Abstract
PACT is a 35-kDa human protein that can directly bind and activate the latent protein kinase, PKR. Here we report that PKR activation by PACT causes cellular apoptosis in addition to PKR autophosphorylation and translation inhibition. We analyzed the structure-function relationship of PACT by measuring its ability to bind and activate PKR in vitro and in vivo. Our studies revealed that among three domains of PACT, the presence of either domain 1 or domain 2 was sufficient for high-affinity binding of PACT to PKR. On the other hand, domain 3, consisting of 66 residues, was absolutely required for PKR activation in vitro and in vivo. When fused to maltose-binding protein, domain 3 was also sufficient for efficiently activating PKR in vitro. However, it bound poorly to PKR at the physiological salt concentration and consequently could not activate it properly in vivo. As anticipated, activation of PKR by domain 3 in vivo could be restored by attaching it to a heterologous PKR-binding domain. These results demonstrated that the structure of PACT is modular: it is composed of a distinct PKR-activation domain and two mutually redundant PKR-interacting domains.
PKR is a serine/threonine protein kinase present, at a low level, in all cells (5, 34). Its cellular abundance is elevated by interferon (IFN) treatment of cells that causes transcriptional induction of the PKR gene. The enzymatic activity of the PKR protein is latent, and it needs to be activated by autophosphorylation. Once activated, PKR can phosphorylate a limited set of cellular proteins, the most well studied of which is the translation initiation factor, eIF-2α. The most potent activator of PKR is double-stranded RNA (dsRNA), a frequent by-product of virus replication in cells. dsRNA binds to PKR with a high affinity and causes a conformational change to the protein, thereby exposing its ATP-binding site (2, 3) which leads to its autophosphorylation (27). Other polyanionic molecules, such as heparin, can also activate PKR in vitro, although their physiological roles remain unclear (23). An alternative route of PKR activation that is more relevant to cells without virus infection was revealed by the discovery of PACT, a protein activator of PKR (20).
The dsRNA-binding domain (DRBD) of PKR has been well characterized. It is located at the amino terminus of the protein and contains two dsRNA-binding motifs (11, 19). Similar motifs are present in other dsRNA-binding proteins as well (30). The higher-order structures of the DRBDs of several proteins, including PKR, have been determined, and they all contain identical α-β-β-β-α structures (17, 28). We have shown that the DRBD of PKR is also involved in protein-protein interaction or dimerization (22). Thus, this dimerization domain (DD) mediates direct homomeric and heteromeric interactions among different members of this family. This property of PKR was exploited in a yeast two-hybrid screen for cloning two new proteins of this family, PACT and DRBP76, the latter being a substrate of PKR (20, 25). Although the same domain of PKR mediates both protein-protein interactions and protein-RNA interactions, the two properties appear to be largely separable because several mutants of PKR are capable of doing one but not the other (22, 24). On the other hand, the two properties may function in a cooperative manner for PKR function since dsRNA-DRBD interaction promotes and stabilizes PKR dimerization that is required for PKR activation (3, 21). In this regard, the dsRNA-DRBD interaction was suggested to expose an additional dimerization site at the C-terminus region of PKR (31); this region and DRBD together may facilitate a stable PKR dimerization required for PKR activation.
PKR's physiological functions were first illuminated in the context of virally infected cells. Activation of PKR, presumably by viral dsRNA, causes eIF-2α phosphorylation which leads to a global inhibition of cellular and viral protein synthesis. To circumvent this PKR-mediated block of virus replication, many viruses encode specific RNAs or proteins which block PKR activation or action (33). In addition to its antiviral role, PKR participates in a broad array of cellular processes such as signal transduction, differentiation, apoptosis, cell growth, and oncogenic transformation (5, 34). PKR has been shown to be an important participant in the transcriptional signaling pathways activated by specific cytokines, growth factors, dsRNA, and extracellular stresses. Although the detailed mechanisms of PKR actions in these cascades of signaling remain to be delineated, the presence of PKR has been shown to be required for the optimal activation of several other protein kinases, such as P38, JNK, and IKK (4, 12, 37), and transcription factors, such as NF-κB, P53, STAT1, ATF, STAT3, and IRF-1 (4, 6, 14, 15, 34, 35). What mediates PKR activation in virally uninfected cells in response to the various extracellular stimuli has remained a mystery, and it is quite likely that PACT plays an important, although as yet undefined, role in this process.
PACT was cloned by virtue of its ability to interact with PKR (20). It is ubiquitously expressed at a low level, and its cellular abundance is not regulated by IFNs or dsRNA. As expected, PACT contains typical domains which are known to mediate protein-protein interactions among the members of the PKR family of dsRNA-binding proteins. Among three such putative domains, domains 1 and 2 have strong sequence conservations with similar domains of PKR and TRBP, whereas domain 3 shows less homology. PACT can bind directly to PKR, and at least a part of this binding is mediated by the DD of PKR. Although both PACT and PKR can bind dsRNA, their mutual interaction does not require dsRNA. Binding of PACT leads to activation of PKR by autophosphorylation. Bacterially expressed PACT that was free of any contaminating RNA could activate PKR in vitro, and PKR mutants which could not be activated by dsRNA could still be activated by PACT (20). Thus, PACT was identified as a true protein activator of PKR. In vivo, the overexpression of PACT caused activation of PKR, phosphorylation of eIF-2α and inhibition of translation in mammalian cells. Similar expression of PACT in yeasts caused a PKR-dependent inhibition of cell growth. RAX, the murine homolog of PACT, has an almost identical primary structure (13). It also activates PKR in vitro and in an interleukin-3-dependent cell line, a variety of stress conditions cause RAX phosphorylation, PKR-RAX association, and activation of PKR. Thus, PACT or RAX functions as a physiological mediator of stress-induced PKR activation.
In the current study, we have analyzed the functional domain structure of PACT. PKR binding and various outcomes of PKR activation were used as markers to delineate a modular structure of PACT. Domain 3 was shown to be both necessary and sufficient for activating PKR, but either domain 1 or domain 2 was needed for a strong binding of PACT to PKR.
MATERIALS AND METHODS
Construction of PACT and PKR mutants.
All internal PACT and PKR deletions (Table 1) were constructed by overlap extension PCR (26). Briefly, two separate PCR reactions were performed to amplify both halves of the coding region of PACT using four primers. An outside-forward primer (P1) was paired with a middle-reverse primer (P2) to synthesize the first half; an outside-reverse primer (P4) was paired with a middle-forward primer (P3) to synthesize the second half. The deletion was introduced by the middle two primers (P2 and P3). The two PCR products were put into the third PCR reaction with the two outside primers (P1 and P4) to produce the desired deletion. The resulting DNA clones were DNA sequenced to confirm the correct deletion and reading frame. For all of these studies a wild-type (wt) PACT variant truncated at amino acid 305 and coding for residues KLCSI at positions 301 to 305 was utilized. A FLAG-epitope tag was added at the N-terminal coding end of all PACT and PKR-PACT hybrid deletion constructs.
TABLE 1.
PACT mutants used
| Protein | Protein description |
|---|---|
| wt | wt PACT, contains residues 1 to 305 |
| Δ1 | PACT missing domain 1 (residues 35 to 99), contains residues 1 to 34 and 100 to 305 |
| Δ2 | PACT missing domain 2 (residues 127 to 192), contains residues 1 to 126 and 193 to 305 |
| Δ3 | PACT missing domain 3 (residues 240 to 305), contains residues 1 to 239 |
| Δ1,2 | PACT missing domains 1 and 2, contains residues 1 to 34, 100 to 126, and 193 to 305 |
| MBP | Maltose-binding protein |
| MBP-3 | MBP tethered to PACT domain 3 residues 240 to 305 |
| DD | PKR DD, contains amino acid residues 1 to 170 |
| DD-Δ1,2 | PKR DD containing residues 1 to 170 tethered to PACT Δ1,2 |
| Δ2DD-Δ1,2 | PKR dimerization domain missing second dimerization motif (residues 100 to 165), tethered to PACT Δ1,2 |
| DD-D3 | PKR tethered to PACT domain 3 |
| Δ2DD-D3 | PKR DD missing second dimerization motif, tethered to PACT domain 3 |
Apoptosis assays. (i) TUNEL.
