Abstract
Huntington’s disease is an autosomal-dominant neurodegenerative disease caused by CAG expansion in exon 1 of the huntingtin (HTT) gene. Since mutant huntingtin (mHTT) protein is the root cause of Huntington’s disease, oligonucleotide-based therapeutic approaches using small interfering RNAs (siRNAs) and antisense oligonucleotides designed to specifically silence mHTT may be novel therapeutic strategies for Huntington’s disease. Unfortunately, the lack of an effective in vivo delivery system remains a major obstacle to realizing the full potential of oligonucleotide therapeutics, especially regarding the delivery of oligonucleotides to the cortex and striatum, the most severely affected brain regions in Huntington’s disease.
In this study, we present a synthetic biology strategy that integrates the naturally existing exosome-circulating system with artificial genetic circuits for self-assembly and delivery of mHTT-silencing siRNA to the cortex and striatum. We designed a cytomegalovirus promoter-directed genetic circuit encoding both a neuron-targeting rabies virus glycoprotein tag and an mHTT siRNA.
After being taken up by mouse livers after intravenous injection, this circuit was able to reprogramme hepatocytes to transcribe and self-assemble mHTT siRNA into rabies virus glycoprotein-tagged exosomes. The mHTT siRNA was further delivered through the exosome-circulating system and guided by a rabies virus glycoprotein tag to the cortex and striatum. Consequently, in three mouse models of Huntington’s disease treated with this circuit, the levels of mHTT protein and toxic aggregates were successfully reduced in the cortex and striatum, therefore ameliorating behavioural deficits and striatal and cortical neuropathologies.
Overall, our findings establish a convenient, effective and safe strategy for self-assembly of siRNAs in vivo that may provide a significant therapeutic benefit for Huntington’s disease.
Keywords: Huntington’s disease, siRNA, exosome, self-assembly, synthetic biology
See Snyder and Grunseich (doi:10.1093/brain/awab398) for a scientific commentary on this article.
Zhang et al. develop a new synthetic biology system that reprograms the host liver to direct the synthesis and self-assembly of mHTT siRNA in secretory exosomes, and facilitates the in vivo delivery of siRNA through circulating exosomes to the cortex and striatum. This approach substantially reduced mHTT expression in three mouse models.
See Snyder and Grunseich (doi:10.1093/brain/awab398) for a scientific commentary on this article.
Introduction
Huntington’s disease, a progressive neurodegenerative disorder inherited as an autosomal-dominant trait, usually develops in midlife and is characterized by movement dysfunction (e.g. hyperkinetic involuntary movements, chorea and dystonia) and cognitive decline.1-3 Huntington’s disease is caused by a CAG repeat expansion in exon 1 of the huntingtin (HTT) gene that leads to expression of mutant HTT (mHTT) protein containing a stretch of 36 or more glutamine residues in the N-terminus.4,5 The mHTT protein aggregates into intracellular inclusion bodies to induce neuronal death primarily in the cortex and striatum, two brain regions that are particularly vulnerable to mHTT-mediated toxicity.6–8 Although the genetic origin of Huntington’s disease has been recognized for >20 years, there is no treatment that can cure or slow the course of Huntington’s disease; the current therapies still aim to ameliorate symptoms.3,9 Given the monogenic nature of Huntington’s disease, application of therapeutics directly targeting the causative gene, mHTT, would likely be a more precise and efficient strategy for Huntington’s disease treatment than the approaches used thus far.
Oligonucleotide-based therapeutic approaches using small interfering RNAs (siRNAs) and antisense oligonucleotides (ASOs) are emerging strategies for silencing of disease-causing genes in gene therapy.10 Because siRNAs and ASOs have unique mechanisms of action—they directly intervene at the mRNA level to reduce production of the causative pathogenic protein—they offer powerful solutions for selective inhibition of disease targets that are notoriously difficult to modulate with traditional small-molecule drugs or antibodies.11–14 Since mHTT protein is an undruggable target due to its large size, complex structure and strong interactions with other proteins,7,15 Huntington’s disease is particularly well suited to alternative oligonucleotide-based therapies. Thus far, preclinical studies in animal models of Huntington’s disease have confirmed that siRNAs and ASOs delivered directly to the CNS successfully reduce toxic mHTT levels, alleviate neuropathologies and prolong survival in animal models of Huntington’s disease,16 and some ASOs designed against HTT mRNA (IONIS-HTTRx and WVE-120101) are currently in pivotal clinical trials involving intrathecal administration.17,18 Although these pioneering studies have provided the groundwork for therapeutic silencing of mHTT with oligonucleotides, some major challenges must be overcome to realize the full potential of oligonucleotide therapeutics in Huntington’s disease. First, the lack of a convenient delivery platform for oligonucleotides remains a major hurdle. Since the blood–brain barrier is still a formidable obstacle limiting access to cells within the CNS, siRNAs and ASOs generally must be directly delivered to the brain via intraparenchymal or intracerebroventricular administration or must be delivered intrathecally into the spinal fluid by lumbar puncture.19,20 These local delivery approaches require invasive surgery and have adverse effects when used for long-term treatment of Huntington’s disease, especially when they are delivered continually through repeated administrations. Second, the poor biodistribution of oligonucleotides to the striatum, a deep brain structure that is the first and most severely affected brain structure in Huntington’s disease, is another key hurdle. Indeed, even for ASOs that are administered by direct intraparenchymal injections, the therapeutic effects are limited to only a small portion of the striatum immediately adjacent to the sites of injection.21 Successful treatment of Huntington’s disease may require an approach that effectively spreads oligonucleotides over long distances in the CNS. Third, since oligonucleotides and their delivery formulations are usually exogenous, the potential risks of side effects, toxicity and immunogenicity should not be underestimated.22 Thus, a convenient, effective and safe delivery strategy is needed to improve the deep brain distribution and neuronal uptake of oligonucleotide therapeutics.
Exosomes have recently emerged as promising delivery vehicles for siRNAs. Exosomes are endogenously produced nano-sized extracellular vesicles that mediate intercellular communication by transferring lipids, proteins and RNAs between cells.23,24 Recently, our group and other groups have discovered that endogenous cells can selectively package miRNAs into exosomes and secrete the exosomes to deliver the miRNAs into recipient cells; the secreted miRNAs robustly block target gene expression at fairly low concentrations.25–27 We have also provided the first evidence that cells can be engineered to produce specific exosomes that carry artificial small RNAs (e.g. miRNA mimics).25 Because exosomes have the innate ability to protect and transport small RNAs across biological barriers in vivo and are more biocompatible with the host immune system than other vehicles,28,29 numerous studies have tried to use exosomes to deliver therapeutic siRNAs and miRNAs in vivo. However, using exosomes to deliver siRNAs remains unfeasible in practice, mainly because not enough exosomes can be harvested for clinical use.30 Additionally, robust and scalable methods for loading siRNA cargos into exosomes are lacking, and complicated exosome purification protocols are not amenable to good manufacturing practice (GMP) standards.
Recent advances in synthetic biology have provided insights for the design of new biomedicines. Synthetic biology aims to achieve new biological functions through the design and assembly of genetic circuits, which are combinations of biological modules that together execute defined functions within a host organism.31,32 The breakthroughs in this field thus far have already shown impressive success in producing energy, food, biosensors, drugs and biomaterials and in creating man-made organisms.32 For instance, new genetic pathways created via assembly of enzymatic cascades or networks in bacteria and yeast have been instrumental for the large-scale economic production of biofuels and pharmaceuticals.33 However, the current practices of synthetic biology are largely confined to micro-organisms, while many of the most pressing problems, particularly those associated with human health and diseases, are inherent to in vivo systems.34 Therefore, a more concerted effort towards advancing synthetic biology in vivo is crucial for the development of next-generation therapeutics. To overcome this bottleneck problem, genetic circuits that are typically developed in cell lines cultured in artificial environments need to be able to be precisely delivered to and rationally assembled in desired body locations, where they can reprogramme endogenous cells to behave in controllable and predictable ways in heterogeneous, dynamic in vivo environments.
Because siRNA therapy and synthetic biology share the key problems of in vivo delivery and assembly of biological components (siRNAs or genetic circuits) in endogenous systems, it is rational to integrate the naturally existing exosome-circulating system with artificial genetic circuits to achieve self-assembly and delivery of siRNAs in vivo. Recently, we developed a convenient, effective and safe synthetic biology strategy that reprograms the host liver as a tissue chassis to direct the self-assembly of siRNAs into secretory exosomes and facilitate the in vivo delivery of siRNAs to desired tissues.35 This strategy is based on the intrinsic capability of the liver to express transgenes introduced by intravenous injection of genetic circuits (in the form of naked DNA plasmids).36,37 In this study, we designed new genetic circuits to engineer mouse liver to transcribe and self-assemble mHTT siRNA into neuron-directed exosomes. The therapeutic value of these genetic circuits was confirmed by efficient delivery of mHTT siRNA to the cortex and striatum in Huntington’s disease mouse models, which resulted in significant mHTT level reduction and symptom alleviation in the model animals.
Materials and methods
Construction of the genetic circuits targeting mHTT
The CMV-siRmHTT circuit was generated by inserting an mHTT siRNA sequence into a 166-bp pre-miR-155 backbone with structurally conserved nucleotide changes to maintain pairing. The inserted sequences are listed in Supplementary Table 1. The CMV-RVG-siRmHTT circuit was generated by fusing the rabies virus glycoprotein (RVG) tag into the extra-exosomal N-terminus of lysosome-associated membrane glycoprotein 2b (Lamp2b); the fused RVG-Lamp2b and pre-miR-155 backbone containing mHTT siRNA were then cloned downstream of the cytomegalovirus (CMV) promoter to generate a circuit that simultaneously encodes mHTT siRNA and the RVG-Lamp2b fusion protein. A circuit designed to express a scrambled RNA was used as the negative control.
