Three interesting ideas about the initiation of translation in eukaryotes have recently emerged in the literature. One is the possibility that internal initiation of translation might occur not only with viral but also with some cellular mRNAs. The second is the possibility of initiating translation without Met-tRNA. The third concerns circumstances under which base pairing might occur between mRNA and rRNA. These ideas, if upheld, would significantly expand our understanding of how eukaryotic ribosomes function. As detailed below, however, there are serious deficiencies in the supporting evidence. Understanding how the present studies fall short might facilitate the design of experiments that are more convincing.
Each of these ideas has gut appeal because certain mRNA sequences appear incompatible with the standard mechanism of initiation. In some cases, “incompatible” means only that the mRNA—e.g., an mRNA with a highly structured, GC-rich leader sequence—would be translated inefficiently by the normal scanning mechanism. As discussed elsewhere (40), however, inefficient translation might be necessary for mRNAs that encode growth regulators, transcription factors, and other potent proteins.
The presumed structures of certain other mRNAs really are incompatible with the normal initiation mechanism. While the scanning mechanism (41) can deal with 5′ untranslated regions (UTRs) that have a few upstream AUG codons, some cDNA sequences predict mRNAs with a dozen or more AUGs before the start of the coding domain (26, 37, 51, 54, 57). One possibility, discussed in the next section, is that ribosomes enter directly at an internal point in these mRNAs. An alternative possibility is that these encumbered cDNA sequences do not reflect the actual structures of mRNAs. As documented in many other cases (42), the problematic cDNAs might derive from incompletely spliced transcripts, in which case the upstream AUG codons could reside in an intron that gets removed from the functional mRNA. Thus, there are alternatives to the ideas explored here.
HOW MANY (IF ANY) CELLULAR MRNAS CONTAIN IRES ELEMENTS?
Internal ribosome entry site (IRES) is the name given to a sequence that allows ribosomes to enter directly at an AUG codon rather than scanning from the capped 5′ end of the mRNA. Putative IRES elements almost always reside at the 5′ end of monocistronic transcripts (33). In theory, however, an IRES should be functional when repositioned to the midpoint—the intercistronic gap—in a dicistronic mRNA. Based on tests with artificially constructed dicistronic transcripts, 26 sequences derived from 25 mammalian mRNAs have been tentatively identified as IRES elements (Table 1; entry numbers cited below refer to this table).
TABLE 1.
Sequences from mammalian mRNAs postulated to function as IRES elements
| Entry no. | mRNAa | Dicistronic test in vivob
|
In vitro translationd of dicistronic or monocistronic mRNA | Comments | |
|---|---|---|---|---|---|
| Translation of 3′ cistronc (increase relative to the stated negative control) | Published analysis of RNA structure | ||||
| 1 | AML1 (57) | 3- to 6-fold (neg ctrl, empty vector; pos ctrl, EMCV [≤20-fold]) | Northern blot (Fig. 6, faint [57]; Fig. 7, extra bands [57]) | Di, 5% as efficient as EMCV control (Fig. 7 [57]); Mono, strongly inhibited (Fig. 3 [57]) | Low efficiency; inadequate RNA analysis |
| 2 | Apaf-1 (7) | 10-fold (neg ctrl, empty vector; pos ctrl, rhinovirus [8-fold]) | Extraneous bands obscure monocistronic position | Mono, untranslatable | Inadequate RNA analysis |
| 3 | BiP (46) | ∼15-fold (neg ctrl, 400-nt Antp insert; pos ctrl, poliovirus); 2.6-fold (with empty vector as neg ctrl [38]) | Polysomal mRNA screened with probe complementary to 5′ but not 3′ cistron | Di, untranslatable (68); poorly translated compared with EMCV (38) | Transfection with dicistronic RNA fails in BHK cells (2); inappropriate negative control (see text) |
| 4 | Cx43 (65) | 46-fold (neg ctrl, empty vector; pos ctrl, EMCV [2.5-fold]) | None | Di, not tested; Mono, inhibited | No RNA data |
| 5 | Cx32 (31) | 2.5- to 5-fold (neg ctrl, empty vector; pos ctrl, EMCV [3- to 4-fold]) | None for dicistronic constructs | Not tested | No RNA data |
| 6 | Cyr61 (35) | 20-fold (neg ctrl, ΔEMCV [440 nt]; pos ctrl, none) | Northern blot | Not tested | Inappropriate vector (ΔEMCV insert might bind factors without which Cyr61 would not score as IRES) |
| 7 | DAP5 (23) | ∼10-fold (neg ctrl, empty vector; pos ctrl, BiP IRES [4-fold]) | Northern blot very faint | Di, barely translatable | Inadequate RNA analysis |
| 8 | FGF-2 (10) | ∼5- to 35-fold (cell type dependent; neg ctrl, hairpin at midpoint; pos ctrl, EMCV) | Amt but not form monitored (10); Northern blot very faint (70) | Di, not tested | Tissue-specific expression is uninterpretable without RNA analysis |
| 9 | eIF4G (16) | 42-fold (neg ctrl, empty vector) | None | Not tested | “IRES” is really an intron |
| 10 | eIF4G (34) | ∼5-fold (neg ctrl, 400-nt Antp insert; pos ctrl, poliovirus [5-fold]) | Northern blot cropped (useless) | Not tested | Low efficiency; inappropriate negative contr (see text) |
| 11 | Gtx (5) | ∼7-fold (increases to 570-fold when 9-base motif is amplified; neg ctrl, empty vector; pos ctrl, poliovirus [32-fold]) | Northern blot shown for construct that stimulates 7-fold but not for high-efficiency constructs | Not tested | Inadequate RNA analysis |
| 12 | IGFII (69) | Barely detectable (not quantified; neg ctrl, empty vector; pos ctrl, EMCV) | None | Di, untranslatable (68) | Low efficiency; no RNA data |
| 13 | IGF-IR (18) | 18-fold (neg ctrl, empty vector; pos ctrl, EMCV [8-fold]) | None | Not tested | No RNA data |
| 14 | KCNA4 (51) | >100-fold using complete 5′ UTR (1,200 nt) or last 200 nt (neg ctrl, empty vector) | None | Die, 50-fold stimulation in sense orientation (but antisense stimulates 20-fold) | No RNA data; AG downstream from Yn might be 3′ splice site |
| 15 | La1′ (4) | 10-fold (neg ctrl, ΔEMCV [440 nt]) | Northern blot | Di, 6 fold stimulation in RRL; 46-fold in HeLa cell extract | Inappropriate vector (see Cyr61) |
| 16 | c-myc (67) | ∼70-fold (neg ctrl, empty vector; pos ctrl, EMCV [14-fold]) | RNase protection assay detects only dicistronic (66) | Di, not tested; Mono, untranslatable | Transfection with dicistronic RNA fails (67) |
| 17 | MYT2 (37) | 5-fold (neg ctrl, empty vector; pos ctrl, BiP IRES [4-fold]) | None | Di, not tested; Mono, untranslatable | Low efficiency; no RNA data; possible splice site YnAG942 |
| 18 | Nkx6.1 (72) | 20-fold (neg ctrl, empty vector) | None | Not tested | No RNA data |