HT1080 or mouse embryonic fibroblasts (MEFs) growing on glass coverslips in six-well dishes were transfected with pcDNA3 vector, pcDNA3-FLAG PACT, or another FLAG-tagged construct. At 6 h after transfection, the cells were treated with 50 ng of actinomycin D per ml. Cells were fixed in 4% methanol-free formaldehyde 24 h after transfection. Thymidine deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) assay using the Apoptosis Detection System (Promega) was performed using the manufacturer protocol. After a 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) wash and rinses in phosphate-buffered saline (PBS) blocking buffer (10% goat serum and 3% bovine serum albumin in Tris-buffered saline-Tween 20) was placed on the cells for 10 min at room temperature (RT). Cells were stained using mouse anti-FLAG (M5 or M2) monoclonal antibody at 1:1,000 dilution for 45 min at RT. After three 5-min washes in PBS, cells were stained with goat anti-mouse immunoglobulin G-Texas red conjugate (Molecular Probes) at 1:1,500 dilution for 45 min at RT. After three more 5-min washes in PBS, the cells were mounted on glass slides in Vectashield with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories) and examined under a fluorescence microscope. Different optical filters were used for screening the same field for detecting blue color for DAPI (all cells in the field), red color for expression of the protein (only the transfected cells), and green color for TUNEL (only the cells undergoing apoptosis). For counting the TUNEL-positive cells among the protein-expressing cells, the red and green colors could be merged, producing a yellow color. For quantitation of apoptosis, at least 300 protein-expressing cells were scored for TUNEL positivity.
(ii) FACS (sub-G1) analysis.
For fluorescence-activated cell sorter (FACS) analysis, HT1080 cells were first transfected with pcDNA3 vector, pcDNA3-FLAG PACT, or mutant with Lipofectamine reagent. At 6 h after transfection, the cells were treated with 50 ng of actinomycin D per ml. Cells were harvested 24 h after transfection as follows. Floating and attached cells were pooled, pelleted, and washed in PBS. The cells were then fixed in 70% ethanol for 30 min, treated with RNase, stained with propidium iodide, and monitored in a flow cytometer.
dsRNA-binding assay.
In vitro-translated, 35S-labeled FLAG epitope-tagged PACT or PACT mutant proteins were synthesized using the TNT T7-coupled reticulocyte system from Promega. The poly(I-C)-agarose binding assay was used to measure dsRNA binding. The translated protein (4 μl) diluted with 25 μl of binding buffer (20 mM Tris-HCl [pH 7.5], 0.3 M NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% NP-40, 10% glycerol) was mixed with 25 μl of poly(I-C)-agarose beads (Amersham-Pharmacia) and incubated at 30°C for 30 min with intermittent shaking. The beads were then washed with 500 μl of binding buffer, four times. The proteins bound to the beads after being washed were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by fluorography (19). Alternatively, bacterially expressed purified proteins were used similarly to measure dsRNA-binding activity and detected by Western blot with anti-PACT polyclonal antibody.
In vitro interaction assay.
In vitro-translated, 35S-labeled PKR (K296R) and FLAG epitope-tagged PACT or PACT mutant proteins were synthesized using the TNT T7-coupled reticulocyte system (Promega) (20). After translation, equal quantities of reticulocyte extracts containing the two proteins to be tested for interaction were mixed and placed at 37°C for 15 min. Then, 5 μl of the mixture was incubated with 20 μl of anti-FLAG (M2)-agarose (Sigma) in immunoprecipitation buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 1 mM EDTA, 1 mM DTT, 100 U of aprotinin per ml, 0.2 mM PMSF, 1% Triton X-100, 20% glycerol) for 30 min at 4°C on a rotating wheel. In some experiments, a low-salt buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 2 mM magnesium acetate, 100 U of aprotinin per ml, 0.2 mM PMSF, 1% Triton X-100, 20% glycerol) or high-salt buffer (immunoprecipitation buffer containing 100 mM NaCl) was used for immunoprecipitation and washing. After binding, the beads were washed six times with 500 μl of immunoprecipitation buffer. The washed beads were then boiled in 2× Laemmli buffer (150 mM Tris-HCl [pH 6.8], 5% SDS, 5% β-mercaptoethanol, 20% glycerol) for 2 min and analyzed by SDS-PAGE on a 12% resolving gel. The radioactive bands were quantitated by densitometry of autoradiograms. In some experiments, PKR or bacterially expressed DD of PKR was immunoprecipitated with matrix-bound antibody. After incubation with purified PACT, maltose-binding protein (MBP), or MBP-3, the beads were centrifuged and washed as described above, and the bound proteins were analyzed by gel electrophoresis followed by Western blotting.
Expression in mammalian cells and coimmunoprecipitation assay.
HT1080 cells were transfected in 100-mm culture dishes with 10 μg of total DNA (5 μg of CMV-PKR [K296R] and 5 μg of FLAG-PACT mutant DNA) using the Lipofectamine reagent (Gibco-BRL). At 24 h after transfection, cells were lysed in immunoprecipitation buffer (20 mM Tris-HCl [pH 7.5], 1 mM DTT, 100 mM NaCl, 2 mM MgCl2, complete protease inhibitors [Roche], 20% glycerol) on ice. The cell extract was used to immunoprecipitate FLAG-PACT with anti-FLAG (M2) agarose as described above for the in vitro interaction assay. The immunoprecipitates were analyzed by Western blotting with anti-PKR (Santa Cruz) and anti-FLAG polyclonal antibodies (Santa Cruz) (20).
Translation inhibition assay.
HT1080 cells were transfected in six-well dishes in triplicate with 200 ng each of pGL2-luciferase reporter, pRSV–β-galactosidase, and pcDNA3-PACT or PACT mutant by Lipofectamine. At 24 h after transfection, the cells were treated with 100 U of IFN-β per ml. Cells were harvested 48 h after transfection and assayed for luciferase activity. The Luciferase Assay System (Promega) was used to prepare and assay the cell extract. Luciferase activity was measured in a Dynatech Laboratories luminometer (20). β-Galactosidase was quantified in liquid assays using ONPG (o-nitrophenyl-β-d-galactopyranoside) as the substrate. Luciferase activities were normalized using the recorded β-galactosidase activities.
Expression and purification of PACT and its mutants from Escherichia coli.
The protein coding region for PACT mutants Δ3 and Δ1,2 were subcloned into pET15b (Novagen) to generate pET15b-PACTΔ3 and pET15b-PACTΔ1,2. This results in an in-frame fusion of correct PACT coding sequence to the histidine tag. The expression vector was transformed into BL21(DE3) cells containing a plasmid overexpressing thioredoxin to enhance protein solubility (36). The bacteria were grown overnight, transferred to a larger culture volume, and grown 3 to 4 h. The culture was shifted to room temperature, with isopropyl-β-d-thiogalactopyranoside (IPTG) then added at a final concentration of 0.5 mM, and grown for 12 h. The culture was harvested at 8,000 rpm for 10 min in a Beckman JA10 rotor. Cells were washed once in ice-cold PBS and then resuspended in 20 ml of lysis buffer (500 mM NaCl, 50 mM NaH2PO4, 20 mM imidazole, 10% glycerol, 0.1% NP-40, complete protein inhibitors, 5 mM β-mercaptoethanol) per liter. Cells were lysed by passing them twice through a French press (1000 lb/in2), and the lysate was cleared by spinning at 15,000 × g for 20 min. The supernatant was mixed with 10 ml of Ni-TA agarose and incubated for 1 h at 4°C on a spinning wheel. After binding, the beads were washed twice in buffer A (500 mM NaCl, 50 mM NaH2PO4, 20 mM imidazole, 10% glycerol; pH 8.0) and packed into a column. The column was washed with 100 ml of buffer A and 100 ml of buffer B (buffer A at pH 6.0). The His-PACT mutant was eluted with 30 ml of elution buffer (150 mM l-histidine in buffer A [pH 6.8]). The eluted protein was concentrated by placing it into dialysis tubing (10,000 molecular weight cut off) covered with dry polyethylene glycol (PEG 20000; Fluka) at 4°C for 1 to 2 h. The protein was further purified by gel filtration (200 mM NaCl) using a Superdex 75 matrix (Pharmacia) at constant flow of 0.5 ml/min. The protein coding region for PACT domain 3 was subcloned into pMAL-c2X (New England Biochemical) to generate pMAL-c2X-PACTD3. This resulted in an in-frame fusion of PACT domain 3 coding sequence to MBP. MBP and MBP-3 were purified by amylose affinity chromatography. After the final elution off of the column, each and every bacterially expressed protein preparation was treated with micrococcal nuclease to remove any contaminating dsRNA (20). To ensure that any and all dsRNA was digested, each protein was heat inactivated by boiling and tested for residual dsRNA in PKR activation assays in vitro. All proteins were stored at −80°C until use.
PKR activation assay in vitro.