The genetic circuits in the form of DNA plasmids were dissolved in 1× PBS without additional formulations. The map and scaffold of the plasmid are shown in Supplementary Fig. 7. The plasmids were transformed into Escherichia coli DH5α competent cells (Tsingke, TSC01), cultured with LB medium (with 50 µg/ml spectinomycin) for 14–16 h in a 37°C shaking incubator and extracted and purified with an EndoFree Maxi Plasmid Kit V2 (Tiangen, DP120) according to the manufacturer’s instructions. The purified plasmids were sequenced to ensure that the sequences of the inserted genetic circuits were correct.
Cell culture
The human embryonic kidney cell line HEK293T was purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences and maintained at 37°C under 5% CO2 in high-glucose (4.5 g/l) Dulbecco's modified Eagle medium (DMEM, Gibco) supplemented with 10% foetal bovine serum (FBS, Gibco), 100 U/ml penicillin and 100 mg/ml streptomycin. Conditionally immortalized wild-type (STHdh-Q7) and mutant (STHdh-Q111) striatal neuronal progenitor cell lines expressing HTT with either 7 or 111 glutamines were derived from a knock-in transgenic mouse containing humanized exon 1 with either 7 or 111 CAG repeats. STHdh-Q7 and STHdh-Q111 cells were cultured in high-glucose (4.5 g/l) DMEM (Gibco) supplemented with 10% FBS (Gibco), Pen-Strep and G418 (0.4 mg/ml) at 33°C in a 5% CO2 atmosphere.
Construction of the HEK293T cell line stably expressing mHTT-Q66-FLAG
A lentiviral plasmid designed to express FLAG-tagged exon 1 of human mHTT (NM_002111.7) with 66 CAG repeats (LV-mHTT-Q66-FLAG) was purchased from HANBIO. The lentivirus was added to HEK293T cells at a multiplicity of infection of 10 together with polybrene (final concentration 5 μg/ml) according to the manufacturer’s instructions. After 48 h, puromycin at a final concentration of 10 μg/ml was added to the infected HEK293T cells three times every 24 h to obtain the HEK293T cell line stably expressing mHTT-Q66-FLAG.
In vitro transfection of the genetic circuits
The genetic circuits (plasmids) were transfected into HEK293T or STHdh-Q7/STHdh-Q111 cells using LipofectamineTM 2000 (Invitrogen) according to the manufacturer’s instructions. A scrambled siRNA was included as a negative control. Total RNA and protein were isolated 48 h post-transfection, and the mHTT mRNA and protein expression levels were assessed using quantitative PCR with reverse transcription (RT-qPCR) and western blotting, respectively.
According to the users’ manual of ImageJ,38 we used the automated counting procedure to quantify the number of aggregated mHTT dots. We chose the green fluorescent protein (GFP) channel to open the original RGB image and then chose the option of ‘Image→Type→16-bit’ to convert the image to greyscale. Once the image was in greyscale, we chose the option of ‘Image→Adjust→Threshold’ and set a fixed threshold of pixel intensity (minimum: 70; maximum: 85) for each image to subtract the background and highlight all the aggregated dots. Subsequently, we chose the option of ‘Analyse→Analyse Particles’ and set a particle size threshold (minimum: 3; maximum: infinity) to exclude small ‘noise’ pixels. Finally, we chose the ‘Display Results’ option, and all counted aggregated dots were shown as numbered outlines in the image. For each group, five immunofluorescent images were analysed.
In vivo injection of the genetic circuits
C57BL/6J mice were purchased from the Model Animal Research Centre of Nanjing University and maintained under specific pathogen-free conditions at Nanjing University. Genetic circuits were formed as naked DNA plasmids and administered to mice at a dose of 5 mg/kg through regular tail vein injection (200 μl of solution was injected in 3 s). After treatment, the mice were killed; blood samples were collected via cardiac puncture; and the liver, cortex and striatum were collected. All animal care and handling procedures were performed in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Review Board of Nanjing University.
RNA extraction and quantitative RT-PCR assay
Total RNA was extracted from cultured cells or mouse tissues using TRIzol® Reagent (Invitrogen) according to the manufacturer’s instructions. Mature mHTT siRNA was quantitated using specific TaqMan™ miRNA probes (Applied Biosystems) according to the manufacturer’s instructions. Briefly, 0.5 µg of total RNA was reverse-transcribed into cDNA using a customized stem-loop reverse transcription primer (Applied Biosystems) and avian myeloblastosis virus (AMV) reverse transcriptase (TaKaRa). The reaction conditions were as follows: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. Real-time PCR was run with a TaqMan™ MicroRNA Assay (Applied Biosystems) and an Applied Biosystems 7300 Sequence Detection System. The reactions were incubated in a 96-well optical plate at 95°C for 5 min and then subjected to 40 cycles at 95°C for 15 s and 60°C for 1 min. All reactions were run in triplicate. After running the reactions, the cycle threshold (CT) values were determined using fixed threshold settings, and the mean CT was determined for PCRs performed in triplicate. For quantitation of the absolute levels of mHTT siRNA, synthetic single-stranded mHTT siRNA was serially diluted to generate a standard curve using RT-qPCR. By referring to the standard curve, the concentrations of mHTT siRNA in various tissues and plasma were calculated and are presented as the absolute amounts of mHTT siRNA in 1 g of total RNA (pmol/g total RNA) or 1 l of plasma (fmol/l).
To quantitate mHTT mRNA, 1 µg of total RNA was reverse-transcribed into cDNA using oligo-d(T) (TaKaRa, 3806) and AMV reverse transcriptase (TaKaRa, 2621) under the following conditions: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. Next, real-time PCR was performed with the reverse transcription product, EvaGreen Dye (Biotium, 31000) and specific primers for the mHTT and β-actin genes. The primer sequences are listed in Supplementary Table 2. The reactions were incubated at 95°C for 5 min and then subjected to 40 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 1 min. After the reactions were completed, the CT values were determined, and the relative levels of mRNA were normalized to the β-actin levels by the 2−ΔΔCT method.
Fluorescence in situ hybridization
To quantitate mHTT siRNA expression and localization, fluorescence in situ hybridization was performed on mouse tissue sections with a digoxygenin (DIG)-labelled probe (miRCURY LNA miRNA detection probe; Qiagen). Detection and analysis were performed by Wuhan Servicebio Technology Co., Ltd.
Western blotting
All cells were rinsed with 1× PBS (pH 7.4) and lysed in RIPA lysis buffer (Beyotime) supplemented with a protease and phosphatase inhibitor cocktail (Thermo Scientific 78440) on ice for 30 min. The tissue samples were frozen solid with liquid nitrogen, ground into a powder and lysed in RIPA lysis buffer containing a protease and phosphatase inhibitor cocktail on ice for 30 min. When necessary, sonication was used to facilitate lysis. Cell lysates or tissue homogenates were centrifuged for 10 min (12 000g, 4°C), the supernatant was collected and the protein concentration was determined using a Pierce BCA protein assay kit (Thermo Scientific). Protein expression levels were analysed using western blotting with corresponding antibodies and were normalized to the α-tubulin level. Briefly, protein lysates (60 μg) were loaded onto 6% SDS-PAGE gels for electrophoretic separation of proteins under denaturing conditions and transferred onto PVDF membranes. The membranes were then blocked for 1 h at room temperature with 5% non-fat dry milk in Tris-buffered saline with 0.1% Tween-20 (TBST), and incubated 3 h at room temperature with primary antibodies. The membranes were washed three times with 1× TBST for 10 min per wash. The membranes were incubated with secondary antibodies for 1 h at room temperature and then subjected to three additional 10-min washes with 1× TBST. The membranes were incubated with enhanced chemiluminescence (ECL) substrate (Thermo Fisher, 46640) according to the manufacturer’s instructions and detected with autoradiography film in a darkroom or read using a chemiluminescence imaging system. The protein bands were analysed using ImageJ. The primary antibodies were as follows: anti-HTT (Millipore, MAB2166), anti-polyglutamine (Millipore, MAB1574), anti-DDDDK tag (Abcam, ab1162), anti-α-tubulin (Abcam, ab18251), anti-GFP (Abcam, ab290), anti-TSG101 (Abcam, ab125011), anti-CD63 (Abcam, ab217345), anti-CD9 (Abcam, ab92726), goat anti-mouse IgG H&L HRP (Abcam, ab205719) and goat anti-rabbit IgG H&L HRP (Abcam, ab205718).
In BACHD mice, human mHTT and mouse endogenous HTT are simultaneously expressed and only differ in a short length. Because the molecular weights of HTT and α-tubulin are quite different (350 kD versus 55 kD), although HTT and α-tubulin could be blotted in the same 6% SDS-PAGE gel, the distance between human mHTT and mouse endogenous HTT would be too small for effectively separating the two. Therefore, we chose to run both a 4% and 10% SDS-PAGE gel, and an equal amount of total protein was loaded in each gel. The 4% gel was used to quantitate human mHTT and mouse endogenous HTT, and the 10% gel was used to quantitate α-tubulin. Thus, we could determine whether a genetic circuit designed to specifically silence human mHTT was able to discriminate between human mHTT and mouse endogenous HTT.
Analysis of exosomes
After tail vein injection of genetic circuits, blood was collected from each mouse and then centrifuged at 3000g for 10 min to collect the plasma. Roughly 200 μl of plasma was obtained from each mouse and was equally divided into two samples: one (∼100 μl) for direct isolation of total RNA and quantitation of total siRNA levels and the other for purification of exosomes. Plasma exosomes were isolated using a Total Exosome Isolation Kit (from plasma) (Invitrogen, 4484450) according to the manufacturer’s instructions. In detail, an equal volume (∼100 μl) of 1× PBS was added to the plasma, and 20% of the total volume (∼40 μl) of Exosome Precipitation Reagent was added. After the sample was incubated for 10 min at room temperature, the exosomes and supernatant were separated by centrifugation at 10 000g for 5 min at room temperature. Finally, the exosome pellet was resuspended in 1× PBS for transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) or lysed in RIPA lysis buffer for western blotting.
