Abstract
Background:
Neonatal sepsis accounts for a large proportion of neonatal deaths in sub-Saharan Africa. The lack of access to diagnostic testing and excessively long turnaround times to result contributes to delays in sepsis identification and initiation of appropriate treatment. This study aims to evaluate the novel InTrays COLOREX Screen and extended-spectrum beta-lactamase for rapid identification of bacterial pathogens causing sepsis and detection of resistance.
Methods:
Neonates with suspected sepsis admitted to the Harare Central Hospital were prospectively enrolled. One blood culture was collected and incubated using the BacT/ALERT automated system. Positive blood cultures with potential pathogens identified by Gram stain were inoculated on the InTray COLOREX Screen and extended-spectrum beta-lactamase culture plates.
Results:
A total of 216 neonates with suspected sepsis were recruited. Pathogens were isolated from blood cultures in 56 (25.9%) neonates of which 54 were Klebsiella pneumoniae. All K. pneumoniae were resistant to ceftriaxone and 53 (98%) were resistant to gentamicin. Sensitivity and specificity for ceftriaxone-resistant K. pneumoniae detection using InTrays were 100%. InTrays results were interpretable as early as 5–10 hours (median 7 hours, interquartile range 7–7) post blood culture positivity enabling rapid identification and notification of result and leading to a 60% reduction in time to result from blood culture collection.
Conclusions:
This study shows that the implementation of a novel culture method was feasible and reduced turnaround times for results by 60% compared with standard microbiologic techniques. An impact on patient outcomes and cost-effectiveness of this method needs to be demonstrated.
Keywords: AMR, antimicrobial resistance, rapid diagnosis, extended-spectrum beta-lactamase, neonatal sepsis
The sustainable developmental goals call for the end of preventable deaths of newborns, with all countries aiming for a neonatal mortality rate of <12 deaths per 1000 live births. The sub-Saharan African region has the highest neonatal mortality globally (27 deaths per 1000 live birth) with 42 of 48 countries at risk of missing the sustainable developmental goal target.1 Globally, 15% of neonatal deaths are attributable to sepsis, but the proportion is likely to be higher in sub-Saharan Africa. A recent study estimated the number of neonatal sepsis cases in sub-Saharan Africa to be 355,500–605,750 annually leading to 177,500–302,870 deaths.2
While neonatal sepsis is a life-threatening condition, nosocomial neonatal sepsis may be reduced by adhering to stringent infection prevention and control practices.3 Effective treatment of neonatal sepsis relies on early and rapid diagnosis and appropriate antibiotic treatment.4 Ideally, empiric antibiotics should take into account the most frequently isolated organisms and antimicrobial susceptibility profiles in a specific setting.5 Targeted antibiotic treatment should be informed by antimicrobial susceptibility of the bacterial isolate cultured from the infant’s blood.6 Unfortunately, antimicrobial susceptibility data are unavailable in most resource-limited settings due to limited laboratory capacity, especially in lower-level facilities such as district and provincial hospitals.7
A systematic review found that the most common Gram-negative bacterial species representing 21% of all blood culture isolates in neonates in sub-Saharan Africa were Klebsiella spp. and that the majority were resistant to ceftriaxone and gentamicin.7 The mechanism of resistance against third-generation cephalosporins is usually extended-spectrum beta-lactamase (ESBL) production. Both the first- and second-line neonatal sepsis treatment regimens recommended by the World Health Organization (WHO) are ineffective in treating ESBL-Klebsiella.8 In the absence of antimicrobial susceptibility results, it is thus not surprising that case fatality rates of neonatal ESBL-Klebsiella sepsis are extremely high exceeding 35%–70% in many low- and middle-income countries.9–12 Even if microbiologic diagnostics are available, results are often delayed or not returned. A recent audit conducted in one of the busiest neonatal care units in Zimbabwe reported that 67% of blood culture results were never received by the responsible clinician. Only 4% of blood culture results arrived in time to impact on clinical decision-making.13 Similar to other settings in sub-Saharan Africa, ESBL-Klebsiella pneumoniae was common both on admission to the unit and during hospitalization. However, the first- and second-line treatment for neonatal sepsis remained aligned with WHO recommendations due to high cost and limited availability of carbapenems.13 InTrays are commercially available, ready-to-use agars containing chromogenic substrates that enable the differentiation between multiple bacterial species based on colony color. InTray COLOREX ESBL plates contain in addition to the chromogenic substrates, antimicrobial compounds for selective identification of ESBL producing organisms. They have a series of advantages over conventional culture media which may lead to shorter turnaround time (TAT) to result. Using conventional microbiology techniques, blood culture results are available after at least 72 hours from blood culture collection.