The kinase activation assay of PKR was performed on PKR purified by monoclonal antibody immobilized on protein G-Sepharose (20). HT1080 cells were treated with 1,000 U of IFN-β per ml for 24 h and lysed in high-salt buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 400 mM NaCl, 1% Triton X-100, 0.2 mM PMSF, 100 U of aprotinin per ml, 20% glycerol). In some experiments, a low-salt buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 1% Triton X-100, 0.2 mM PMSF, 100 U of aprotinin per ml, 20% glycerol) was used in place of the high-salt buffer. HT1080 lysate was mixed with 1 μl of PKR monoclonal antibody 71/10 (Ribogene) in high-salt buffer and placed on a spinning wheel for 30 min at 4°C. Then, 25 μl of protein G-Sepharose was added, and the mixture was spun an additional 30 min at 4°C. The protein G-Sepharose beads were washed four times in 500 μl of high-salt buffer and two times in 500 μl of activity buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 2 mM magnesium acetate, 7 mM β-mercaptoethanol, 20% glycerol). The activation assay was performed on immobilized PKR in activity buffer containing 1 to 100 nmol of purified protein activator, 2.5 mM MnCl2, 0.1 mM ATP, and 10 μCi of [γ32-P]ATP for 20 min at 30°C. The labeled protein was analyzed by SDS-PAGE on an 11% resolving gel. Autoradiography was performed at RT.
RESULTS
PKR-mediated cellular apoptosis by PACT.
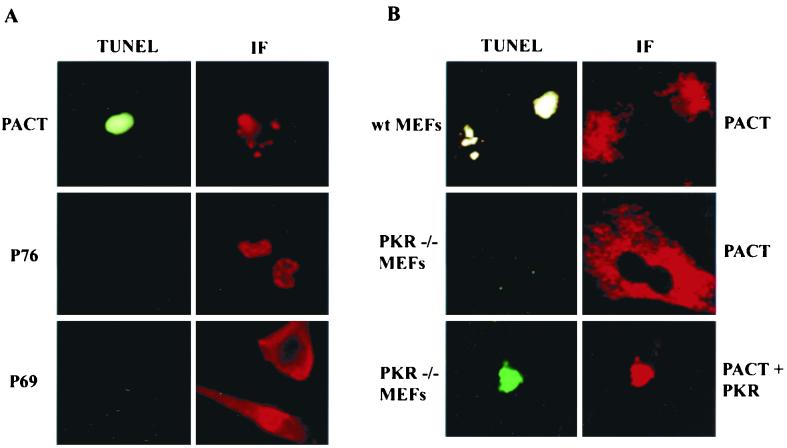
Because PKR activation in vivo has been shown to cause apoptosis, we investigated whether PACT overexpression could cause the same (32). Human HT1080 cells were transfected with expression vectors for PACT or two other dsRNA-binding proteins, P76 and P69. P76 is a nuclear protein that is a substrate of PKR and P69 is an isozyme of 2–5(A) synthetase (9, 25). All three proteins contained a FLAG tag for easy detection and were expressed at the same level in the transfected cells. As shown by immunofluorescence, PACT was expressed mostly in the cytoplasm, P76 was exclusively nuclear, and P69 was exclusively cytoplasmic (Fig. 1A). The transfected cells were stressed by treating them with a low dose of actinomycin D. This treatment did not cause apoptosis in vector-transfected cells (data not shown). Similarly, apoptosis was not observed in P76- or P69-expressing cells as well (Fig. 1A). In contrast, PACT-expressing cells were undergoing apoptosis as revealed by TUNEL assay (Fig. 1A). The TUNEL-positive cells appeared green which, during the long exposures required for photography, was sometimes bleached to gray. The morphology of the cells undergoing apoptosis was also quite distinct. Although only one cell is shown here in a high-resolution image, the majority of PACT-expressing cells were TUNEL positive (Table 2). Apoptosis was also observed when PACT was expressed in wild-type MEFs (upper panel in Fig. 1B). In contrast, similar expression of PACT in PKR−/− fibroblasts did not cause apoptosis (middle panel in Fig. 1B), demonstrating the need of PKR for this effect of PACT. As expected, when PKR was cotransfected with PACT into the same PKR−/− cells, the apoptotic response was restored (lower panel in Fig. 1B). These results demonstrated that PACT can cause PKR-mediated apoptosis in stressed cells.
FIG. 1.
PKR-dependent apoptosis by PACT. (A) PACT, but not two other dsRNA-binding proteins, causes apoptosis. TUNEL and immunofluorescence (IF) assays in HT1080 cells expressing FLAG-PACT (top panels), P76-FLAG (middle panels), and FLAG-P69 2–5(A) synthetase (bottom panels). (B) PKR is required for apoptosis by PACT. TUNEL and IF assays in wt MEFs and PKR−/− MEFs. wt MEFs (top panels) and PKR−/− MEFs (middle panels) were transfected with FLAG-PACT. Bottom panels show PKR−/− MEFs that were transfected with both FLAG-PACT and PKR. The panels labeled TUNEL show green immunofluorescence, some of which were bleached to gray during photography. The panels labeled IF show red immunofluorescence detecting the presence of the FLAG-tagged proteins. Independent photographs of the same cells, with different filters, are shown.
TABLE 2.
Quantitation of apoptotic cells expressing PACT mutants
| Expressed protein | No. of TUNEL-positive cellsa |
|---|---|
| wt PACT | 237 |
| Δ1 | 240 |
| Δ2 | 240 |
| Δ3 | 0 |
| Δ1,2 | 29 |
| DD | 0 |
| DD-Δ1,2 | 198 |
| DD-D3 | 0 |
| Δ2DD-Δ1,2 | 0 |
| Δ2DD-D3 | 0 |
For each protein, 300 protein-expressing cells were scored for each TUNEL signal. The nonspecific background was 18 cells, a amount which has been subtracted from all values.
PKR and dsRNA-binding by PACT deletion mutants.
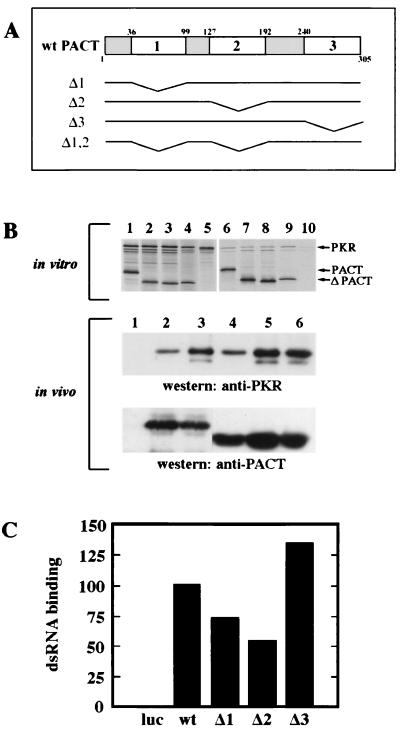
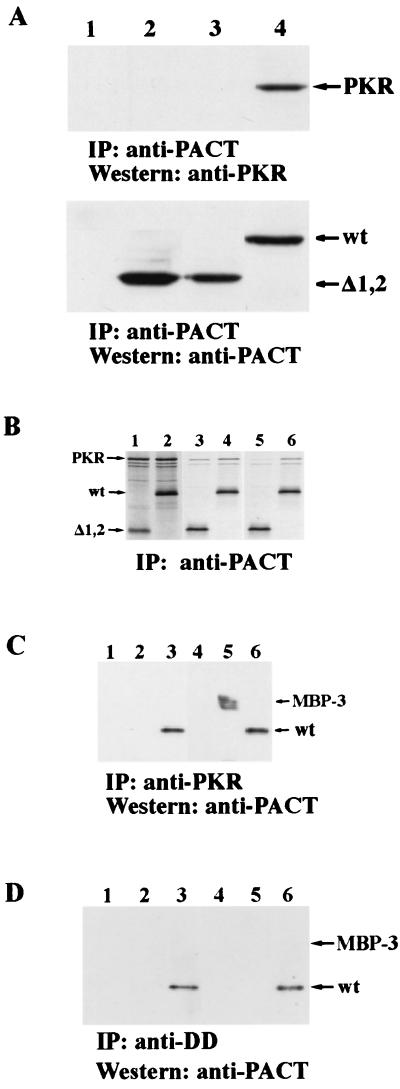
To begin a series of investigations on PACT structure and function, we generated three deletion mutants, Δ1, Δ2, and Δ3, that are missing domains 1, 2, and 3, respectively (Fig. 2A). These three domains were previously identified by comparing the amino acid sequence of PACT with those of other dsRNA-binding and PKR-interacting proteins (20). Domains 1 and 2 of PACT have strong sequence homology with the dsRNA-binding motif present in those proteins. We have previously shown that the same motif of PKR also mediates homomeric and heteromeric protein-protein interactions. Domain 3 of PACT, however, has only weak homology with this motif. The functions of the deletion mutants of PACT and the wt protein were tested in a series of experiments. To test their ability to bind to PKR, the proteins were translated in vitro and mixed with in vitro-translated PKR (upper panel in Fig. 2B). When PACT or its mutants was immunoprecipitated from these mixtures, PKR was coprecipitated with them, as shown by the appearance of the uppermost band in lane 6 to 9. There was no PKR band in lane 10, confirming the specificity of the immunoprecipitation procedure (upper panel in Fig. 2B). Quantitation of the data revealed that 22% of the input PKR was bound to wt PACT, 12% was bound to Δ1, 15% was bound to Δ2, and 30% was bound to Δ3. These results demonstrated that all three deletion mutants of PACT were capable of interacting with PKR in vitro. Similar interactions in vivo were confirmed by cotransfecting PACT or its mutant with an enzymatically inactive PKR mutant (middle panel in Fig. 2B). In this experiment, PACT was immunoprecipitated using its FLAG tag, and coimmunoprecipitation of PKR was detected by Western blotting of the precipitates. When only PKR was transfected, nothing was precipitated by the Flag-antibody (lane 1, middle panel, in Fig. 2B). But when only PACT was transfected, endogenous PKR coprecipitated with transfected PACT (lane 2). More PKR was precipitated with wt PACT when PKR was cotransfected (lane 3). But most importantly, PKR was coprecipitated with all three deletion mutants of PACT (lanes 4 to 6, middle panel, in Fig. 2B). That similar levels of PACT and its mutants were expressed in the cells and immunoprecipitated was confirmed by Western blotting of the precipitates with anti-FLAG antibody (lower panel in Fig. 2B).
