Abstract
Current serological tests for Babesia bigemina use semipurified merozoite antigens derived from infected erythrocytes. One of the major drawbacks of these tests is that antigen quality can vary from batch to batch. Since the quality of the antigen contributes to the sensitivity and specificity of serological tests, the use of standardized recombinant antigens should ensure consistency in assay quality. Previously, a 200-kDa merozoite antigen (p200) was identified as a candidate diagnostic antigen for use in a serological assay for the detection of B. bigemina antibodies in infected cattle. In this study, we have cloned, characterized, and expressed p200. A 3.5-kbp cDNA clone encoding p200 was isolated and shown to be almost full length, lacking approximately 300 bp at the 5′ end. The predicted amino acid sequence shows that p200 consists of a long, highly charged central repeat region of an uninterrupted α helix, indicative of a fibrous protein. Immunoelectron microscopy localized p200 to the merozoite cytoplasm, suggesting that the antigen may be a structural protein involved in forming filament structures within the cytoskeleton. The 3.5-kbp cDNA was expressed in bacteria as a fusion protein with glutathione S-transferase (GST), but the yield was poor. To improve the yield, cDNA fragments encoding antigenic domains of p200 were expressed as fusions with GST. One of these fusion proteins, C1A-GST, is composed of a 7-kDa fragment of the p200 repeat region and contains epitopes that react strongly with sera from cattle experimentally infected with B. bigemina. Recombinant C1A-GST should permit the development of an improved enzyme-linked immunosorbent assay for the detection of antibodies against B. bigemina.
Babesia bigemina is a tick-borne protozoan parasite of cattle that causes a disease variously referred to as Texas fever, redwater fever, or cattle tick fever. The disease is characterized by fever, hemolytic anemia, hemoglobinuria and, in acute cases, death (8). The parasite is widely distributed throughout Africa, southern Europe, southern Asia, southeastern Asia, Australia, Central America, and South America, coincident with its main tick vectors Boophilus decoloratus, Boophilus microplus, and Boophilus annulatus (2). Economically, it is most important as a cause of heavy losses in susceptible cattle, particularly in imported taurine breeds.
The classical diagnosis of animals acutely infected with Babesia is made by the light microscopic demonstration of intraerythrocytic parasites in Giemsa-stained blood smears (7). However, when infections are subclinical or latent, parasites may not always be demonstrable by microscopy because of low levels of parasitemia (13). Alternatively, infection of an animal by Babesia can be determined directly by PCR-based tests (4, 21) or indirectly by measurement of the humoral response using serological tests (27). While PCR can provide good sensitivity and specificity and is able to detect current, carrier infections, such tests are complex and time-consuming, requiring specialized laboratory equipment and highly trained personnel. As such, PCR-based tests are currently not applicable for use in many of the regions where babesiosis causes high economic losses. Serodiagnostic methods, however, are generally much simpler to perform and can provide important information for implementing control measures and for epidemiological studies. Several serological tests for the detection of antibodies to B. bigemina have been developed, including complement fixation, passive hemagglutination, capillary tube agglutination, card agglutination, indirect immunofluorescence test, and enzyme-linked immunosorbent assay (ELISA) (17, 27). The indirect immunofluorescence test and the ELISA are most widely used because of their superior sensitivity, robustness, and ease of use. These tests, however, use either whole parasites or semipurified antigens, whose qualities can vary from batch to batch. Also, the production of antigens for these tests requires experimentally infected cattle, making production time-consuming and expensive.
A merozoite antigen of approximately 200 kDa (p200) in B. bigemina is a candidate diagnostic antigen (18), and it was shown that 98% of sera collected from cattle in areas in which B. bigemina is endemic recognized this antigen (J. M. Katende, unpublished data). Monoclonal antibodies (MAbs) to p200 were used to immobilize native antigen in an indirect antibody ELISA. This ELISA was shown to be specific for B. bigemina antibodies, lacking cross-reactivities with sera from cattle infected with Babesia bovis, Theileria parva, Theileria taurotragi, or Anaplasma marginale (18). A major improvement in this assay would be obtained through the use of standardized recombinant p200.
In this study, the expression of recombinant p200 in bacteria was undertaken to facilitate the production of large quantities of standardized antigen for the development of a serodiagnostic test. Major bovine B-cell epitopes were identified within p200, and these were expressed as a recombinant 7-kDa fragment. This recombinant p200 fragment is a strong candidate diagnostic antigen that should facilitate the development of an improved antibody ELISA for B. bigemina.
MATERIALS AND METHODS
Parasite stocks.
B. bigemina (Kikuyu stock) from Kiambu District, Kenya, was provided by A. Kelly, National Veterinary Laboratories, Kabete, Kenya. The Pongola strain of B. bigemina was obtained from D. T. de Waal, Onderstepoort Veterinary Institute, Onderstepoort, South Africa.
Purification of B. bigemina merozoites.
