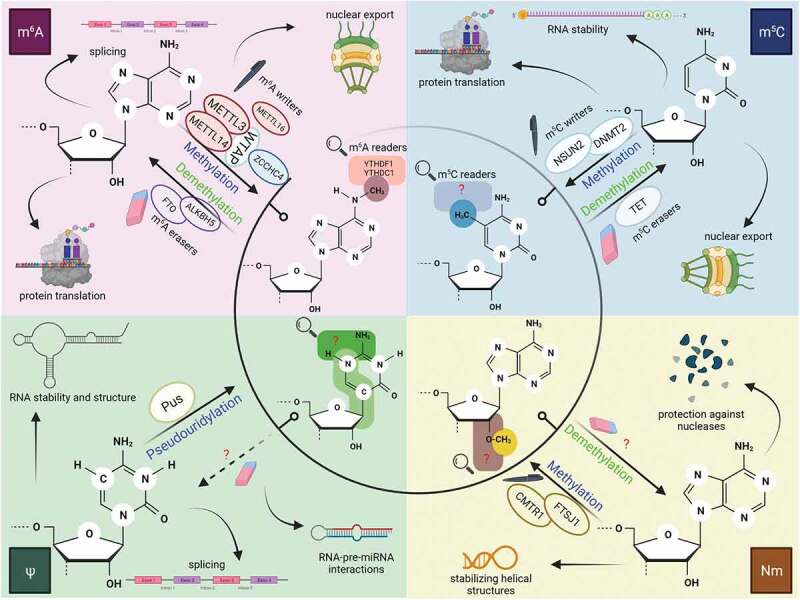
Epitranscriptomic modifications which occur in RNA species and their possible roles in RNA metabolism including splicing, nuclear export, and translation
Writers are responsible for the generation of modified sites in the RNA structure, erasers reverse the chemically modified sties and finally, readers recognize RNA modifications. METTL – methyltransferase-like; FTO – Fe2+/α-KG-dependent enzymes, namely the mass- and obesity-associated protein; ALKBH5 – α-ketoglutarate-dependent dioxygenase alkB homolog 5; ZCCHC4 – zinc finger CCHC-type containing 4; YTHD – members of a family of proteins with a YT521-B homology (YTH) domain; NSUN2 – NOP2/SUN RNA methyltransferase; DNMT2 – DNA methyltransferase-2; TET – ten-eleven family demethylases; Pus – pseudouridine synthases; CMTR1 – cap methyltransferase 1; FTSJ1 – putative ribosomal RNA methyltransferase 1. Image created with BioRender.com m5C deposition (Fig. 1) patterns on mRNA are enriched at CG dinucleotides near transcription initiation sites [21]. Among various eukaryotic m5C methyltransferases, the roles of two major writer proteins, DNA methyltransferase-2 (Dnmt2) and NOP2/SUN RNA methyltransferase (Nsun2), have been studied individually [22]. Although there is no known protein that can entirely ‘erase’ m5C to cytosine, the 10–11 family demethylases (TET) that also control DNA demethylation can change m5C into 5-hydroxymethylcytosine (hm5C), 5-formylcytosine and 5-carboxycytosine [23–25].
Pseudouridine (also known as 5-ribosyluracil or ψ) (Fig. 1) is generated by the isomerization of uracil, more precisely the C1 of the ribose binds to the C5 of uracil, thus freeing N1 and leading to a more rigid structure [26]. Pseudouridylation has been found to have a significant influence on cellular processes such as translation, splicing, telomere maintenance and gene expression regulation [27]. The writers catalysing this RNA modification are pseudouridine synthases (Pus enzymes), which commonly share the same fold and require aspartate, as an active site for catalysis [28]. They can act in two different manners: (a) RNA-dependent guided-pseudouridylation, via the family of ribonucleoproteins which consists of the box H/ACA RNA and four core proteins (dyskerin, Nhp2, Nop10 and Gar 1). This complex is responsible for guiding base paring with the RNA substrate and ensuring pseudouridylation [29]; (b) the RNA-independent pseudouridylation which command stand-alone Pus enzymes that are able to generate ψ modifications in targeted molecules [30]. Pus enzymes are classified into six groups based on their consensus sequences: TruA, TruB, TruD, RluA and RsuA [31]. Nevertheless, no direct eraser or reader proteins have been found thus far, leaving its reversibility and downstream pathways unknown.
Nm motif (Fig. 1), unlike other modifications, does not need a particular nucleotide and may occur on any base by adding a methyl group to the 2ʹ-hydroxyl of ribose [32]. Nm has a variety of effects on RNAs, including stabilizing helical structures, protecting against nucleases, increasing hydrophobicity, and affecting interactions between modified RNAs and proteins [33]. Just a few Nm methyltransferases have been discovered so far and they usually target sites in the tRNA anticodon loop [34]. For instance, cap methyltransferase 1 (CMTR1) has been identified as a Nm methyltransferase which acts on the first transcribed nucleotide of mRNA structures [35], whereas putative ribosomal RNA methyltransferase 1 (FTSJ1) targets sites in tRNAs anticodon loops [36]. Many gaps still need to be filled regarding this RNA modification, as it is unknown whether Nm motif is reversible or if it can be detected by specific proteins.
Based on the current knowledge available on the RNA modifications found in different RNA spices, in this review we aim to address the implication of epitranscriptomic plasticity in the neural differentiation of stem cells and finally, correlate the interplay between RNA signalling dysfunction and epitranscriptomic modifications during the development of severe neurological disorders.
2. Epitranscriptomic marks in stem cell fate towards the neural lineage
Differentiation of embryonic stem cells in neural stem cells
Over the past few years, many studies have focused on understanding the role of epitranscriptomic signatures in regulating the differentiation state of embryonic stem cells (ESCs) to neural stem/progenitor cells (NSCs). Even though there are many processes that need to be elucidated, significant progress has been made in this direction, due to the use of modern high-throughput sequencing techniques that allow the identification of RNA-modified sites.