FIG. 2.
PKR and dsRNA binding by deletion mutants of PACT. (A) Maps of human wt PACT protein and deletion constructs. wt PACT contains three putative dsRNA-protein or protein-protein interaction domains indicated by the large numbers 1, 2, and 3. Small numbers indicate the amino acid residue number. Δ1 is missing PACT amino acid residues 35 to 99; Δ2 is missing PACT amino acid residues 127 to 192; Δ3 is missing PACT amino acid residues 240 to 305; Δ1,2 is missing PACT amino acid residues 35 to 99 and 127 to 192. (B) PACT and its mutant proteins each interact with PKR in vitro and in vivo. In vitro and in vivo coimmunoprecipitation of PKR with FLAG-tagged PACT and its mutants. (In vitro) 35S-labeled PKR (K296R), FLAG-wt PACT, and FLAG-PACT mutants were synthesized independently. A total of 3 μl of the reticulocyte lysate containing PKR (K296R) was mixed with 3 μl of the lysates containing FLAG-wt PACT, FLAG-Δ1, FLAG-Δ2, FLAG-Δ3. PACT was immunoprecipitated using anti-FLAG (M2) agarose, and the proteins coimmunoprecipitating with it were analyzed. Lanes 1 to 5 show all proteins in the mixture before immunoprecipitation, and lanes 6 to 10 represent immunoprecipitated proteins. All lanes contain PKR (K296R). Lanes 1 and 6, wt PACT; lanes 2 and 7, Δ1, lanes 3 and 8, Δ2; lanes 4 and 9, Δ3; lanes 5 and 10, only PKR. (In vivo) Coimmunoprecipitation of PKR in transfected HT1080 cells with anti-FLAG agarose as described in Materials and Methods. A total of 5 μg each of CMV-PKR (K296R) and FLAG-tagged PACT construct was transfected unless indicated otherwise below. Lane 1, PKR (K296R) alone (10 μg, transfected); lane 2, wt PACT alone (10 μg, transfected); lane 3, PKR and wt PACT; lane 4, PKR and Δ1; lane 5, PKR and Δ2; lane 6, PKR and Δ3. (C) PACT and its mutant proteins bind dsRNA. wt PACT and its mutant proteins were tested for poly(I-C) agarose binding activity as described in Materials and Methods. Equal amounts of the total translation mix were loaded for all samples. PhosphorImager analysis was done to quantify the binding activity. The fraction of bound protein was calculated as the radioactivity in the bound protein band/total radioactivity assayed. The dsRNA binding of wt PACT was considered 100%, and values for other PACT mutant proteins or luciferase are presented as a percentage of that value. Forty percent of input wt PACT bound to the resin.
Because PACT can also bind dsRNA, we were interested in measuring the abilities of the different in vitro-translated deletion mutants to bind dsRNA. As shown in Fig. 2C, all three mutants could efficiently bind dsRNA under conditions in which no luciferase bound to it. The Δ1 and Δ2 mutants were slightly deficient in binding compared to the wt protein, whereas the Δ3 protein was more efficient. These results suggested that the presence of none of the three domains is absolutely required for dsRNA binding and that domains 1 and 2 may both mediate this process.
Functional analysis of the deletion mutants.
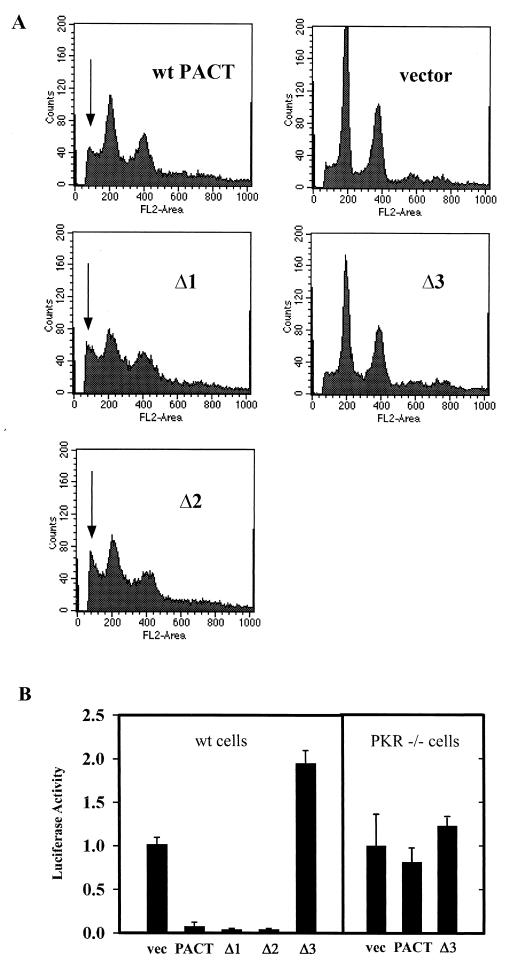
Although the three deletion mutants of PACT could all bind PKR or dsRNA, there were surprising differences in their functional properties. Their cellular effects were tested by measuring apoptosis upon transfecting them (Fig. 3A). Apoptosis was measured by analyzing the whole population of cells by FACS according to cellular DNA contents. In vector-transfected cells, most of the cells were in G0/G1 and G2 peaks with diploid or tetraploid DNA contents. But in wt PACT transfected cells, these peaks were reduced, and many cells with subdiploid DNA contents, because of apoptotic DNA degradation, appeared (as indicated by the arrow in Fig. 3). Even more enhanced apoptosis was observed with the Δ1 and Δ2 mutants. In contrast, the Δ3 mutant was totally inert and did not cause apoptosis (Fig. 3A). The same conclusions were confirmed by TUNEL assays (Table 2). These data indicated that the Δ3 mutant was nonfunctional, although it could bind to PKR (Fig. 2).
FIG. 3.
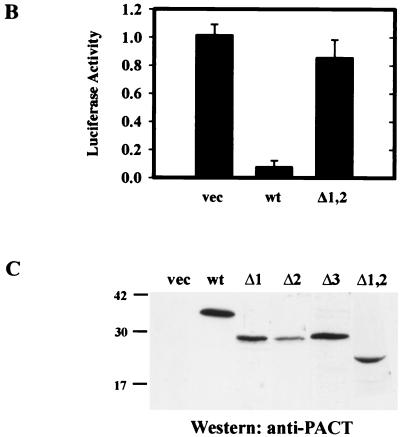
Induction of apoptosis and translation inhibition by PACT deletion mutants. (A) Effect of PACT and its mutants on apoptosis. HT1080 cells transfected with wt PACT, Δ1, Δ2, Δ3, or pcDNA3 vector were analyzed for presence of sub-G1 peak by fluorescence monitoring in a flow cytometer. The x axis represents the fluorescence intensity; the y axis represents the cell number. The arrows point to cells in sub-G1, indicating cells undergoing apoptosis. (B) Effect of PACT deletion mutants on inhibition of translation in vivo. HT1080 cells were transfected with normalizing β-galactosidase reporter, luciferase reporter, and different PACT expression constructs, as indicated. Cell extracts were assayed for β-galactosidase and luciferase activities. Normalized luciferase activities are presented. The error bars represent the standard error calculated from six independent values. vec, pcDNA vector; PACT, wt PACT. The assays in the left block were done in wt cells, and those in the right block were done in PKR−/− MEFs.
