Abstract
Background
Vancomycin-resistant enterococci (VRE) are a major cause of morbidity and mortality in immunocompromised patients. Tracking the dissemination of VRE strains is crucial to understand the dynamics of emergence and spread of VRE in the hospital setting.
Methods
Whole genome sequencing (WGS) and phylogenetic analyses were performed to identify dominant VRE strains and potential transmission networks between 35 patients with VRE-positive rectal swabs and their rooms (main rooms and bathrooms) on the leukemia (LKM) and the hematopoietic cell transplant (HCT) floors. Sequence types (STs), drug resistance genes, and patients’ outcomes were also determined.
Results
A total of 89 VRE strains grouped into 10 different STs, of which newly described STs were isolated from both floors (ST736, ST494, ST772, and ST1516). We observed highly genetically related strains transmitted between rooms, floors, and time periods in an average period of 39 days (ranging from 3 to 90 days). Of 5 VRE bacteremia events, 3 strains were lacking the pili operon fms14–17–13 (ST203) and the remaining 2 were resistant to daptomycin (DAP; ST736, ST664). Of 10 patients harboring DAP-resistant strains, only 2 were exposed to DAP within 4 months before strain recovery.
Conclusions
Our comparisons of VRE strains derived from the environment and immunocompromised patients confirmed horizontal transfer of highly related genetic lineages of multidrug-resistant (particularly to DAP) VRE strains between HCT and LKM patients and their room environment. Implementing WGS can be useful in distinguishing VRE reservoirs where interventions can be targeted to prevent and control the spread of highly resistant organisms.
Keywords: vancomycin-resistant enterococci (VRE), immunocompromised, daptomycin (DAP), whole genome sequencing, transmission
We observed highly genetically related strains transmitted vancomycin-resistant enterococci (VRE) strains between distinct rooms, floors, and time periods in an average period of 39 days. Ten patients had daptomycin (DAP)-resistant strains, but only 2 were exposed to DAP.
Vancomycin-resistant enterococci (VRE) constitute a major cause of morbidity and mortality in patients with underlying malignancies [1]. In allogeneic hematopoietic cell transplant (allo-HCT) recipients, subjected to high antibiotic administration, over one third of the population is colonized with VRE [1]. VRE infections range from 3.6% to 22% within the first year after transplantation, with 30-day mortality rates of 38% and sometimes up to 88% [2]. In these patients, loss of mucosal immunity and disruption of the gastrointestinal barrier contribute to the translocation of VRE strains into the bloodstream after colonization [3, 4]. Of interest, VRE-colonized HCT recipients also experience higher rates of norelapse mortality compared to noncolonized patients [5].
Daptomycin (DAP) has been used for treatment of VRE infections since 2003 [6]. This drug has bactericidal activity against VRE with low toxicity potential [7]. However, emergence of DAP-resistant VRE causing invasive infections seems to be on the rise [6]. In fact, a study showed an increase in DAP-resistant VRE bacteremic isolates from 3.4% in 2007 to 15.2% in 2009, with 83% occurring in patients with hematologic malignancies [6]. DAP exposure, along with immunosuppression, might be associated with the development of resistance in enterococci. In a recent study, colonization or infection with DAP non-susceptible enterococci was detected in 60% of patients who had exposure to DAP therapy for about 14 days [8]. Of these patients, 68% had DAP nonsusceptible enterococci infections and 36% experienced bacteremia. In another study, patients with DAP-resistant enterococcal bacteremia were more likely to be HCT recipients (74%) and to be exposed to high-dose (>6mg/kg per day) DAP (71%) than patients with other hematologic malignancies [2]. Nonetheless, development of DAP resistance in the absence of DAP exposure has also been documented [6, 9, 10].
Considering the serious public health threat associated with the emergence of DAP-resistant VRE and VRE in general, studying the clinical epidemiology and transmission dynamics of these organisms are the initial steps to understand their emergence and spread in the hospital setting. In this study, using whole genome sequencing (WGS) and single nucleotide polymorphism (SNP) analyses, we identified VRE dynamics and transmission networks in leukemia (LKM) and allo-HCT patients at the University of Texas MD Anderson Cancer Center, Houston, Texas, USA. In addition, we studied the transmission and influence of DAP-resistant strains in patients’ outcomes.
