Abstract
This scientific commentary refers to ‘Therapeutic reversal of Huntington’s disease by in vivo self-assembled siRNAs’ by Zhang et al. (doi:10.1093/brain/awab354).
This scientific commentary refers to ‘Therapeutic reversal of Huntington’s disease by in vivo self-assembled siRNAs’ by Zhang et al. (doi:10.1093/brain/awab354).
Advances in next-generation sequencing technology have enabled the development of interventions specific to an underlying disease mechanism. This has resulted in improvements in the fields of precision oncology and pharmacogenomics and treatment for some forms of Mendelian disease.1 For neurological disorders, strategies have been developed that target the source of the disease by modifying the expression of abnormal RNA transcripts. One approach to achieving this specificity has been through the use of small interfering RNAs (siRNAs) containing a guide strand complementary to the target RNA. In this issue of Brain, Zhang and co-workers2 demonstrate the utility of using the naturally existing system of circulating exosomes to both assemble and deliver siRNA in Huntington’s disease models.
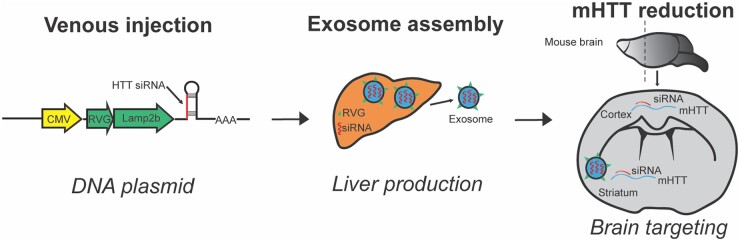
Zhang et al.2 engineered a DNA plasmid which, when incorporated into host liver tissue, allows for the production of secretory exosomes containing siRNA (Fig. 1). Specifically, siRNAs targeting mutant human huntingtin (mHTT) mRNA were generated in vivo and delivered to the brain via exosomes expressing a short peptide of rabies virus glycoprotein fused to the N-terminus of lysosome-associated membrane glycoprotein 2b (Lamp2b). After first validating the efficacy of siRNA-mediated knockdown in human embryonic kidney cells, Zhang et al.2 used fluorescence labelling to confirm that the exosomes were able to cross the blood–brain barrier and reach striatal and cortical neurons. They then tested the therapeutic application of this approach in three mouse models of Huntington’s disease, and showed a reduction in target tissue mHTT levels as well as improvements in motor performance and neuropathology.
Figure 1.
Schematic overview of exosome delivery of siRNA targeting huntingtin mRNA. The plasmid administered by venous injection contains a CMV promoter, neuron-steering RVG and huntingtin siRNA.2 When the plasmid reaches the liver the transcription of huntingtin siRNA and RVG fused to Lamp2b results in the production of RVG-labelled exosomes containing huntingtin siRNA. RVG-tagged exosomes are targeted to the brain where siRNA reduces expression of mHTT mRNA.
This technique of delivering siRNA is an exciting development for the field of precision therapeutics and provides a potential new strategy for targeting specific tissues within the body. The efficacy of siRNA administration can be reduced by processes such as nuclease-mediated degradation, renal clearance, and poor cellular uptake; the use of exosomes as a shuttle for siRNA may help mitigate these issues. The potential for exosomes to serve as vehicles for delivery of siRNA to the brain has previously been demonstrated using harvested dendritic cells3 or reprogramming of host liver tissue.4 The assembly of exosomes in vivo may allow for a more ‘natural’ system for shuttling siRNA that will be less likely to trigger the induction of inflammatory immune responses.5
Oligonucleotides do not readily cross the blood–brain barrier and require lumbar puncture for intrathecal delivery. Shuttling of exosomes to specific target tissues will allow more focused therapeutic delivery. The use of rabies virus glycoprotein to help achieve delivery of siRNA to the brain may reduce the risk of off-target effects in tissues that are not pertinent to the disease and may potentially improve the pharmacological effect in the target tissue. Moreover, this approach could facilitate dosing by allowing for peripheral administration of therapeutic agents, thereby improving patient access to care.
In the case of Huntington’s disease, targeting multiple brain regions will be important, particularly the striatum and cortex. Previous approaches using antisense oligonucleotides (ASOs) to mediate the degradation of mHTT mRNA in non-human primates resulted in lower efficiency of target reduction in the striatum compared to the cortex and spinal cord.6 Zhang et al.2 report in mouse models that the exosome delivery of siRNA resulted in similar distribution to both the cortex and striatum with comparable mHTT protein reductions observed in both locations. Additional studies will be needed to determine if exosome delivery results in robust targeting of the disease-relevant brain regions in other animal models.