B. bigemina merozoites were prepared from blood collected at peak parasitemia from experimentally infected, splenectomized Friesian calves. Infected blood was collected into an equal volume of heparinized Alsever's solution. The blood was centrifuged at 2,500 × g for 30 min at 4°C, and the packed cells were washed four times with Alsever's solution by centrifugation as before. For RNA preparation, an aliquot of the packed infected erythrocyte pellet was lysed with an equal volume of 1% cold acetic acid in water, and unlysed cells were removed by centrifugation at 2,500 × g for 10 min at 4°C. The supernatant was centrifuged at 9,000 × g for 10 min at 4°C. The pellet was resuspended in an equal volume of Dulbecco's phosphate-buffered saline (DPBS) and centrifuged again. The resulting pellet of B. bigemina merozoites and erythrocyte ghost membranes was immediately snap frozen as droplets in liquid nitrogen and stored at −70°C. For genomic DNA extraction, packed infected erythrocytes were lysed in an equal volume of 1 mg of saponin per ml in water for 10 s. Lysis was stopped by the addition of four volumes of 20 mM Tris-HCl (pH 8.0)–10 mM EDTA–100 mM NaCl (TEN buffer) and followed by centrifugation at 1,000 × g for 15 min at 4°C to remove leukocyte nuclei and unlysed erythrocytes. The supernatant was recovered and further centrifuged at 8,000 × g for 30 min at 4°C to pellet the merozoites. The pellet of merozoites was washed with TEN buffer by further centrifugation until it was free of hemoglobin and stored at −70°C.
Genomic DNA preparations.
DNA was prepared from purified merozoites by standard methods of proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation (22). The B. bigemina DNA preparations from Argentina (pathogenic strain S2P), Brazil (wild-type strain B.68Cpo6dc), and Mexico (cloned strain J629) were obtained from the Department of Veterinary Microbiology and Pathology, Washington State University, Pullman. DNA from B. bovis (strain KR) was obtained from A. Lew and W. Jorgensen, Animal Research Institute, Yeerongpilly, Queensland, Australia.
Production of antibodies.
Hyperimmune polyclonal antiserum BJ28 was prepared by intravenously inoculating a Boran steer with B. bigemina (Kikuyu) blood stabilate. Three booster inoculations were given subcutaneously at 2-week intervals after the primary inoculation. Serum was collected 2 weeks after the final inoculation. Bovine immunoglobulins (immunoglobulin G1 [IgG1] and IgG2) were purified by DE52 chromatography by standard procedures (11) from sera pooled from 28 cattle that had recovered from B. bigemina infection. Rat polyclonal antisera to p200 was raised by inoculation with p200 purified by preparative sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). MAbs Bb F4/86.6, Bb F4/86.11, Bb F4/86.19, and Bb F4/86.34, with specificity for the B. bigemina p200 antigen, were derived from BALB/c mice inoculated with B. bigemina (Kikuyu) merozoite lysate by methods described previously (19). Bb F4/86.6, Bb F4/86.11, and Bb F4/86.19 are isotype IgM, while Bb F4/86.34 is isotype IgG1. For sequential bovine infection sera, Boran steer BJ26 was infected with B. bigemina sporozoite stabilate 3899, and sera were collected at approximately 5-day intervals over a 228-day period.
ELISA, SDS-PAGE, and immunoblotting.
The indirect antibody ELISA was carried out essentially as described previously (14). Bound antibodies were detected with horseradish peroxidase (HRP)-conjugated secondary antibodies and ABTS [2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)] as a chromogen. Optical densities (OD) were measured at 414 nm. Results were expressed as OD values or as percent positivity (PP) values. PP was calculated as follows: (OD of test serum/OD of a strong positive serum) × 100 (13). SDS-PAGE under reducing conditions and immunoblotting were performed as detailed previously (11). For immunoblotting, secondary antibodies were conjugated to HRP (14) or 125I (Amersham International, Aylesbury, United Kingdom). For blots incubated with HRP-labeled conjugates, antibody binding was visualized by the addition of a substrate-chromogen (hydrogen peroxide-diaminobenzidine) solution. For filters incubated with 125I-labeled conjugates, antibody binding was visualized by exposure to X-ray film.
Construction and immunoscreening of a B. bigemina cDNA expression library.
Total RNA was isolated from B. bigemina (Kikuyu) merozoites by the hot phenol-SDS method (25). mRNA isolated by oligo(dT)-cellulose chromatography was used to construct an expression library in λgt11 by methods described previously (22). Immunoscreening of λgt11 phage plaques propagated on Escherichia coli strain Y1090 was carried out by standard methods (22). The library was screened with MAb Bb F4/86.6 diluted to 10 mg/ml, bovine recovery serum immunoglobulins at 20 μg/ml, hyperimmune antiserum BJ28 diluted to 1:100, and polyvalent rat antisera to p200 diluted to 1:50. Phage plaques that were positive with all the sera were selected for further study.
Southern and Northern hybridization.
Southern hybridization was carried out by standard methods (22). Five micrograms of Babesia genomic DNA and 15 μg of bovine lymphocyte DNA were digested with EcoRI, electrophoresed through a 1% agarose gel, transferred to a Hybond-N nylon membrane (Amersham), and hybridized with an [α-32P]dCTP-labeled probe. Blots were washed at a high stringency (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% SDS at 65°C). Northern hybridization was carried out by published methods (20). Two micrograms of B. bigemina merozoite mRNA was electrophoresed through a 1% agarose gel and transferred to Hybond-N. Hybridization and washing of Northern blots were as described for Southern blots.
Sequencing of the cDNA encoding p200.