ESCs are unique stem cells that live in the early embryo blastocyst and give rise to all tissues, thus possessing a pluripotent character. These cells can self-renew and can be kept in their pluripotent condition in vitro for an undetermined period. ESCs are often separated into two states: ‘naive’, which are found in the blastocyst inner cell mass, and ‘primed’, which are in the epiblast and are primed to differentiate [37]. Several molecular criteria separate naive and primed cells, including X chromosomal inactivation, OCT4 enhancer activity, and the signalling necessary for in vitro maintenance [38]. A group of key transcription factors are responsible for orchestrating the pluripotent state of ESCs, among them OCT4, SOX2, NANOG and KLF4 [39].
Early studies have highlighted the controversial role of m6A modifications. Wang et al. reported in 2014 that knockdown of METTL3 and METTL14 reduces m6A motifs, thus leading to impaired stem cell self-renewal and inhibition of gene expression of key pluripotency genes, like DPPA3, NANOG and SOX2 [40]. On top of that they found that knocking down these methyltransferases resulted in the promotion of developmental regulators in mouse ESCs [41]. On the other hand, another study observed that METTL3 knockdown in mouse ESCs supported self-renewal but inhibited cell differentiation [42]. Yet, both groups looked at in vitro mouse ESCs, which makes it difficult to fully understand the functions of m6A in vivo stimulation during embryonic development. Later, Geula et al. have addressed this matter and looked at m6A modifications in the naïve state of ESCs. When knocking down METTL3, their findings indicated that compared to more mature, ‘primed’ stem cells, naïve ESCs are in a different molecular state. They also concluded that m6A is a critical regulator for naive state termination and entrance into the primed state, which is required for appropriate lineage differentiation. On top of that, their research brought to light the fact that naïve ESCs can be separated from ‘primed’ cells, with the help of m6A modifications. The consequences of poor differentiation are so severe that m6A deficiency can result in early cell death [43]. Ultimately, they concluded that m6A modifications are responsible for controlling gene expression in ESCs’ fate regulation, but the exact molecular events that govern lineage fate remain unknown [44].
As most studies have focused on m6A modifications, little is known about other RNA modifications in this specific field of research, but studies have confirmed the presence of m5C sites in mRNA molecules of mouse ESCs [45]. Moreover, double knockdown of Dnmt2 and Nsun2 [46] led to underdeveloped phenotype and impaired cell differentiation in mice, demonstrating that m5C modifications are essential for normal embryonic cellular differentiation and proliferation. It has been found that methylated RNA supports protein synthesis, which can also provide an explanation for the poor differentiation exhibited in Dnmt2- and Nsun2-lacking tissues, as protein synthesis has been correlated with differentiation in mESC [47]. Interestingly, when knocking down only Dmnt2 no severe changes were observed, because its loss was compensated by m5C methyltransferase Nsun2 [48]. However, their exact function and the way they modulate ESCs’ activity in developing neural progenitors still needs to be further investigated.
The exact functions of ψ are still not fully understood, but there is some evidence that indicates its implications in regulation of cellular processes in the nervous system. PUS3 transcripts, for example, are found only in the nervous system of developing mouse embryos, while TruA family proteins are found throughout the body [49]. In human patients suffering from severe intellectual disability (ID) and with a history of global development delay, mutations in Pus3 gene were identified coupled with loss of pseudouridine sites in their tRNA [50].
Dyskerin, an H/ACA box-mediated pseudouridine synthase, showed increased expression levels of transcripts restricted to the neuronal tissues in the telencephalon region, indicating that pseudouridyation is likely to play an important role in neurodevelopment [51]. Additionally, it has been found that mutations in dyskerin may affect stem cell fate in mice and humans, as it is highly expressed in ESCs and a key regulator of telomere elongation, controlling the expression of important pluripotency factors, among them OCT4 and SOX2 [52,53]. Revealing the molecular events involved in these depositions’ appearance would be groundbreaking for a better understanding of how these significant changes, which exhibit control over important biological processes, could be targeted to develop therapeutic strategies.
Neural stem cells – the road to mature neurons and glial cells
Neural stem/progenitor cells (NSCs) form a specific cell population, which has self-renewal properties and is responsible for giving birth to mature cell types in the nervous system, namely neurons, astrocytes, oligodendrocytes [54]. These transformations are highly coordinated processes that need to occur smoothly during developmental stage, to avoid neurological disorders, but also during the regeneration phase, following an injury. Probably, the most studied RNA modification in this context is m6A, which has been demonstrated to have a severe impact on the proliferation, embryonic and adult stem cell differentiation, as well as the maintenance of neural progenitor proficiency [43,55,56], being in general very abundant in the nervous system. All the findings on the regulation of m6A in neurodevelopment will highly impact the development of stem cell-based therapies or gene therapies to avoid disease or as treatment for current affections within the nervous system.
As mentioned earlier, it has been proven that epitranscriptomic mechanisms are responsible for governing the gene expression programme, which is responsible for the commitment towards neural and glial lineage and the maintenance of their differentiated state [55]. The transcription factor network that maintains the stem cell in the pluripotent state is repressed as the cell differentiates from an embryonic state to a specific lineage, due to key molecular switches [57,58]. It has been demonstrated that transcripts responsible for maintaining the pluripotent state are m6A methylated and thus, transcript turnover is affected during lineage commitment [41–43]. In response to a differentiation signal, it is hypothesized that two major events occur: (a) the transcription of genes involved in early cell fate decisions is switched off (via an unrelated m6A mechanism) and (b) m6A is responsible for ensuring the exit from the pluripotent state, by breaking up the corresponding set of transcripts [59,60].
Additionally, significant evidence has been found over the past few years concerning the role of m6A in controlling and regulating stem cell proliferation and differentiation during neural lineage commitment. It has been shown that METTL3-METTL4 writer complex is essential for proper nervous system development [61,62]. Furthermore, one study demonstrated that in YTHDF2−/− mice embryos, neural progenitor cells’ proliferation and differentiation decreased significantly, and the derived-neurons were not able to form proper neurites [63], thus pointing out the importance of each element involved in m6A complex, each with its specific, crucial role during differentiation and development.