In vivo activation of PKR can be measured by a translation inhibition assay as well. We have previously shown by this assay that PACT can block the translation of a cotransfected reporter protein, luciferase. When the deletion mutants were tested by this assay for their functions, the Δ1 and Δ2 mutants were slightly more efficient than the wt protein in blocking luciferase synthesis (Fig. 3B). In contrast, the Δ3 mutant enhanced its translation, probably by functioning as a dominant-negative mutant. That these effects were mediated by PKR was established by marginal effects of wt or Δ3 PACT on luciferase translation in PKR−/− cells (Fig. 3B). The levels of luciferase mRNA were the same in cells transfected with vector, wt PACT, or mutant PACT (data not shown), indicating that the observed effects were at the level of translation of the luciferase protein. Similar levels of the transfected proteins were expressed (see Fig. 5C).
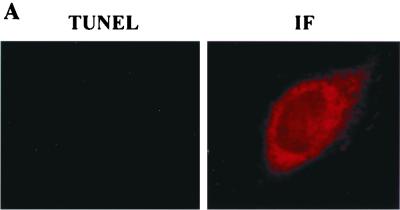
FIG. 5.
Failure of PKR activation in vivo by domain 3. (A) Δ1,2 cannot cause apoptosis in vivo. TUNEL or immunofluorescence (IF) assay in HT1080 cells expressing FLAG-Δ1,2. (B) Δ1,2 cannot inhibit translation in vivo. Procedures were as described in Fig. 3B. The error bars represent the standard error calculated from six independent values. (C) PACT-related protein levels in the transfected cells of Fig. 3B and 5B.
Activation of PKR in vitro by domain 3 alone.
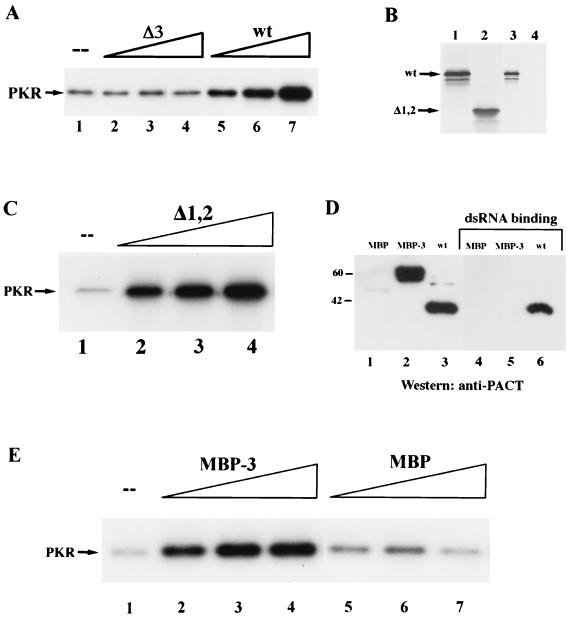
The above in vivo data indicate that domain 3 of PACT was required for its ability to activate PKR. This conclusion was directly tested in the experiment shown in Fig. 4A. wt PACT and Δ3 PACT were expressed as polyhistidine-tagged proteins in E. coli and purified to homogeneity by affinity chromatography. The ability of these proteins to activate PKR in vitro was tested by monitoring PKR autophosphorylation. The addition of increasing amounts of the wt protein activated PKR increasingly, whereas the same amounts of Δ3 protein had no effect on PKR autophosphorylation. These results, combined with those shown in Fig. 3, demonstrated conclusively that the presence of domain 3 was necessary for PACT-mediated PKR activation.
FIG. 4.
Activation of PKR in vitro by PACT and its mutants. (A) Effects of purified bacterially expressed wt PACT and Δ3 on PKR activation in vitro. Lane 1, activity buffer; lanes 2 and 5, 1 nmol of purified protein; lanes 3 and 6, 10 nmol of purified protein; lanes 4 and 7, 100 nmol of purified protein. (B) Δ1,2 cannot bind dsRNA. wt PACT and its mutant protein Δ1,2 were tested for poly(I-C)-agarose binding activity as described in Materials and Methods. Lanes 1 and 2 show the in vitro-translated proteins before poly(I-C) binding; lanes 3 and 4 represent dsRNA-binding proteins. Lanes 1 and 3, wt PACT; lanes 2 and 4, Δ1,2. (C) Δ1,2 activates PKR. The effect of increasing concentrations of purified bacterially expressed Δ1,2 on PKR activation in vitro was tested. Lane 1, activity buffer; lane 2, 1 nmole of Δ1,2 protein; lane 3, 10 nmol of Δ1,2 protein; lane 4, 100 nmol of Δ1,2 protein. (D) MBP-3 cannot bind dsRNA. MBP, MBP-3, and wt PACT were tested for poly(I-C)-agarose binding activity. Equal amounts of purified MBP, MBP-3, or wt PACT were bound to poly(I-C)-agarose beads. The input proteins and the proteins which remained bound to the beads after washing were analyzed by SDS-PAGE followed by Western blotting with anti-PACT polyclonal antibody. Lanes 1 to 3 show proteins before dsRNA binding, and lanes 4 to 6 represent proteins which bound dsRNA. Lanes 1 and 4, MBP (1 μg); lanes 2 and 5, MBP-3 (1 μg); lanes 3 and 6, wt PACT (1 μg). (E) MBP-3, but not MBP, activates PKR. The effect of purified bacterially expressed MBP and MBP-3 on PKR activation in vitro was tested. Lane 1, activity buffer, lanes 2 to 4, MBP-3; lanes 5 to 7, MBP; lanes 2 and 5, 1 nmol of purified protein; lanes 3 and 6, 10 nmol of purified protein; lanes 4 and 7, 100 nmol of purified protein.
Once the need for domain 3 in PACT functions was established, we asked whether domain 3, by itself, is sufficient for activating PKR. Because domain 3 consists of only 66 residues, we expressed it as fusion proteins. One such protein, Δ1,2, was a PACT derivative devoid of domains 1 and 2 (see Fig. 2A). It contained not only domain 3 but also the end and the linker regions in between the different domains. The Δ1,2 protein was translated in vitro and tested for dsRNA binding. As anticipated from Fig. 2C, Δ1,2 did not bind dsRNA (Fig. 4B). The Δ1,2 protein was then expressed in E. coli and purified to homogeneity. This protein was very efficient in activating PKR in a dose-dependent manner (Fig. 4C), demonstrating that under the in vitro activation conditions, the presence of domains 1 and 2 was not necessary for PACT's activity. The sufficiency of domain 3 was more rigorously tested by transplanting it to a heterologous context. An MBP-domain 3 fusion protein containing only the 66 residues of PACT domain 3 (MBP-3), was expressed in E. coli and purified. This protein, but not parental MBP, reacted with PACT antibody (Fig. 4D, lanes 1 to 3). However, it did not bind to dsRNA as the wt PACT had done (Fig. 4D, lanes 4 to 6). The MBP-3 protein, however, activated PKR very efficiently, whereas MBP by itself did not (Fig. 4E, lanes 1 to 7). The experiments shown in Fig. 4 demonstrated that domain 3 of PACT is not only necessary but is also sufficient for activating PKR in vitro.
Failure of PKR activation in vivo by domain 3.