The 3.5-kbp cDNA encoding p200 (designated Bb3.5) was subcloned into pUC18 for sequencing. Sequencing was performed with the fmol (Promega Corp., Madison, Wis.) and Sequenase (United States Biochemicals, Cleveland, Ohio) sequencing systems according to the manufacturers' instructions and with primers to the vector and primers based upon acquired sequences. Additional sequences were obtained from fragments of Bb3.5 cDNA that were generated by PvuI, Sau3AI, and BAL 31 digestions, subcloned into pUC18, pBluescript II KS(+) (Stratagene, La Jolla, Calif.), or pNEB193 (New England Biolabs), and sequenced with vector primers. Sequence analysis was performed with DNASIS V2.5 for Windows (Hitachi Software Engineering America Ltd., San Bruno, Calif.) and PC/gene 6.80 software (IntelliGenetics Inc., Mountain View, Calif.). The prediction of protein secondary structure and hydrophobicity was done with published algorithms (8, 11). The BLAST facility of the National Center for Biotechnology Information was used for sequence homology searches (http://www.ncbi.nlm.nih.gov/blast/blast.cgi).
Bacterial expression of the p200 antigen.
Recombinant proteins were produced in bacteria as fusion proteins with 26-kDa glutathione S-transferase (GST) by use of the pGEX1λT expression system (Pharmacia Biotech, Uppsala, Sweden). Recombinant proteins were produced in E. coli strain XL1-Blue (Stratagene) and affinity purified on glutathione-Sepharose (Pharmacia) according to the manufacturer's instructions. Parental GST was produced from nonrecombinant vector pGEX1N (24).
Construction and immunoscreening of a subfragment library of the p200 3.5-kbp cDNA.
Random DNA fragments of approximately 100 to 500 bp were created by digestion of the 3.5-kbp cDNA in pUC18 with a DNase Shotgun Cleavage Kit (Novagen Inc., Madison, Wis.) according to the manufacturer's instructions. The fragments were cloned into λgt11, and approximately 10,000 recombinant plaques were immunoscreened with hyperimmune antiserum BJ28. Immunopositive clones were also screened with MAbs Bb F4/86.6, Bb F4/86.11, Bb F4/86.19, and Bb F4/86.34. Inserts from selected clones were subcloned into pGEX1λT and expressed as GST fusion proteins.
Epitope mapping with synthetic peptides.
A set of 11 overlapping peptides on pins was synthesized by Chiron Mimotopes (Clayton, Victoria, Australia) to represent the fragment of p200 encoded by clone C1A (see below). Each peptide was 12 residues long and overlapped the preceding and subsequent peptides by 7 residues, except for the last peptide, which overlapped the preceding peptide by 10 residues. A 12-residue peptide with an irrelevant amino acid sequence was used as a control. The peptides were tested in an ELISA with hyperimmune antiserum BJ28 diluted to 1:100, naive bovine serum diluted to 1:100, and MAb Bb F4/86.6 at a concentration of 10 μg/ml. For reuse, the pins were treated in a sonication bath with 1% SDS–0.1% 2-mercaptoethanol in 0.1 M phosphate buffer (pH 7.2) for 10 min at 60°C, followed by rinsing in distilled water at 60°C.
Localization of the B. bigemina p200 antigen in merozoites by immunoEM.
A 200-μl aliquot of blood from a calf that had 10% parasitemia with B. bigemina (Kikuyu stock) was collected directly into 2% parafomaldehyde in 0.1 M phosphate buffer (pH 7.3) and fixed in suspension for 1 h at room temperature. Fixed cells were centrifuged at 13,000 × g for 5 min, and the cell pellet was processed into Lowicryl K4M resin (Chemische Werke Lowi GmbH & Co., Waldkraiburg, Germany) by the method described previously (1). Cells were prepared for immunogold electron microscopy (immunoEM) by the method described previously (3). Briefly, 60-nm sections of embedded cells were cut and collected on Parlodion-coated copper grids. The sections were preincubated by flotation on 20-μl droplets of 5% bovine serum albumin (BSA) in DPBS in a humidified chamber for 30 min. The sections were then incubated for 1 h with MAb Bb F4/86.11 diluted 1:50 in 5% BSA–DPBS. After being washed in 5% BSA–DPBS, the sections were incubated for 1 h with goat antimouse IgG–5-nm colloidal gold (British Biocell, Cardiff, United Kingdom) diluted 1:10 in 5% BSA–DPBS. The sections were washed twice with DPBS, followed by two washes with distilled water. Sections were then contrasted for 2 min with 2% aqueous osmium tetroxide, washed, and stained for 20 min with 2% aqueous uranyl acetate. The sections were examined with a Zeiss EM 10A electron microscope (Carl Zeiss, Oberkochen, Germany) operating at 80 kV.
Nucleotide sequence accession number.
Nucleotide sequence data reported in this paper are available in the GenBank database under accession number AF142406.
RESULTS
Characterization of antibodies to the B. bigemina p200 antigen.
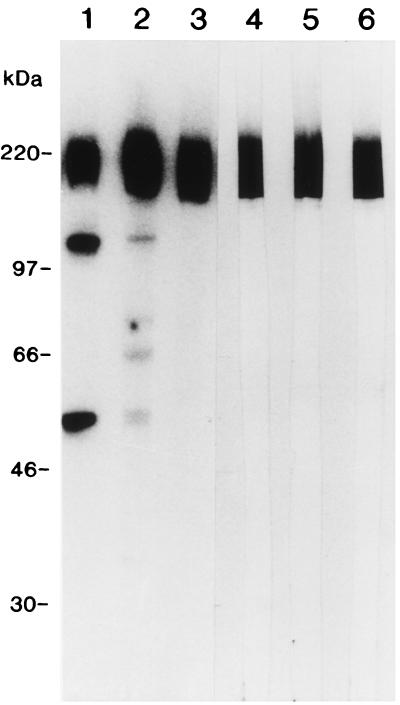
Anti-p200 antibodies that were used for immunoscreening an expression library were characterized by immunoblotting. Antigens recognized by bovine hyperimmune serum BJ28, bovine recovery serum immunoglobulins, rat antisera, and MAbs to the p200 antigen are shown in Fig. 1. All sera recognized the p200 antigen as a diffuse band extending from approximately 150 to 250 kDa. Hyperimmune and recovery sera also recognized other proteins between 58 and 125 kDa, but p200 appeared as the most immunodominant. Rat polyvalent antisera and MAbs were specific for p200. None of the antibodies reacted with bovine erythrocyte proteins (data not shown).