Exosome vesicles were visualized using TEM (HT7700, Japan). Briefly, exosomes resuspended in PBS were fixed in 2% paraformaldehyde (PFA). Each fixed sample was adsorbed onto a copper mesh coated with formaldehyde in a dry environment for 20 min. The sample was fixed in 1% glutaraldehyde for 5 min. After rinsing in distilled water, the sample was dyed with uranyl oxalate for 5 min and then dyed with uranyl acetate for 10 min on ice. Excess liquid was removed from the mesh with filter paper, and the mesh was stored at room temperature until imaging.
The size distribution and quantity of exosomes were measured with a NanoSight NS 300 system (NanoSight Technology) equipped with a blue laser. The nanoparticles were illuminated with the laser, and their Brownian motion was captured for 60 s. At least five videos were collected from each individual sample to provide representative concentration measurements. The size distribution curves were evaluated with NTA software and were averaged within each sample from the video repetitions and then averaged between repetitions to provide a representative size distribution.
Exosome labelling and incubation
Exosomes were labelled with PKH26 (Sigma) according to the manufacturer’s instructions. Briefly, each exosome pellet (60 μg total protein content) was resuspended in 1 ml of Diluent C and then mixed with 4 μl of PKH26 and incubated for 4 min. Next, an equal volume of 1% FBS was added to stop the labelling reaction. Then, the exosomes were isolated, washed with 1× PBS and cocultured with recipient cells for 6 h. After incubation, the cells were washed, fixed and observed under confocal microscopy.
GFP fluorescence intensity detection
GFP-transgenic mice were purchased from the Model Animal Research Centre of Nanjing University and maintained under specific pathogen-free conditions at Nanjing University. After GFP-transgenic mice were intravenously injected with PBS, CMV-scrR, CMV-siRGFP or CMV-RVG-siRGFP seven times, their cortices and striata were sequentially soaked in 4% PFA and in 15% and 30% sucrose solutions for sectioning. Subsequently, cryostat sections (20 μm) of the cortex and striatum were fixed in 4% PFA for 10 min, and the sections were incubated in 1× PBS containing 5% BSA and 0.3% TritonTM X-100 for 60 min at room temperature. The sections were then incubated overnight with an anti-NeuN antibody (Abcam, ab177487) at 4°C. After three rinses with 1× PBS and incubation with the appropriate secondary antibodies (CST, 4413) for 1 h at room temperature, the sections were washed with 1× PBS and then placed in DAPI staining solution for 10 min. After washing with 1× PBS, the sections were ready for examination of GFP fluorescence intensity using a Nikon TI-SH-J confocal microscope.
Evaluation of the therapeutic effects of the genetic circuits in mouse models of Huntington’s disease
Three mouse models of Huntington’s disease , including N171-82Q (JAX: 003627), BACHD (JAX: 008197) and YAC128 (JAX: 004938), were bred under standard conditions of temperature (22 ± 1°C) and humidity (30%) in a pathogen-free facility and exposed to a 12-h light/dark cycle. N171-82Q mice were maintained on a B6C3 background, and BACHD and YAC128 mice were maintained on the congenic FVB/N background. The Huntington’s disease mice of each type were randomly divided into two groups and intravenously injected with 5 mg/kg CMV-scrR or CMV-RVG-siRmHTT circuit. N171-82Q and BACHD mice were injected once every 2 days for a total of seven times (duration 2 weeks), while YAC128 mice were injected twice a week for a total of 16 times (duration 8 weeks).
A rotarod test was performed as previously reported with slight modifications.39 Before the experiment, all Huntington’s disease mice were trained three times a day for 3 days on a rotarod device (TECHMAN, ZB-200) at an accelerating speed (5–40 rpm, 5 min). Wild-type littermates were trained in the same way to serve as a control. On the testing day, the Huntington’s disease and wild-type mice were tested on the rotarod device at an accelerating speed (5–40 rpm, 5 min), and the latency to fall was measured for each mouse. Rotarod data were collected from three trials with a 10-min recovery time between trials. The retention time data for each group were averaged and plotted. Investigators performing all behavioural tests were blinded to group assignment in behavioural experiments.
Immunofluorescence staining was performed as previously reported.40 Briefly, tissues were sequentially soaked in 4% PFA, 15% and 30% sucrose solution to make sections. The cryostat sections (15 μm) were fixed in 4% PFA for 10 min, incubated in 1× PBS containing 5% BSA and 0.3% TritonTM X-100 for 60 min at room temperature, and incubated overnight with primary antibodies at 4°C. After three rinses with 1× PBS and incubation with the appropriate secondary antibodies for 1 h at room temperature, the sections were washed with 1× PBS and then placed in DAPI staining solution for 10 min. After washing with 1× PBS, the sections were ready for examination using a Nikon TI-SH-J or Lei TCS SP8-MaiTai MP confocal microscope. The primary antibodies included anti-EM48 (Millipore, MAB5374) and anti-NeuN (Millipore, MAB377). The secondary antibodies included anti-rabbit IgG (H+L), Alexa Fluor 555 (CST, 4413), anti-mouse IgG (H+L), Alexa Fluor® 488 (CST, 4408) and Alexa Fluor® 594 (Life Technologies, A11042).
Analyses of biochemical indexes and tissue damage
C57BL/6J mice were intravenously injected with PBS or with 5 mg/kg CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT every 2 days for a total of seven times. Twelve hours after the last injection, the mice were euthanized to collect peripheral blood and tissues. Fresh peripheral blood was divided into two samples. One sample was subjected to separation of serum, and representative biochemical indexes, including alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, albumin, lactic dehydrogenase, total bilirubin, creatinine and blood urea nitrogen, were measured in the serum. These biochemical indexes were examined according to the instructions of an automatic biochemical analyser (Hitachi, 7100). The other sample was collected by vacuum blood sampling (with EDTA anticoagulant). Then, the numbers of red blood cells, white blood cells and platelets in peripheral blood were detected with an automatic blood cell diagnostic instrument (ADVIA 2120i) according to the manufacturer’s instructions.
Liver, spleen, lung and kidney tissues were extracted for haematoxylin and eosin staining to evaluate tissue damage. Every tissue was fixed with 4% neutral formaldehyde, embedded in paraffin and cut into 5-μm sections. The sections were stained with haematoxylin for 3 min, rinsed with running water for 10 s and colour-separated with 1% hydrochloric acid ethanol briefly after dewaxing and rehydration. Next, the sections were rinsed with distilled water for 60 s, stained with eosin for 60 s and dehydrated with 95% and 100% ethanol twice (1 min per time) after washing with distilled water for 10 s. All sections were then permeabilized with xylene and mounted with neutral balsam. Finally, the haematoxylin and eosin-stained sections were observed under a microscope camera (Nikon, DS-Ri2).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7 software. All data are presented as the mean ± standard error of the mean (SEM) from at least three independent experiments. Comparison between two groups was performed by using Student’s t-test. Multiple group comparisons were analysed by using one-way ANOVA followed by Bonferroni’s multiple comparisons test. Significance was accepted as follows: *P < 0.05; **P < 0.01; ***P < 0.001 and ****P < 0.0001. Details on the statistics are indicated in the figure legends.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.
Results
Design and construction of the genetic circuits
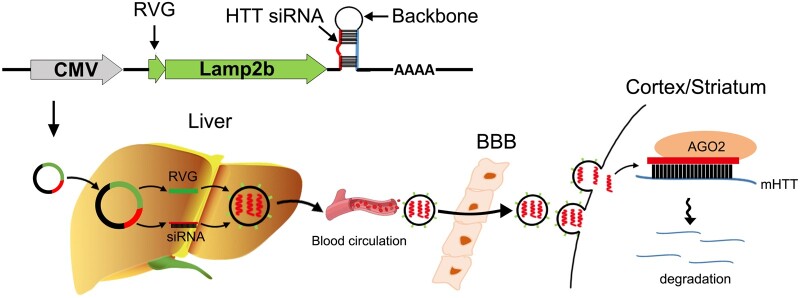
We first designed a genetic circuit allowing combination of three different functional modules: (i) an siRNA-expressing backbone that encodes an siRNA targeting specifically mHTT; (ii) a guiding tag that modifies the membrane-anchored proteins of exosomes to enable neuron-targeting capability; and (iii) a co-driving promoter that drives a dual expression cassette for the mHTT siRNA and guiding tag (Fig. 1).
Figure 1.
Schematic description of the architecture of the genetic circuit. The genetic circuit contains three different functional modules: a co-driving CMV promoter, a neuron-guiding RVG tag and an mHTT siRNA-expressing backbone. When the genetic circuit is placed in a tissue chassis such as the liver, the CMV promoter drives the transcription of mHTT siRNA and facilitates loading of the payload of mHTT siRNA into exosomes as cargo; simultaneously, the CMV promoter directs the localization of the RVG guidance tag onto the exosome surface to confer neuron-targeting properties on the exosomes. After being released into the circulation and delivered through the exosome-circulating system, the mHTT siRNA enclosed in RVG-tagged exosomes is guided by the RVG tag to penetrate the blood–brain barrier (BBB) and arrive at the cortex and striatum, ultimately resulting in mRNA degradation and decreased protein expression of the mHTT gene.
The CMV promoter was selected as the co-driving promoter because it has been demonstrated to be an excellent choice for transcription of both guiding proteins and siRNAs.35 For the guiding tag, a sequence encoding a short peptide of RVG fused to the N-terminus of Lamp2b (a canonical exosome membrane protein) was inserted downstream of the CMV promoter to confer a neuron-targeting property on exosomes, because the RVG tag anchored onto the exosome surface via Lamp2b has been shown to facilitate the delivery of exosomal cargos across the blood–brain barrier and into neuronal cells.41 For the siRNA-expressing backbone, mHTT siRNA was embedded in a pre-miR-155 backbone and inserted downstream of the expression cassette for the RVG-Lamp2b fusion protein. By integrating the three modules together, a CMV-directed genetic circuit encoding both an RVG-Lamp2b fusion protein and an mHTT siRNA was constructed (hereafter, the CMV-RVG-siRmHTT circuit). Theoretically, when the CMV-RVG-siRmHTT circuit is taken up and processed by the liver after intravenous injection, the CMV promoter will drive the transcription of mHTT siRNA and the RVG-Lamp2b fusion protein and direct the siRNA payload into RVG-tagged exosomes; after being delivered through the exosome-circulating system, the exosomes will be guided by an RVG tag to penetrate the blood–brain barrier and enable the release of mHTT siRNA into neuronal cells (Fig. 1).