Although Δ1,2 could activate PKR in vitro efficiently, its expression in vivo did not cause massive apoptosis (Fig. 5A). Quantitation revealed that only 10% of cells were TUNEL positive, as against 80% positivity by wt PACT (Table 2). Similarly, it did not appreciably inhibit the translation of luciferase reporter protein (Fig. 5B). We wondered whether this failure was due to an inefficient binding of Δ1,2 to PKR under physiological conditions. In vivo interaction of the two proteins was measured by their coimmunoprecipitation from extracts of cells expressing transfected proteins. As expected, PKR was precipitated, along with wt PACT (lane 4 in Fig. 6A), but neither endogenous PKR (lane 2 in Fig. 6A) nor transfected PKR (lane 3 in Fig. 6A) precipitated along with Δ1,2 PACT, demonstrating the lack of interaction between PKR and Δ1,2 in vivo. In vitro interaction assays (Fig. 6B and C) established that binding of Δ1,2 PACT or MBP-3 to PKR is weak and that it cannot withstand physiological salt concentrations. Under low-salt conditions similar to those used for PKR activation assays in vitro, Δ1,2 bound to PKR but less well than wt PACT (Fig. 6B, lanes 3 and 4). Quantitation of the bands showed that 35% of input PKR was bound to wt PACT and 22% to Δ1,2 under these conditions. Under physiological salt conditions, virtually no PKR was bound to Δ1,2, although 25% of it still bound to wt PACT (note the PKR bands in lanes 5 and 6 in Fig. 6B). Similar results were obtained with bacterially expressed MBP-3 mixed with human cell extracts containing PKR. wt PACT, but not MBP-3, coimmunoprecipitated with PKR under physiological salt conditions (Fig. 6C, lanes 1 to 3), whereas under low-salt conditions, both wt PACT and MBP-3 coimmunoprecipitated with PKR (Fig. 6C, lanes 5 and 6). These results demonstrated that although domain 3 could bind and activate PKR under low-salt conditions used for PKR-activating assays in vitro, it activated PKR poorly in vivo because of its failure to bind to PKR strongly under isotonic conditions. Our previous results indicated that wt PACT interacted with the DD of PKR (20). When interactions of purified DD with wt PACT and MBP-3 were tested, even under low-salt conditions only the full-length PACT bound to DD (Fig. 6D), indicating that the weak interaction of domain 3 with PKR is mediated by a region other than DD of PKR.
FIG. 6.
Weak interactions of domain 3 with PKR at a physiological salt concentration. (A) Failure of Δ1,2 to interact with PKR in vivo. Coimmunoprecipitation of PKR in transfected HT1080 cells with anti-FLAG agarose as described in Materials and Methods. Lane 1, PKR; lane 2, Δ1,2; lane 3, PKR and Δ1,2; lane 4, PKR and wt PACT. (B) Δ1,2 interacts weakly with PKR under physiological salt concentrations in vitro. 35S-labeled PKR and FLAG-Δ1,2 mutant or FLAG-wt PACT were synthesized independently in vitro. A total of 3 μl of the reticulocyte lysate containing PKR was mixed with 3 μl of the lysates containing wt PACT or Δ1,2. PACT was immunoprecipitated from the lysate using anti-FLAG (M2) agarose in high (physiological)-salt buffer or low-salt buffer, and the proteins coimmunoprecipitating with it were analyzed. Lanes 1 and 2 show all proteins in the mixture before immunoprecipitation, and lanes 3 to 6 represent immunoprecipitated proteins. Proteins in lanes 3 and 4 were immunoprecipitated and washed in low-salt buffer, while proteins in lanes 5 and 6 were immunoprecipitated and washed in high-salt buffer. Lanes 1, 3, and 5, PKR and Δ1,2; lanes 2, 4, and 6, PKR and wt PACT. (C) MBP-3 can be coimmunoprecipitated with PKR in low-salt but not in high-salt conditions. MBP, MBP-3, and wt PACT were tested for PKR binding in conditions of low or high salt. Equal amounts of purified MBP, MBP-3, or wt PACT were added to an extract from HT1080 cells treated with 1,000 U of IFN β per ml for 24 h. PKR was immunoprecipitated with anti-PKR monoclonal antibody and washed in conditions of high or low salt. The proteins which remained bound to the beads after washing were analyzed by Western blotting with anti-PACT polyclonal antibody. Lanes 1 to 3 show immunoprecipitated proteins in high salt conditions, and lanes 4 to 5 show immunoprecipitated proteins in low-salt conditions. Lanes 1 and 4, MBP (1 μg); lanes 2 and 5, MBP-3 (1 μg); lanes 3 and 6, wt PACT (1 μg). (D) MBP-3 cannot bind to DD. The conditions were the same as for panel C, except that purified DD was used instead of cell extracts containing PKR. The same PKR antibody immunoprecipitated DD and its associated proteins.
Restoration of PKR activation in vivo by domain 3 upon its attachment to a heterologous PKR-interacting domain.
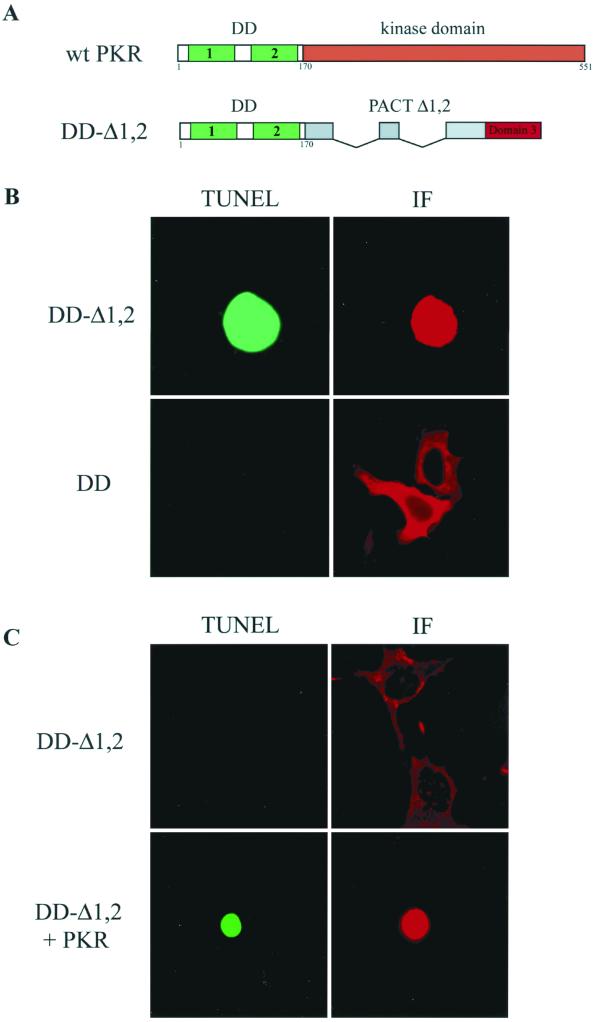
The experimental results presented above suggested that domain 3 can activate PKR but can do so in vivo only if domain 1 or 2 mediates a strong binding to PKR. We argued that if this were the case, a heterologous PKR-interacting domain, when attached to domain 3, should be able to activate PKR in vivo. To test this idea, a fusion protein containing the DD of PKR and Δ1,2 of PACT was generated (Fig. 7A). The dimerization domain of PKR located at its amino terminus is known to mediate PKR-PKR interaction in a dsRNA-independent fashion (21, 22). The DD-Δ1,2 protein, when expressed in HT1080 cells, caused strong apoptosis, while the DD protein itself was ineffective (Fig. 7B and Table 2). These data, taken together with results presented previously, demonstrated that the two portions of the fusion protein could not cause apoptosis separately, but when expressed together as parts of the same protein, they were effective in vivo (Table 2, Fig. 5 and 7). The apoptotic effect of DD-Δ1,2 was mediated by PKR because it was ineffective in PKR−/− cells unless PKR was cotransfected with it (Fig. 7C).
FIG. 7.
Apoptosis by DD-Δ1,2. (A) Maps of wt PKR and PKR-PACT hybrid. DD-Δ1,2 contains PKR amino acid residues 1 to 170 tethered to PACT Δ1,2. PKR contains two dsRNA-dimerization motifs indicated by the large numbers 1 and 2. The small numbers indicate the amino acid residue number. (B) TUNEL or immunofluorescence (IF) assay was performed in HT1080 cells expressing FLAG-DD-Δ1,2 (top panels) and FLAG-DD (bottom panels). (C) TUNEL or immunofluorescence (IF) assay in PKR−/− MEFs transfected with FLAG-DD-Δ1,2 (top panels) and FLAG-DD-Δ1,2 plus PKR (bottom panel).