Interestingly, very recent research studies provide proof of an extensive crosstalk between epigenetic and epitranscriptomic pathways during the regulation of m6A deposit and removal. In case of m6A, it has been identified that this specific signature controls stem cell differentiation into mature neurons and glial cells via chromatin modifications. In this respect, Wang et al. [64] conducted an extensive study in which they have deleted METTL14 in mouse NSCs and found that m6A motif in RNA is essential for the regulation of NSCs self-renewal. Further, this study has evidenced that m6A modifications regulate the levels of some histone modifications, such as H3K27ac, H3K27me3 and H3K4me3. Thus, they have observed for the first time one mechanism by which m6A on RNA transcripts may impact histone modifications. Namely, this mechanism is responsible for pluripotency regulation of NSCs takes place via destabilizing transcripts encoding for H3K27 histone acetyltransferases P300/CBP, but not for polycomb repressive complex 2 (PRC2), suggesting that these findings required further in-depth investigations.
Even though most studies strived to elucidate the role of m6A modification in regulating stem cell proliferation and differentiation, few studies have also focused on investigating the possible regulation mechanism of m5C chemical RNA modification. In this regard, m5C has been found to regulate the proliferation, motility, and differentiation of neural progenitors. A while ago, it has been found that the wrong deposition of m5C and mutations in the nsun2 gene can lead to severe neuronal deficiency, retardation, and microcephaly in both human and mouse models [65] (Fig. 2). Also, it has been observed that fragmentation of tRNA is affected in both mouse models and human cells lacking Nsun2 protein, thus leading to neurological abnormalities [66]. Further investigations on mice have elucidated that the lack of m5C methylated tRNA fragments lead to impaired neurogenesis and underdeveloped brain structures [67]. All this evidence suggests the importance of m5C methyltransferases in regulating cellular differentiation and stresses upon the fact that failure to properly methylate RNA molecules can have devastating effects on the nervous system.
Figure 2.
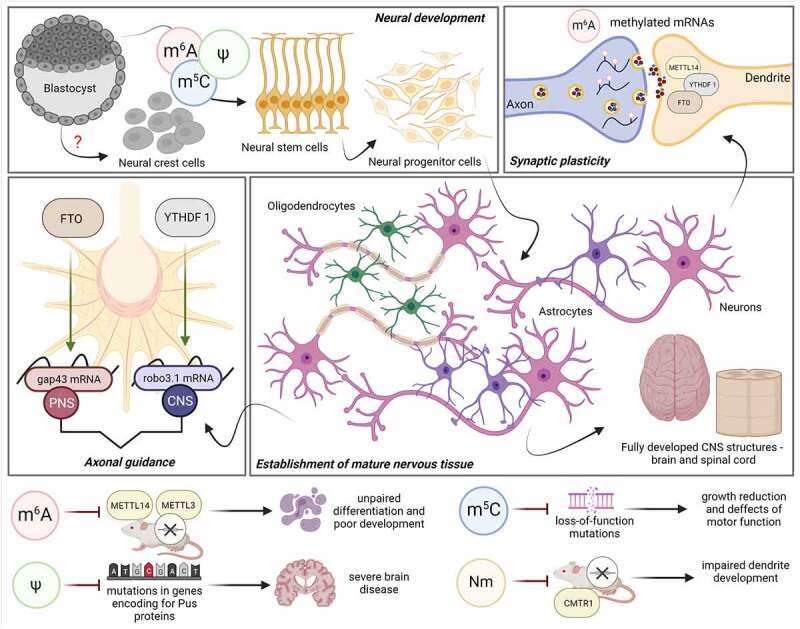
Role of epitranscriptomic marks in the cellular differentiation and development of neural structures
Epitranscriptomic marks impact different processes which are necessary for the establishment of neural tissues: during early ESC differentiation towards the neural progenitors and later on in mature neurons and glial cells; in synaptic transmission, m6A enzymes have been found to be present in dendrites of cortical and hippocampal neurons [68] and during axonal growth of the PNS, FTO acts on gap43 [69], whereas in the CNS, YTHDF1 stimulates translation of axon guidance-related transcript robo3.1 [70]. Defects or knockdown of important compounds responsible for epitranscriptomic modifications may lead to underdeveloped nervous tissue. METTL14 – methyltransferase-like 14; YTHDF1 – protein with a YT521-B homology (YTH) domain; FTO – Fe2+/α-KG-dependent enzymes; gap43 – growth-associated protein 43; robo3.1 – roundabout 3.1; METTL3 – methyltransferase-like 3; CMTR1 – cap methyltransferase 1; CNS – central nervous system; PNS – peripheral nervous system. Image created with BioRender.com
Furthermore, Nm methylation has also been found to act upon tRNA and rRNA in the nervous system, thus being related to neurodevelopment. In this context, it has been reported that tRNA 2-O-methyltransferase FTSJ1 is significantly increased in human foetal nervous tissue in comparison to others, and that mutations in FTSJ1 are a risk for the development of non-syndromic X-linked ID [71] (Fig. 2). Lee et al. [72] have demonstrated that knockdown of CMTR1 leads to impaired dendrite development in mice brain development revealing that Nm may act as an important epitranscriptomic mark in gene regulation, dendritic growth and brain development (Fig. 2).
The m6A modification has also been identified to play an essential role in the differentiation of NSCs towards glial cells, such as astrocytes and oligodendrocytes, during development. Several studies have presented the importance of m6A writers [62] and m6A readers [73] in glial lineage commitment and their alteration, which can lead to the failure of proper glial development and subsequent axon myelination.
iPSCs – exploring their potential for neural lineage commitment
Ever since the success of safely obtaining induced pluripotent stem cells (iPSCs) has been reported, groundbreaking discoveries regarding many biological processes in various tissues have been made. In this context, it has been widely studied how RNA modifications regulate the pluripotency of iPSCs on one hand, and on the other how they act during their commitment to the neuronal lineage [74]. Using iPSCs as a study model, to fully understand the mechanisms underlying the differentiation process of neural progenitor cells to mature neurons, can serve as a useful tool for disease exploration and drug testing.