Characterization of the genetic circuits in vitro
Next, we examined whether the three modules can functionally integrate to form a genetic circuit that has the ability to silence mHTT in vitro. HEK293T cells were stably transfected with a lentiviral vector expressing FLAG-tagged exon 1 of mHTT with 66 CAG repeats (LV-mHTT-Q66-FLAG) and then transfected with the CMV-RVG-siRmHTT circuit. Two CMV-RVG-siRmHTT circuits designed to target different sites of mHTT exon 1 were compared to optimize the silencing potency, and a CMV-directed genetic circuit encoding a scrambled RNA (CMV-scrR circuit) served as the control. Both circuits mediated significant suppression of mHTT protein and mRNA expression in HEK293T cells (Supplementary Fig. 1A–C), and the circuit with the greatest interference efficiency, CMV-RVG-siRmHTT-2, was selected. Furthermore, a CMV-directed genetic circuit encoding only an mHTT siRNA (the CMV-siRmHTT circuit) was compared with the CMV-RVG-siRmHTT circuit; equivalent silencing of mHTT protein and mRNA expression was detected after transfection of HEK293T cells with either circuit (Supplementary Fig. 1D–F), indicating that modification with the genetic circuit including the guiding tag module did not alter the silencing efficiency of the siRNA-expressing module. Likewise, since polyglutamine expansion in exon 1 of mHTT leads to misfolding of mHTT and formation of aggregates,5 we determined whether toxic protein aggregates could be removed by the genetic circuits. The CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT circuit was transfected into HEK293T cells stably transfected with a lentiviral vector coexpressing mHTT-Q66 and GFP. CMV-scrR-treated cells exhibited large nuclear aggregates and small cytoplasmic aggregates, whereas CMV-RVG-siRmHTT- or CMV-siRmHTT-treated cells displayed only diffuse nuclear and cytoplasmic fluorescence (Supplementary Fig. 1G and H). In addition, immortalized striatal progenitor cell lines expressing either wild-type HTT (STHdh-Q7) or mHTT with 111 CAG repeats (STHdh-Q111) were used. Transfection of three different CMV-RVG-siRmHTT circuits into STHdh-Q111 cells resulted in significant reductions in mHTT protein and mRNA levels in the cells, and the CMV-RVG-siRmHTT-2 circuit still had the highest interference efficiency (Supplementary Fig. 1I–K). Overall, these results establish the genetic circuit as an organic combination of multiple modules that direct the generation of functional mHTT siRNA in vitro.
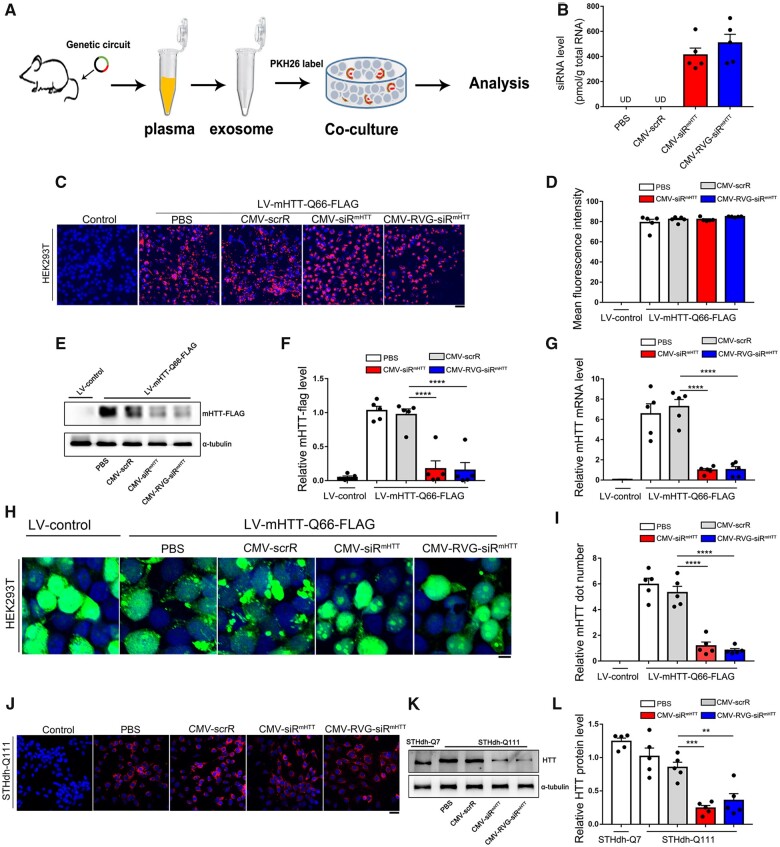
Genetic circuits direct the self-assembly and release of functional mHTT siRNA in an ex vivo model
To ensure that the genetic circuits developed in vitro work well in the complex in vivo environment and have an innate ability to automatically assemble siRNA-encapsulating, neuron-directed exosomes, we established an ex vivo model to examine the production, assembly and release of exosome-enclosed mHTT siRNA. In this model, PBS, CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT was intravenously injected into C57BL/6J mice seven times. The exosomes were then purified from mouse plasma and incubated with HEK293T cells for further analyses (Fig. 2A). First, we confirmed proper enrichment of exosomes from mouse plasma. NTA showed that the exosome size in each group peaked at ∼100 nm, and the exosomes in the different groups had similar concentrations (Supplementary Fig. 2A). TEM confirmed that the purified exosomes had the characteristic morphology and size of exosomes (Supplementary Fig. 2B). Moreover, enrichment of exosomal markers (CD63, TSG101 and CD9) was detected only in purified exosomes but not in exosome-depleted plasma (Supplementary Fig. 2C). These results validate that the genetic circuits do not affect the size, structure or physical properties of endogenous exosomes. Second, we examined whether the genetic circuits direct efficient loading of siRNAs into exosomes. Notable and equivalent amounts of mHTT siRNA were detected in the exosomes derived from the mice injected with CMV-siRmHTT or CMV-RVG-siRmHTT (Fig. 2B). Third, the purified exosomes were labelled with PKH26, a lipophilic fluorescent dye and the fluorescently labelled exosomes were added to the culture medium of HEK293T cells. The intracellular fluorescence intensity was monitored by confocal microscopy. Exosomes derived from mice injected with PBS, CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT were equally taken up by HEK293T cells (Fig. 2C and D). These results validate that the incorporation of siRNA cargo does not significantly interfere with exosome uptake. Fourth, we evaluated whether the formation of toxic protein aggregates can be reduced by the genetic circuits. When mHTT-Q66-expressing HEK293T cells were incubated with exosomes derived from mice injected with CMV-siRmHTT or CMV-RVG-siRmHTT, efficient knockdown of mHTT protein and mRNA expression was achieved (Fig. 2E–G). Similarly, when HEK293T cells coexpressing mHTT-Q66 and GFP were incubated with exosomes derived from mice injected with different types of genetic circuits, extensive nuclear and cytoplasmic protein aggregation was observed in the cells incubated with the control exosomes from PBS- or CMV-scrR-injected mice; however, significantly less protein aggregation was observed in the cells incubated with exosomes from CMV-siRmHTT- or CMV-RVG-siRmHTT-injected mice (Fig. 2H and I). Fifth, we evaluated the rescue of mHTT toxicity by genetic circuits in striatal cell lines. PKH26-labelled exosomes derived from mice injected with CMV-siRmHTT or CMV-RVG-siRmHTT were readily incorporated by STHdh-Q111 cells (Fig. 2J), which further resulted in apparent reductions in mHTT protein levels in the STHdh-Q111 cells (Fig. 2K and L). Overall, these results reveal that the genetic circuits can direct the self-assembly of mHTT siRNA into exosomes, which can be further internalized by recipient cells to block mHTT protein expression, alleviate protein aggregation and prevent cytotoxicity.
Figure 2.
Characterization of self-assembled mHTT siRNA in an ex vivo model. (A) Schematic of the experimental design. C57BL/6J mice were intravenously injected with PBS or with 5 mg/kg CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT circuit every 2 days for a total of seven times, and then the exosomes were purified from mouse plasma and incubated with HEK293T cells. Next, uptake of self-assembled mHTT siRNA by HEK293T cells and the subsequent suppression of mHTT expression and aggregation by self-assembled mHTT siRNA were examined in this ex vivo model. (B) Quantitative RT-PCR analysis of mHTT siRNA levels in purified exosomes (n = 5 in each group). (C) Purified exosomes were fluorescently labelled with PKH26, and PKH26-labelled exosomes were incubated with HEK293T cells for 6 h. The levels of intracellular fluorescence intensity were monitored by confocal microscopy. (D) Quantitation of the fluorescence intensity shown in C (n = 5 in each group). (E) Western blot analysis of mHTT-FLAG protein levels in mHTT-Q66-expressing HEK293T cells after incubation with purified exosomes. HEK293T cells were stably transfected with LV-mHTT-Q66-FLAG ahead of incubation. (F) Quantitation of the mHTT-FLAG protein levels shown in E (n = 5 in each group). (G) Quantitative RT-PCR analysis of mHTT mRNA levels in mHTT-Q66-expressing HEK293T cells after incubation with purified exosomes (n = 5 in each group). (H) HEK293T cells coexpressing mHTT-Q66 and GFP were incubated with purified exosomes. Fluorescence microscopy was used to assess mHTT aggregates (bright green puncta). Scale bar = 5 μm. (I) Quantitation of the aggregated mHTT dots shown in H (n = 5 in each group). (J) Purified exosomes were fluorescently labelled with PKH26, and PKH26-labelled exosomes were incubated with STHdh-Q111 cells for 6 h. The levels of intracellular fluorescence intensity were monitored by confocal microscopy. Scale bar = 20 μm. (K) Western blot analysis of mHTT protein levels in STHdh-Q111 cells after incubation with purified exosomes. STHdh-Q7 cells expressing wild-type HTT served as a control. (L) Quantitation of the HTT protein levels shown in K (n = 5 in each group). Values are presented as mean ± SEM. Significance was determined using one-way ANOVA followed by Bonferroni's multiple comparisons in F, G, I and L. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns = not significant.