MATERIALS AND METHODS
Sampling and Selection of VRE Isolates
Rectal swabs are performed routinely as part of an infection control surveillance program for HCT recipients and LKM patients on admission and once a week during their stay until discharge at MD Anderson Cancer Center, Houston, Texas. Environmental samples were collected from rooms on 2 consecutive floors that housed LKM and HCT patients who had VRE-positive rectal swabs between April and August 2012 (clinical and environmental samples). The environmental samples from the main room and bathroom of VRE-positive colonized patients were obtained using sterile swabs, after patient discharge, but before terminal cleaning using standard sampling practices. Sampled high-touch surfaces included bedrails, tables, telephones, doorknobs, and call button in the room, and toilet seat, doorknobs, and handrails in the bathroom. All samples were evaluated qualitatively for VRE presence by inoculating swabs in Enterococcosel medium (Beckton Dickinson Company, Franklin Lakes, New Jersey, USA). A total of 133 clinical and environmental VRE strains were isolated. Eighty-nine of the 133 VRE isolates were selected based on pairing VRE-positive environmental swabs (28 bathroom/26 main room samples) from the same rooms of patients with VRE-positive rectal swabs (35 of 64 rectal swabs).
Data Collection
Data were retrieved retrospectively from infection control and pharmacy databases. The variables collected included length of stay, dates of VRE positive cultures, time to acquisition of VRE colonization or infection, room number and unit (HCT/LKM floors), admission and discharge dates, exposure to DAP, and development of bacteremia as a main clinical outcome.
Whole Genome Sequencing and Analyses
DNA extraction was performed on the 89 selected VRE isolates using QIAamp DNA Stool Kit (Qiagen, Germantown, Maryland, USA). WGS was performed using Illumina MiSeq (250 bp paired-end reads) [11]. Sequencing adapters and low-quality bases were removed with Trimmomatic v0.36 [12], genomes were assembled using SPAdes [13], and annotated using Prokka [14]. Core genomes were identified using Roary [15]. VRE sequence types (STs) were determined using the MLST database [16]. Genes conferring resistance to the major antibiotics against enterococci were identified using Abricate (ResFinder) [17]. Amino-acid substitutions in predominant LiaFSR proteins were identified using BLAST+ v2.6.0 [18] for DAP resistance (threonine to alanine in amino-acid position 120 of the LiaS histidine-kinase sequence and trytophan to cystein in position 73 of the LiaR response regulator) [19]. The identification of highly related VRE isolates and VRE transmission networks between patients and rooms on both floors was conducted according to the number of SNPs found within the core genome of VRE isolates. Briefly, the core genome alignment file was processed with Snippy software to identify the number of SNPs between all isolates [14, 15]. We defined highly related strains as those differing by 5 SNPs or less. The 5 SNP threshold was selected as 62% of genomes were within 0–5 SNPs of at least 1 other isolate to a maximum of 5 other isolates [20, 21]. Phylogenetic trees were constructed using filtered core genome SNPs, and a maximum likelihood tree search algorithm RAxML [22] and iTOL softwares were used to generate and visualize the tree [23]. To further assess the SNPs called within this analysis, we sequenced one ST736 isolate (33-S-51112) on an Oxford Nanopore GridION X5 sequencer using an R9.4 flow cell and the SQK-RBK004 rapid barcoding kit. A consensus assembly was generated using long-read sequencing and the short-read Illumina data using Flye hybrid assembly pipeline (version 0.6; https://github.com/wshropshire/flye_hybrid_assembly_pipeline). This consensus assembly was used as the reference for reference-based SNP calling of all ST736 within this study using Snippy and adjusting for recombination using Gubbins [24].
Ultraviolet-C Device Disinfection Devices
An ultraviolet-C pulsed-field xenon device (PX-UV) was tested for its efficacy to control the spread of nosocomial VRE infections [25] in a controlled prospective study. This device generates a broad-spectrum, high-intensity ultraviolet light through pulsed xenon flash lamps to deactivate and kill bacteria, spores, and viruses on high-touch surfaces in 5 minutes or less [25, 26]. A disinfection step using the PX-UV device versus no intervention was performed after discharge of VRE-positive patients and after terminal cleaning of rooms on the LKM and HCT floors. Information on the acquisition of nosocomial VRE infection by the subsequent room’s occupant was captured as an outcome measure.