A significant increase of m6A has been reported when mouse embryonic fibroblasts have been efficiently reprogrammed to iPSCs, by altering the expression of pluripotency transcription factors SOX2, NANOG and OCT4 [74]. The same study evidenced iPSCs’ colony reduction upon METTL3 knockdown [75]. A more recent study demonstrated that knockdown of METTL3 inhibits the proliferation rate of iPSCs and leads to impaired expression of pluripotency genes [76]. Further work indicates that the m6A reader, YTHDF2 is upregulated in iPSCs, but downregulated during their neural differentiation. A series of m6A modified transcripts associated with development have been found to be directly regulated by YTHDF2 and during depletion of YTHDF2 it has been indicated that iPSCs lose their pluripotency state and induces neural gene expression [77].
These studies evidence that m6A controlling mechanisms on pluripotency and reprogramming strongly depend on cellular context; as such more in-depth analysis should be carried out to elucidate the functions of epitranscriptomic modifications and their role in regulating iPSCs’ neural lineage commitment.
3. Discussion
Correlation between RNA modifications and their role during axon regeneration and degenerative diseases of the nervous system
The constant challenge that preoccupies the mind of researchers are the injuries and lesions that occur in the adult nervous system and the lack of concrete methods that can be used to avoid or heal them. This is also because the two systems, CNS and PNS, differ significantly in their response upon injuries. Whereas neurons in the PNS can respond and act when axons are injured, those in the CNS are unable to respond and undergo apoptosis, thus leading to severe tissue damage and thereby neurological disorders. During this event, the synaptic contact between neurons or between neurons and the target tissue is lost and this has a severe impact on the patient’s life. The axonal regeneration and final re-establishment of the synaptic connections request a specific set of translated proteins, which are responsible for the correct occurrence of these events. Some other proteins are also required to act as retrograde signals and trigger injury signals in the soma of the neurons [78].
Various stimuli, such as guidance cues, injury signals and growth factors are responsible for local translation of specific mRNAs in axons [79,80]. Usually, this same situation is also found in dendritic extensions, and they must respond simultaneously to the different signals [79]. Interestingly, it has been suggested that RNA modifications hold some control over the regulation of specific mRNA translocations upon extracellular signal in both cellular structures. Thus, is has been demonstrated that in axons, dynamic demethylation of mRNA is catalysed by FTO [69] and that inhibition or knockdown of FTO can lead to increase of axonal m6A-modified mRNA and reduction in axonal growth [81]. This can have a subsequent effect on generating alterations in the methylation state of the mRNA encoding for growth-associated protein 43 (gap43) which is known for its beneficial effects on promoting axon growth during development and regeneration [82]. With this evidence, it may be that mRNA demethylation by FTO is one epitranscriptomic mark which controls the local translation in axons, thus having important effects in different processes that occur in the nervous tissue, such as regeneration and axonal guidance.
The healing process concerning injured axons of adult neurons in the dorsal root ganglia relies on the transcription and translation of regeneration-associated genes (RAGs) (Fig. 3A) [83]. More exactly, specific molecules which are activated at the injury site transport information to the cell body, thus activating RAGs expression. This event is also known as retrograde signalling [84]. For efficient axonal regrowth the epigenetic machinery is also responsible via activation and regulation of RAGs [85]. Lately, it has been established that genes in peripheral nerve injuries are regulated by an epitranscriptomic mechanism. Namely, Weng et al. observed that in injured mice the m6A levels of mRNA encoding for RAGs are highly present. Moreover, the same group revealed that m6A also targets transcripts of retrograde injury signal molecules, demonstrating that m6A response is required for proper retrograde signalling, which is significant during efficient axon regeneration. In line with this idea, knockout of METTL14 impairs functional sensory axon regeneration in adult mice, resulting in poor axonal extension and decreased epidermal innervation. The same study observed that deletion of YTHDF1 reduces axonal growth following injury, which is an m6A reader responsible for promoting translation. The m6A writer complex is thought to be recruited by the same epigenetic processes and transcription factors that trigger RAGs transcription, according to theory [86]. Following these findings, a model for the contribution of m6A to sensory axon regeneration is established: injury results in transcription and increased methylation of RAGs, and YTHDF1-mediated, m6A-dependent enhanced translation of these transcripts generates the proteome machinery necessary for efficient axonal regeneration (Fig. 3A) [77]. It is possible that deficiencies in this post-transcriptional coordinating process may result in impaired axon regeneration and functional recovery. This scenario emphasizes the importance of methylation in driving stimulus-induced changes: whereas YTHDF1 contributes only in a limited way to the enhancement of transcript translation, it is critical for the recovery of the body after an injury. The varied ability to use this epitranscriptomic regulatory mechanism to quickly switch on a regeneration programme, may explain the variations in the regenerative capacity of various neurons and more generally organisms [77].
Figure 3.