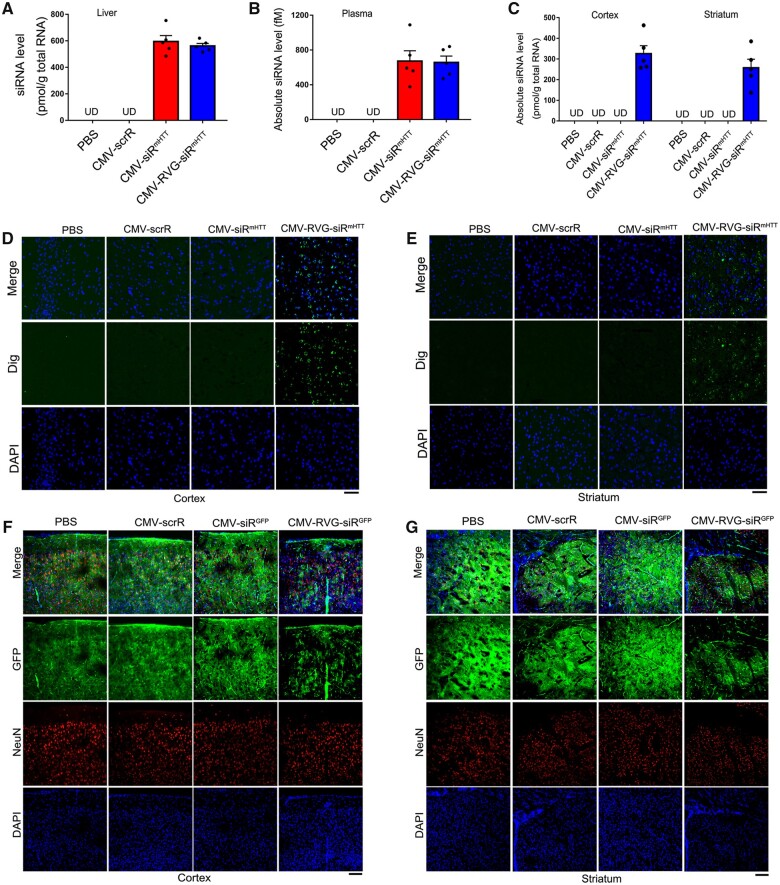
Tracking and visualization of the delivery of self-assembled mHTT siRNA into the cortex and striatum
Since achieving an appropriate concentration of mHTT siRNA in the cortex and striatum is critical for therapeutic efficacy,42 we next determined whether self-assembled mHTT siRNA was efficiently delivered to striatal and cortical neurons via endogenous exosome circulation. First, we evaluated the delivery of exosomes to the cortex and striatum. PBS, CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT was intravenously injected into C57BL/6J mice; the resultant blood exosomes were purified and labelled with PKH26, and then the fluorescently labelled exosomes were injected into another group of C57BL/6J mice. While the four types of exosomes all generated apparent fluorescent signals (shown in red) in the liver (Supplementary Fig. 3A), only the exosomes derived from CMV-RVG-siRmHTT-injected mice generated fluorescent signals in the cortex and striatum (Supplementary Fig. 3B and C). These results suggest that only the CMV-RVG-siRmHTT circuit successfully endows exosomes with the ability to target the cortex and striatum in the brain.
Second, we examined siRNA accumulation in the cortex and striatum. In C57BL/6J mice injected with CMV-siRmHTT or CMV-RVG-siRmHTT, similar amounts of mHTT siRNA were produced in the liver (Fig. 3A). Accordingly, equivalent amounts of mHTT siRNA were detected in the plasma (Fig. 3B). These results are consistent with the findings of our previous study 35 and support the idea that the liver can take up exogenous circuits and continuously release exosome-enclosed siRNA into the circulation. Only the CMV-RVG-siRmHTT circuit resulted in notable accumulation of mHTT siRNA in the cortex and striatum, whereas no mHTT siRNA was detected in these tissues after CMV-siRmHTT circuit injection (Fig. 3C). Furthermore, the tissue distribution of mHTT siRNA was confirmed by fluorescence in situ hybridization. Apparent hybridization signals of mHTT siRNA were detected in the livers of mice injected with CMV-siRmHTT or CMV-RVG-siRmHTT (Supplementary Fig. 4A). While no signals were detected in the cortex and striatum in mice injected with CMV-siRmHTT, strong hybridization signals were present in these brain regions after CMV-RVG-siRmHTT injection (Fig. 3D and E). These results reveal that the RVG guiding tag successfully endows exosomes with the ability to transport their cargo siRNAs to striatal and cortical neurons, bypassing the blood–brain barrier.
Figure 3.
Tracking and visualization of the delivery of self-assembled mHTT siRNA into the cortex and striatum. (A–C) Quantitative RT–PCR analysis of mHTT siRNA levels in the liver, plasma and cortex/striatum after intravenous injection of C57BL/6J mice with PBS or with 5 mg/kg CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT circuit for a total of three times (n = 5 in each group). Values are presented as the means ± SEM. (D and E) In situ detection of mHTT siRNA in the mouse cortex and striatum. C57BL/6J mice were intravenously injected with PBS or with 5 mg/kg CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT circuit for a total of three times. Positive in situ hybridization signals in the cortex and striatum are shown in green, and DAPI-stained nuclei are shown in blue. Scale bar = 50 μm. (F and G) Direct visualization of the suppression of GFP fluorescence levels in vivo by self-assembled GFP siRNA. GFP-transgenic mice were intravenously injected with PBS or with 5 mg/kg CMV-scrR, CMV-siRGFP or CMV-RVG-siRGFP circuit every 2 days for a total of seven times. After treatment termination, mice were killed and GFP fluorescence levels were assessed in frozen sections of the cortex and striatum. Representative fluorescence microscopy images are shown. Positive GFP signals are shown in green, NeuN-stained neurons are shown in red and DAPI-stained nuclei are shown in blue. Scale bar = 100 μm.
Third, to directly visualize the in vivo distribution of the neuron-directed, self-assembled siRNAs, we intravenously injected a CMV-directed circuit expressing a GFP-silencing siRNA with or without an RVG-Lamp2b fusion protein (CMV-RVG-siRGFP versus CMV-siRGFP) into a transgenic mouse model ubiquitously expressing GFP. GFP fluorescence was significantly decreased in the livers of GFP-transgenic mice treated with CMV-RVG-siRGFP or CMV-siRGFP (Supplementary Fig. 4B), indicating that functional GFP siRNA was generated in the liver. Only the CMV-RVG-siRGFP circuit led to marked reductions in GFP levels in the cortex and striatum in GFP-transgenic mice (Fig. 3F and G), again validating the importance of the RVG guiding tag for the delivery of siRNAs to the brain. Accordingly, both GFP protein and GFP mRNA levels in the cortex and striatum were significantly suppressed by the CMV-RVG-siRGFP circuit in GFP-transgenic mice (Supplementary Fig. 5). These findings support the idea that the liver, through spontaneous organization and continuous release of siRNA-encapsulating exosomes, enables the delivery of functional siRNA into the cortex and striatum despite the presence of the blood–brain barrier, which limits the ability of siRNAs to access these brain regions.
Silencing of mHTT with self-assembled mHTT siRNA attenuates striatal and cortical neuropathology and behavioural deficits
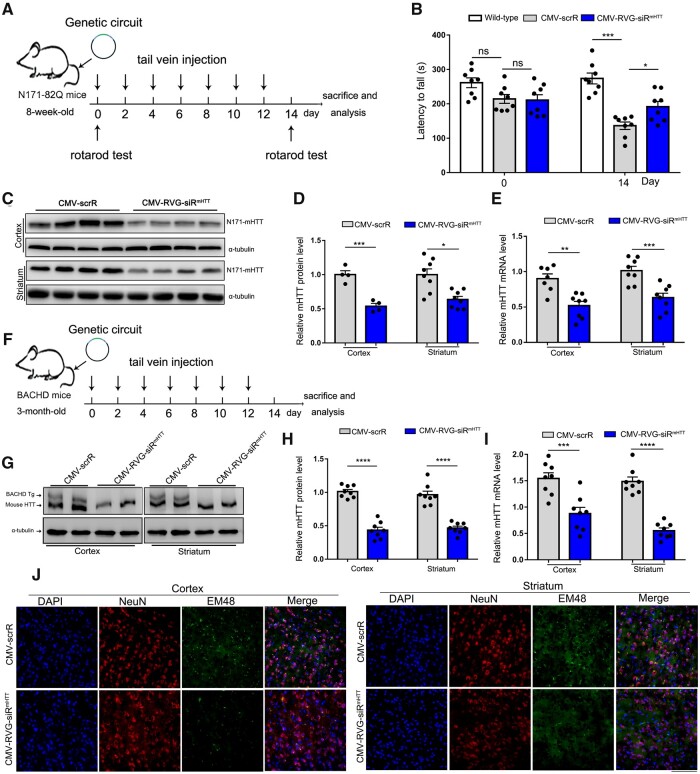
Next, we evaluated the therapeutic potential of the genetic circuits in three mouse models of Huntington’s disease.43–45 First, 8-week-old N171-82Q transgenic mice were intravenously injected with 5 mg/kg CMV-scrR or CMV-RVG-siRmHTT circuit every 2 days for a total of seven injections (Fig. 4A). N171-82Q mice express a fragment of human mHTT containing 82 CAG repeats and develop a progressive phenotype of Huntington’s disease including motor incoordination and neuropathology.10 As expected, motor deficit, which develops in N171-82Q mice beginning at 8 weeks of age, was clearly exacerbated in the N171-82Q mice after 2 weeks of treatment with the CMV-scrR circuit. This deficit aggravation manifested as an apparent latency to fall in the rotarod task for the CMV-scrR-treated N171-82Q mice compared with wild-type normal mice (Fig. 4B). In contrast, the N171-82Q mice treated with the CMV-RVG-siRmHTT circuit displayed significantly improved motor skill (Fig. 4B). Accordingly, treatment with the CMV-RVG-siRmHTT circuit led to significant reductions in mHTT protein and mRNA levels in the cortex and striatum in N171-82Q mice (Fig. 4C–E). These results indicate that intravenous injection of the CMV-RVG-siRmHTT circuit facilitates suppression of mHTT expression in the cortex and striatum and improves motor performance in N171-82Q Huntington’s disease mice.