We conducted a study to evaluate the novel InTrays COLOREX Screen and ESBL for the work-up of positive neonatal blood cultures in Zimbabwe and investigated whether an algorithm including the chromogenic agar would reduce time to results.
METHODS
Study Design and Setting
This study included babies admitted to the neonatal unit at Harare Central Hospital (HCH) between March 4th and June 29th, 2020 in Zimbabwe. HCH is a tertiary teaching hospital. The neonatal care unit has the capacity to admit 100 neonates, but often operates at 150% capacity leading to unit overcrowding and difficulties in implementing effective infection prevention and control measures. Monthly admissions before the COVID-19 pandemic averaged 400. The HCH guidelines for managing neonatal sepsis were adapted from the NICE guidelines and the WHO Pocket book of hospital care for children on management of neonatal sepsis.6,14 The first-line treatment for neonatal sepsis is benzyl-penicillin and gentamicin and ceftriaxone is used as second-line.
Any neonate with suspected sepsis based on the presence of one major risk factor or two or more minor risk factors for sepsis was eligible for inclusion (Supplemental Digital Content 1, http://links.lww.com/INF/E366). Extremely low birth weight neonates (birth weight < 1000 g) were excluded. Mothers of eligible neonates provided written informed consent. Skin disinfection before blood culture collection was performed using 70% alcohol wipes. Blood cultures aiming for a volume of 2 mL were collected and sent to the laboratory at the Biomedical Research and Training Institute within 4 hours of the blood draw. Sample volume was assessed by weighing the blood culture bottles before and after filling. Only one sample was included per neonate. Repeat samples were not analyzed in the study.
An interviewer administered questionnaire was used to collect information about the antenatal and perinatal period.
Microbiologic Investigations
Blood culture bottles were incubated using the automated BacT/ALERT (bioMerieux, Marcy-l’Etoile, France) system with continuous monitoring for growth. Positive blood cultures were processed using conventional methods. Samples showing Gram-negative bacilli were also inoculated on the InTray COLOREX Screen and ESBL chromogenic media (Biomed Diagnostic, White City, OR; Supplemental Digital Content 2, http://links.lww.com/INF/E366). underwent Gram staining, one drop of the positive blood culture was inoculated on conventional media (MacConkey, blood and chocolate agar (HiMedia, Bombay, India)) on day 0. If the Gram stain showed Gram-negative bacilli or Gram-positive cocci in chains, the sample was also inoculated on the InTray COLOREX Screen and ESBL chromogenic media (Biomed Diagnostic; Supplemental Digital Content 3, http://links.lww.com/INF/E366). InTray cultures were incubated at 37 °C and read at the end of the working day (6–8 hours postinoculation) and at 24 hours postinoculation. For Gram-negative bacilli, direct antimicrobial susceptibility testing (AST) from the positive blood culture was performed and read on day 1. Enterobacteriaceae was identified using biochemical tests on subcultured isolates (Analytical Profile Index, API 20E, bioMerieux) with results available on day 2. Other organisms were identified by colony morphology and additional conventional microbiology tests including catalase, tube coagulation and appearance on bile-aesculin agar. AST (“indirect AST”) was repeated using a 0.5 McFarland inoculum for all isolates which underwent AST directly from the sample. AST was performed by disc diffusion for ampicillin, ceftriaxone, imipenem, ciprofloxacin, gentamicin, amikacin, chloramphenicol and by determining minimum inhibitory concentration using Etests (bioMerieux) for vancomycin. AST results were interpreted using the EUCAST standards.15 Figure 1 shows the testing algorithm for positive blood cultures using the InTray system. Quality control of laboratory tests for media growth support, bacterial identification and AST was performed using ATCC strains.