FIG. 1.
Immunoblotting of B. bigemina merozoite lysate with antibodies to the p200 antigen. B. bigemina merozoite proteins were separated by SDS-PAGE through a 7.5 to 17.5% gradient gel, transferred to a nitrocellulose membrane, and probed with B. bigemina hyperimmune serum BJ28 diluted to 1:100 (lane 1), 20 μg of bovine recovery serum immunoglobulins per ml (lane 2), polyclonal rat antisera to p200 diluted to 1:50 (lane 3), and 10 μg each of anti-p200 MAbs Bb F4/86.6 (lane 4), Bb F4/86.34 (lane 5), and Bb F4/86.11 (lane 6) per ml. The positions of molecular mass standards are indicated to the left.
Isolation of cDNA encoding the p200 antigen.
Approximately 80,000 PFU from a B. bigemina (Kikuyu) cDNA expression library in λgt11 were immunoscreened for the p200 antigen. Thirty-five plaques were positive with all screening sera: MAb Bb F4/86.6, bovine hyperimmune serum BJ28, bovine recovery serum immunoglobulins, and rat antisera to p200. The clone that gave the strongest signals on immunoscreening was selected for further study. The cDNA in this clone was estimated to be 3.5 kbp (data not shown) and was designated Bb3.5.
Southern and Northern hybridization.
In Southern blotting, Bb3.5 cDNA hybridized to a single band of approximately 20 kbp in EcoRI-digested DNA of B. bigemina isolated from Kenya, South Africa, Australia, Brazil, Argentina, and Mexico (Fig. 2A), indicating that the p200 gene is conserved among B. bigemina stocks and strains from different geographical regions. There was no hybridization of Bb3.5 cDNA to B. bovis DNA (Fig. 2A) or to bovine lymphocyte DNA (data not shown). Bb3.5 cDNA hybridized to a single band of 3.8 kb in a Northern blot of B. bigemina (Kikuyu) mRNA (Fig. 2B). A transcript of 3.8 kb is sufficient to encode a protein of approximately 140 kDa.
FIG. 2.
Southern and Northern hybridization. (A) Southern hybridization. Conservation of the gene encoding the p200 antigen among stocks of B. bigemina from different geographical locations. Genomic DNA (between 5 and 10 μg) was digested with EcoRI, electrophoresed through a 1% agarose gel, transferred to a nylon membrane, and probed with [α-32P]dCTP-labeled Bb3.5 cDNA. The DNA samples were Kikuyu stock from Kenya (lane 1), Pongola strain from South Africa (lane 2), strain S2P from Argentina (lane 3), strain J629 from Mexico (lane 4), wild-type strain B.68Cpo6dc from Brazil (lane 5), and B. bovis strain KR from Australia (lane 6). The positions of DNA size markers are indicated on the left. (B) Northern hybridization. Two micrograms of B. bigemina (Kikuyu) mRNA was electrophoresed through a 1% agarose gel, transferred to a nylon membrane, and probed with [α-32P]dCTP-labeled Bb3.5 cDNA (lane 1). RNA size markers (lane M) are indicated to the left.
Sequence analysis of the p200 Bb3.5 cDNA.
Sequence analysis of the p200 Bb3.5 cDNA showed that it is composed of 3,464 bp (DNA data not shown) with a large open reading frame from bases 1 to 3324, sufficient to encode a protein of 133 kDa. A TAG stop codon at position 3325 and a poly(A) tail at positions 3446 to 3464 demonstrated that the 3′ end of Bb3.5 cDNA was complete. There was no ATG initiation codon near the 5′ end of the sequence (the first ATG codon occurred at bp 253), indicating that Bb3.5 is a partial-length cDNA. Because the p200 transcript is 3.8 kb (Fig. 2B), it was deduced that Bb3.5 could be lacking ∼300 bases from the 5′ end. The p200 amino acid sequence deduced from the open reading frame in Bb3.5 cDNA is shown in Fig. 3. The amino acid sequence could be divided into three distinct regions: N- and C-terminal regions flanking a large central region containing complex repeating peptide sequences.
FIG. 3.
Partial-length amino acid sequence of B. bigemina (Kikuyu) p200 as deduced from Bb3.5 cDNA. Amino acids 186 to 966 are vertically aligned to show the structure of repeat sequences. The left and middle columns (amino acids 186 to 741) are composed of R1 and R2 repeats, while the right column is composed of R3, R4, and R5 repeats. Potential sites for N-linked glycosylation are indicated by N. The boxed and circled amino acid residues indicate potential sites for protein kinase C phosphorylation and casein kinase II phosphorylation, respectively. The sequence marked by single underlining represents overlapping regions (amino acids 213 to 270 and 257 to 314) which have 100% identity to the p200 fragment encoded by clone C1A. The sequence marked by double underlining (amino acids 883 to 960) represents a region which has 100% identity to the p200 fragment encoded by clone C3A.