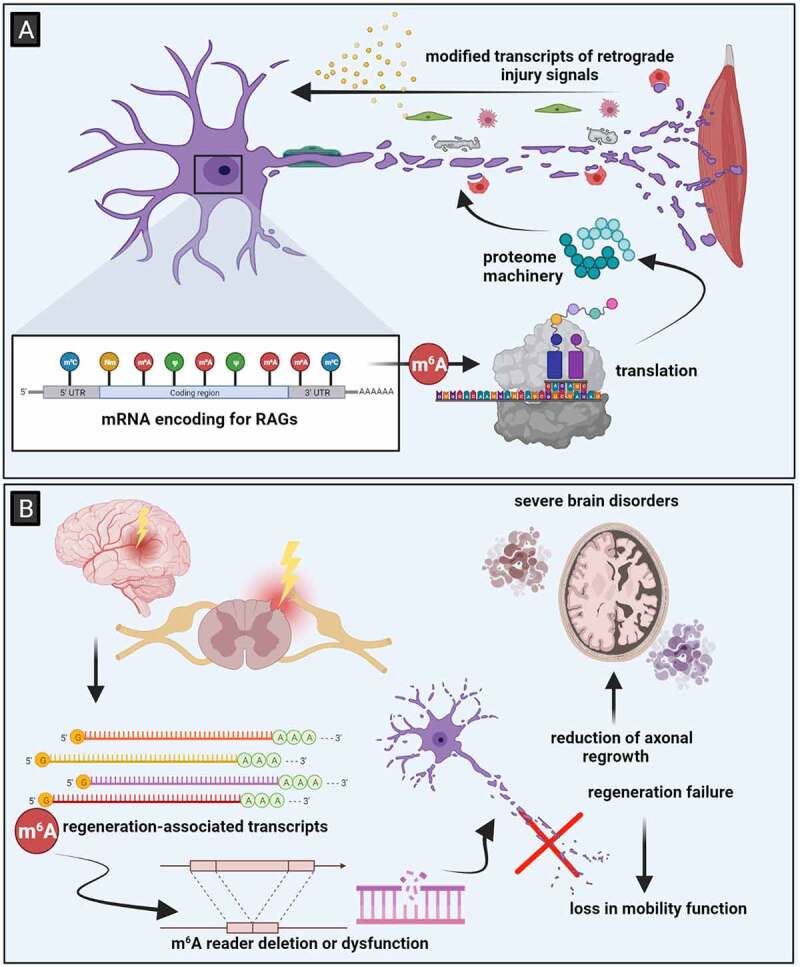
Epitranscriptomic regulation in injured nervous tissue. (A) The response to injuries in the PNS relies on the activation of RAGs, which have been found to be methylated, thus indicating that RNA modifications are required for proper regeneration proteome machinery responsible for ensuring regeneration. (B) Upon injuries in the CNS, neurons fail to regenerate, leading to degenerative tissue which is commonly for several severe brain disorders. Image created with BioRender.com
Unfortunately, research reports concerning other RNA modifications, aside from m6A, are unavailable, thus leaving a significant gap in the understanding of epitranscriptomic regulation during PNS injuries.
In the case of CNS, even though it is most likely that regeneration will not occur following a lesion, one study has offered new insights regarding spinal cord injuries (SCI). Xing et al. [87] have demonstrated for the first time, that following SCI most regeneration-related genes are highly m6A methylated. Also, they have found an increase in the expression level of METTL3 in both zebra fish and mice following SCI, demonstrating that the regeneration process is at least partly regulated by epitranscriptomic modifications. Moreover, it has been demonstrated that pten deletion-induced axon regeneration of retinal neurons in adult mammalian CNS is reduced upon METTL14 knockdown, suggesting that m6A methylation has a significant role in the responses generated by injury signals in the adult nervous system (Fig. 3B) [86].
The fact that mature neurons in the CNS are not capable of undergoing regeneration processes can further lead to the degradation of neurons, which can result in neurological disorders, such as Alzheimer’s (AD) or Parkinson’s (PD). The possible role of epitranscriptomic regulation has been recently investigated in case of degenerative disorders and it has been correlated with m6A and m5C modifications which occurred in mRNAs encoding for apoptosis-related proteins [88,89] in the nervous system. Thus, leading to the conclusion that failure of regeneration associated to neurodegenerative disorders may be also due to m6A dysregulation.
AD and PD are two of the most aggressive human degenerative diseases that occur in the ageing brain, leading to progressive memory loss and impaired cognitive function [90], tremor and rigidity [91], respectively. Both disorders have been widely studied over the past decade, but the pathogenesis remains unclear. Significant steps have been made towards understanding the regulatory mechanism underlying the development of these disorders and so it has been identified that RNA modifications contribute to neurological disorders. In this context, studies on AD mouse models have demonstrated the presence of increased m6A methylation in the hippocampus and cortex, and simultaneously it has been detected that expression of METTL3 was upregulated, whereas fto expression was downregulated [92,93] (Fig. 3B). Moreover, studies on human genetics have identified different variants of FTO which have been associated with AD [94]. On top of this, aside from m6A that has been identified as a potent regulator, a study highlighted that CMTR1, a Nm writer, is significantly increased in ageing AD mouse models [95]. In case of PD, which is a disease characterized by loss of dopaminergic innervation [96], depletion of FTO increased m6A methylation and reduced the translation of dopaminergic-related transcripts [97]. Moreover, upregulation of m6A demethylases ALKBH5 and FTO, as well as low levels of m6A motifs have been identified in PD rat models [98].
Targeting stem cell reprogramming and RNA modifications as a potential therapeutic approach for nervous system disorders
All the above-mentioned evidence suggests that the field of epitranscriptomics has a significant contribution to the proper development and regeneration of injuries in the nervous system, but the key question here is how these modifications can be targeted to act as therapeutic agents or as treatment agents for all the affections of the nervous system. Over the years, several methods have been developed to use RNA modification or to target them with the hopes of finding new tools which will provide satisfying recovery of the damaged tissue.
Chemically modified RNA (cmRNA) has been proposed as a safe tool for cell programming (aka differentiation) and reprograming (aka dedifferentiation) and tissue engineering applications [99]. So far, cmRNA-based therapy has been investigated in case of cancer immunotherapy [100], mRNA vaccines [101 112], gene editing [102 113] and regenerative medicine [103]. In the same manner that fibroblasts have been reprogrammed to become iPSCs, cmRNA methods could potentially dedifferentiate specific neuronal subpopulations in case of injury and severe diseases or to improve stem cells response upon injuries. In this context, human mesenchymal stem cells (hMSCs) have been reprogrammed to the state of induced NSCs by sox2 cmRNA [104], therefore proposing a more reliable source of stem cells which have successfully responded to neural stimulation, thus developing a potential treatment for neurological disorders. Moreover, it has been reported that integrin a4 mRNA transfected-MSCs have successfully migrated to the brain ischaemic area, where MSCs can exhibit their anti-inflammatory effects and attenuate the inflammation site [105]. Even though, these studies expose significant steps in therapeutic application, a lot of challenges, such as control release of cmRNA or developing proper delivery vehicles still need to be overcome to fully apply these methods on brain disorder-affected patients.