FIGURE 1.
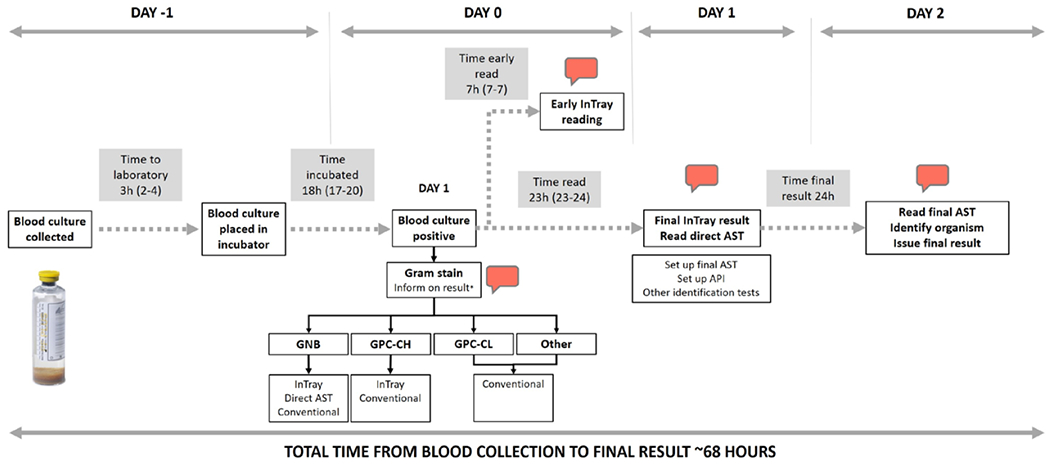
Laboratory procedures and result reporting for positive blood cultures. Call outs represent results which were notified in real-time using messaging platforms. The times presented in the shaded boxes represent median times and interquartile ranges. *The Gram stain results were only notified for GNB and GPC in chains. Conventional methods included inoculation on MacConkey, blood and chocolate agar. InTrays read were InTray COLOREX Screen and ESBL. Time to early bacterial identification and ESBL-detection (“Early InTray reading”): 7 hours from culture positivity. Time to full pathogen identification and AST results using conventional methods (“Read final AST, Identify organism, Issue final result”): 47 hours from culture positivity (68 hours from culture collection). API indicates analytical profile index; AST, antimicrobial susceptibility testing; GNB, Gram-negative bacilli; GPC-CH, Gram-positive cocci in chains; GPC-CL, Gram-positive cocci in clusters.
Result Reporting
Positive results were reported directly to the attending clinicians using an encrypted smartphone-based messaging platform as soon as they were available. These included Gram stain results for potential pathogens, growth of suspected resistant organisms on the InTray ESBL plates at 6–8 hours postinoculation, preliminary bacterial identification and AST results from the directly inoculated sample at 24 hours, and final bacterial identification and AST for pathogens at 48 hours postinoculation.
Data Entry and Analysis
Statistical analyses were performed in STATA v.15 (Stata-Corp, TX). Descriptive analysis was performed to characterize the study population and laboratory findings. A Kaplan–Meier analysis was performed to compare time to death from blood culture collection in neonates with and without K. pneumoniae sepsis. Babies were censored at 28 days or at discharge for those without a day-28 follow-up visit.
Permission and Ethics
Permission to conduct the study was obtained from the HCH ethics committee.
Institutional Review Board approval was obtained from the Medical Research Council Zimbabwe (MRCZ/A/2547).
RESULTS
Between March 1 and June 30, 2020, 972 neonates were admitted to the unit of which 451 (46.4%) had an episode of suspected sepsis during their admission (Fig. 2). A total of 216 neonates with suspected sepsis were recruited into the study.
FIGURE 2.