| 19 | Notch2 (45) | ∼10-fold (neg ctrl, FeLV leader; pos ctrl, poliovirus [3-fold]) | Northern blot | Not tested | Requires better RNA analysis; translation via IRES does not fit with the biologyf |
| 20 | NRF (54) | 30- to 130-fold (ctrl, poliovirus) | Northern blot | Not tested | Requires better RNA analysis; possible splice site YnAG498 |
| 21 | ODC (58) | 2.5-fold (neg ctrl, empty vector) | Northern blot | Di, barely translatable; Mono, strongly inhibited (48) | Low efficiency |
| 22 | PITSLRE (8) | 25-fold (neg ctrl, deletion mutant; pos ctrl, monocistronic) | Northern blot (Fig. 4E, faint [8]; Fig. 4F, strong [8]) | Di, untranslatable | Strong candidate (see text) |
| 23 | Pim-1 (35) | 50-fold (neg ctrl, ΔEMCV [440 nt]; pos ctrl, none) | Northern blot | Mono, inhibited (29) | Inappropriate vector (see Cyr61) |
| 24 | c-sis (3) | 2.5-fold (in differentiated cells; neg ctrl, empty vector; pos ctrl, EMCV [3-fold]) | Northern blot | Di, untranslatable as control (57); Mono, inhibited (60) | Low efficiency |
| 25 | VEGF (1) | ∼20-fold (reduced to 4-fold when internal promoter is deleted; neg ctrl, empty vector; pos ctrl, EMCV [60-fold]) | Vector produces dicistronic and monocistronic mRNAs | Not tested | “IRES” functions as strong transcriptional promoter |
| 26 | XIAP (26, 27) | 150-fold (neg ctrl, empty vector) | None | Die, detectable translation of 3′ cistron but no control to assess efficiency (27) | No RNA data; cryptic promoter ruled out (26), but splicing is possible |
The number in parentheses indicates the primary reference for each entry. All candidate IRES elements derive from the 5′ UTR of the stated mRNA except for PITSLRE and Notch2, for which the putative IRES resides in the coding domain. Two different sequences from translation initiation factor eIF4G are postulated to have IRES activity (entries 9 and 10). Genes from which the candidate IRES elements derive are abbreviated as follows: AML1, runt domain transcription factor activated in acute myeloid leukemia; Apaf-1, apoptotic protease activating factor; BiP, immunoglobulin heavy-chain binding protein; Cx43 and Cx32, connexin-43 and -32 gap junction proteins; Cyr61, extracellular matrix-associated signaling protein; DAP5, death-associated protein 5; FGF-2, fibroblast growth factor 2; Gtx, homeodomain protein; IGFII, insulin-like growth factor II; IGF-IR, insulin-like growth factor I receptor; KCNA4, cardiac voltage-gated potassium channel Kv1.4; MYT2, myelin transcription factor 2; Nkx6.1, β-cell transcription factor; NRF, NF-κB repressing factor; ODC, ornithine decarboxylase; VEGF, vascular endothelial growth factor; XIAP, X-linked inhibitor of apoptosis.
In vivo refers to experiments carried out by transient transfection of cultured cells with plasmid DNA, except for FGF-2 (entry 8), which was tested in mice. Source references are indicated in parentheses and/or brackets where applicable.
Positive control (pos ctrl) and negative control (neg ctrl) constructs have the stated sequence inserted between the 5′ and 3′ cistrons. The empty vector is a control construct with no expansion of the intercistronic sequence.
The mRNAs used for in vitro translation have the putative IRES sequence at the midpoint of a dicistronic (Di) transcript or at the 5′ end of a monocistronic (Mono) transcript. For dicistronic transcripts, the stated efficiency pertains to translation of the 3′ cistron. For monocistronic transcripts, “inhibited” means there is a strong reduction in translation when a simple 5′ UTR is replaced by the IRES. Source references are indicated in parentheses or brackets where applicable.
It is unlikely that the input dicistronic mRNA would have remained intact during incubation in reticulocyte lysates for 1.5 or 2 h (entries 14 and 26). Even the “standard” 1-h incubation at 30°C is much too long when the initiation step of translation is the focus of the study. A better protocol for in vitro experiments might be to add edeine after the first 5 min at 25°C, which is adequate time and temperature for initiation, followed by incubation for another 20 min to allow polypeptide elongation.
The short form of the protein postulated to be translated via an IRES in the coding domain of Notch2 (45) was shown in the case of Notch1 to be produced via proteolysis (49). Whereas the proteolytic mechanism shows the expected dependence on ligand binding, there is no evidence that the putative IRES in Notch2 is regulated. Studies of a recombinant retrovirus that carries the pertinent region of Notch2 did not strongly support the IRES hypothesis. The majority (70%) of Notch2 expression from the provirus apparently resulted from reinitiation of translation (45). The residual expression, which was taken as evidence for IRES activity, might have resulted instead from leaky scanning. Mutations that should have excluded leaky scanning were tested separately and shown to have a small effect, but those mutations were not combined with the mutations that exclude reinitiation.
The dicistronic test has been proclaimed the “gold standard for detecting IRES activity” (62), but whether it is “gold” or “tin” depends on how the test is carried out. For reasons explained below—reasons having to do with the low efficiency of translation in many cases and incomplete data in other cases—only one of the 26 entries in Table 1 emerges as a strong candidate IRES. If deficiencies in the data pertaining to other entries are not recognized, the list will continue to grow, but it will never prove anything. Only when genuine IRES elements are distinguished from sequences that function in other ways will it be possible to recognize common features that might explain how internal initiation is accomplished.
Relative to what?
Dicistronic vectors are often constructed with chloramphenicol acetyltransferase (CAT) or luciferase (LUC or LucF) as the 3′ cistron. Because the assays for these enzymes are so sensitive, low-level expression of the 3′ cistron is detectable even with the empty vector control (a construct that carries a short, nonspecific sequence in the intercistronic gap). In reticulocyte lysates and similar in vitro systems, this background translation probably results from cleavage of the dicistronic mRNA in a way that brings the 3′ CAT or LUC sequence close enough to the 5′ end to be translated via the conventional scanning mechanism. The background expression seen when dicistronic control vectors are tested in vivo might result from mRNA cleavage or from a more physiological mechanism, such as a cryptic promoter or splice site that generates a small amount of monocistronic mRNA. In some cases, low-level production of monocistronic mRNA from an empty dicistronic vector has actually been detected (14).