Defective RNA modifications sites, such as the functional duplicity of writers, erasers and readers could be responsible for failure of normal cell functioning and thus leading to unwanted neurological disorders. Fine and modern molecular biology technologies have led to the discovery of the Cas13 family of proteins which have high affinity for endogenous RNA, thus opening new doors in the field of gene transcript editing [106]. Association of clustered regularly interspaced short palindromic repeats (CRISPR) with the catalytically inactive form of Cas13 (dCas13) leads to the generation of the programmable CRISPR-dCas13 system, which can target a specific nucleic acid site within RNA molecules [107]. Very recent work has investigated the achievement of m6A editing by the CRISPR-dCas13 system and it has been reported that m6A erasers, ALKBH5 and FTO can be included in the CRISPR-dCas13 system and therefore target hypermethylated regions or control hypomethylation, thus making mRNAs more stable [108]. Also, transcripts that lose m6A modifications due to dysfunctions in METTL3 or METTL14 can be edited and repaired by the CRISPR-dCas13 system. Wilson et al. [109] have associated dCas13 with a modified METTL3/METTL14 complex, thus resulting in a CRISPR-dCas13 construct that was able to include site-specific m6A motifs on mRNA encoding for sox2 in human cells. Based on these findings, the CRISPR-dCas13 system could serve as an efficient tool for RNA editing in case of brain damage and disorders. Addressing high-resolution single nucleotides techniques will be crucial in identification of abnormal RNA modifications which are correlated with neuropathological affections. Therefore, more future investigations are necessary to specifically reveal the downstream mechanism underlying the nervous tissue damage caused by dysfunction in RNA modifications and help identify a transcript editing-based therapeutic strategy.
4. Concluding remarks
To sum up, significant steps have been made in understanding how epitranscriptomic marks govern to complex process of stem cell differentiation towards the neural lineage. It has been established that RNA modifications are strongly correlated with proper cellular functions such as splicing, nuclear transport and gene expression. Thus, it is crucial for these events to occur smoothly, as it has been also highlighted in this review. In case of mutations occurrence in the genes encoding for enzymes controlling RNA modifications, it is most likely that neurological pathologies appear and have a terrific impact on one patient’s life. Therefore, it is crucial to fully understand the molecular mechanism underlying RNA modifications in stem cell differentiation towards the neural progeny and the development of healthy neural tissue. Next, epitranscriptomic marks could be used as therapeutic agents as treatment for damaged brain tissue and injured peripheral nerves or to use them as prevention against neurodegenerative affections.
Acknowledgments
This work was supported by a grant of the Romanian Ministry of Education and Research, CNCS-UEFISCDI, project number PN-III-P1-1.1-TE-2019-1191, within PNCDI III. The publication of this review was also possible in the frame of COST Action CA16120 (EPITRAN).
Funding Statement
This work was supported by the European Cooperation in Science and Technology [CA16120 (EPITRAN)]; Unitatea Executiva pentru Finantarea Invatamantului Superior, a Cercetarii, Dezvoltarii si Inovarii [PN-III-P1-1.1-TE-2019-1191].
Conflict of Interest
No potential competing interest was reported by the authors.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Kumar S, Mohapatra T.. Deciphering epitranscriptome: modification of mRNA bases provides a new perspective for post-transcriptional regulation of gene expression. Front Cell Dev Biol. 2021;9:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20(6):303–322. [DOI] [PubMed] [Google Scholar]
- [3].Schaefer M, Kapoor U, Jantsch MF. Understanding RNA modifications: the promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017;7(5):170077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang B, Jiang H, Dong Z, et al. The critical roles of m6A modification in metabolic abnormality and cardiovascular diseases. Genes Dis. 2020. 1;(8)6:746-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang Y, Sun BF, Xiao W, et al. Dynamic m6A modification and its emerging regulatory role in mRNA splicing. Sci Bull. 2015;60(1):21–32. [Google Scholar]
- [6].Yang C, Hu Y, Zhou B, et al. The role of m 6 A modification in physiology and disease. Cell Death Dis. 2020;11(11):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hsu PJ, Shi H, He C. Epitranscriptomic influences on development and disease. Genome Biol. 2017;18(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Du K, Zhang L, Lee T, et al. m6A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56(3):1596–1606. [DOI] [PubMed] [Google Scholar]
- [9].Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Horiuchi K, Kawamura T, Iwanari H, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288(46):33292–33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Van Tran N, Ernst FGM, Hawley BR, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47(15):7719–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ren W, Lu J, Huang M, et al. Structure and regulation of ZCCHC4 in m 6 A-methylation of 28S rRNA. Nat Commun. 2019;10(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brown JA, Kinzig CG, DeGregorio SJ, et al. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc Nat Acad Sci. 2016;113(49):14013–14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bokar JA, Rath-Shambaugh ME, Ludwiczak R, et al. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269(26):17697–17704. [PubMed] [Google Scholar]
- [16].Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem Sci. 2002;27(10):495–497. [DOI] [PubMed] [Google Scholar]
- [17].Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kretschmer J, Rao H, Hackert P, et al. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. Rna. 2018;24(10):1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao YL, Liu YH, Wu RF, et al. Understanding m 6 A function through uncovering the diversity roles of YTH domain-containing proteins. Mol Biotechnol. 2019;61(5):355–364. [DOI] [PubMed] [Google Scholar]
- [20].Lasman L, Krupalnik V, Viukov S, et al. Context-dependent functional compensation between Ythdf m6A. Genes Dev. 2020;34(19-20):1373-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xue C, Zhao Y, Li L. Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer. Biomark Res. 2020;8(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31(5):458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30(7):733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Trixl L, Lusser A. The dynamic RNA modification 5‐methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10(1):e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 2019;10(2):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49(5):341–351. [DOI] [PubMed] [Google Scholar]