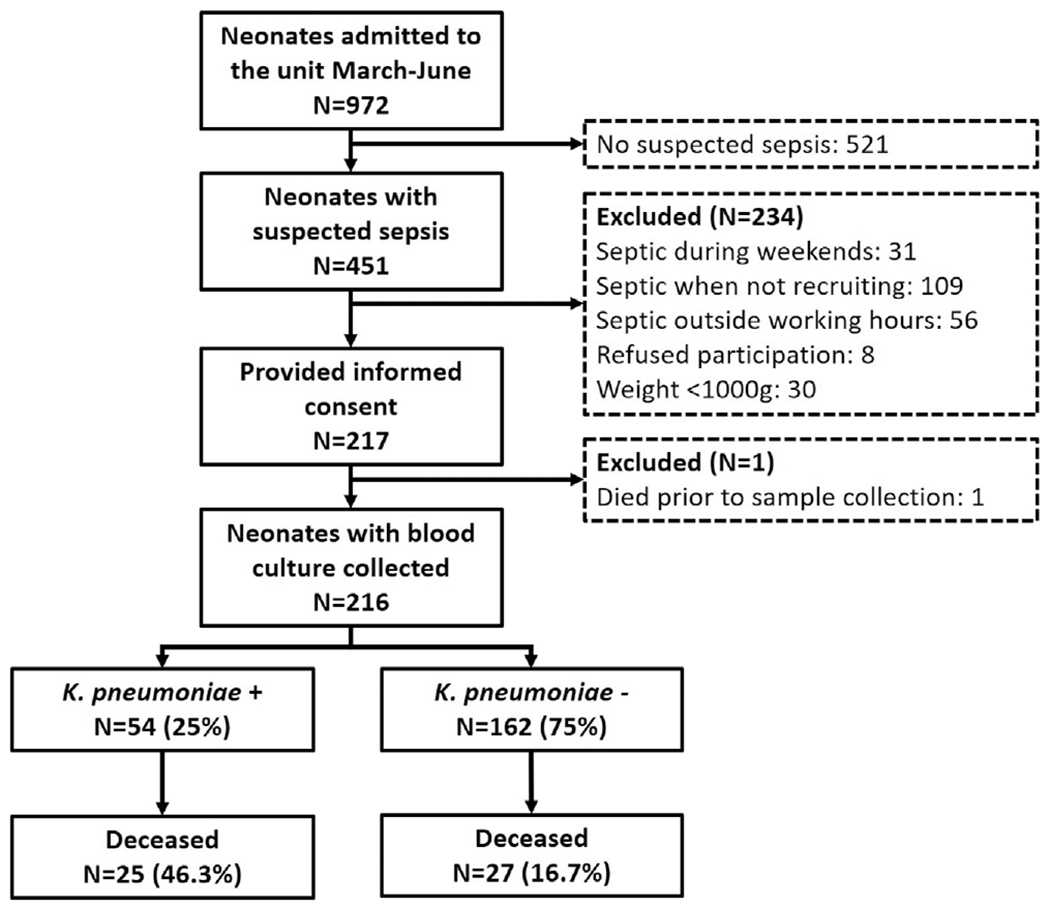
Flowchart of study participants. Septic when not recruiting: neonates were diagnosed with sepsis during the national lockdown (April–May) or before the study started recruitment in March and after the study completion in June (total number of neonatal admissions were only available for the complete month). Neonates who were septic outside regular working hours (weekend and at night) could not be included into the study. For these neonates, blood cultures were collected and sent to the routine laboratory. For one neonate who was severely ill, the mother provided informed consent, but the baby died before a sample could be collected.
Maternal and Birth Characteristics
Median age for the mothers was 25 years (interquartile range [IQR] 22–31), 22 (12.8%) were HIV infected and 90 (41.7%) were primigravidae. The place of delivery was HCH in 136 (63%), another healthcare facility in 51 (23.6%) and 23 (10.6%) babies were born before arrival to health facilities. For 6 babies, the place of delivery was not documented. The majority of babies (164/216, 75.9%) were delivered by normal vertex delivery, while 46 (21.3%) were born by cesarean section. Maternal and neonatal characteristics are described in Table 1.