Deduced amino acids 1 to 185 of the N terminus do not contain peptide repeats. Of these, 19% are charged, 44% are hydrophilic, and 37% are hydrophobic. Two asparagines at amino acid positions 118 and 141 are potential sites for N-linked glycosylation. Serines at positions 59 and 120 and a threonine at position 107 are potential sites for protein kinase C phosphorylation, while the serine at position 111 is a potential site for casein kinase II phosphorylation.
The central repeat region of p200 is composed of 781 amino acids (positions 186 to 966), of which 74% are charged, 20% are hydrophobic, and 6% are hydrophilic. Fifty-one percent of the charged residues are glutamic acid, and the remainder are arginine, histidine, lysine, and aspartic acid. The central region lacks potential sites of posttranslational modification.
Vertical alignment of the amino acids using a backbone of EKAERE identified five types of related repeating sequences, designated R1 to R5 (Fig. 3). These repeats can be divided into two domains: amino acids 186 to 741, containing R1 and R2 repeats, and amino acids 742 to 966, containing R3, R4, and R5 repeats.
The R1 repeat has 8 amino acids with the consensus sequence EKAEREAK/R, while the R2 repeat has 10 amino acids with the consensus sequence EKAEREQRER. There are 37 R1 repeats and 26 R2 repeats. The arrangement of amino acids in R1 and R2 is that of alternating negatively and positively charged residues interrupted by a hydrophobic or hydrophilic residue at positions 3 and 7. The sequences of R1 and R2 are highly conserved, with only eight R1 repeats and one R2 repeat having amino acid substitutions. R1 alternates with R2 imperfectly, since there are one, two, or three R1 repeats after every R2 repeat.
The R3 repeat has the consensus sequence EEAERLAREQ, which is similar to that of R2 except for substitutions at positions 2, 6, 7, and 10. The consensus sequence of the R4 repeat, AEREAREK, is similar to that of amino acids 2 to 8 of R2 except for substitutions at positions 5 and 8. The five-amino-acid R5 repeat sequence AEREA is identical to amino acids 3 to 7 of R1. The amino acid arrangement of charged, hydrophobic, and hydrophilic residues of the R3, R4, and R5 repeats is similar to that of R1 and R2. Seven of 13 R3 repeats and 9 of 10 R4 repeats have at least one amino acid substitution. The three R5 repeats are conserved. The repeats are arranged such that an R3 repeat alternates with an R4 or R5 repeat.
The C terminus of p200 (amino acids 967 to 1108) is composed of nonrepeating amino acid sequences, of which 35% are charged, 25% are hydrophilic, and 40% are hydrophobic. The asparagine at position 1029 is a candidate for N-linked glycosylation, while the serines at positions 986, 1006, and 1020 are potential sites for casein kinase II phosphorylation.
From a search of protein databases, the predicted amino acid sequence of p200 showed similarities with several proteins that were also rich in glutamic acid. These proteins included troponin T, trichohyaline, calreticulin, and Plasmodium falciparum glutamic acid-rich protein, although the overall level of homology with these proteins was insignificant (data not shown).
Predicted secondary structure of the p200 antigen.
Computer analysis predicted that the highly charged central repeat region of p200 has a secondary structure of an uninterrupted α helix (data not shown). All of the α helix was predicted to be hydrophilic. In contrast, the N and C termini were predicted to be composed of short fragments of coil, β-sheet, and α-helix conformations, forming several regions of hydrophobicity (data not shown).
ImmunoEM localization of the p200 antigen in B. bigemina merozoites.
The p200 antigen is highly labile (data not shown) and therefore required mild fixation with 2% paraformaldehyde. These mild conditions, however, resulted in poor fixation, obscuring some detail of the parasite ultrastructure. Labeling was predominantly cytosolic, with no labeling of rhoptries or the nucleus. The spheroid body appeared to be labeled to a similar degree as the cytosol (Fig. 4).
FIG. 4.
Immunogold localization of the p200 antigen in B. bigemina merozoites by electron microscopy. The electron micrograph shows a longitudinal section through a merozoite in the erythrocyte cytoplasm. The spheroid body (S), rhoptries (R), and nucleus (N) of the merozoite are indicated. MAb Bb F4/86.11 immunogold labeling was mainly located in the merozoite cytoplasm, although some was also present in the spheroid body. Bar = 0.5 μm.
Expression of the p200 Bb3.5 cDNA.
Bb3.5 cDNA was cloned into the EcoRI site of the pGEX1λT bacterial expression plasmid. The yield of GST-p200 fusion protein (Bb3.5-GST) was poor, equivalent to only 0.5 μg/ml of bacterial culture. SDS-PAGE analysis demonstrated that the Bb3.5-GST preparation contained a diffuse band at approximately 150 to 250 kDa, a nonrecombinant GST-sized protein of 26 kDa, and several other bands of from 20 to 100 kDa (Fig. 5A). On immunoblotting with MAb Bb F4/86.6, there was a very strong signal extending over approximately 150 to 250 kDa, indicating successful expression of recombinant Bb3.5-GST, albeit at low levels (Fig. 5B). Bb3.5-GST also reacted similarly with bovine recovery sera (data not shown). Several bands of 20 to 100 kDa in the Bb3.5-GST preparation were immunologically nonreactive and may have been of bacterial origin.
FIG. 5.