A serious problem is that this background level of translation from control vectors varies. In one in vitro study, expression of the 3′ cistron was increased severalfold simply by inserting a synthetic sequence to lengthen the intercistronic gap (15). In another study, insertion of the antisense version of a putative IRES increased translation of the 3′ cistron almost as much (40%) as the correctly oriented sequence (51). Should the negative control in that case be the original empty vector or the one with the antisense insert? Similar complications occur in vivo, as when insertion of certain test sequences caused translation of the 3′ cistron to fall substantially below (to <10%) the level obtained with the empty vector (see Fig. 7 in reference 57).
To create the negative control for studies with BiP and eIF4G (entries 3 and 10), a 400-bp inverted segment of the Drosophila Antp gene was inserted between the 5′ CAT and 3′ LUC cistrons. Rather than being neutral, the Antp insert might have depressed background expression of the 3′ cistron, in which case the test sequences might have scored as IRES elements simply because they replaced an inhibitory sequence. This could explain why the IRES activity of the BiP sequence was considerably lower than originally reported when it was retested against an empty vector (38; see also the positive control for entries 7 and 17 in Table 1). The dicistronic vector used as a starting point for testing IRES activity in Cyr61, La1′, and Pim-1 mRNAs (entries 6, 15, and 23) is problematic for a different reason. A 440-nucleotide (nt) structured sequence from encephalomyocarditis virus (EMCV) inserted at the midpoint of this vector— purportedly to block readthrough from the first cistron—might make it too easy for sequences appended downstream to score as IRES elements. Although a small deletion in the EMCV insert prevents it from functioning independently as an IRES, it might still bind protein factors without which the appended test sequence would not score. The EMCV insert could also function in a less physiological way by attracting RNases (13). The use of such an unnecessarily complicated vector invites misinterpretation.
If IRES activity were routinely judged by comparison to a strong positive control, such as a monocistronic transcript that carries the globin 5′ UTR, the validity of the negative control would be less of an issue. Of the 26 studies described in Table 1, however, only one (entry 22) tested an appropriate monocistronic construct alongside the dicistronic vector. Many studies used a dicistronic construct containing the EMCV IRES as a positive control, but the EMCV sequence, which supports efficient translation under some conditions, is weak under other conditions (5, 57, 65). Comparison with EMCV might be used in addition to, but should not be used instead of, a monocistronic control. Use of other “proven” IRES elements as positive controls sets the bar much too low, as when BiP (which stimulated translation fourfold above background) was used to identify MYT2 (fivefold stimulation; entry 17) as an IRES.
The variability in background expression from dicistronic vectors constitutes a warning against interpreting every small change as evidence of IRES activity. An IRES has to support translation well enough to be physiologically relevant. Because insertion of sequences derived from AML1, BiP, Cx32, eIF4G, IGFII, MYT2, ODC, and c-sis (entries 1, 3, 5, 10, 12, 17, 21, and 24) only slightly improved expression of the 3′ cistron, it is doubtful that those sequences qualify as candidate IRES elements.
RNA analyses shape the story.
Some sequences in Table 1 scored strongly when tested for ability to support expression of the 3′ cistron from a dicistronic vector, but the underlying mechanism turned out to be something other than internal initiation of translation. In the case of VEGF (entry 25), for example, RNA analyses revealed a transcriptional promoter within the putative IRES, meaning that translation of the 3′ cistron actually occurred from an unanticipated monocistronic mRNA (1). Because a marginal level of activity persisted after the promoter element had been deleted, the authors continue to call the VEGF sequence an IRES; but the residual activity is too close to background to be convincing.
In the case of eIF4G (entry 9), the putative IRES turned out to be part of an intron (19). The portion of the eIF4G sequence required for “internal initiation” was mapped to the 3′ splice junction (17), and the most reasonable interpretation is that translation occurs from a monocistronic mRNA generated by splicing rather than from the intended dicistronic transcript. The presence of a possible splice-junction motif (Yn closely followed by AG) near the 3′ end of the putative IRES elements from KCNA4, MYT2, NRF, and XIAP mRNAs (entries 14, 17, 20, and 26) raises the possibility of splicing in those cases as well.
Even without the foregoing examples of dicistronic vectors that turned out to function by mechanisms other than internal initiation of translation, the need to determine whether a vector produces only the intended dicistronic mRNA seems obvious. Reports that include no RNA analyses do not merit further consideration (entries 4, 5, 9, 12, 13, 14, 17, 18 and 26). Reports in which Northern blot analyses are very faint (entries 7 and 8), closely cropped (entry 10), or contaminated with extraneous bands (entries 1 and 2) attempt to address the issue without settling anything.
The sequence derived from c-myc mRNA (entry 16) appears, at first glance, to be one of the strongest candidates for a cellular IRES. Insertion of the c-myc sequence into a dicistronic DNA vector stimulated expression of the 3′ LucF cistron by 70-fold in vivo, and RNA analyses failed to uncover alternative transcripts (66). However, the observed inability to translate the 3′ cistron when cells were transfected directly with dicistronic RNA (67), rather than with the DNA vector, is strong evidence that the dicistronic mRNA is not the functional template for translation. Because the in vivo level of expression from the dicistronic DNA vector varied considerably among cell lines, Stonely et al. (67) postulated that cell-specific protein factors might be required for the c-myc IRES to function. But an alternative possibility—consistent with the aforementioned failure of RNA transfection experiments—is that a cell-specific promoter or pattern of splicing generates a monocistronic transcript in cell lines that allow LucF translation. In patients with multiple myeloma, the detection of a point mutation in the 5′ UTR of c-myc (6) is of considerable interest, but the slight (1.5- to 4-fold) augmentation of downstream translation when the mutated sequence was tested in a dicistronic vector is not compelling evidence that the mutation increases IRES activity.
Control experiments are sometimes designed to rule out leaky scanning, which is an unlikely explanation when so many AUG codons intervene before the start of the second cistron, and to rule out reinitiation, which can occur in eukaryotes following the translation of a short upstream open reading frame (ORF) but not after translation of a full-length 5′ cistron (41). These experiments thus test unlikely mechanisms, while the most likely alternative explanation—that the 3′ cistron is actually translated from a monocistronic mRNA produced via splicing or an internal promoter—is rarely tested in a convincing way. The fact that translation of the second cistron persists when a hairpin structure is introduced before the first cistron is often cited as proof that the second cistron is translated independently of the first (3, 5, 7, 8, 18, 23, 45, 46, 50, 65). Since the hairpin barrier could be circumvented by splicing or transcription from an internal promoter, however, the hairpin test—unless accompanied by careful RNA analyses—is not proof of IRES activity.