RESULTS
Characteristics of the VRE Strains
All 89 VRE strains belonged to the species Enterococcus faecium and were classified by their source of isolation and the inpatient floors they were isolated from (LKM and HCT) (Table 1). Percentages of VRE recovery were very similar and equally distributed on both LKM and HCT floors according to the type and source of VRE isolation. When comparing the MLST profile of these isolates, 10 different STs were identified (Table 1). Most of the isolates (80%) had known STs, whereas the rest exhibited potential recombination events in 1or more of the housekeeping genes used for MLST profiling and had unknown STs (Table 1 and Supplementary Table 1). Among those with known STs, 75% found on both floors grouped under STs 18 (n = 25), 584 (n = 16), and 736 (n = 12) (Table 1). One strain belonging to ST772 was found on the HCT floor. Moreover, only 1 strain found on the LKM floor belonged to ST494. This VRE strain was the only isolate harboring the vanB gene cluster. All other VRE isolates contained the vanA cluster. Finally, 1 strain on the LKM floor belonged to ST1516, a new identified ST in March 2019 according to the MLST website that did not group under any clonal complex yet.
Table 1.
Characteristics of the Selected 89 Strains
| HCT Floor (n = 63) | LKM Floor (n = 26) | ||||
|---|---|---|---|---|---|
| DAP-R | Others | DAP-R | Others | ||
| Type | |||||
| Environmental | Bathroom, n (%)a | 4 (6) | 17 (27) | 0 | 7 (27) |
| Main room, n (%) | 5 (8) | 12 (19) | 2 (8) | 7 (27) | |
| Patient | Rectal swabs, n (%) | 6 (10) | 19 (30) | 2 (8) | 8 (30) |
| VRE bacteremia | 1 (2) | 1 (2) | 1 (4) | 2 (8) | |
| Sequence types | |||||
| Unknown, n (%)b | 5 (8) | 7 (11) | 0 | 6 (23) | |
| Known, n (%) | |||||
| ST18, n (%) | 0 | 19 (30) | 0 | 6 (23) | |
| ST584, n (%) | 0 | 12 (19) | 0 | 4 (15) | |
| ST736, n (%) | 8 (13) | 0 | 4 (15) | 0 | |
| ST203, n (%) | 0 | 4 (6) | 0 | 2 (8) | |
| ST412, n (%) | 0 | 4 (6) | 0 | 0 | |
| ST664, n (%) | 2 (3) | 0 | 0 | 0 | |
| ST17, n (%) | 0 | 1 (2) | 0 | 2 (8) | |
| ST772, n (%) | 0 | 1 (2) | 0 | 0 | |
| ST494, n (%) | 0 | 0 | 0 | 1 (4) | |
| ST1516, n (%) | 0 | 0 | 0 | 1 (4) | |
Abbreviations: DAP-R, daptomycin resistant strains; HCT, hematopoietic cell transplant; LKM, leukemia; n, number; STs, sequence types; VRE, vancomycin-resistant enterococci.
a Percentages are in relation to either the HCT or the LKM total patient numbers.
b Due to recombination events in one or more of the 7 housekeeping genes selected for VRE MLST.
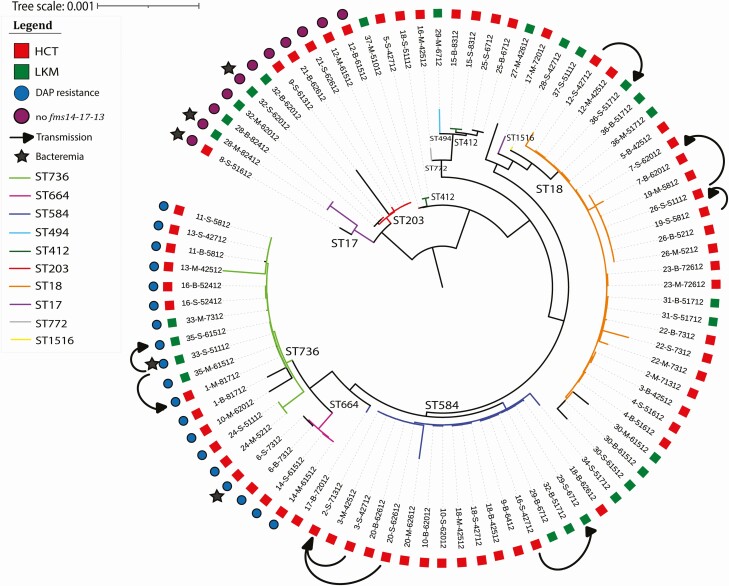
DAP-resistant strains were compared to the other VRE strains on both the LKM and the HCT floors (Table 1). All DAP-resistant strains grouped either under ST736 or ST664. These latter STs were not found for any of the other VRE strains (Figure 1 and Supplementary Figure 1). Interestingly, DAP-resistant strains were responsible of around an equal number of bacteremia event on both floors when compared to the other VRE strains; 12.5% of VRE strains collected in this cohort were DAP-resistant with 3% conferring bacteremia. In fact, 2 of these VRE strains recovered from blood cultures were DAP-resistant, and the remaining 3 were lacking the putative pilus operon fms14-17-13 (Figure 1). Most of the 89 VRE strains were resistant to macrolides, lincomide, streptogramin B (97%), and aminoglycosides (81%) (Table 2). An overview of the 89 VRE strains clustered according to SNPs found on their core genome, their sources, STs, and any additional information is shown in the phylogenetic trees in Figure 1 and Supplementary Figure 1.