SDS-PAGE and immunoblot analyses of p200 recombinant antigen. (A) One microgram of Bb3.5-GST fusion protein preparation was analyzed by SDS-PAGE and stained with Coomassie blue (lane 1). The bracket indicates a small amount of a high-molecular-weight protein. Five micrograms of GST was also electrophoresed (lane 2). (B) Immunoblot of gel arranged as in panel A and probed with MAb Bb F4/86.6. (C) Two micrograms of GST (lane 1) and 10 μg each of C1A-GST (lane 2) and C3A-GST (lane 3) were analyzed by SDS-PAGE and stained with Coomassie blue. (D) Immunoblot of gel arranged as for panel C and probed with hyperimmune serum BJ28. (E) Immunoblot of gel arranged as for panel C and probed with MAb Bb F4/86.6. The sizes of molecular mass standards (lanes M) are shown.
Identification and expression of antigenic regions of p200.
A subfragment library of Bb3.5 cDNA was created in λgt11. Ten thousand plaques were immunoscreened with hyperimmune serum BJ28, and 34 positive clones were identified; 5 of these also reacted with MAbs Bb F4/86.6, Bb F4/86.11, Bb F4/86.19, and Bb F4/86.34. Two clones were selected for further study: clone C1A, which gave the strongest signal on immunoscreening with hyperimmune serum BJ28, and clone C3A, which reacted weakly with serum BJ28 but gave the strongest signal on immunoscreening with MAbs. The inserts from C1A and C3A were subcloned into pGEX1λT, sequenced, and expressed. The protein encoded by the insert in C1A has identity with two overlapping regions in p200 at positions 213 to 270 and 257 to 314 (Fig. 3) and is therefore composed of R1 and R2 repeats. The protein encoded by the C3A insert has identity with residues 883 to 960 of the p200 antigen (Fig. 3) and is therefore composed of R3, R4, and R5 repeats. Yields of C1A-GST and C3A-GST were equivalent to 20 μg/ml of bacterial culture. On SDS-PAGE, C1A-GST and C3A-GST appeared as stable fusion proteins of 33 and 39 kDa, respectively (Fig. 5C). This result demonstrates that the p200 portion of C1A-GST is 7 kDa, while for C3A-GST, it is 11 kDa. On immunoblotting, C1A-GST reacted strongly with hyperimmune serum BJ28 but showed no reactivity with MAb Bb F4/86.6 (Fig. 5D and E). In contrast, C3A-GST reacted strongly with MAb Bb F4/86.6 and weakly with hyperimmune serum BJ28 (Fig. 5D and E). These results demonstrate that the major epitopes for these bovine and murine antibodies are located within different regions of p200.
Antibody epitope mapping with synthetic peptides.
Eleven overlapping peptides representing the C1A fragment of p200 all reacted strongly with hyperimmune serum BJ28 (data not shown). C1A peptides showed no reactivity with a control bovine serum or with MAb Bb F4/86.6 (data not shown). A control peptide with an irrelevant amino acid sequence showed no reactivity with BJ28 serum, control bovine serum, or MAb Bb F4/86.6 (data not shown).
Antibody responses in an experimentally infected animal.
Steer BJ26 infected with a B. bigemina sporozoite stabilate developed a significant antibody response to p200 by day 14, as determined by an ELISA with C1A-GST as the antigen. The antibody response was maintained at high levels until day 179, after which the response fell to approximately 60% its maximal level by day 228 (Fig. 6). These results indicate that soon after infection an animal can develop a detectable antibody response which is then maintained over a long period.
FIG. 6.
Antibody responses to C1A-GST in sequential sera from a steer experimentally infected with B. bigemina. Boran steer BJ26 was infected with B. bigemina sporozoites, and sequential sera were collected over a period of 228 days. Sera were diluted to 1:100 and tested by an ELISA with C1A-GST as the antigen. ELISA OD values were expressed as PP.
DISCUSSION
Screening of a B. bigemina λgt11 cDNA expression library with MAbs and polyclonal antibodies to the p200 antigen identified an immunoreactive clone that contained a 3.5-kbp insert (Bb3.5). Hybridization to DNA of B. bigemina that originated from Kenya, South Africa, Australia, Mexico, Argentina, and Brazil demonstrated that the gene encoding the p200 antigen is geographically conserved. It was shown by Northern hybridization that the mRNA encoding p200 is 3.8 kb, indicating that the Bb3.5 cDNA was missing approximately 300 bases from the 5′ end. A transcript of 3.8 kb would encode a protein with a predicted mass of approximately 140,000, in contrast to the mass of 150 to 250 kDa on immunoblots of merozoite p200. The discrepancy in molecular mass is most likely due to the presence of highly charged repeating amino acid residues, which may have disrupted the binding of SDS to the protein (10). Similar migration patterns have been observed for a highly charged, glutamic acid-rich protein encoded by the Pf332 gene of P. falciparum and for which the predicted molecular mass of 700,000 is in marked contrast to the actual mass of 2,500 kDa (16).
A striking feature of the p200 protein is the presence of a central, highly charged, glutamic acid-rich region composed of five types of related repeat sequences, designated R1 to R5. Unlike repetitive proteins of Plasmodium and Trypanosoma, where repeats are arranged tandemly (6, 15), the repeats within p200 alternate imperfectly. The N-terminal end of the central repeat region is composed of highly conserved R1 and R2 repeats, suggesting that this region may be critical for the function of the protein. In contrast, the C terminus of the central repeat region contains divergent R3 and R4 repeats.