How careful is careful? If the 3′ cistron is translated with only 5% efficiency compared to a monocistronic control (the control that almost everyone omits), RNA analyses must be able to detect—to rule out—an adventitious monocistronic transcript produced at 1/20 the level of the dicistronic form. Northern blot assays usually cannot meet that standard. A carefully quantified study carried out with a synthetic IRES (12) underscores the point. When the synthetic IRES was inserted into a dicistronic vector, translation of the 3′ cistron was stimulated 10-fold relative to the empty-vector control, and this downstream translation was calculated to be 5% as efficient as cap-mediated translation from the 5′ end. For many of the candidate IRES elements in Table 1, the observed stimulation was ≤10-fold above background, and therefore meaningful RNA analyses must be sensitive enough to rule out a monocistronic mRNA present at ≤5% the level of the dicistronic mRNA.
Regulated IRES function?
A recent report (10) describes transgenic mice produced by injecting eggs with a dicistronic construct in which the putative IRES from FGF-2 mRNA precedes the 3′ LucF cistron. This is entry 8 in Table 1. The observation that high-level expression of LucF was restricted to the adult brain is interesting, but because the study was published without analyzing RNA structure in tissues that allow or disallow translation of LucF, it is premature to call the FGF-2 sequence a tissue-specific IRES. An alternative explanation is that a tissue-specific promoter located in the FGF-2 insert might generate an efficiently translated monocistronic mRNA only in the brain. Another recent report claims that radiation resistance in certain cell lines results from gamma-radiation-induced activation of the XIAP IRES (28) but, again, the results were published without RNA analyses. Before claiming regulation of IRES activity, one first has to show that the sequence is an IRES, and that cannot be done without analyzing RNA.
The strongest candidate IRES in Table 1 (entry 22) is unusual in that it derives from the interior rather than the 5′ end of the mRNA. A 219-nt sequence from the middle of the coding domain for p110PITSLRE directs initiation of a truncated form of the protein kinase (p58PITSLRE) which is upregulated during the G2/M phase of the cell cycle. For testing, the 219-nt putative IRES was inserted into a dicistronic vector and translation of the 3′ cistron was compared against a monocistronic control. Careful RNA analyses (Fig. 4F in reference 8) appear to rule out a shorter form of mRNA, although this would have been more convincing had an adequate exposure been shown also for the Northern blot in Fig. 4E, where active and inert constructs were compared side by side. The inability to translate the synthetic dicistronic mRNA in vitro makes it important to confirm the activity of the PITSLRE IRES by liposome-mediated RNA transfection, which has not yet been done. Because the p110PITSLRE gene generates multiple protein isoforms via alternative splicing (75) (a splice junction sequence is present 20 nt upstream from the start codon for p58PITSLRE), it is not clear why an alternative mode of expression, via the internal entry of ribosomes, is also employed. Nevertheless, the data from this study appear convincing.
That is not the case for ODC, another putative cell-cycle-dependent IRES (entry 21). In contrast to the >25-fold stimulation produced by the PITSLRE sequence, insertion of the ODC sequence into a dicistronic vector stimulated translation only 2.5-fold above that of the empty-vector control. Even if it is true that cell cycle regulation of the endogenous ODC gene occurs at the level of translation rather than by posttranslational proteolysis (the 20-min duration used for pulse-labeling was really too long to rule out proteolysis; see reference 71), there is no justification for calling this cell cycle regulation of internal initiation. The outcome of the dicistronic mRNA test is simply too weak to be counted as evidence for internal initiation.
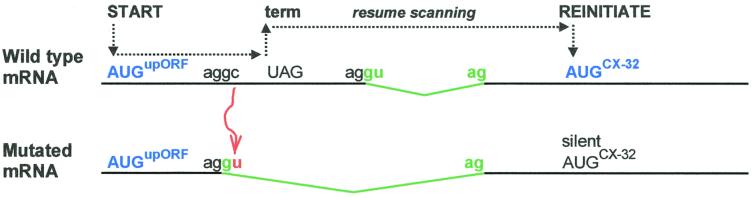
A naturally occurring C-to-U mutation in the 5′ UTR of connexin-32 mRNA was postulated to impair IRES function (31), but again the weak results of the dicistronic test—and absence of RNA analyses—do not justify classifying this sequence as an IRES. The reason why the C-to-U mutation impairs translation of the natural (monocistronic) form of connexin 32 mRNA might be because the mutation creates an upstream splice site. As diagrammed in Fig. 1, the old-fashioned scanning mechanism could explain how the resulting change in structure of the 5′ UTR prevents translation of connexin 32 in patients with Charcot-Marie-Tooth neuropathy (32).
FIG. 1.
Proposed mechanism to explain silencing of connexin 32 translation by a naturally occurring C-to-U mutation in the 5′ UTR. An AUG codon near the 5′ end of the wild-type mRNA creates a small upstream ORF (upORF) which terminates before the connexin 32 start site (AUGCX-32). This makes it likely that connexin 32 is translated normally by a reinitiation mechanism, as indicated in the top line. The C-to-U mutation (shown in red) creates a potential splice donor motif (AG/GU) within the upORF. Splicing from the new upstream site would enlarge the intron (green) and eliminate the UAG codon that normally terminates translation of the upORF. In the resulting mRNA, the elongated upORF overlaps the connexin 32 start site, thereby precluding reinitiation. This could explain the inability to translate connexin 32 in patients with Charcot-Marie-Tooth neuropathy (32).
What's missing?
What's missing are biological clues: natural examples of dicistronic cellular mRNAs that require internal initiation of translation. There are well-characterized dicistronic mRNAs that contain overlapping cistrons which are translated by leaky scanning (40), and there are a few dicistronic mRNAs that translate a small upstream ORF and a second, nonoverlapping ORF by reinitiation (41). But no cellular mRNA of proven function contains two full-length, nonoverlapping cistrons, the second of which requires direct internal initiation of translation. A dicistronic mRNA that encodes enzymes involved in molybdopterin synthesis looked initially like a promising candidate. The original cDNA revealed an ORF for MOCSIA upstream from the ORF for MOCS1B, with an intercistronic gap of 18 nt (61). Follow-up studies, however, suggest that this mRNA produces only the upstream MOCS1A protein and that MOCS1B is translated as a fusion protein from a spliced transcript (21). Another potential candidate is the dicistronic SNRPN mRNA (20). In this case, the rather small size of the first cistron might allow translation of the second cistron by reinitiation. Splicing is also a possible explanation, inasmuch as a second transcript was evident in some tissues. Further studies are needed to determine whether SNRPN mRNA actually functions as a dicistronic transcript and, if so, whether internal initiation is involved.
With regard to the putative IRES elements in Table 1, what's missing is a common sequence or structure that might explain how the elements function. Complementarity to 18S rRNA is sometimes invoked, but with no experimental justification (see below). Computer-generated secondary structures that have been postulated to define IRES elements differ from case to case, and they have no experimental foundation. The presence of an oligopyrimidine tract is often mentioned as a hallmark of IRES elements, but it is not present in every case. An often overlooked fact is that the paradigmatic EMCV IRES retains 70% of its activity when the oligopyrimidine tract near the putative ribosome entry site is deleted (36).