Figure 1.
Phylogenetic tree showing the genetic relatedness according to SNPs found in the whole genome of the 89 VRE isolates. Transmissions between patients are shown on the figure. Abbreviations: B, bathroom; DAP, daptomycin; HCT, hematopoietic cell transplant; LKM, leukemia; M, main room; S, rectal swabs; SNP, single nucleotide polymorphism; ST, sequence type; VRE, vancomycin-resistant enterococci.
Table 2.
Antibiotic Resistance Genes Present in the 89 Vancomycin-resistant Enterococci (VRE) Strains
| Total | HCT (n = 63) | LKM (n = 26) | ||
|---|---|---|---|---|
| Vancomycin | vanA, n (%) | 88 (99) | 63 (72) | 25 (28) |
| vanB, n (%) | 1 (1) | 0 | 1 (100) | |
| Daptomycina | n (%) | 19 (21) | 15 (79) | 4 (21) |
| Macrolide | erm(B), n (%) | 56 (63) | 39 (70) | 17 (30) |
| Lincosamide | Inu(B), n (%) | 26 (29) | 19 (73) | 7 (27) |
| Macrolide, lincosamide and streptogramin B | msr(C), n (%) | 84 (94) | 58 (69) | 26 (31) |
| Tetracycline | 65 (73) | 46 (71) | 19 (29) | |
| Tetracycline only | tet(L), n (%) | 36 (40) | 24 (67) | 12 (33) |
| Minocycline and tetracycline | tet(M), n (%) | 31 (35) | 23 (74) | 8 (26) |
| All | tet(L) and tet(M), n (%) | 21 (24) | 15 (71) | 6 (29) |
| Aminoglycoside | 72 (81) | 52 (72) | 20 (28) | |
| Streptomycin | ant(6)-Ia, n (%) | 6 (7) | 3 (50) | 3 (50) |
| Kanamycin, neomycin | aph(3′)-III, n (%) | 23 (26) | 17 (74) | 6 (26) |
| All | ant(6)-Ia and aph(3’)-III, n (%) | 43 (48) | 32 (74) | 11 (26) |
| Trimethoprim | dfrG, n (%) | 26 (29) | 17 (65) | 9 (35) |
Abbreviations: HCT, hematopoietic cell transplant; LKM, leukemia; n, number.
aDaptomycin resistance was conferred by the presence of amino-acid substitutions in predominant LiaFSR proteins; threonine to alanine on position 120 of the LiaS protein sequence and trytophan to cystein on position 73 the LiaR sequence.