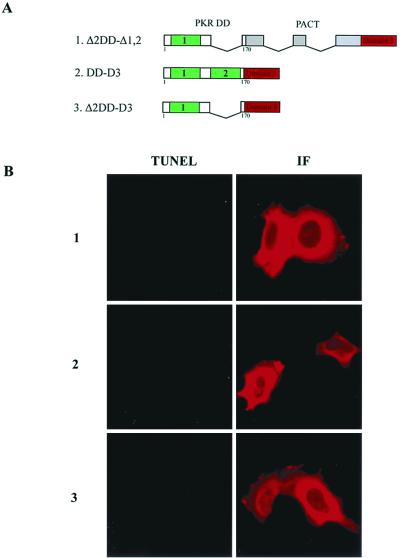
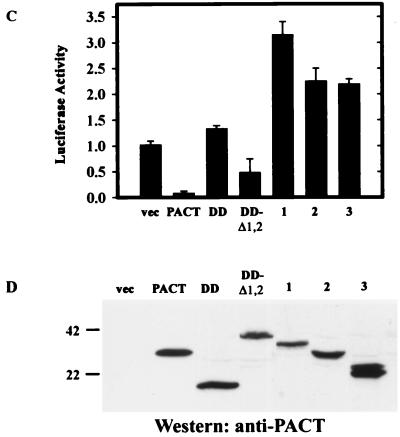
The DD of PKR contains two motifs, both of which are needed for the functional property of this domain (11, 21, 24). As anticipated, when motif 2 of the DD was deleted, the resulting fusion protein, Δ2DD-Δ1,2, did not cause apoptosis (Fig. 8B, Table 2). Surprisingly, elimination of the linker regions of PACT from the DD-Δ1,2 (see Fig. 2A) also inactivated the protein: when the DD was attached to only domain 3, the resulting protein, DD-D3, did not cause apoptosis (Fig. 8B, Table 2). Similarly, Δ2DD-D3 was also inactive (Fig. 8B, Table 2). When tested in the translation inhibition assay, DD was neutral, but DD-Δ1,2 was strongly inhibitory (Fig. 8C). In contrast, the three mutants, 1, 2, and 3 (Fig. 8A), stimulated translation (Fig. 8C). The levels of expression of the different proteins were comparable (Fig. 8D). Thus, it is conceivable that mutants 1, 2, and 3 had dominant-negative effects on the inhibition of luciferase synthesis. These results suggest that, for the appropriate functioning of domain 3, not only does the domain require an additional strong PKR-interacting domain but the spacing of the two domains is also critical for PKR activation.
FIG. 8.
Failure of PKR activation in vivo by PKR-domain 3 mutants. (A) Maps of PKR-PACT hybrid mutants. Δ2DD-Δ1,2 contains PKR amino acid residues 1 to 99 and 166 to 170 tethered to PACT Δ1,2. DD-D3 contains PKR amino acid residues 1 to 170 tethered to PACT domain 3. Δ2DD-D3 contains PKR amino acid residues 1 to 99 and 166 to 170 tethered to PACT domain 3. (B) Failure of PKR-domain 3 mutants to cause apoptosis in vivo. TUNEL or immunofluorescence (IF) assay in HT1080 cells expressing FLAG-Δ2DD-Δ1,2 (top panels), FLAG-DD-D3 (middle panels), and FLAG-Δ2DD-D3 (bottom panels). (C) Dominant inhibitor effects of PKR-domain 3 mutants on translation in vivo. HT1080 cells were transfected with luciferase reporter and different DD constructs, as indicated, and assayed for luciferase activity after normalizing for β-galactosidase activities. The error bars represent the standard error calculated from six independent values. vec, pcDNA3 vector; PACT, wt PACT; DD, the DD of PKR (residues 1 to 170); DD-Δ1,2, Δ1,2 attached to DD; 1, mutant Δ2DD-Δ1,2; 2, mutant DD-D3; 3, mutant Δ2DD-D3. (D) Expression levels of PACT related proteins.
DISCUSSION
PACT was discovered as a PKR-interacting protein and shown to activate PKR in vitro and in vivo (13, 20). Although both PKR and PACT contain typical dsRNA-binding domains, this activation is dsRNA independent. One of the consequences of PKR activation in vivo is the triggering of cellular apoptosis (7, 32). Extracellular stimuli such as virus infection, dsRNA, the addition or removal of specific growth factors, lipopolysaccharide, or Ca2+ have been shown to activate cellular PKR and cause apoptosis. That PKR is the crucial mediator of the observed apoptosis was established by using dominant-negative mutants of PKR and more convincingly by demonstrating that the apoptotic effects of the extracellular stimuli were abrogated in PKR−/− cells. Moreover, overexpression of PKR, either by transfection or by a vaccinia virus vector, caused apoptosis without any deliberate stimulus (7, 16). However, controlled PKR expression using an inducible promoter required a stimulus for causing apoptosis (1, 8). The exact mechanism of PKR-mediated induction of apoptosis remains to be elucidated. Although translational inhibition caused by PKR-mediated eIF-2α phosphorylation and altered gene expression as a result of NFκB activation by PKR may participate in this process, their specific contributions to apoptosis have yet to be evaluated (10, 29). In one study, PKR activation of the FADD-mediated pathway has been demonstrated, and cells deficient in this pathway were protected from PKR-induced apoptosis (1).
In many of the experimental systems cited above, what activates PKR in the cells has remained an enigma, especially in the cases where synthetic or viral dsRNA was not involved. In this context, PACT is an attractive candidate for cellular activator of PKR because both proteins are present in all cells at low levels. Thus, overexpression of either partner may trigger their interaction. This was indeed the case as shown here. Overexpression of PACT, but not other dsRNA-binding proteins, caused cellular apoptosis (Fig. 1). One of these proteins, P76, also interacts with PKR and gets phosphorylated by it (25). However, unlike PACT, P76 cannot activate PKR. Overexpression of PACT by itself, however, did not cause apoptosis (data not shown). It required application of additional stress to the cells, achieved in this study by a low level of actinomycin D treatment. The PACT-mediated apoptosis was observed in many human and mouse normal and tumor cell lines which express PKR. In the absence of PKR, PACT did not cause apoptosis, demonstrating that PKR activation is the crucial cellular function of PACT (Fig. 1). Our results established that, in our experimental setting, PACT, PKR, and extracellular stress were all required for apoptosis.
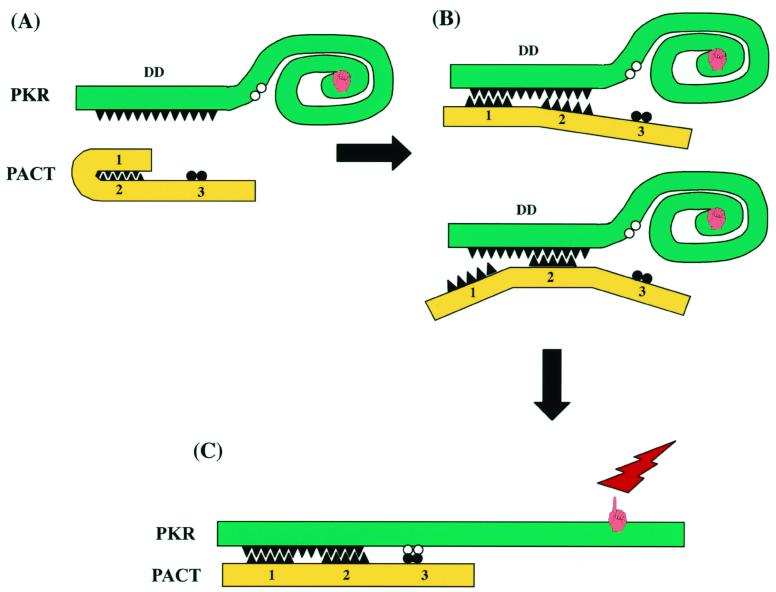
The main focus of this investigation was to define the domains of PACT that are required for PKR interaction and activation. It was already suspected that just the binding of another protein to the DD of PKR is not sufficient for its activation because the DD of PKR itself, TRBP, or P76, all of which bind to PKR, does not activate PKR (18, 25). Thus, it was anticipated that PACT contains an additional PKR activation domain. This domain has now been identified. We previously suggested, as indicated by sequence homology, that domains 1 and 2 of PACT mediate both protein-protein and protein-dsRNA interactions (20). The role of domain 3, which has limited homology with the dsRNA-binding motif, was unclear. The same was true for the intervening spacer regions in between the three domains (Fig. 2A). The results presented here confirmed that the absence of either domain 1 or domain 2 by itself does not inhibit the strong interactions with PKR in vitro and in vivo. The same was true for dsRNA binding. Domain 3, on the other hand, could not bind dsRNA (Fig. 4B and D). It could bind PKR, but only weakly (Fig. 6). Unlike domains 1 and 2, however, domain 3 was absolutely required for PKR activation (Fig. 4) and for causing translation inhibition and apoptosis (Fig. 3). Domain 3 was also sufficient for activating PKR in vitro (Fig. 4). Its failure to properly activate PKR in vivo (Fig. 5) was attributed to its inability to bind PKR under physiological salt concentrations (Fig. 6). When PKR binding was restored by attaching domain 3 to the DD of PKR, its ability to activate PKR in vivo was restored (Fig. 7). These results have led us to formulate a working model for PKR activation by PACT (Fig. 9). Inactive PKR contains the DD at its amino terminus. Binding of dsRNA to this region is known to change the conformation of the protein so that the ATP-binding site is exposed and the protein acquires enzymatic activity (2, 3). We propose that a similar activation process requires two sets of interactions in the case of PACT. Either domain 1 or domain 2 can direct PACT's binding to the DD of PKR (Fig. 9B). However, this binding in itself is not sufficient for PKR activation. Domain 3 needs to interact with PKR as well to change its conformation (Fig. 9C). The interaction of domain 3 with PKR is not mediated by DD, as shown in Fig. 6D, but by an as-yet-unidentified locus in the PKR kinase domain. Although the PKR-domain 3 interaction is sufficient for PKR activation under conditions that allow weak interaction of proteins, at physiological salt concentrations, the domain 3-PKR interaction is not strong enough, and some other domain has to provide the PKR binding function. However, note that even the weak interaction of Δ1,2 with PKR in vivo caused some, albeit minor level of apoptosis (Table 2) and inhibition of translation (Fig. 5B).