Figure 4.
Evaluation of the therapeutic efficacy of self-assembled mHTT siRNA in N171-82Q and BACHD Huntington’s disease models. (A) Schematic of the experimental design. At 8 weeks of age, N171-82Q mice were intravenously injected with 5 mg/kg CMV-scrR or CMV-RVG-siRmHTT circuit every 2 days for a total of 2 weeks. After treatment termination, the mice were monitored to evaluate behavioural performance and mHTT accumulation. (B) Motor performance in a rotarod test. Non-transgenic littermates were included as behavioural controls (n = 8 in each group). (C) Western blot analysis of mHTT protein levels in the cortex and striatum. α-Tubulin served as the internal loading control. (D) Quantitation of the mHTT protein levels shown in C (n = 4 in cortex; n = 8 in striatum). (E) Quantitative RT-PCR analysis of mHTT mRNA levels in the cortex and striatum (n = 7–8 in each group). (F) Schematic of the experimental design. At 3 months of age, BACHD mice were intravenously injected with 5 mg/kg CMV-scrR or CMV-RVG-siRmHTT circuit every 2 days for a total of 2 weeks. After treatment termination, the mice were monitored to evaluate mHTT accumulation and neuropathology. (G) Western blot analysis of mHTT protein levels in the cortex and striatum. The more slowly migrating band is the human mHTT protein, and the more rapidly migrating band is mouse endogenous HTT. Endogenous HTT and α-tubulin served as the internal loading control. (H) Quantitation of the mHTT protein levels shown in G (n = 8 in each group). (I) Quantitative RT–PCR analysis of mHTT mRNA levels in the cortex and striatum (n = 8 in each group). (J) Immunofluorescence staining of aggregated mHTT (EM48, green), neurons (NeuN, red) and nuclei (DAPI, blue) in striatal and cortical sections. Scale bar = 50 μm. Values are presented as the means ± SEM. Significance was determined using two-sided t-test in D, E, H and I, or using one-way ANOVA followed by Bonferroni's multiple comparisons in B. *P < 0.05; ***P < 0.001; ****P < 0.0001; ns = not significant.
Second, the therapeutic efficacy of the CMV-RVG-siRmHTT circuit was evaluated in BACHD mice, which recapitulate the characteristics of human Huntington’s disease due to expression of a full-length human mHTT gene under the control of the endogenous HTT promoter.46 Since human mHTT and mouse endogenous HTT are simultaneously expressed in BACHD mice, this Huntington’s disease model is especially suitable for determining whether a genetic circuit designed to specifically silence human mHTT can discriminate between expanded human mHTT and mouse endogenous HTT transcripts in vivo.16,47 BACHD mice at 3 months of age were intravenously injected with the CMV-scrR or CMV-RVG-siRmHTT circuit seven times within 2 weeks (Fig. 4F). Selective inhibition of human mHTT protein rather than mouse endogenous HTT protein was observed in the cortex and striatum in CMV-RVG-siRmHTT-treated BACHD mice compared to their CMV-scrR-treated littermates (Fig. 4G and H). Likewise, significant reductions in human mHTT mRNA levels were detected in the cortex and striatum in BACHD mice after treatment with the CMV-RVG-siRmHTT circuit (Fig. 4I). More importantly, the amounts of neuropil aggregates, which are formed by abnormal folding and aggregation of mHTT protein, were significantly reduced in striatal and cortical neurons of BACHD mice by treatment with CMV-RVG-siRmHTT circuit; these reductions manifested as reduced EM48-staining intensity with an EM48 polyclonal antibody that preferentially recognizes and binds to aggregated mHTT protein (Fig. 4J). These results reveal that the CMV-RVG-siRmHTT circuit can cause efficient silencing of the mHTT gene, thereby preventing striatal and cortical neuropathology in BACHD mice.
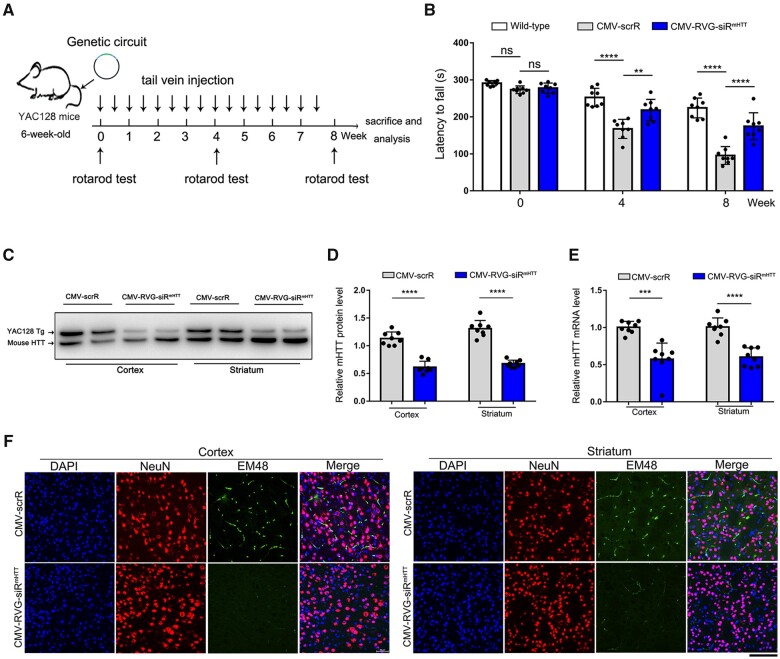
Third, to assess the long-term benefits of reductions in mHTT levels for Huntington’s disease therapy, the CMV-scrR or CMV-RVG-siRmHTT circuit was intravenously injected into 6-week-old YAC128 mice twice a week for a total of 8 weeks (Fig. 5A). YAC128 mice express a full-length human mHTT transgene with ∼120 CAG repeats and display many hallmarks of Huntington’s disease, including motor dysfunction and neuropathologies in the cortex and striatum.43,48 While the motor performance of the CMV-scrR-treated YAC128 mice progressively worsened with increasing age, treatment with CMV-RVG-siRmHTT significantly improved the motor coordination of YAC128 mice in the rotarod test (Fig. 5B). Moreover, remarkable reductions in mHTT protein and mRNA levels were observed in the cortex and striatum in CMV-RVG-siRmHTT-treated YAC128 mice (Fig. 5C–E). Consequently, mHTT aggregation in cortical and striatal neurons was largely ameliorated by the CMV-RVG-siRmHTT circuit (Fig. 5F). These results verify the longevity and beneficial effect of the CMV-RVG-siRmHTT circuit and suggest that brain-directed, self-assembled mHTT siRNA may be useful therapeutic tools for Huntington’s disease.
Figure 5.
Evaluation of the long-term therapeutic efficacy of self-assembled mHTT siRNA in the YAC128 Huntington’s disease model. (A) Schematic of the experimental design. At 6 weeks of age, YAC128 mice were intravenously injected with 5 mg/kg CMV-scrR or CMV-RVG-siRmHTT circuit twice a week for a total of 8 weeks. After treatment termination, the mice were monitored to evaluate behavioural performance, mHTT accumulation and neuropathology. (B) Motor performance on a rotarod test. Non-transgenic littermates were included as behavioural controls (n = 8 in each group). (C) Western blot analysis of mHTT protein levels in the cortex and striatum. The more slowly migrating band is the human mHTT protein, and the more rapidly migrating band is mouse endogenous HTT. Endogenous HTT served as the internal loading control. (D) Quantitation of the mHTT protein levels shown in (C) (n = 8 in each group). (E) Quantitative RT–PCR analysis of mHTT mRNA levels in the cortex and striatum (n = 8 in each group). (F) Immunofluorescence staining of aggregated mHTT (EM48, green), neurons (NeuN, red) and nuclei (DAPI, blue) in striatal and cortical sections. Scale bar = 50 μm. Values are presented as mean ± SEM. Significance was determined using two-sided t-test in D and E, or using one-way ANOVA followed by Bonferroni's multiple comparisons in B. ***P < 0.001; ****P < 0.0001; ns = not significant.
Confirmation of the safe and non-toxic features of the self-assembled mHTT siRNA in vivo
To confirm that mHTT siRNA can be used in vivo without causing toxic effects and tissue damage, C57BL/6J mice were intravenously injected seven times with PBS, CMV-scrR, CMV-siRmHTT or CMV-RVG-siRmHTT. Representative serum biochemical indexes, including alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, albumin, lactic dehydrogenase, total bilirubin, creatinine and blood urea nitrogen, were examined. No significant alterations in serum biochemical indexes were observed after treatment with the genetic circuits (Supplementary Fig. 6A). Similarly, the counts of red blood cells, white blood cells and platelets in peripheral blood were not affected by the genetic circuits (Supplementary Fig. 6B). In addition, repeated injection of the genetic circuits caused negligible tissue toxicity, as confirmed by histological examination of the livers, spleens, lungs and kidneys of genetic circuit-treated mice and by the absence of any noticeable irregularities or overt tissue damage in these tissues (Supplementary Fig. 6C). Overall, these results suggest that the genetic circuits can generate brain-directed, self-assembled mHTT siRNA in a safe, non-toxic and biocompatible manner.