VRE Transmission Network
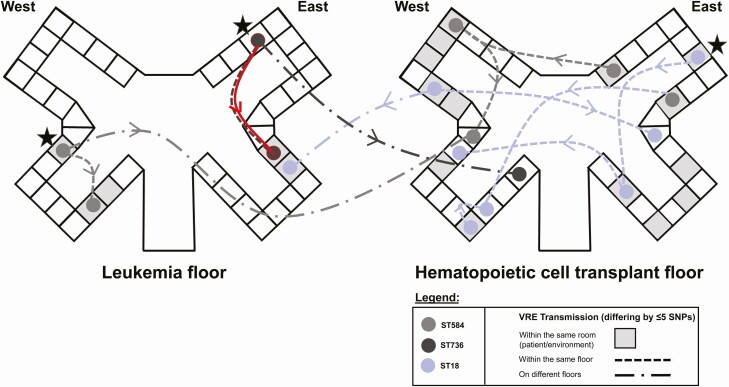
The core genome length used in our phylogenetic analysis and SNP distance assessments consisted of 728 056 bases, with the entire pan-genome consisting of 5 549 736 bases. The VRE strains isolated from patients and their respective room environment (pairs) were found to be highly genetically related. We also found highly related clusters of VRE isolates recovered from different rooms and floors, including 5 on the HCT and 3 on the LKM units (Figure 2). The transmission events occurred within a mean of 39 days (ranging from 3 to 90 days). The transmitted VRE strains on the LKM floor belonged to ST736 and ST584. Those transmitted on the HCT floor belonged to ST18 and the VRE strains found on both floors belonged to ST584, ST18, and ST736 (Figures 1 and 2). Additionally, a heatmap representing all clusters of the 89 strains can be found in the Supplementary material (Supplementary Figure 2).
Figure 2.
Transmission networks of VRE isolates differing by ≤5 SNPs in their core genomes on stem cell transplant and leukemia floors. Potential index cases of VRE are marked with a star. Possible daptomycin-resistant VRE strain transmission between rooms is highlighted in red. Transmission time period range from 3 to 90 days with an average of 39 days. Abbreviations: SNP, single nucleotide polymorphism; VRE, vancomycin-resistant enterococci.
We also further investigated the SNP distances between isolates within ST736 using the hybrid assembly of 33-S-51112 as a whole-genome reference. We observed a slight increase in SNPs called between isolates (max—70, mean—27.98) but observed the same clustering as completed with the full data set using a core-gene based method (Supplementary Table 2).
Daptomycin Exposure
A total of 19 strains were found to be resistant to DAP and harbored chromosomally encoded LiaRW73C and LiaST120A substitutions (Figure 1) [19]. Of the 19 DAP-resistant VRE strains, 8 were isolated from patients’ rectal swabs and 11 from their respective rooms. Interestingly, only 2 of the 8 patients with DAP resistant-VRE were exposed to DAP for 5 and 150 days, respectively. The remaining 6 patients had no prior exposure to DAP raising the possibility of transmission of these highly resistant strains between patients or their environment. In fact, 1 patient on the northeast side of the LKM floor was treated with DAP for VRE bacteremia and was still positive for VRE after 1 month. A possible transmission of this DAP-resistant VRE strain may have occurred 1 month later to another patient on the southeast side of the LKM floor (0 SNP difference between both patients’ strains) (Figure 2, highlighted in red). A total of 4 of the 8 patients with DAP-resistant strains died within a median of 2 months after strain recovery (range of 1–5 months). Deaths were mainly attributed to respiratory failure or cancer relapse.
PX-UV Study
After confirming transmission of VRE strains between our patients’ floors, and the likely contribution of environmental contamination in the transmission network, we sought to evaluate the efficacy of a PX-UV device versus no intervention after sampling each patient’s room. Half of the rooms on the LKM and the HCT floors were disinfected with the PX-UV devices (12/29 rooms on HCT floor and 6/13 rooms on the LKM floor). Subsequent patients admitted in the same disinfected rooms were monitored for potential acquisition of VRE or VRE infections. One patient had a hospital-acquired VRE colonization 6 days after admission to a room that did not receive a PX-UV intervention where VRE ST18 was detected previously (data not shown). No VRE acquisition or infection was identified in subsequent patients who were admitted to rooms disinfected with the PX-UV devices during the study period.
DISCUSSION
The dominance of VRE in the gastrointestinal tract, the use of broad-spectrum antibiotic therapy, and VRE transmission through direct contact or via the surrounding environment are all factors contributing to the spread of VRE in the hospital setting [27–29]. Due to the lack of therapeutic options for VRE infections, understanding VRE transmission networks within the hospital environment is of paramount importance to control and contain the emergence and the propagation of these pathogens.