One challenge in the development of targeted therapeutics is the ability to generate allele-specific knockdown that will selectively target the mutant allele without altering the normal wild-type copy of the gene.7 Strategies to achieve this may include targeting of single nucleotide polymorphisms that are frequently detected on the mutant allele.8 Zhang et al.2 show that the wild-type copy of huntingtin is unaffected in their animal studies; however, this is a result of their siRNA being targeted against the human transcript instead of the endogenous mouse transcript, suggesting that this specificity may be lost when translating the approach to humans.
In addition to allele targeting, it is also useful to consider if the therapy will target a specific cell type such as neurons, astrocytes, or oligodendrocytes. In their study, Zhang et al.2 made use of a neuron-targeting rabies virus glycoprotein tag to direct assembled exosomes to neurons expressing the nicotinic acetylcholine receptor. To assess the delivery of siRNA to neurons, the authors used the exosome strategy to deliver GFP-silencing siRNA in a transgenic mouse model ubiquitously expressing GFP. Although the knockdown of GFP should in theory be restricted to the NeuN-expressing neurons, knockdown was also observed in regions deficient of NeuN staining. This calls into question the cell-specificity of the model and raises concerns about possible off-target effects. While the delivery of siRNA to NeuN-negative cells may be helpful in targeting non-neuronal cells affected in disease, understanding the mechanism by which this occurs will be helpful in evaluating the application of this technology to other diseases in which more precise delivery is desired.
Other considerations in the development of an exosome-based therapy will be the optimization of dosing frequency and the evaluation of safety. One advantage of ASO therapies in Huntington’s disease is the stability of ASOs within the CSF and the relatively long interval between doses.9 Zhang et al.2 dose their siRNA vector using a more frequent schedule of at least once per week. They evaluated biochemical studies, peripheral blood cell counts, and tissue histology to assess the safety of their approach in mice treated with their vector every 2 days for a total of seven administrations. However, the long-term consequences of repeated administration on liver function will need to be determined. A larger volume of distribution, differences in hepatic and renal metabolism, and tissue-specific half-lives are all challenges that will have to be overcome in translation.
In conclusion, the results presented by Zhang et al.2 show the utility of exosome-guided delivery of siRNA in several mouse models of Huntington’s disease. The paradigm of using the host liver tissue to direct exosomes to the nervous system is intriguing and may be applicable to other neurological diseases. The use of exosomes to deliver therapy is also being developed in other fields of medicine including cancer immunotherapy.10 This work highlights a new strategy for delivering siRNA to the brain and a potential additional therapy to be tested in Huntington’s disease. Success of this approach would open the door for delivery of a diverse array of therapies to the nervous system.
Funding
This work was supported by intramural research funding from the National Institute of Neurological Disorders and Stroke, NIH.
Competing interests
The authors report no competing interests.
References
- 1. Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17(9):507–522. [DOI] [PubMed] [Google Scholar]
- 2. Zhang L, Wu T, Shan Y, et al. Therapeutic reversal of Huntington’s disease by in vivo self-assembled siRNAs. Brain. 2021;144(11):3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. [DOI] [PubMed] [Google Scholar]
- 4. Fu Z, Zhang X, Zhou X, et al. In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res. 2021;31(6):631–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alshaer W, Zureigat H, Al Karaki A, et al. siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur J Pharmacol. 2021;905:174178. [DOI] [PubMed] [Google Scholar]
- 6. Kordasiewicz HB, Stanek LM, Wancewicz EV, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74(6):1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reiner A, Dragatsis I, Zeitlin S, Goldowitz D.. Wild-type huntingtin plays a role in brain development and neuronal survival. Mol Neurobiol. 2003;28(3):259–275. [DOI] [PubMed] [Google Scholar]
- 8. Skotte NH, Southwell AL, Ostergaard ME, et al. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: Providing a therapeutic option for all Huntington disease patients. PLoS One. 2014;9(9):e107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, et al. ; Phase 1–2a IONIS-HTTRx Study Site Teams. Targeting huntingtin expression in patients with Huntington’s disease. N Engl J Med. 2019;380(24):2307–2316. [DOI] [PubMed] [Google Scholar]
- 10. Syn NL, Wang L, Chow EK-H, et al. Exosomes in cancer nanomedicine and immunotherapy: Prospects and challenges. Trends Biotechnol. 2017;35(7):665–676. [DOI] [PubMed] [Google Scholar]