Abstract
This scientific commentary refers to ‘Predictors of functional outcomes in patients with facioscapulohumeral muscular dystrophy’ by Katz et al. (doi:10.1093/brain/awab326).
This scientific commentary refers to ‘Predictors of functional outcomes in patients with facioscapulohumeral muscular dystrophy’ by Katz et al. (doi:10.1093/brain/awab326).
Facioscapulohumeral muscular dystrophy (FSHD), inherited in an autosomal dominant fashion, is the most common adult-onset muscular dystrophy. The disease is caused by a deletion on the long arm of chromosome 4,1 in a highly methylated region containing a motif of D4Z4 repeats. The size of the D4Z4 repeat contraction due to this deletion influences disease behaviour: longer deletions result in earlier onset of disease and more severe weakness. This suggests that the disorder is caused by an epigenetic disturbance, with the size of the deletion influencing the extent of epigenetic dysregulation. The discovery that hypomethylation of the D4Z4 repeats leads to reduced transcriptional regulation of developmentally regulated genes, and that double homeobox 4 (DUX4)—a developmentally regulated gene in muscle—shows persistent expression in FSHD, provided further confirmation of this.2 In this issue of Brain, Katz and co-workers analyse the relationship between the D4Z4 allele size and disease characteristics and confirm that smaller allele sizes correlate with earlier onset of disease, earlier diagnosis, more severe weakness and earlier need for wheelchair assistance.3
The authors divided their patient cohort into three groups: (i) 1–3 D4Z4 repeats; (ii) 4–7 D4Z4 repeats; and (iii) 8–10 D4Z4 repeats. Surprisingly there were more female FSHD patients than male in the 1–3 D4Z4 repeat cohort. This was unexpected because females were thought to be less severely affected than males and to be affected at an older age.4 However, there were no differences in age at diagnosis or age at symptom onset between males and females with 1–3 D4Z4 repeats. The majority of the patients had 4–7 D4Z4 repeats. Males were more likely to report upper extremity or proximal weakness at onset of symptoms, while females had more facial weakness (and were thus more likely to be misdiagnosed). Males were more likely than females to complain of breathing difficulty. This is an important area for future investigation. Reduced lung capacity and diaphragmatic and intercostal muscle weakness may result in significant morbidity and may contribute to higher mortality. Complications such as sleep-disordered breathing and ineffective airway secretion clearance are treatable and should be screened for.
One of the surprising findings by Katz et al.3 was the higher incidence of wheelchair use and shorter time from diagnosis to wheelchair use in females, independent of allele length. This did not seem to be related to breathing issues, as discussed above. Earlier studies, by contrast, had suggested that females were less severely affected than males and that oestrogens have a protective role in FSHD.5 Under-reporting by females of muscle weakness, especially in the limbs, is one possible explanation. The authors also suggest that females may be more accepting of a wheelchair than males. However, this still does not satisfactorily explain the earlier loss of ambulation in females. This should be explored further in prospective studies, especially in the context of patterns of muscle involvement clinically and on MRI, and how these may relate to changes in ambulatory functions. Such changes should be considered in relation to genotype and other covariates, such as pregnancy, which has been reported to worsen progression of weakness in other neuromuscular cohorts.6
In rare neuromuscular diseases, such as FSHD, having too much data tends not to be a concern. However, parsing cohort-level data at the electronic medical record level or identifying novel variants at the genomic level may require the use of artificial intelligence (AI) algorithms. But what is the difference between AI, machine learning, and deep learning?
AI, simply put, is when a computer does something humans do using ‘intelligence’. Machine learning is a large subset of AI that uses various methods to create algorithms that learn from data. Deep learning is a smaller subset of machine learning that utilizes neural networks to learn, and solve, even more complex problems.
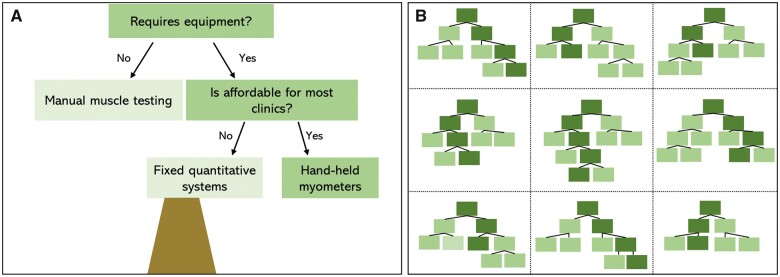
Katz et al.3 utilized random forest modelling, a machine learning method, to analyse clinical data gathered in the United States National Registry for FSHD Patients and Family members.7 Random forest modelling is built on decision trees (Fig. 1).8 Decision trees apply simple logic to depict possible outcomes for a given variable. The example in Fig. 1A shows a decision tree and possible outcomes for selection of strength testing methods when considering specific variables (e.g. need for equipment, affordability). While this decision tree is useful in logically arriving at a decision, the result could be biased by the features selected. Random forest modelling uses multiple decision trees based on ‘bootstrapped’ or random sampling of the full dataset and a subset of variables for each tree. Each instance adds a new tree to the forest. Bagging, or aggregating, the results across the training dataset and trees creates a more accurate and stable prediction than could be achieved with a single decision tree (Fig. 1B).
Figure 1.
Depiction of how a strength testing method can be selected via (A) a decision tree versus (B) a random forest model. The decision tree only considers a certain number of features or factors (e.g. need for equipment, affordability), whereas a random forest model will randomly select variables from across a dataset (e.g. equipment, affordability, reliability, sensitivity to change, ease of standardization, etc) to evaluate which features have the greatest influence on the outcome.