WGS is increasingly used as an epidemiologic tool to distinguish strains that would appear identical by conventional typing, identify potential transmission networks, and accurately stratify the different clusters of VRE as well as any predominant strain circulating in multiple rooms and persisting in the hospital environment. Among the attributable STs, 3 were recognized as most predominant in our hospital setting (ie, ST18, ST584, and ST736). ST17, thought to be the leading ST in clonal complex 17, is being replaced by the newest STs such as ST18, ST203, ST412, ST584, ST736, and ST664. This shift was observed worldwide in Canada, [30], the United States, [31], Australia, Germany, and China [32]. Interestingly, newly identified STs belonging to clonal complex 17 were isolated in our institution (ie, ST494 and ST736). ST736 is a novel clone exhibiting DAP nonsusceptibility [31]. In this study, 13% and 15% of the VRE ST736 were found on the HCT and the LKM floors, respectively. ST736 was previously identified in 76% of DAP nonsusceptible enterococci [31] and the 2012 DAP-resistant VRE cohort in a New York hospital [33]. Similarly, we showed that all strains belonging to ST736 and ST664 isolated in our study were DAP-resistant, providing evidence of a potential association between these genetic lineages and DAP resistance. Moreover, 1 VRE strain belonging to ST494 was isolated from the LKM floor and harbored the vanB gene cluster, whereas all other strains possessed vanA [34, 35]. ST494 was only reported once in a Peruvian hospital in 2010 [36]. However, both vanA and vanB determinants reside on mobile elements that can be transferred to other enterococci strains as well as other gram-positive organisms posing a major public health challenge [37]. The propagation of newly identified genetic lineages in different regions and countries demonstrates the significance of VRE evolutionary dynamics and genome malleability [36]. Interestingly, a cluster of 11 VRE isolates lacked the fms 14-17-13 pili operon and were responsible for 60% of the bacteremia cases in this study. It has been previously shown that the absence of a pilus protein can result in increased interactions between alternative pilus proteins and human cells, causing subsequent invasion and meningitis in group B Streptococcus [38].
The phylogenetic analyses of 89 VRE isolates suggest that potential transmission events seem to have occurred within the LKM and the HCT floors between the patient and the room environment and among patients on the same floor or on different floors, highlighting the high potential for spread in VRE. Likely, environmental sources of VRE were located on the west and east wing of the LKM floor and were associated with the spreading of VRE ST584 and ST736, respectively. Additionally, the west wing of the HCT floor was another potential environmental source of VRE ST18 to rooms on different floors. These results are in agreement with other studies showing ST18, ST584, and ST736 as predominant VRE STs currently circulating in hospitals worldwide [30–32].
An important focus of our study was the dynamics of transmission of DAP resistance in our patients [8]. As such, prior exposure to DAP may be an important factor to select for DAP nonsusceptible strains. Indeed, a recent study showed that the majority of patients with DAP-resistant strains (60%) had a prior exposure to daptomycin [8]. In contrast, other studies have documented de novo emergence of DAP resistance in the absence of prior exposure to DAP. Indeed, Kamboj et al found that 89% of their patients with DAP-resistant VRE (mostly hematologic malignancy patients) had no documented DAP exposure [6], a finding supported by other studies [9, 10]. Here we showed that 75% of our patients harboring DAP-resistant VRE had no prior exposure to DAP, suggesting possible nosocomial transmission of DAP-resistant VRE strains in the hospital environment. This assumption is supported by our findings that some of the DAP-resistant isolates are highly genetically related.
The presence of VRE clonal transmission in the hospital setting underscores the importance of infection control measures and antimicrobial stewardship interventions to control the emergence and transmission of highly resistant pathogen between rooms and patients, including DAP-resistant strains. These measures include wearing appropriate personnel protective equipment when needed and compliance with hand hygiene, in addition to enhanced cleaning protocols such as the use of UV-C devices [39]. The routine and real-time use of WGS for identifying new or unrecognized transmission networks and potential reservoirs of predominant pathogens within a hospital has proven to be a reliable method of infection control surveillance and may aid in limiting the transmission of pathogens, especially multidrug-resistant organisms within the hospital.
This study has several limitations. In total, 34 rooms were sampled on both floors as only VRE-positive patients were included. Half of these were disinfected with PX-UV devices. This constitute a small sample size; however, the efficacy of PX-UV in disinfecting hospital rooms has been widely tested in other studies [26, 40]. Additionally, single colonies were used in the analyses, which cannot capture the full diversity of strains within a patient and has the potential to miss some transmission events. One potential limitation is the lack of sampling of healthcare workers monitoring of infection control practices. A social network analysis might reveal additional information on transmission events occurring within the hospital. Finally, the rooms were only sampled following patient discharge; as such, we cannot be fully certain about the direction of transmission.