Discussion
Oligonucleotide approaches based on siRNAs and ASOs provide promising new therapeutic strategies for direct intervention in Huntington’s disease through silencing of the causative protein, mHTT.10 Unfortunately, in vivo delivery of siRNAs and ASOs remains a major obstacle to translational success.49,50 In this study, we reprogrammed a native exosome-circulating system with artificial genetic circuits to facilitate the delivery of siRNAs in vivo. Our delivery strategy has several advantages over conventional approaches in three main areas: safety, efficiency and convenience. First, our strategy is safe. Current siRNA transfer techniques use viruses and synthetic agents as delivery vehicles. These foreign formulations may cause immune responses and side effects when used in vivo.51–53 Given the limitations of the current delivery vehicles, borrowing the body’s own small RNA transport machinery may be an optimal solution. Hence, we exploited the intrinsic properties of exosomes to functionally deliver siRNAs in vivo. The exosomes are derived from host cells, and the siRNA cargo loading is performed by endogenous systems; thus, this natural formulation is fully compatible with the immune system and has no toxicity or adverse effects. Second, our strategy is efficient. Because the cortex and striatum are among the most severely affected brain areas in Huntington’s disease,6 optimal Huntington’s disease therapeutics may require targeting of mHTT in both striatal and cortical neurons. Unfortunately, the poor biodistribution of siRNAs and ASOs to the striatum remains a major obstacle to successful treatment of Huntington’s disease. For example, it has been shown that intrathecal administration of ASOs can reduce HTT levels by 70% in the cortex but only by 35% in the striatum.54 Likewise, a bolus cisternal injection of ASOs reduces HTT levels by 60% in the cortex and by 36% in the striatum.54 Another study also revealed that injection of ASOs into the CSF of non-human primates can reduce HTT mRNA levels by 53% in the anterior cortex but only 25% in the striatum.16 These data generally support a preferential ASO distribution in the cortex rather than in the striatum when ASOs are applied with a local delivery strategy. In contrast, our strategy resulted in efficient accumulation of similar amounts of siRNA (∼325.77 pmol siRNA per gram of total RNA in the cortex versus ∼257.52 pmol siRNA per gram of total RNA in the striatum) and equivalent mHTT protein silencing efficiency in the cortex and striatum (∼50% reduction in the cortex versus ∼45% reduction in the striatum). Thus, our innovative strategy for siRNA delivery may provide an improved translational paradigm to optimize the dose of siRNA in deep brain regions. Since toxic protein aggregation in deep brain is a hallmark of many neurodegenerative diseases, in vivo self-assembled siRNAs are expected to accelerate the development of therapies for other neurodegenerative disorders, such as Parkinson’s disease and Alzheimer’s disease. Third, our strategy is convenient. In this study, functional delivery of mHTT siRNA to the cortex and striatum was achieved simply through intravenous injection of the genetic circuit; then, the siRNA was spontaneously produced by liver cells and transferred through the exosome-circulating system. This non-invasive strategy avoids the surgical procedures, high costs and intensive labour associated with conventional delivery techniques. Overall, our delivery strategy compensates for the shortcomings of conventional siRNA delivery systems and may open up new avenues for future treatment of Huntington’s disease.
Synthetic biology holds great potential to shape new biomedicine practices and to combat hitherto intractable diseases, including Huntington’s disease. However, most genetic networks are deployed in microbes or immortalized cell lines32; since clinical problems are usually correlated with the complex and dynamic endogenous environment, there exists a critical need to move synthetic biology towards in vivo systems. Therefore, the central hurdle concerning the therapeutic application of synthetic biology is the efficient delivery of genetic circuits into the human body and the rational assembly of genetic circuits for programming and control of host cell functions.55 In this study, we conceptualized genetic circuits in a new way as ‘medicines’ instead of ‘agents’. This strategy is advantageous because of its intelligence, controllability and composability. With regard to intelligence, one of the pursuits of synthetic biology is to redesign existing natural biological systems. One approach to ensure that synthetic biology systems work well in the complex natural environment is to gradually step through increasing levels of complexity in an attempt to best approximate the natural system, while a more elegant solution might be to design systems that can evolve and adapt to the natural environment.31 Here, we converted the host system, engineering it to produce ‘living therapeutics’: once they took up the intravenous genetic circuits, liver cells expressed the siRNA payloads and coordinated the self-assembly of the siRNA into secretory exosomes. The secretory siRNA was spontaneously transferred by the native exosome-circulating system to target cells. Thus, our synthetic biology design borrowed the body’s own workshops and reprogrammed host systems to perform user-defined behaviours. This intelligent strategy is beyond the paradigm of current applications of synthetic biology in biomedicine, such as implantation of ex vivo-engineered cells (which involves the major challenge of ensuring that there is no interference between implanted cells and host homeostasis) or virus-mediated delivery of genetic circuits in vivo.56 Our system is also controllable. In vivo applications of synthetic biology are often limited by the sizes of genetic circuits, which often exceed the capacity of delivery vehicles. More modules are needed to realize complex biological functions, which will inevitably increase the sizes of genetic circuits, leading to decreased stability and less controllability. By combining the naturally existing small RNA assembly and transportation machinery with artificial genetic circuits, our design helps to minimize the sizes of genetic circuits so that the circuits are small enough for stability and easy delivery but complex enough to carry out biological functions in vivo. Finally, our approach is composable. A key challenge of genetic circuit engineering is composability (the ability to connect any two modules and yield predictable behaviours).57 We have demonstrated here that the genetic circuit is composable and allows the free combination of three different functional modules. Because the sequence in the siRNA-expressing backbone and the guidance tag are switchable, our strategy theoretically provides a personalized treatment approach that can be tailored for different genes in different tissues. In summary, we have developed a synthetic biology strategy that allows precise control of siRNA distribution and target gene expression in a complex, dynamic in vivo environment. With such a toolbox, gene networks can be engineered and remodelled to target multiple different genes, cell populations and symptoms, thereby addressing a broad range of needs in biomedicine.
In this study, RVG-tagged exosomes had the ability to transport their cargo siRNAs to striatal and cortical neurons, bypassing the blood–brain barrier. Indeed, RVG peptide has been frequently used to confer a neuron-targeting property on exosomes, liposomes, micelles and polymers, because this ligand can specifically recognize the nicotinic acetylcholine receptor (nAChR),58–60 which is widely located on the extracellular surface of microvascular endothelial cells and neurons and on the surface of the blood–brain barrier. Therefore, RVG-coupled materials are more likely to localize to nerve cells through nAChR-mediated cellular transduction. However, although the brain-specific RVG peptide recombinantly expressed on exosomal surfaces is necessary to guide exosomes towards nerve cells, the RVG peptide itself is not necessarily involved in the transcytosis process of exosomes across the blood–brain barrier, and the influence of RVG peptide on the biodistribution of exosome-enclosed siRNAs remains to be fully elucidated. According to previous studies, exosomes can be taken up by recipient cells through several ways, including direct membrane fusion and endocytosis/phagocytosis.61 It is still not clear which pathway is the main one used by RVG-tagged exosomes. One study found that exosomes can be internalized into multivesicular bodies (MVBs) of recipient cells and then released again to be reinternalized into MVBs of secondary recipient cells; thus, by moving from cell to cell via the MVB compartment, exosomes could cross the multiple cell layers in the blood–brain barrier.62 Likewise, another study revealed that tumour-derived extracellular vesicles could cross the blood–brain barrier by modulating the endocytic pathway in brain endothelial cells to increase the efficiency of their transcellular transport.63 In agreement with these observations, we also found that PKH26-labelled RVG exosomes could penetrate the blood–brain barrier and generate apparent fluorescent signals in the cortex and striatum. These findings suggest that absorption of whole exosomes via phagocytosis or endocytosis is one of the mechanisms for exosomes and its cargos to enter nerve cells. For the siRNAs that successfully get into nerve cells through direct membrane fusion, they might be repackaged into new exosomes within cytoplasm, which are difficult to track and visualized using current methods. Moreover, these repackaged exosomes, because of the rearrangement of surface proteins, may exhibit high affinity for cells others than neurons, including microglia and astrocytes.64,65 In agreement with this hypothesis, when we injected the CMV-siRGFP into GFP-transgenic mice, we also observed an apparent reduction of GFP fluorescence in the regions deficient of NeuN staining, indicating that other CNS cell types such as microglia and astrocytes could also take up some siRNA. The related molecular mechanisms underlying these phenotypes need to be further explained to provide insights that can guide the design of better brain delivery systems for Huntington’s disease and other neurological diseases.
Admittedly, there is still much room for improvement with our strategy. First, its safety, dosing and durability need to be evaluated more systematically and accurately. In particular, the biodistribution of siRNA to the cortex and striatum needs to be adequately fine-tuned. Second, patients with Huntington’s disease express both mutant and wild-type HTT alleles.15 In this study, a sequence targeting human mHTT was designed in a non-selective way; therefore, our approach may not allow for targeting of the mutant allele with high selectivity while preserving wild-type allele expression at a normal level. Since wild-type HTT has numerous physiological activities that are important for neuronal function, suppression of both mutant and wild-type alleles may not be appropriate and allele-specific silencing of mutant alleles via targeting of the associated single-nucleotide polymorphisms (SNPs) is a promising alternative. Third, regular injection of genetic circuits is currently indispensable for long-term Huntington’s disease treatment because the siRNA effect is not permanent. The problems associated with repeated injection should be dealt with properly. Furthermore, since the injected circuits are mainly processed in the liver, it is quite important to examine whether the physiological functions and status of the liver are impaired after long-term therapy. Finally, the therapeutic potential has only been verified in mouse models of Huntington’s disease. This approach should be further investigated in models with larger mammals, such as pig models of Huntington’s disease.
In summary, we have developed a synthetic biology strategy that reprograms the host liver to direct the synthesis and self-assembly of mHTT siRNA into secretory exosomes and facilitate the in vivo delivery of siRNA through circulating exosomes to the cortex and striatum. This strategy results in substantial reductions in mHTT expression and symptom alleviation in mouse models of Huntington’s disease. Our strategy overcomes the main barrier to the clinical applications of siRNAs in Huntington’s disease therapy and may provide a promising solution for the treatment of Huntington’s disease.