Secondary structure analysis of p200 predicted a long α-helical central domain flanked by short N- and C-terminal regions each composed of alternating short segments of coil, β-sheet, and α-helix conformations. The high propensity for an α-helical conformation in the repeats is due to the high proportion of helix-stabilizing amino acid residues, such as glutamic acid, alanine, lysine, and glutamine (29). The α-helix of the central region of p200 is predicted to form an elongated, uninterrupted, inflexible rod.
Unlike the central repeat region, the N- and C-terminal regions contained consensus sites for posttranslational modification. Three sites for N-linked glycosylation, with typical NXS/T consensus sequences, were identified. Also present were seven sites for casein kinase II and protein kinase C phosphorylation. Restriction of posttranslational modification sites to the C and N termini is typical of proteins containing central regions of repeated amino acids. A good example is the intermediate filaments (IF) of the cytoskeleton, which contain consensus sites for phosphorylation at the nonrepeating “head” and “tail” domains of the molecule (23). For IF, phosphorylation sites close to the repeat region appear to affect filament assembly, while sites further away appear to be important for interactions with other cellular components. It has been shown that phosphorylation plays an important role in the regulation of IF dispersal throughout the nucleus and the cytoplasm, especially during cell division (5). It is tempting to speculate that phosphorylation of the p200 antigen may be necessary for the molecule to perform its cellular functions. Immunogold localization of the p200 antigen to the merozoite cytoplasm supports the idea that this protein may function as a cytoskeletal structural protein.
The p200 Bb3.5 cDNA was expressed as a fusion with GST. On immunoblotting, Bb3.5-GST appeared as a diffuse band that extended from approximately 150 to 250 kDa, similar to native p200. Bb3.5-GST reacted strongly with MAb Bb F4/86.6 and with bovine recovery serum, indicating that it contained immunodominant epitopes. A major drawback in the use of Bb3.5-GST as a potential diagnostic antigen was the very low yield of fusion protein. It has been observed that fusions of GST with proteins that are larger than 100 kDa are insoluble (24). It is possible that the low yields of Bb3.5-GST were due, at least in part, to poor solubility resulting from the large size of the p200 fragment being expressed. To improve yields of recombinant p200, we expressed small fragments of the antigen that contained the critical diagnostic epitopes. We produced two smaller constructs, C1A-GST and C3A-GST, containing 7- and 11-kDa fragments of p200, respectively. C1A-GST reacted strongly with bovine sera, whereas C3A-GST reacted strongly with MAbs to p200. Since C1A was derived from the region containing R1 and R2 repeats and C3A was derived from the region containing R3, R4, and R5 repeats, it appears that the epitopes for the bovine and murine antibodies used in this study are located within different regions of p200. ELISA analysis of synthetic peptides representing the C1A protein showed that bovine epitopes are distributed throughout this fragment. C1A peptides did not react with MAb Bb F4/86.6, confirming that bovine and murine epitopes in p200 are distinct. Since the R1 and R2 repeats are well conserved in p200, it is assumed that the entire R1 and R2 repeat region (amino acids 186 to 741 of the p200 antigen) is immunodominant in cattle.
We have shown that an ELISA with C1A-GST as an antigen can detect a significant antibody response in an animal soon after experimental infection with B. bigemina (day 14). Also, this antibody response was still detectable on day 228 postinfection, indicating that this assay can detect antibodies during both early and late stages of infection. Furthermore, it has been demonstrated that ELISAs with C1A-GST or native p200 are equally efficient at detecting antibodies in B. bigemina-infected cattle (26), indicating that C1A contains major p200 epitopes recognized by bovine infection sera. Detailed descriptions of the field and laboratory validations of an ELISA with C1A-GST for the detection of antibodies to B. bigemina in cattle will be published elsewhere.
ACKNOWLEDGMENTS
We sincerely thank W. Mwangi and P. Pandit for assistance with sequencing, C. Nkonge and L. Gichuru for assistance with the protein analysis, and D. Lugo, T. Njoroge, A. Tonui, and S. Njuguna for monitoring infected animals. For artwork we thank D. Elsworth, F. Shikhubari, and J. Mwaura.
Footnotes
ILRI publication 99/0164.