CONCLUSION
The efficacy of infection control measures such as hand hygiene, use of recommended barrier precautions, and UV-C surface disinfection to limit the dissemination of pathogens can be evaluated by surveillance and routine use of WGS. Our data suggest the presence and the transfer of highly related and invasive DAP-susceptible and DAP-resistant VRE isolates between HCT recipients, LKM patients, and their room environment. Our findings underscore the significance of infection control strategies and the importance of implementing WGS to early detect, prevent, and control the spread of this opportunistic organism in the hospital setting.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author Contributions. L. E. H., C. P. H., and S. S. G. conducted the experiments. L. E. H., B. H., and R. F. C. analyzed the results. R. F. C. designed the study, and M. S. provided assistance with the study design. L. E. H., B. H., M. S., C. A. A., and R. F. C. wrote and edited the manuscript. All authors approved the manuscript preparation.
Acknowledgment. The authors thank the MD Anderson Biospecimen Extraction and the Sequencing and Microarray Facilities for DNA extraction and sequencing of the VRE strains. They also thank Dr Glen Otero and Dr Samuel V. Scarpino for their valuable input provided for this study.
Financial support. Xenex Disinfection Services provided funding for laboratory analysis. L. E. H.’s research was funded by the American Cancer Society Postdoctoral award number 131044-PF-18-191-01-LIB. The work was also supported by the National Institutes of Health/National Cancer Institute (NIH/NCI) under award number P30CA016672. C. A. A. is supported by NIH/National Institute of Allergy and Infectious Diseases (NIAID) grant numbers R01 AI134637, R21 AI143229, and K24 AI121296.
Potential conflicts of interest. R. F. C. has received research grants from and personal fees as a consultant to Xenex Disinfection Services. M. S. is an employee/shareholder with Xenex Disinfection Services. C. A. A. has received grant support from Merck, Entasis Pharmaceuticals, and MeMed Diagnostics (money paid to institution). C. A. A. also report personal fees as an employee with University of Texas Health Science Center, royalties/personal fees from UptoDate, Harrison Principles of Internal Medicine, Mandell Principles and Practice of Infectious Diseases (chapters); personal fees from serving as study section member/grant reviewer for NIH/NIAID, reimbursement for traveling to IDWeek and ID program committee meetings as IDWeek Chair from the Infectious Diseases Society of America (IDSA), and reimbursement for traveling to ASM Microbe and Antimicrobial Agents and Chemotherapy Editor’s stipend (paid to institution) from American Society for Microbiology. All other authors report no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
REFERENCES
- 1. Ubeda C, Taur Y, Jenq RR, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120:4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chong PP, van Duin D, Bangdiwala A, et al. Vancomycin-resistant enterococcal bloodstream infections in hematopoietic stem cell transplant recipients and patients with hematologic malignancies: impact of daptomycin MICs of 3 to 4 mg/L. Clin Ther 2016; 38:2468–76. [DOI] [PubMed] [Google Scholar]
- 3. CDC. VRE in healthcare settings. Available at: https://www.cdc.gov/hai/organisms/vre/vre.html. Accessed 10 May 2011.