If and when genuine cellular IRES elements are identified, the next step will be to determine how they function. For putative IRES elements derived from picornaviruses, the common belief is that binding of one or another initiation factor to the mRNA guides ribosomes to that site (62), but in no case have the factor-mRNA complexes been shown to function as chase-able intermediates. The first real evidence that prebinding of eIF4G to mRNA can mediate internal initiation comes from an unheralded study in which a synthetic IRES was created by inserting, at the midpoint of a dicistronic transcript, the iron response element derived from ferritin mRNA. This sequence was shown to mediate translation of the 3′ cistron when an eIF4G-IRP fusion protein was provided in trans (12). The synthetic IRES takes advantage of the high affinity of the iron regulatory protein (IRP) for its target sequence. Whether eIF4G unlinked to a carrier protein can bind natural IRES elements with sufficiently high affinity to mediate ribosome entry has yet to be demonstrated.
The fact that the 5′ UTR from ODC mRNA can support translation in extracts wherein cleavage of eIF4G prevents interaction with the cap binding factor eIF4E (Fig. 4A in reference 58), but the 5′ UTR from ODC mRNA does not perform credibly in the dicistronic test—the gold standard for defining IRES elements—constitutes a warning against equating cap-independent translation with internal initiation. The same false equation was made with regard to AML1, BiP, eIF4G, and IGFII (34, 57, 69), all of which are insensitive to cleavage of eIF4G but function poorly when tested directly in dicistronic vectors (entries 1, 3, 10, and 12). The consequences of perturbing the amount or interaction of eIF4G and/or eIF4E in vivo are more complicated (44, 52) and perhaps more interesting than simply allowing mRNAs that contain IRES elements to emerge.
CAN TRANSLATION INITIATE IN THE A-SITE AND WITHOUT MET-TRNA?
The genomic RNA of cricket paralysis virus (CrPV) has an unusual structure. The first ORF which encodes nonstructural proteins is punctuated by normal start and stop codons, but the downstream ORF which encodes viral capsid proteins lacks an AUG (or a standard alternative) start codon. A synthetic transcript that contains the CrPV intercistronic sequence and the start of the downstream ORF (with a reporter gene in place of the capsid coding sequence) supports measurable translation in vitro, showing that—at least in vitro—ribosomes can initiate without a standard start codon. This is postulated to occur via a totally unconventional mechanism (73) in which neither Met-tRNA nor initiation factors are required—an initiation mechanism that starts with binding of Ala-tRNA in the A-site of the ribosome, directed by the sequence CCU-GCU(6214–6219) in the viral mRNA. The CCU “initiator codon” that occupies the P-site is postulated to pair, not with tRNA, but with an upstream sequence in the viral mRNA. As explained below, the experiments on which this model is predicated have serious deficiencies.
Some other insect viruses that display the same peculiar coding properties as CrPV, but for which there is less experimental evidence, have been discussed elsewhere (59).
Sedimentation analyses.
Sucrose gradient analyses showed that ribosomes could bind to CrPV mRNA in the absence of initiation factors and that the complexes were insensitive to standard inhibitors of initiation such as GMPPNP, edeine, and l-methioninol (73). This insensitivity to inhibitors could mean either that CrPV uses a radically different mechanism of initiation or that the ribosome-mRNA complexes are nonfunctional aggregates. No attempt was made to assess functionality. (EDTA-induced dissociation of the complexes is not proof of authenticity, inasmuch as EDTA disrupts a variety of RNP aggregates.) An easy, routine test for distinguishing functional initiation complexes from inert aggregates is to show that radiolabeled Met-tRNA cosediments with the complexes (43, 47). The parallel test called for with CrPV would be to show that radiolabeled Ala-tRNA binds to and cosediments with 80S ribosome-CrPV mRNA complexes. Indeed, it should be possible not only to show binding of Ala-tRNA but also to synthesize a dipeptide simply by adding tRNAs and elongation factors. Without such tests, there is no reason to think that the complexes formed in the absence of initiation factors are functional.
Edeine resistance.
Wilson et al. (73) contend that the edeine-resistant complexes detected by sucrose gradient analysis are genuine because translation of luciferase, when directed by an mRNA that carries CrPV sequences at the 5′ end, was also resistant to edeine. But this was true only at extremely low concentrations of the antibiotic (0.25 to 0.5 μM in Fig. 3K in reference 73). In the same experiment, translation of CrPV mRNA was inhibited by 80% in the presence of 1 μM edeine. The concentration of edeine routinely used to inhibit eukaryotic mRNAs ranges from 1 to 10 μM (9, 25, 43, 55, 63). Thus, rather than CrPV translation being unusually resistant to edeine, the EMCV-derived mRNA used as the control in Fig. 3K in reference 73 might be unusually sensitive. It seems strange that, after making the point that 0.25 μM edeine is sufficient to inhibit EMCV translation, these authors consistently used a much higher concentration of edeine to disrupt EMCV initiation complexes (2.5 μM in Fig. 3D in reference 73 and 10 μM in Fig. 5A in reference 73).
Toeprinting assays.
Complexes detected using primer-extension inhibition (toeprinting) assays with CrPV mRNA also showed no requirement for initiation factors (Fig. 2B in reference 73). Indeed, the toeprint attributed to binding of a 40S ribosomal subunit to CrPV mRNA was strongly suppressed when initiation factors were added in vitro. (How the viral mRNA gets translated in vivo, despite the presence of initiation factors, was not addressed.)
As with the sucrose gradient assays, it is not clear what the complexes detected by toeprinting mean. Certain bands (primer extension pauses) are cited as evidence that a particular sequence in the mRNA occupies the A- or P-site of the ribosome, but no functional test supports these assignments. When binding was carried out with purified ribosomes in the absence of factors, the position of the major primer extension stop at C6226 was anomalous, since it mapped only 13 nt downstream from the CCU start codon rather than the customary 15- to 18-nt distance. When binding was carried out using an unfractionated reticulocyte lysate supplemented with cycloheximide to inhibit elongation, the C6226 stop was greatly reduced and a strong primer extension stop appeared instead at A6232 (Fig. 5B). Since the latter stop is the right distance downstream from the start codon, the most straightforward interpretation might be that only the cycloheximide-dependent stop near A6232 represents an authentic initiation complex. In other words, an authentic initiation complex forms only in the complete system.
Wilson et al. (73) pick and choose which toeprint bands are important and which can be ignored. They discuss at length a band at G6229 which they attribute (without evidence) to Ala-tRNA having entered the A-site; but no band is visible at G6229 in Fig. 5B in reference 73. (The authors say the band is weak because “pseudotranslocation occurs rapidly,” but if this key intermediate exists, it could have and should have been demonstrated by adding Ala-tRNA to purified ribosomes in the absence of eEF2.) In the same figure, the prominent extraneous bands seen in the presence of edeine are not explained convincingly. No explanation is offered as to why the binding of purified 40S subunits to CrPV mRNA gives a strong toeprint at A6161 in addition to the “authentic” toeprint at C6226 or why the addition of 60S ribosomal subunits produces yet another extraneous, upstream toeprint at G6182 (Fig. 2D in reference 73). The story is created by picking and choosing.