In random forest models, the computer selects random samples to determine which features of the dataset result in the most trees and highest accuracy of prediction. The random selection of data and variables in the random forest model allows it to be more generalizable as it is not conditioned or biased by specific data features.2 In our strength testing example, other features such as sensitivity to change, ease of use, variability of measurement, standardization of scoring and training, or time to complete testing, could be considered across various trees and data samples to arrive at our outcome.
Overfitting a model to a dataset is a concern with machine learning and important to consider. This can occur if a large portion of the data is used to train a model; in this instance, the model may be highly accurate within the dataset but too specific to be applied to another. Katz et al.3 used a small subset of 15 records to train the model and then applied it to the full cohort, lending credibility to their findings as overfitting is not likely.
Machine learning algorithms succeed with large datasets as there is sufficient data to both train the model and deliver accurate results. The authors report several relationships between data features and risk of wheelchair use, but these relationships should not necessarily be considered causative. For example, consider whether disease duration, presence of comorbidities (specifically breathing difficulty), and use of medications may influence wheelchair use. Disease duration was most predictive, which is not entirely surprising as weakness tends to progress and the likelihood of wheelchair use will therefore increase with time. The authors correctly posit that while breathing difficulty strongly influenced progression to wheelchair use, it may be that difficulty breathing is a medical condition that suggests worsening disease status and will thus be seen more frequently in patients using wheelchairs. This may also be true for medication use.
The FSHD Registry, even though prospective in nature, is heavily dependent on recall and thus subject to recall bias. The data that are collected are primarily subjective or binary. To minimize in-person visits, and to capture a larger group of patients, objective measures such as muscle strength or pulmonary function, are not part of the registry data. There is also participation bias, with more severely affected patients more likely to participate than their less severely affected counterparts. Registries are great for creating a snapshot of a disease but are not designed per se to measure disease progression. Though much cheaper to run and less burdensome for patients and caregivers, registries are significantly different from natural history studies, which although limited in terms of the number of subjects they can capture, provide a more objective and detailed characterization of disease behaviour and progression.
There are currently several prospective natural history studies ongoing that will help further define motor outcomes in FSHD (NCT04635891), and to validate clinical outcome measures through repeated objective assessments (NCT03458832).9 Efforts to validate additional biomarkers, such as MRI changes over time (NCT01671865),10 as well as changes on electrical impedance myography (EIM) (NCT03458832) are also underway. Such efforts are directly linked to clinical trial readiness, allowing for more accurate capture of disease progression during therapeutic interventions.
As technology improves and the integration of electronic medical records with growing registries yields ever larger datasets, use of AI models to understand relationships within datasets, predict patient outcomes, and identify novel genes or genetic modifiers influencing patient trajectories will intensify. In FSHD and other rare patient cohorts, collaboration and data sharing will be required to enable effective use of AI. Data quality should be prioritized over data quantity, with standardized outcomes gathered by trained evaluators and well-planned registries collecting data on meaningful, interpretable end points.
Funding
L.N.A. was supported by professional service agreements from Sarepta Therapeutics and Audentes Therapeutics through the Nationwide Children’s Hospital, grant support from Cure VCP Disease and Novartis Gene Therapies. T.M. was supported by National Institutes of Health grants RO1AR078340 and U24NS107210.
Competing interests
T.M. received research support from ATyr and Fulcrum for phase 2 clinical trials in FSHD. T.M. served on the data safety monitoring board for FSHD clinical trials for Acceleron. L.N.A. reports in-kind support from the Microsoft Philanthropies Artificial Intelligence for Health initiative for a study of infant movement.
References
- 1. Wijmenga C, Hewitt JE, Sandkuijl LA, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2(1):26–30. [DOI] [PubMed] [Google Scholar]
- 2. Lemmers RJLF, van der Vliet PJ, Klooster R, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329(5999):1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katz NK, Hogan J, Delbango R, Cernik C, Tawil R, Statland JM.. Predictors of functional outcomes in patients with facioscapulohumeral muscular dystrophy. Brain. 2021;144(11):3451–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zatz M, Marie SK, Cerqueira A, Vainzof M, Pavanello RC, Passos-Bueno MR.. The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severely and more frequently than females. Am J Med Genet. 1998;77(2):155–161. [PubMed] [Google Scholar]
- 5. Teveroni E, Pellegrino M, Sacconi S, et al. Estrogens enhance myoblast differentiation in facioscapulohumeral muscular dystrophy by antagonizing DUX4 activity. J Clin Invest. 2017;127(4):1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moore UJ, Jacobs M, Mayhew A, et al. The clinical outcome study for dysferlinopathy: pregnancy in dysferlinopathy. Neuromuscul Disord. 2019;29:S104. [Google Scholar]
- 7. Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 8. Verdú-Díaz J, Alonso-Pérez J, Nuñez-Peralta C, et al. Accuracy of a machine learning muscle MRI-based tool for the diagnosis of muscular dystrophies. Neurology. 2020;94(10):e1094–e1102. [DOI] [PubMed] [Google Scholar]
- 9. Wang LH, Friedman SD, Shaw D, et al. MRI-informed muscle biopsies correlate MRI with pathology and DUX4 target gene expression in FSHD. Hum Mol Genet. 2019;28(3):476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dahlqvist JR, Andersen G, Khawajazada T, Vissing C, Thomsen C, Vissing J.. Relationship between muscle inflammation and fat replacement assessed by MRI in facioscapulohumeral muscular dystrophy. J Neurol. 2019;266(5):1127–1135. [DOI] [PubMed] [Google Scholar]