- 4. Dubin K, Pamer EG. Enterococci and their interactions with the intestinal microbiome. Microbiol Spectr 2014; 5:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scheich S, Lindner S, Koenig R, et al. Clinical impact of colonization with multidrug-resistant organisms on outcome after allogeneic stem cell transplantation in patients with acute myeloid leukemia. Cancer 2018; 124:286–96. [DOI] [PubMed] [Google Scholar]
- 6. Kamboj M, Cohen N, Gilhuley K, Babady NE, Seo SK, Sepkowitz KA. Emergence of daptomycin-resistant VRE: experience of a single institution. Infect Control Hosp Epidemiol 2011; 32:391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benamu E, Deresinski S. Vancomycin-resistant Enterococcus infection in the hematopoietic stem cell transplant recipient: an overview of epidemiology, management, and prevention. F1000Res 2018; 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Storm JC, Diekema DJ, Kroeger JS, Johnson SJ, Johannsson B. Daptomycin exposure precedes infection and/or colonization with daptomycin non-susceptible Enterococcus. Antimicrob Resist Infect Control 2012; 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fraher MH, Corcoran GD, Creagh S, Feeney E. Daptomycin-resistant Enteroccoccus faecium in a patient with no prior exposure to daptomycin. J Hosp Infect 2007; 65:376–8. [DOI] [PubMed] [Google Scholar]
- 10. Wudhikarn K, Gingrich RD, de Magalhaes Silverman M. Daptomycin nonsusceptible enterococci in hematologic malignancy and hematopoietic stem cell transplant patients: an emerging threat. Ann Hematol 2013; 92:129–31. [DOI] [PubMed] [Google Scholar]
- 11. Kwong JC, McCallum N, Sintchenko V, Howden BP. Whole genome sequencing in clinical and public health microbiology. Pathology 2015; 47:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30:2068–9. [DOI] [PubMed] [Google Scholar]
- 15. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31:3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belén A, Pavón I, Maiden MCJ. Multilocus sequence typing. Methods Mol Biol (Clifton, NJ) 2009; 551:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67:2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 1999; 174:247–50. [DOI] [PubMed] [Google Scholar]
- 19. Diaz L, Tran TT, Munita JM, et al. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 2014; 58:4527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brodrick HJ, Raven KE, Harrison EM, et al. Whole-genome sequencing reveals transmission of vancomycin-resistant Enterococcus faecium in a healthcare network. Genome Med 2016; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raven KE, Gouliouris T, Brodrick H, et al. Complex routes of nosocomial vancomycin-resistant Enterococcus faecium transmission revealed by genome sequencing. Clin Infect Dis 2017; 64:886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 2016; 44:W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El Haddad L, Ghantoji SS, Stibich M, et al. Evaluation of a pulsed xenon ultraviolet disinfection system to decrease bacterial contamination in operating rooms. BMC Infect Dis 2017; 17:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chemaly RF, Simmons S, Dale C Jr, et al. The role of the healthcare environment in the spread of multidrug-resistant organisms: update on current best practices for containment. Ther Adv Infect Dis 2014; 2:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 2012; 10:266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Hal SJ, Ip CL, Ansari MA, et al. Evolutionary dynamics of Enterococcus faecium reveals complex genomic relationships between isolates with independent emergence of vancomycin resistance. Microb Genom 2016; 2:e000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Schaik W, Top J, Riley DR, et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 2010; 11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCracken M, Wong A, Mitchell R, et al. ; Canadian Nosocomial Infection Surveillance Program . Molecular epidemiology of vancomycin-resistant enterococcal bacteraemia: results from the Canadian Nosocomial Infection Surveillance Program, 1999–2009. J Antimicrob Chemother 2013; 68:1505–9. [DOI] [PubMed] [Google Scholar]
- 31. Wang G, Kamalakaran S, Dhand A, et al. Identification of a novel clone, ST736, among Enterococcus faecium clinical isolates and its association with daptomycin nonsusceptibility. Antimicrob Agents Chemother 2014; 58:4848–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lam MM, Seemann T, Tobias NJ, et al. Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium. BMC Genomics 2013; 14:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang G, Yu F, Lin H, et al. Evolution and mutations predisposing to daptomycin resistance in vancomycin-resistant Enterococcus faecium ST736 strains. PLoS One 2018; 13:e0209785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hill CM, Krause KM, Lewis SR, et al. Specificity of induction of the vanA and vanB operons in vancomycin-resistant enterococci by telavancin. Antimicrob Agents Chemother 2010; 54:2814–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eliopoulos GM, Gold HS. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin Infect Dis 2001; 33:210–9. [DOI] [PubMed] [Google Scholar]
- 36. Panesso D, Reyes J, Rincón S, et al. Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals. J Clin Microbiol 2010; 48:1562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev 2000; 13:686–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maisey HC, Hensler M, Nizet V, Doran KS. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J Bacteriol 2007; 189:1464–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hayden MK, Blom DW, Lyle EA, Moore CG, Weinstein RA. Risk of hand or glove contamination after contact with patients colonized with vancomycin-resistant Enterococcus or the colonized patients’ environment. Infect Control Hosp Epidemiol 2008; 29:149–54. [DOI] [PubMed] [Google Scholar]
- 40. Ghantoji SS, Stibich M, Stachowiak J, et al. Non-inferiority of pulsed xenon UV light versus bleach for reducing environmental Clostridium difficile contamination on high-touch surfaces in Clostridium difficile infection isolation rooms. J Med Microbiol 2015; 64:191–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.