No subgenomic mRNA?
While initiation at an unconventional codon does appear to occur (somehow) when CrPV mRNA is translated in vitro, that is not necessarily the case in vivo. If a subgenomic mRNA were produced in infected cells, the mRNA might acquire an AUG start codon via splicing or editing or discontinuous transcription. The Northern blot of infected cell RNA shown by Wilson et al. (Fig. 1 in reference 74) reveals only genome-sized RNA, but that RNA preparation, extracted at a single unstated time point, might have derived primarily from infecting virions or progeny virus particles. It would seem useful to search for subgenomic mRNAs in infected cells by careful analysis of transcripts derived from polysomes that are actually engaged in synthesizing capsid proteins. (Cauliflower mosaic virus [CaMV], long cited as a rare example in which nonoverlapping cistrons are translated from a polycistronic mRNA, illustrates how careful analysis of transcripts can change a story: sensitive techniques recently uncovered spliced mRNAs that had been missed by Northern analysis [39], and additional splicing has not been ruled out.)
If there is no subgenomic mRNA and CrPV capsid proteins are indeed translated from an ORF positioned at the 3′ end of the genomic RNA, the intercistronic sequence (nt 6025 to 6216) must function as an IRES. Wilson et al. (74) attempted to show this by constructing a synthetic dicistronic vector into which the CrPV intercistronic sequence was inserted (5′-lucR[CrPV]lucF-3′). But all the questions raised above regarding the efficiency of putative cellular IRES sequences apply again here. For translation in vitro, uncapped dicistronic transcripts were used (Fig. 2C in reference 73), thus minimizing translation from the 5′ cistron and making the modest yield from the 3′ cistron look better. In one case where the 5′ cistron was preceded by the EMCV IRES and the 3′ cistron was preceded by the CrPV IRES, the yield of the 5′ product greatly exceeded the yield of the 3′ product (Fig. 6 in reference 74). Even when cells were transfected directly with the dicistronic mRNA (Fig. 4 in reference 74), the result is inconclusive because of the structure of the vector (the intercistronic region included a mutated version of the EMCV IRES along with the CrPV sequence; see the foregoing discussion of this vector) and because IRES function was scored only by monitoring the lucF/lucR ratio. What we need to know is whether the absolute yield from the 3′ end of a synthetic dicistronic mRNA is anywhere near the yield obtainable from a normal monocistronic mRNA. Is it anywhere close to the efficiency required to produce CrPV capsid proteins in infected cells?
BASE PAIRING BETWEEN MRNA AND 18S RRNA?
The 5′ UTR of Gtx mRNA contains the sequence CCGGCGGGU which is complementary to bases 1124 to 1132 in 18S rRNA. Insertion of this 9-nt sequence near the 5′ end of a monocistronic test transcript was shown to inhibit translation (30). These authors attributed the inhibition to base pairing with rRNA, although a simpler explanation might be that insertion of a GC-rich sequence creates a secondary structure that restricts the entry or movement of ribosomes. In contrast to the effect on monocistronic mRNAs, insertion of a single copy of the CCGGCGGGU motif at the midpoint of a dicistronic mRNA slightly stimulated translation of the 3′ cistron, and translation of the 3′ cistron was stimulated several hundredfold when 10 copies were inserted (5). Unfortunately, mRNA produced by the high-expressing construct was not analyzed to rule out the possibility that the GC-rich intercistronic insert might have functioned as a transcriptional promoter, producing a monocistronic mRNA from which the 3′ cistron was actually translated. Without having ruled out this and other alternative explanations, the authors concluded from the dicistronic test that the Gtx-derived sequence is an IRES. They postulate that IRES activity results from base pairing between mRNA and rRNA, citing as evidence the ability of the CCGGCGGGU element to be photochemically cross-linked to 18S rRNA (30). The cross-linking, however, might be an artifact, unrelated to function, since it did not require formation of a ribosome-mRNA initiation complex. Indeed, cross-linking occurred even when the Gtx-derived sequence was incubated with deproteinized 18S rRNA.
Complementarity to 18S rRNA has also been invoked to explain some instances of ribosome shunting. Shunting, or discontinuous scanning by 40S ribosomal subunits, has been suggested in cases where a stable hairpin structure inserted into the 5′ UTR fails to inhibit translation. Shunting has been postulated to occur with heat shock protein 70 (hsp70) and adenovirus late mRNAs (76), as well as with CaMV mRNA (see below). Because the adenovirus and hsp70 experiments were carried out only in vivo, the possibility that the hairpin might be bypassed by splicing or some other nontranslational mechanism has not been rigorously excluded. Rudimentary mapping of the sequences in adenovirus mRNA required for (what appears to be) ribosome shunting identified a sequence in the 5′ UTR with extensive complementarity to bases 1841 to 1867 near the 3′ end 18S rRNA (76). There are several reasons, however, to be cautious about concluding that base pairing between mRNA and rRNA promotes shunting: (i) large deletions, rather than point mutations, were used to map the required sequences in adenovirus mRNA; (ii) hsp70 mRNA shows far less complementarity to 18S rRNA (e.g., the boxed sequence in hsp70 mRNA postulated to pair with CGGAAGG is complementary to that rRNA sequence in only three of seven positions); and (iii) the implicated sequence in 18S rRNA forms a stable hairpin structure (bases 1843 to 1866) which should make it unavailable to interact with mRNA.
Schärer-Hernández and Hohn have studied other 5′ UTRs in which hairpin structures that normally inhibit translation are somehow evaded. The residual translation was too low (∼5%) for confident interpretation when the full 5′ UTR from CaMV was tested in vivo (64), but the ∼20% efficiency observed when partially synthetic 5′ UTR sequences were tested in vitro was adequate to allow analysis of the mechanism. The most notable finding from these studies is that bypassing of the hairpin barrier requires that it be preceded by a small upstream ORF (22, 56). The authors' interpretation is that, after translating the upstream ORF and penetrating the base of the hairpin, the ribosome dissociates from the mRNA and then reenters downstream. It is easy to envision how reformation of the hairpin might nudge the ribosome off the mRNA; what is missing, however, is a mechanism for directing reentry of the ribosome—a mechanism such as the one that allows discontinuous translation of bacteriophage T4 gene 60 in Escherichia coli (24).