- [27].Borchardt EK, Martinez NM, Gilbert WV. Regulation and function of RNA pseudouridylation in human cells. Annu Rev Genet. 2020;54(1):309–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rintala-Dempsey AC, Kothe U. Eukaryotic stand-alone pseudouridine synthases–RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;14(9):1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Duan J, Li L, Lu J, et al. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell. 2009;34(4):427–439. [DOI] [PubMed] [Google Scholar]
- [30].Mueller EG, Ferré-D’Amaré AR. Pseudouridine formation, the most common transglycosylation in RNA. In: Grosjean H, editor. DNA and RNA modification enzymes: structure, mechanism, function and evolution. Madame Curie Bioscience Database [Internet] Landes Bioscience, Austin (Texas); 2009. p. 363–376. [Google Scholar]
- [31].Hamma T, Ferré-D’Amaré AR. Pseudouridine synthases. Chem Biol. 2006;13(11):1125–1135. [DOI] [PubMed] [Google Scholar]
- [32].Ayadi L, Galvanin A, Pichot F, et al. RNA ribose methylation (2′-O-methylation): occurrence, biosynthesis and biological functions. Biochimica Et Biophysica Acta (Bba)-gene Regulatory Mechanisms. 2019;1862(3):253–269. [DOI] [PubMed] [Google Scholar]
- [33].Abou Assi H, Rangadurai AK, Shi H, et al. 2′-O-Methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucleic Acids Res. 2020;48(21):12365–12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Funk HM, DiVita DJ, Sizemore HE, et al. (2021). Identification of a motif in Trm732 required for 2′-O-methylation of the tRNA anticodon loop by Trm7. bioRxiv. [DOI] [PMC free article] [PubMed]
- [35].Inesta‐Vaquera F, Cowling VH. Regulation and function of CMTR1‐dependent mRNA cap methylation. Wiley Interdiscip Rev RNA. 2017;8(6):e1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].He Q, Yang L, Gao K, et al. FTSJ1 regulates tRNA 2ʹ-O-methyladenosine modification and suppresses the malignancy of NSCLC via inhibiting DRAM1 expression. Cell Death Dis. 2020;11(5):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Takahashi S, Kobayashi S, Hiratani I. Epigenetic differences between naïve and primed pluripotent stem cells. Cell Mol Life Sci. 2018;75(7):1191–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–492. [DOI] [PubMed] [Google Scholar]
- [39].Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang X, Lu Z, Gomez A, et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505(7481):117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang Y, Li Y, Toth JI, et al. N 6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16(2):191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Batista PJ, Molinie B, Wang J, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. [DOI] [PubMed] [Google Scholar]
- [44].Zhao BS, He C. Fate by RNA methylation: m 6 A steers stem cell pluripotency. Genome Biol. 2015;16(1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Amort T, Rieder D, Wille A, et al. Distinct 5-methylcytosine profiles in poly (A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tuorto F, Liebers R, Musch T, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19(9):900–905. [DOI] [PubMed] [Google Scholar]
- [47].Sampath P, Pritchard DK, Pabon L, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448–460. [DOI] [PubMed] [Google Scholar]
- [48].Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311(5759):395–398. [DOI] [PubMed] [Google Scholar]
- [49].Angelova MT, Dimitrova DG, Dinges N, et al. The emerging field of epitranscriptomics in neurodevelopmental and neuronal disorders. Front Bioeng Biotechnol. 2018;6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shaheen R, Han L, Faqeih E, et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet. 2016;135(7):707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Heiss NS, Bächner D, Salowsky R, et al. Gene structure and expression of the mouse dyskeratosis congenita gene, dkc1. Genomics. 2000;67(2):153–163. [DOI] [PubMed] [Google Scholar]
- [52].Agarwal S, Loh YH, McLoughlin EM, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464(7286):292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fong YW, Ho JJ, Inouye C, et al. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. Elife. 2014;3:e03573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tang Y, Yu P, Cheng L. Current progress in the derivation and therapeutic application of neural stem cells. Cell Death Dis. 2017;8(10):e3108–e3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yao B, Christian KM, He C, et al. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17(9):537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Boles NC, Temple S. Epimetronomics: m6A marks the tempo of corticogenesis. Neuron. 2017;96(4):718–720. [DOI] [PubMed] [Google Scholar]
- [57].Atlasi Y, Stunnenberg HG. The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet. 2017;18(11):643–658. [DOI] [PubMed] [Google Scholar]
- [58].Weinberger L, Ayyash M, Novershtern N, et al. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17(3):155–169. [DOI] [PubMed] [Google Scholar]
- [59].Ke S, Pandya-Jones A, Saito Y, et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31(10):990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Frye M, Harada BT, Behm M, et al. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wang CX, Cui GS, Liu X, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16(6):e2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yoon KJ, Ringeling FR, Vissers C, et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171(4):877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Li M, Zhao X, Wang W, et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang Y, Li Y, Yue M, et al. N 6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Frye M, Blanco S. Post-transcriptional modifications in development and stem cells. Development. 2016;143(21):3871–3881. [DOI] [PubMed] [Google Scholar]
- [66].Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of t RNA s links cellular stress to neuro‐developmental disorders. EMBO J. 2014;33(18):2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Reports. 2017;8(1):112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Merkurjev D, Hong WT, Iida K, et al. Synaptic N 6-methyladenosine (m 6 A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci. 2018;21(7):1004–1014. [DOI] [PubMed] [Google Scholar]