REFERENCES
- 1.Berryman M A, Rodewald R D. An enhanced method for post-embedding immunocytochemical staining which preserves cell membranes. J Histochem Cytochem. 1990;38:159–170. doi: 10.1177/38.2.1688894. [DOI] [PubMed] [Google Scholar]
- 2.Brown C G D, Hunter A G, Luckins A G. Diseases caused by protozoa. In: Sewell M M H, Brocklesby D W, editors. Handbook on animal diseases in the tropics. 4th ed. London, England: Baillière Tindall; 1990. pp. 161–226. [Google Scholar]
- 3.Burleigh B A, Wells C W, Clarke M W, Gardiner P R. An integral membrane glycoprotein associated with an endocytic compartment of Trypanosoma vivax: identification and partial characterization. J Cell Biol. 1993;120:339–352. doi: 10.1083/jcb.120.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calder J A, Reddy G R, Chieves L, Courtney C H, Littell R, Livengood J R, Norval R A, Smith C, Dame J B. Monitoring Babesia bovis infections in cattle by using PCR-based tests. J Clin Microbiol. 1996;34:2748–2755. doi: 10.1128/jcm.34.11.2748-2755.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou Y H, Risevear E, Goldman R D. Phosphorylation and disassembly of intermediate filaments in mitotic cells. Proc Natl Acad Sci USA. 1989;86:1885–1889. doi: 10.1073/pnas.86.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotrim P C, Paranhos-Baccala G, Santos M R, Mortensen C, Cano M I, Jolivet M, Camargo M E, Mortara R A, Da Silveira J F. Organization and expression of the gene encoding an immunodominant repetitive antigen associated to the cytoskeleton of Trypanosoma cruzi. Mol Biochem Parasitol. 1995;71:89–98. doi: 10.1016/0166-6851(95)00036-z. [DOI] [PubMed] [Google Scholar]
- 7.de Vos A J, Jorgensen W K. Manual of recommended diagnostic techniques and requirements for biological products. Paris, France: Office International des Epizootics; 1991. Bovine babesiosis; pp. 1/14–14/14. [Google Scholar]
- 8.de Vos A J, Potgieter F T, De Waal D T, van Heerden J. Babesioses. In: Coetzer J A W, Thompson G R, Tustin R C, editors. Infectious diseases of livestock with special reference to southern Africa. Oxford, United Kingdom: Oxford University Press; 1994. pp. 278–308. [Google Scholar]
- 9.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 10.Hames B D. One-dimensional polyacrylamide gel electrophoresis. In: Hames B D, Rickwood D, editors. Gel electrophoresis of proteins. A practical approach. Oxford, United Kingdom: Oxford University Press; 1990. pp. 1–147. [Google Scholar]
- 11.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 12.Janin J. Surface and inside volumes in globular proteins. Nature. 1979;277:491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- 13.Jorgensen W, de Vos A J, Dalgleish J R. Methods for diagnosis of babesiosis in developing countries. Present and future trends. In: Uilenberg G, Permin A, Hansen J W, editors. Use of applicable biotechnological methods for diagnosing haemoparasites. Rome, Italy: FAO; 1994. pp. 65–84. [Google Scholar]
- 14.Katende J, Morzaria S, Toye P, Skilton R, Nene V, Nkonge C, Musoke A. An enzyme-linked immunosorbent assay for detection of Theileria parva antibodies in cattle using a recombinant polymorphic immunodominant molecule. Parasitol Res. 1998;84:408–416. doi: 10.1007/s004360050419. [DOI] [PubMed] [Google Scholar]
- 15.Kemp D J, Coppel R L, Anders R F. Repetitive proteins and genes of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- 16.Mattei D, Scherf A. The Pf322 gene of Plasmodium falciparum codes for a giant protein that is translocated from the parasite to the membrane of infected erythrocytes. Gene. 1992;110:71–79. doi: 10.1016/0378-1119(92)90446-v. [DOI] [PubMed] [Google Scholar]
- 17.Montenegro S, James M A, Levy M G, Preston M D, Esparza H, Ristic M. Utilization of culture derived soluble antigen in the latex agglutination test for bovine babesiosis and anaplasmosis. Vet Parasitol. 1981;8:291–297. [Google Scholar]
- 18.Morzaria S, Katende J, Kairo A, Nene V, Musoke A. New methods for the diagnosis of Babesia bigemina infection. Mem Inst Oswaldo Cruz. 1992;87:201–205. doi: 10.1590/s0074-02761992000700033. [DOI] [PubMed] [Google Scholar]
- 19.Pearson T W, Pinder M, Roelants G E, Kar S K, Lundin L B, Mayor-Withey K S, Hewett R S. Methods for derivation and detection of anti-parasite monoclonal antibodies. J Immunol Methods. 1980;34:141–154. doi: 10.1016/0022-1759(80)90168-4. [DOI] [PubMed] [Google Scholar]
- 20.Pellé R, Murphy N B. Northern hybridization: rapid and simple electrophoretic conditions. Nucleic Acids Res. 1993;21:2783–2784. doi: 10.1093/nar/21.11.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salem G H, Liu X, Johnsrude J D, Dame J B, Roman Reddy G. Development and evaluation of an extra chromosomal DNA-based PCR test for diagnosing bovine babesiosis. Mol Cell Probes. 1999;13:107–113. doi: 10.1006/mcpr.1998.0223. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Shaw G. Identification of previously unrecognised sequence motifs at the extreme carboxyterminus of the neurofilament. Biochem Biophys Res Commun. 1989;162:294–299. doi: 10.1016/0006-291x(89)91995-5. [DOI] [PubMed] [Google Scholar]
- 24.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione-S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 25.Taylor D W, Cordingley J S, Butterworth A E. Immunoprecipitation of surface antigen precursors from Schistosoma mansoni messenger RNA in vitro translation products. Mol Biochem Parasitol. 1984;10:305–318. doi: 10.1016/0166-6851(84)90029-x. [DOI] [PubMed] [Google Scholar]
- 26.Tebele N. Ph.D. thesis. Uxbridge, United Kingdom: Brunel University; 1996. [Google Scholar]
- 27.Weiland G, Reiter I. Methods for the measurement of the serological response to Babesia. In: Ristic M, editor. Babesiosis of domestic animals and man. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 143–162. [Google Scholar]
- 28.Wright I G. Immunodiagnosis of and immunoprophylaxis against the haemoparasites Babesia sp. and Anaplasma sp. in domestic animals. Rev Sci Tech. 1990;9:345–356. doi: 10.20506/rst.9.2.508. [DOI] [PubMed] [Google Scholar]
- 29.Zubay G. Biochemistry. New York, N.Y: MacMillan Publishing Company; 1988. Methods for characterization and purification of proteins; pp. 100–130. [Google Scholar]