Instead of the postulated shunting mechanism (22, 56), the CaMV results might be explained by an “augmented linear scanning” mechanism whereby the 40S subunit or 80S ribosome, perhaps aided by helicases that enter at the terminator codon of the upstream ORF (11), penetrates the hairpin structure and then reinitiates somewhere downstream. Because the closest AUG codons are often bypassed when ribosomes scan in the reinitiation mode, this linear scanning mechanism is not ruled out by the observed failure to initiate at some AUG codons positioned within the hairpin. Further studies, perhaps employing a covalent cross-link to preclude disruption of the hairpin, should clarify whether the base-paired structure in CaMV-derived transcripts is surmounted by discontinuous scanning (shunting) or by augmented linear scanning. Either way, the interesting mechanism emerging from the studies of Hohn and his colleagues might shed light on why small upstream ORFs occur with high frequency in cellular mRNAs that have highly structured 5′ noncoding sequences (40). Of note here is that, when the upstream ORF in CaMV-derived transcripts was replaced with the aforementioned adenovirus sequence that is complementary to 18S rRNA, the resulting mRNA could not be translated (22).
When considering claims of base pairing between eukaryotic mRNAs and 18S rRNA, it is useful to recall a lesson from prokaryotes. For many years, it was fashionable to postulate base pairing between 16S rRNA and an mRNA sequence located downstream from the AUG codon in cases where an upstream Shine-Dalgarno sequence was absent. But when the downstream box hypothesis was finally tested by mutating the postulated target sequence in 16S rRNA, it was firmly ruled out (53). The point is that it is easy to spot sequences in mRNAs complementary to one or another rRNA sequence, but even when changes in the mRNA sequence perturb translation, that is a long way from proving that the mRNA and rRNA sequences interact.
ACKNOWLEDGMENT
Research in my laboratory is supported by grant GM33915 from the National Institutes of Health.
REFERENCES
- 1.Akiri G, Nahari D, Finkelstein Y, Le S-Y, Elroy-Stein O, Levi B-Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay P K, Wang C, Lipton H L. Cap-independent translation by the 5′ untranslated region of Theiler's murine encephalomyelitis virus. J Virol. 1992;66:6249–6256. doi: 10.1128/jvi.66.11.6249-6256.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein J, Sella O, Le S-Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-liked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 4.Carter M S, Sarnow P. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J Biol Chem. 2000;275:28301–28307. doi: 10.1074/jbc.M004657200. [DOI] [PubMed] [Google Scholar]
- 5.Chappell S A, Edelman G M, Mauro V P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell S A, LeQuesne J P C, Paulin F E M, deSchoolmeester M L, Stonely M, Soutar R L, Ralston S H, Helfrich M H, Willis A E. A mutation in the c-myc IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 7.Coldwell M J, Mitchell S A, Stoneley M, MacFarlane M, Willis A E. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 9.Craig N, Kostura M. Inhibition of protein synthesis in CHO cells by actinomycin D: lesion occurs after 40S initiation complex formation. Biochemistry. 1983;22:6064–6071. doi: 10.1021/bi00295a004. [DOI] [PubMed] [Google Scholar]
- 10.Créancier L, Morello D, Mercier P, Prats A-C. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czaplinski K, Majlesi N, Banerjee T, Peltz S W. Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA. 2000;6:730–743. doi: 10.1017/s1355838200992392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Gregorio E, Preiss T, Hentze M W. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elgadi M M, Smiley J R. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkelstein Y, Faktor O, Elroy-Stein O, Levi B-Z. The use of bi-cistronic transfer vectors for the baculovirus expression system. J Biotechnol. 1999;75:33–44. doi: 10.1016/s0168-1656(99)00131-5. [DOI] [PubMed] [Google Scholar]
- 15.Gallie D R, Ling J, Niepel M, Morley S J, Pain V M. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan W, Rhoads R E. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 17.Gan W, La Celle M, Rhoads R E. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- 18.Giraud, S., A. Greco, J.-J. Diaz, and P. Delafontaine. Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem., in press. [DOI] [PubMed]
- 19.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray T A, Saitoh S, Nicholls R D. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray T A, Nicholls R D. Diverse splicing mechanisms fuse the evolutionarily conserved bicistronic MOCS1A and MOCS1B open reading frames. RNA. 2000;6:928–936. doi: 10.1017/s1355838200000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemmings-Mieszczak M, Hohn T, Preiss T. Termination and peptide release at the upstream open reading frame are required for down stream translation on synthetic shunt-competent mRNA leaders. Mol Cell Biol. 2000;20:6212–6223. doi: 10.1128/mcb.20.17.6212-6223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henis-Korenblit S, Strumpf N L, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbst K L, Nichols L M, Gesteland R F, Weiss R B. A mutation in ribosomal protein L9 affects ribosomal hopping during translation of gene 60 from bacteriophage T4. Proc Natl Acad Sci USA. 1994;91:12525–12529. doi: 10.1073/pnas.91.26.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiremath L S, Hiremath S T, Rychlik W, Joshi S, Domier L L, Rhoads R E. In vitro synthesis, phosphorylation, and localization on 48S initiation complexes of human protein synthesis initiation factor 4E. J Biol Chem. 1989;264:1132–1138. [PubMed] [Google Scholar]
- 26.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk R G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 27.Holcik M, Korneluk R G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holcik M, Yeh C, Korneluk R G, Chow T. Translational upregulation of X-linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene. 2000;19:4174–4177. doi: 10.1038/sj.onc.1203765. [DOI] [PubMed] [Google Scholar]
- 29.Hoover D S, Wingett D G, Zhang J, Reeves R, Magnuson N S. Pim-1 protein expression is regulated by its 5′-untranslated region and translation initiation factor eIF-4E. Cell Growth Differ. 1997;8:1371–1380. [PubMed] [Google Scholar]
- 30.Hu M C-Y, Tranque P, Edelman G M, Mauro V P. rRNA-complementarity in the 5′ untranslated region of mRNA specifying the Gtx homeodomain protein: evidence that base-pairing to 18S rRNA affects translational efficiency. Proc Natl Acad Sci USA. 1999;96:1339–1344. doi: 10.1073/pnas.96.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudder A, Werner R. Analysis of a Charcot-Marie-Tooth disease mutation reveals an essential internal ribosome entry site element in the connexin-32 gene. J Biol Chem. 2000;275:34586–34591. doi: 10.1074/jbc.M005199200. [DOI] [PubMed] [Google Scholar]
- 32.Ionasescu V V, Searby C, Ionasescu R, Neuhaus I M, Werner R. Mutations of the noncoding region of the connexin32 gene in X-linked dominant Charcot-Marie-Tooth neuropathy. Neurology. 1996;47:541–544. doi: 10.1212/wnl.47.2.541. [DOI] [PubMed] [Google Scholar]