- [69].Yu J, Chen M, Huang H, et al. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018;46(3):1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhuang M, Li X, Zhu J, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3. 1 expression. Nucleic Acids Res. 2019;47(9):4765–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Guy MP, Shaw M, Weiner CL, et al. Defects in tRNA anticodon loop 2′‐O‐methylation are implicated in nonsyndromic X‐linked intellectual disability due to mutations in FTSJ1. Hum Mutat. 2015;36(12):1176–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lee YL, Kung FC, Lin CH, et al. CMTR1-Catalyzed 2′-O-Ribose Methylation Controls Neuronal Development by Regulating Camk2α Expression Independent of RIG-I Signaling. Cell Rep. 2020;33(3):108269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wu R, Li A, Sun B, et al. A novel m 6 A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29(1):23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Aguilo F, Walsh MJ. The N6-methyladenosine RNA modification in pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Chen T, Hao YJ, Zhang Y, et al. m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16(3):289–301. [DOI] [PubMed] [Google Scholar]
- [76].Wu R, Liu Y, Zhao Y, et al. m 6 A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 2019;10(3):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Livneh I, Moshitch-Moshkovitz S, Amariglio N, et al. The m 6 A epitranscriptome: transcriptome plasticity in brain development and function. Nat Rev Neurosci. 2020;21(1):36–51. [DOI] [PubMed] [Google Scholar]
- [78].Hanz S, Fainzilber M. Retrograde signaling in injured nerve–the axon reaction revisited. J Neurochem. 2006;99(1):13–19. [DOI] [PubMed] [Google Scholar]
- [79].Spaulding EL, Burgess RW. Accumulating evidence for axonal translation in neuronal homeostasis. Front Neurosci. 2017;11:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Cioni JM, Koppers M, Holt CE. Molecular control of local translation in axon development and maintenance. Curr Opin Neurobiol. 2018;51:86–94. [DOI] [PubMed] [Google Scholar]
- [81].Walters BJ, Mercaldo V, Gillon CJ, et al. The role of the RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology. 2017;42(7):1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Donnelly CJ, Park M, Spillane M, et al. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013;33(8):3311–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Mahar M, Cavalli V. Intrinsic mechanisms of neuronal axon regeneration. Nat Rev Neurosci. 2018;19(6):323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Abe N, Cavalli V. Nerve injury signaling. Curr Opin Neurobiol. 2008;18(3):276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhou FQ. Genetic study of axon regeneration with cultured adult dorsal root ganglion neurons. J Vis Exp. 2012;66(66):e4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Weng YL, Wang X, An R, et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97(2):313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Xing L, Cai Y, Yang T, et al. Epitranscriptomic m6A regulation following spinal cord injury. J Neurosci Res. 2021;99(3):843–857. [DOI] [PubMed] [Google Scholar]
- [88].Casella G, Tsitsipatis D, Abdelmohsen K, et al. mRNA methylation in cell senescence. Wiley Interdiscip Rev RNA. 2019;10(6):e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Min KW, Zealy RW, Davila S, et al. Profiling of m6A RNA modifications identified an age‐associated regulation of AGO 2 mRNA stability. Aging Cell. 2018;17(3):e12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sengoku R. Aging and Alzheimer’s disease pathology. Neuropathology. 2020;40(1):22–29. [DOI] [PubMed] [Google Scholar]
- [91].Kouli A, Torsney KM, Kuan WL. Parkinson’s disease: etiology, neuropathology, and pathogenesis. In: Stoker TB, Greenland JC, editors. Parkinson's Disease: Pathogenesis and Clinical Aspects, Exon Publications, Brisbane (AU); 2018;p. 3–26. [Google Scholar]
- [92].Han M, Liu Z, Xu Y, et al. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front Neurosci. 2020;14:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Shafik AM, Zhang F, Guo Z, et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021;22(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Li H, Ren Y, Mao K, et al. FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem Biophys Res Commun. 2018;498(1):234–239. [DOI] [PubMed] [Google Scholar]
- [95].Boza-Serrano A, Yang Y, Paulus A, et al. Innate immune alterations are elicited in microglial cells before plaque deposition in the Alzheimer’s disease mouse model 5xFAD. Sci Rep. 2018;8(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Grosch J, Winkler J, Kohl Z. Early degeneration of both dopaminergic and serotonergic axons–a common mechanism in Parkinson’s disease. Front Cell Neurosci. 2016;10:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hess ME, Hess S, Meyer KD, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16(8):1042–1048. [DOI] [PubMed] [Google Scholar]
- [98].Chen X, Yu C, Guo M, et al. Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem Neurosci. 2019;10(5):2355–2363. [DOI] [PubMed] [Google Scholar]
- [99].Badieyan ZS, Evans T. Concise review: application of chemically modified mRNA in cell fate conversion and tissue engineering. Stem Cells Transl Med. 2019;8(8):833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Van Hoecke L, Verbeke R, Dewitte H, et al. mRNA in cancer immunotherapy: beyond a source of antigen. Mol Cancer. 2021;20(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang Y, Zhang Z, Luo J, et al. mRNA vaccine: a potential therapeutic strategy. Mol Cancer. 2021;20(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Tang Q, Liu J, Jiang Y, et al. Cell-selective messenger RNA delivery and CRISPR/Cas9 genome editing by modulating the interface of phenylboronic acid-derived lipid nanoparticles and cellular surface sialic acid. ACS Appl Mater Interfaces. 2019;11(50):46585–46590. [DOI] [PubMed] [Google Scholar]
- [103].Patel S, Athirasala A, Menezes PP, et al. Messenger RNA delivery for tissue engineering and regenerative medicine applications. Tissue Eng Part A. 2019;25(1–2):91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kim BE, Choi SW, Shin JH, et al. Single-factor SOX2 mediates direct neural reprogramming of human mesenchymal stem cells via transfection of in vitro transcribed mRNA. Cell Transplant. 2018;27(7):1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang S, Lachance BB, Moiz B, et al. Optimizing stem cell therapy after ischemic brain injury. J Stroke. 2020;22(3):286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Cox DB, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-targeting CRISPR–Cas systems and their applications. Int J Mol Sci. 2020;21(3):1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Mo J, Chen Z, Qin S, et al. TRADES: targeted RNA demethylation by suntag system. Adv Sci. 2020;7(19):2001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Wilson C, Chen PJ, Miao Z, et al. Programmable m 6 A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol. 2020;38(12):1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]