Supplementary Material
Acknowledgements
We would like to thank Dr Xiaoyang Zhang (School of Life Sciences, Nanjing University) for her help in analysing the results of brain sections.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 32022015 and 31871295), the National Key Research and Development Program of China, Stem Cell and Translational Research (No. 2017YFA0105104), the Natural Science Foundation of Jiangsu Province (No. BE2016737), the Guangdong Provincial Science and Technology Plan Project (No. 2016B030230002), the Guangdong Provincial Key Laboratory for Diagnosis and Treatment of Major Neurological Diseases (2017B030314103) and the Fundamental Research Funds for the Central Universities (020814380162).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- ASO
antisense oligonucleotide
- CMV
cytomegalovirus
- GFP
green fluorescent protein
- Lamp2b
lysosome-associated membrane glycoprotein 2b
- mHTT
mutant huntingtin
- RVG
rabies virus glycoprotein
- siRNA
small interfering RNA
References
- 1. Novak MJ, Tabrizi SJ.. Huntington's disease. BMJ. 2010;340:c3109. [DOI] [PubMed] [Google Scholar]
- 2. Ross C, Aylward E, Wild E, et al. Huntington disease: Natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10(4):204–216. [DOI] [PubMed] [Google Scholar]
- 3. Tabrizi S, Flower M, Ross C, Wild E.. Huntington disease: New insights into molecular pathogenesis and therapeutic opportunities. Nat Rev Neurol. 2020;16(10):529–546. [DOI] [PubMed] [Google Scholar]
- 4. MacDonald ME, Duyao MP, Myers RH, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72(6):971–983. [DOI] [PubMed] [Google Scholar]
- 5. Sathasivam K, Neueder A, Gipson T, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A. 2013;110(6):2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ross CA, Tabrizi SJ.. Huntington's disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10(1):83–98. [DOI] [PubMed] [Google Scholar]
- 7. Bates G. Huntingtin aggregation and toxicity in Huntington's disease. Lancet. 2003;361(9369):1642–1644. [DOI] [PubMed] [Google Scholar]
- 8. Walker FO. Huntington's disease. Lancet. 2007;369(9557):218–228. [DOI] [PubMed] [Google Scholar]
- 9. McColgan P, Tabrizi SJ.. Huntington's disease: A clinical review. Eur J Neurol. 2018;25(1):24–34. [DOI] [PubMed] [Google Scholar]
- 10. Sah D, Aronin N.. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121(2):500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davidson B, McCray P.. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12(5):329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kole R, Krainer A, Altman S.. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11(2):125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rinaldi C, Wood M.. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat Rev Neurol. 2018;14(1):9–21. [DOI] [PubMed] [Google Scholar]
- 14. Xia H, Mao Q, Paulson HL, Davidson BL.. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20(10):1006–1010. [DOI] [PubMed] [Google Scholar]
- 15. Yu D, Pendergraff H, Liu J, et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150(5):895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kordasiewicz H, Stanek L, Wancewicz E, et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 2012;74(6):1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Svrzikapa N, Longo K, Prasad N, et al. Investigational assay for haplotype phasing of the huntingtin gene. Mol Ther Methods Clin Dev. 2020;19:162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, et al. Targeting huntingtin expression in patients with Huntington's disease. N Engl J Med. 2019;380(24):2307–2316. [DOI] [PubMed] [Google Scholar]
- 19. Gomes M, Martins S, Sarmento B.. siRNA as a tool to improve the treatment of brain diseases: Mechanism, targets and delivery. Ageing Res Rev. 2015;21:43–54. [DOI] [PubMed] [Google Scholar]
- 20. Pardridge WM. shRNA and siRNA delivery to the brain. Adv Drug Deliv Rev. 2007;59(2-3):141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caron NS, Dorsey ER, Hayden MR.. Therapeutic approaches to Huntington disease: From the bench to the clinic. Nat Rev Drug Discov. 2018;17(10):729–750. [DOI] [PubMed] [Google Scholar]
- 22. Roberts T, Langer R, Wood M.. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. 2020;19(10):673–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kalluri R, LeBleu V.. The biology function and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Brien K, Breyne K, Ughetto S, Laurent L, Breakefield X.. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21(10):585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–144. [DOI] [PubMed] [Google Scholar]
- 26. Li J, Zhang Y, Liu Y, et al. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem. 2013;288(32):23586–23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yin Y, Cai X, Chen X, et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014;24(10):1164–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y, Zhou G, Tian C, et al. Exosome-mediated small RNA delivery for gene therapy. Wiley Interdiscip Rev RNA. 2016;7(6):758–771. [DOI] [PubMed] [Google Scholar]
- 29. van Niel G, D'Angelo G, Raposo G.. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. [DOI] [PubMed] [Google Scholar]
- 30. Syn N, Wang L, Chow E, Lim C, Goh B.. Exosomes in cancer nanomedicine and immunotherapy: prospects and challenges. Trends Biotechnol. 2017;35(7):665–676. [DOI] [PubMed] [Google Scholar]
- 31. Kitada T, DiAndreth B, Teague B, Weiss R.. Programming gene and engineered-cell therapies with synthetic biology. Science. 2018;359(6376):eaad1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weber W, Fussenegger M.. Emerging biomedical applications of synthetic biology. Nat Rev Genet. 2011;13(1):21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Atsumi S, Hanai T, Liao J.. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. [DOI] [PubMed] [Google Scholar]
- 34. Elani Y. Interfacing living and synthetic cells as an emerging frontier in synthetic biology. Angew Chem Int Ed Engl. 2021;60(11):5602–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu Z, Zhang X, Zhou X, et al. In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res. 2021;31(6):631–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu F, Song Y, Liu D.. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6(7):1258–1266. [DOI] [PubMed] [Google Scholar]
- 37. Zhang G, Budker V, Wolff J.. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Therapy. 1999;10(10):1735–1737. [DOI] [PubMed] [Google Scholar]
- 38. Tiago Ferreira WR. ImageJ User Guide. https://imagej.nih.gov/ij/docs/guide/
- 39. Marco S, Giralt A, Petrovic M, et al. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington's disease models. Nat Med. 2013;19(8):1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fox L, Kim K, Johnson C, et al. Huntington's disease pathogenesis is modified in vivo by Alfy/Wdfy3 and selective macroautophagy. Neuron. 2020;105(5):813–821.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood M.. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. [DOI] [PubMed] [Google Scholar]
- 42. DiFiglia M, Sena-Esteves M, Chase K, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104(43):17204–17209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hodgson J, Agopyan N, Gutekunst C, et al. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23(1):181–192. [DOI] [PubMed] [Google Scholar]
- 44. Menalled LB, Chesselet M-F.. Mouse models of Huntington's disease. Trends Pharmacol Sci. 2002;23(1):32–39. [DOI] [PubMed] [Google Scholar]
- 45. Beal MF, Ferrante RJ.. Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat Rev Neurosci. 2004;5(5):373–384. [DOI] [PubMed] [Google Scholar]
- 46. Wang N, Gray M, Lu X, et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington's disease. Nat Med. 2014;20(5):536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gu X, Cantle J, Greiner E, et al. N17 Modifies mutant huntingtin nuclear pathogenesis and severity of disease in HD BAC transgenic mice. Neuron. 2015;85(4):726–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pouladi MA, Morton AJ, Hayden MR.. Choosing an animal model for the study of Huntington's disease. Nat Rev Neurosci. 2013;14(10):708–721. [DOI] [PubMed] [Google Scholar]
- 49. Kim B, Park J, Sailor M.. Rekindling RNAi therapy: Materials design requirements for in vivo siRNA delivery. Adv Mater. 2019;31(49):e1903637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McKenzie LK, El-Khoury R, Thorpe JD, Damha MJ, Hollenstein M.. Recent progress in non-native nucleic acid modifications. Chem Soc Rev. 2021;50(8):5126–5164. [DOI] [PubMed] [Google Scholar]
- 51. Vogel G. Gene therapy. FDA moves against Penn scientist. Science. 2000;290(5499):2049–2051. [DOI] [PubMed] [Google Scholar]
- 52. Zuckerman J, Gritli I, Tolcher A, et al. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc Natl Acad Sci U S A. 2014;111(31):11449–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gabisonia K, Prosdocimo G, Aquaro G, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569(7756):418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu T, Su F, Feng Y, et al. Mesenchymal stem cells alleviate AQP-4-dependent glymphatic dysfunction and improve brain distribution of antisense oligonucleotides in BACHD mice. Stem Cells. 2020;38(2):218–230. [DOI] [PubMed] [Google Scholar]
- 55. Khalil A, Collins J.. Synthetic biology: Applications come of age. Nat Rev Genet. 2010;11(5):367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tan X, Letendre J, Collins J, Wong W.. Synthetic biology in the clinic: Engineering vaccines, diagnostics, and therapeutics. Cell. 2021;184(4):881–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Glass D, Alon U.. Programming cells and tissues. Science. 2018;361(6408):1199–1200. [DOI] [PubMed] [Google Scholar]
- 58. Lentz T, Wilson P, Hawrot E, Speicher D.. Amino acid sequence similarity between rabies virus glycoprotein and snake venom curaremimetic neurotoxins. Science. 1984;226(4676):847–848. [DOI] [PubMed] [Google Scholar]
- 59. Kumar P, Wu H, McBride J, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448(7149):39–43. [DOI] [PubMed] [Google Scholar]
- 60. Lentz T, Burrage T, Smith A, Crick J, Tignor G.. Is the acetylcholine receptor a rabies virus receptor? Science. 1982;215(4529):182–184. [DOI] [PubMed] [Google Scholar]
- 61. Mulcahy LA, Pink RC, Carter DRF.. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014. Aug 4;3:10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. El Andaloussi S, Lakhal S, Mäger I, Wood MJA.. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65(3):391–397. [DOI] [PubMed] [Google Scholar]
- 63. Morad G, Carman CV, Hagedorn EJ, et al. Tumor-derived extracellular vesicles breach the intact blood-brain barrier transcytosis. ACS Nano. 2019;13(12):13853–13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rodrigues G, Hoshino A, Kenific CM, et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat Cell Biol. 2019;21(11):1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Men Y, Yelick J, Jin S, et al. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat Commun. 2019;10(1):4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.