- 33.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 34.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johannes G, Carter M S, Eisen M B, Brown P O, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaminski A, Belsham G J, Jackson R J. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J G, Armstrong R C, Berndt J A, Kim N W, Hudson L D. A secreted DNA-binding protein that is translated through an internal ribosome entry site (IRES) and distributed in a discrete pattern in the central nervous system. Mol Cell Neurosci. 1998;12:119–140. doi: 10.1006/mcne.1998.0701. [DOI] [PubMed] [Google Scholar]
- 38.Kim Y K, Hahm B, Jang S K. Polypyrimidine tract-binding protein inhibits translation of Bip mRNA. J Mol Biol. 2000;304:119–133. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- 39.Kiss-László Z, Blanc S, Hohn T. Splicing of cauliflower mosaic virus 35S RNA is essential for viral infectivity. EMBO J. 1995;14:3552–3562. doi: 10.1002/j.1460-2075.1995.tb07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 42.Kozak M. Do the 5′ untranslated domains of human cDNAs challenge the rules for initiation of translation (or is it vice versa)? Genomics. 2000;70:396–406. doi: 10.1006/geno.2000.6412. [DOI] [PubMed] [Google Scholar]
- 43.Kozak M, Shatkin A. Migration of 40S ribosomal subunits on messenger RNA in the presence of edeine. J Biol Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 44.Lai H-K, Borden K L B. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene. 2000;19:1623–1634. doi: 10.1038/sj.onc.1203473. [DOI] [PubMed] [Google Scholar]
- 45.Lauring A S, Overbaugh J. Evidence that an IRES within the Notch2 coding region can direct expression of a nuclear form of the protein. Mol Cell. 2000;6:939–945. doi: 10.1016/s1097-2765(05)00084-5. [DOI] [PubMed] [Google Scholar]
- 46.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 47.Maiti T, Das S, Maitra U. Isolation and functional characterization of a temperature-sensitive mutant of the yeast Saccharomyces cerevisiae in translation initiation factor eIF5: an eIF5-dependent cell-free translation system. Gene. 2000;244:109–118. doi: 10.1016/s0378-1119(99)00570-3. [DOI] [PubMed] [Google Scholar]
- 48.Manzella J M, Blackshear P J. Regulation of rat ornithine decarboxylase mRNA translation by its 5′-untranslated region. J Biol Chem. 1990;265:11817–11822. [PubMed] [Google Scholar]
- 49.Mumm J S, Schroeter E H, Saxena M T, Griesemer A, Tian X, Pan D J, Ray W J, Kopan R. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 50.Nanbru C, Lafon I, Audigier S, Gensac M-C, Vagner S, Huez G, Prats A-C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 51.Negulescu D, Leong L E-C, Chandy K G, Semler B L, Gutman G A. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J Biol Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- 52.Novoa I, Carrasco L. Cleavage of eukaryotic translation initiation factor 4G by exogenously added hybrid proteins containing poliovirus 2Apro in HeLa cells: effects on gene expression. Mol Cell Biol. 1999;19:2445–2454. doi: 10.1128/mcb.19.4.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Connor M, Asai T, Squires C L, Dahlberg A E. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc Natl Acad Sci USA. 1999;96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oumard A, Hennecke M, Hauser H, Nourbakhsh M. Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol Cell Biol. 2000;20:2755–2759. doi: 10.1128/mcb.20.8.2755-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson D T, Safer B, Merrick W C. Role of eukaryotic initiation factor 5 in the formation of 80S initiation complexes. J Biol Chem. 1979;254:7730–7735. [PubMed] [Google Scholar]
- 56.Pooggin M M, Hohn T, Fütterer J. Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J Biol Chem. 2000;275:17288–17296. doi: 10.1074/jbc.M001143200. [DOI] [PubMed] [Google Scholar]
- 57.Pozner A, Goldenberg D, Negreanu V, Le S-Y, Elroy-Stein O, Levanon D, Groner Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol Cell Biol. 2000;20:2297–2307. doi: 10.1128/mcb.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 59.RajBhandary U L. More surprises in translation: initiation without the initiator tRNA. Proc Natl Acad Sci USA. 2000;97:1325–1327. doi: 10.1073/pnas.040579197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao C D, Pech M, Robbins K C, Aaronson S A. The 5′ untranslated sequence of the c-sis/platelet-derived growth factor 2 transcript is a potent translational inhibitor. Mol Cell Biol. 1988;8:284–292. doi: 10.1128/mcb.8.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reiss J, Cohen N, Dorche C, Mandel H, Mendel R R, Stallmeyer B, Zabot M-T, Dierks T. Mutations in a polycistronic nuclear gene associated with molybdenum cofactor deficiency. Nat Genet. 1998;20:51–53. doi: 10.1038/1706. [DOI] [PubMed] [Google Scholar]
- 62.Sachs A B. Cell cycle-dependent translation: IRES elements prevail. Cell. 2000;101:243–245. doi: 10.1016/s0092-8674(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 63.Safer B, Kemper W, Jagus R. Identification of a 48S preinitiation complex in reticulocyte lysate. J Biol Chem. 1978;253:3384–3386. [PubMed] [Google Scholar]
- 64.Schärer-Hernández N, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA in transgenic tobacco plants. Virology. 1998;242:403–413. doi: 10.1006/viro.1998.9038. [DOI] [PubMed] [Google Scholar]
- 65.Schiavi A, Hudder A, Werner R. Connexin43 mRNA contains a functional internal ribosome entry site. FEBS Lett. 1999;464:118–122. doi: 10.1016/s0014-5793(99)01699-3. [DOI] [PubMed] [Google Scholar]
- 66.Stoneley M, Paulin F E M, Le Quesne J P C, Chappell S A, Willis A E. c-myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 67.Stoneley M, Subkhankulova T, Le Quesne J P C, Coldwell M J, Jopling C L, Belsham G J, Willis A E. Analysis of the c-myc IRES: a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teerink H, Kasperaitis M A M, De Moor C H, Voorma H O, Thomas A A M. Translation initiation on the insulin-like growth factor II leader 1 is developmentally regulated. Biochem J. 1994;303:547–553. doi: 10.1042/bj3030547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teerink H, Voorma H O, Thomas A A M. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim Biophys Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 70.Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Daalen Wetters T, Macrae M, Brabant M, Sittler A, Coffino P. Polyamine-mediated regulation of mouse ornithine decarboxylase is post-translational. Mol Cell Biol. 1989;9:5484–5490. doi: 10.1128/mcb.9.12.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watada H, Mirmira R G, Leung J, German M S. Transcriptional and translational regulation of β-cell differentiation factor Nkx6.1. J Biol Chem. 2000;275:34224–34230. doi: 10.1074/jbc.M004981200. [DOI] [PubMed] [Google Scholar]
- 73.Wilson J E, Pestova T V, Hellen C U T, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 74.Wilson J E, Powell M J, Hoover S E, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiang J, Lahti J M, Grenet J, Easton J, Kidd V J. Molecular cloning and expression of alternatively spliced PITSLRE protein kinase isoforms. J Biol Chem. 1994;269:15786–15794. [PubMed] [Google Scholar]
- 76.Yueh A, Schneider R J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]