FIG. 9.
Model of PACT binding and activation of PKR. (A) PKR contains the DD at its amino terminus and the kinase domain at its carboxyl terminus. PACT has three domains: DDs 1 and 2 can possibly interact with each other. (B) Either domain 1 or domain 2 can interact with PKR DD by itself. This strong interaction of PACT with PKR is needed in vivo. (C) Domain 3 of PACT binds to an undefined site of PKR weakly and changes its conformation, leading to PKR activation.
Domains 1 and 2 of PACT have the potential to interact with each other (Fig. 9A). Such an intramolecular interaction should decrease the ability of PACT to bind PKR and activate it. If this were true, removal of either domain 1 or 2 from PACT should increase its ability for PKR activation. There is a hint that this could be the case: both Δ1 and Δ2 proteins were slightly more potent than wt PACT in inhibiting translation in vivo and causing apoptosis (Fig. 3). The in vitro properties of these proteins could not be tested because the proteins were not expressed in bacteria in a soluble form (data not shown). It is conceivable that the putative interaction between domains 1 and 2 in vivo can be disrupted by posttranslational modification of PACT. Indeed, Ito et al. (13) have reported that stress-kinase-mediated phosphorylation of RAX, the mouse PACT, increases its affinity for PKR. Thus, we hypothesize that domains 1 and 2 are not only required for PACT binding to PKR in vivo but may also modulate the efficiency of this process through their mutual interactions.
How domain 3 activates PKR remains a major question. The first step in that understanding will require mapping the domain of PKR with which it interacts. In Fig. 9 we have placed this hypothetical site in the C-terminal domain of PKR because MBP-3 failed to bind DD (Fig. 6D). This scenario is supported by the fact that heparin can bind and activate PKR deletion mutants which cannot bind dsRNA (23). Thus, it is conceivable that PKR can be activated upon binding of activators, such as heparin or domain 3 of PACT, to a site distinct from the dsRNA-binding site. In vivo such binding, however, needs to be stabilized by the strong binding of domain 1 or domain 2. Because the DD of PKR could substitute functionally for domain 1 or domain 2 (Fig. 7), we concluded that mediating strong PKR interaction is the only function of PACT domains 1 and 2. As expected, deletion of motif 2 of the DD of PKR in the hybrid DD-DD-Δ1,2 protein eliminated its function because such a deletion is known to affect the dimerization property negatively. More surprising was the observation that DD-D3 was inactive, whereas DD-Δ1,2 was not (Fig. 8). This indicates that the spacer regions of PACT (Fig. 2A) have functional roles. As shown by the results in Fig. 4E with MBP-3, which does not contain the spacer regions, these regions are not required for the ability of domain 3 to activate PKR in vitro. It is conceivable that in vivo they serve as linkers and separate the two PKR-interacting domains, domains 1 and 2 and domain 3, appropriately so that each can interact with the two cognate sites of the PKR protein (Fig. 9). When they are too close, as in DD-D3, although the protein has the potential to interact with either site of PKR, it is spatially constrained to interact with both at the same time. Future experiments with additional mutants of DD-Δ1,2 or Δ1 PACT can test this space-constraining model.
ACKNOWLEDGMENTS
We thank Judy Drazba for advice on fluorescence microscopy, Theresa Rowe and Fahima Rahman for experimental assistance, and Karen Toil for secretarial help.
This work was supported by National Institutes of Health grants CA-62220 and CA-68782 to G.C.S. G.A.P. was supported, in part, by Training Grant DK-07678.
REFERENCES
- 1.Balachandran S, Kim C N, Yeh W C, Mak T W, Bhalla K, Barber G N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff J R, Samuel C E. Mechanism of interferon action. The interferon-induced phosphoprotein P1 possesses a double-stranded RNA-dependent ATP-binding site. J Biol Chem. 1985;260:8237–8239. [PubMed] [Google Scholar]
- 3.Carpick B W, Graziano V, Schneider D, Maitra R K, Lee X, Williams B R G. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J Biol Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 4.Chu W M, Ostertag D, Li Z W, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 5.Clemens M J, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 6.Cuddihy A R, Wong A H, Tam N W, Li S, Koromilas A E. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene. 1999;18:2690–2702. doi: 10.1038/sj.onc.1202620. [DOI] [PubMed] [Google Scholar]
- 7.Der S D, Yang Y L, Weissmann C, Williams B R. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donze O, Dostie J, Sonenberg N. Regulatable expression of the interferon-induced double-stranded RNA dependent protein kinase PKR induces apoptosis and fas receptor expression. Virology. 1999;256:322–329. doi: 10.1006/viro.1999.9618. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh A, Sarkar S, Sen G. Cell growth regulatory and anti-viral effects of the P69 isozyme of 2–5(A) synthetase. Virology. 2000;266:319–328. doi: 10.1006/viro.1999.0085. [DOI] [PubMed] [Google Scholar]
- 10.Gil J, Alcami J, Esteban M. Induction of apoptosis by double-stranded-RNA-dependent protein kinase (PKR) involves the alpha subunit of eukaryotic translation initiation factor 2 and NF-κB. Mol Cell Biol. 1999;19:4653–4663. doi: 10.1128/mcb.19.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green S R, Mathews M B. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 12.Iordanov M S, Paranjape J M, Zhou A, Wong J, Williams B R, Meurs E F, Silverman R H, Magun B E. Activation of p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito T, Yang M, May W S. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J Biol Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 14.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Yang Y L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S B, Esteban M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 17.Nanduri S, Carpick B W, Yang Y, Williams B R, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park H, Davies M V, Langland J O, Chang H W, Nam Y S, Tartaglia J, Paoletti E, Jacobs B L, Kaufman R J, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci USA. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel R C, Sen G C. Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J Biol Chem. 1992;267:7671–7676. [PubMed] [Google Scholar]
- 20.Patel R C, Sen G C. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel R C, Sen G C. Requirement of PKR dimerization mediated by specific hydrophobic residues for its activation by double-stranded RNA and its antigrowth effects in yeast. Mol Cell Biol. 1998;18:7009–7019. doi: 10.1128/mcb.18.12.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel R C, Stanton P, McMillan N M, Williams B R, Sen G C. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel R C, Stanton P, Sen G C. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J Biol Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 24.Patel R C, Stanton P, Sen G C. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J Biol Chem. 1996;271:25657–25663. doi: 10.1074/jbc.271.41.25657. [DOI] [PubMed] [Google Scholar]
- 25.Patel R C, Vestal D J, Xu Z, Bandyopadhyay S, Guo W, Erme S M, Williams B R, Sen G C. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 26.Pogulis R J, Vallejo A N, Pease L R. In vitro recombination and mutagenesis by overlap extension PCR. In: Trower M K, editor. In vitro mutagenesis protocols. Vol. 57. Totowa, N.J: Humana Press, Inc.; 1996. pp. 167–176. [DOI] [PubMed] [Google Scholar]
- 27.Romano P R, Garcia-Barrio M T, Zhang X, Wang Q, Taylor D R, Zhang F, Herring C, Mathews M B, Qin J, Hinnebusch A G. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol Cell Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryter J M, Schultz S C. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava S P, Kumar K U, Kaufman R J. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- 30.St. Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan S L, Gale M J, Jr, Katze M G. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol Cell Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan S L, Katze M G. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J Interferon Cytokine Res. 1999;19:543–554. doi: 10.1089/107999099313677. [DOI] [PubMed] [Google Scholar]
- 33.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 375–399. [Google Scholar]
- 34.Williams B R. PKR, a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 35.Wong A H, Tam N W, Yang Y L, Cuddihy A R, Li S, Kirchhoff S, Hauser H, Decker T, Koromilas A E. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasukawa T, Kanei-Ishii C, Maekawa T, Fujimoto J, Yamamoto T, Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J Biol Chem. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]
- 37.Zamanian-Daryoush M, Mogensen T H, DiDonato J A, Williams B R. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol Cell Biol. 2000;20:1278–1290. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]