Table 1.
Characteristics of Mothers and Neonates Enrolled into the Study
| Neonatal Characteristics | N =216 |
|---|---|
| Female sex, n (%) | 90 (41.7) |
| Birth weight, n (%) | |
| <1500 g | 54 (25.4) |
| 1500–2499 g | 65 (31.5) |
| ≥2500 g | 94 (44.1) |
| Product of multiple pregnancies, n (%) | 23 (10.7) |
| Maternal characteristics | |
| Age in years, median (IQR) | 25 (22–31) |
| No. antenatal clinic visits, n (%) | |
| None | 65 (30.1) |
| 1 | 55 (25.5) |
| 2 | 34 (15.7) |
| 3 | 27 (12.5) |
| ≥4 | 35 (16.2) |
| HIV positive*, n (%) | 22 (12.8) |
| Parity, n (%) | |
| Primigravida | 90 (41.7) |
| Multigravida | 126 (58.3) |
| Prolonged rupture of membranes (18 h or more), n (%) | 23 (19.2) |
| Prolonged labor (24 h or more), n (%) | 7 (3.8) |
| Cesarean delivery, n (%) | 46 (21.3) |
| Birth outside a healthcare facility, n (%) | 23 (10.7) |
| Meconium-stained amniotic fluid, n (%) | 42 (19.4) |
| Neonatal care variables | |
| NG/OG-feed, n (%) | 86 (39.8) |
| Oxygen, n (%) | 84 (38.9) |
| CPAP, n (%) | 35 (16.2) |
| Invasive ventilation, n (%) | 4 (1.9) |
| Required surgery, n (%) | 8 (3.7) |
| Duration of hospital stay in days, median (IQR) | 8 (5–14) |
The following variables had missing data: birth weight (n = 3), HIV status (n = 44), prolonged rupture of membranes, excluding neonates born by cesarean section and outside a healthcare facility (n = 50); prolonged labor, excluding neonates born outside a healthcare facility (n = 23).
CPAP indicates continuous positive airway pressure; NG, nasogastric; OG, orogastric.
Neonatal Characteristics
This study included 125 (57.9%) boys, 90 (41.7%) girls and one baby with ambiguous genitalia; 23 (10.6%) babies were from multiple pregnancies. Birth weight was very low (1000–1499 g) in 54 (25.4%), low (1500—2499 g) in 65 (30.5%) and normal (≥2500 g) in 87 (40.8%). Seven (3.3%) babies had a birth weight of >4000 g.
Microbiologic Diagnosis
A blood culture was collected within the first 72 hours of life in 142 (65.1%), between 4 and 7 days in 49 (22.5%) and after the first week in 27 (12.4%; Fig. 3). The median filling volume of the blood culture bottles was 1.08 mL (IQR 0.56–1.75). Pathogens were isolated from blood cultures in 56 (25.9%) neonates and further 29 (13.4%) blood cultures were categorized as contaminated (Fig. 2). K. pneumoniae was isolated from 54 and Enterococcus spp. from 3 neonates (Fig. 3). One baby had 2 pathogens isolated from the blood culture. Coagulase-negative staphylococci were the most common contaminants (n = 24) followed by Bacillus spp. (n = 4) and nonpathogenic viridans streptococci (n = 1). The prevalence of resistance in K. pneumoniae was 54/54 (100%) for ceftriaxone, 53/54 (98%) for gentamicin, 50/53 (94%) for amikacin, 9/54 (17%) for chloramphenicol, 6/54 (11%) for ciprofloxacin and 0/54 (0%) for imipenem. Appropriate treatment with meropenem was initiated in neonates with ESBL-K. pneumoniae as soon as the microbiology results were available. All K. pneumoniae isolates showed growth of blue colonies on the InTray Screen and ESBL culture indicating ceftriaxone resistance. Enterococcus spp. isolates only had growth on the InTray Screen plate and were all ampicillin-resistant. Sensitivity and specificity for ESBL-K. pneumoniae detection using InTrays were 100%. Risk factors for K. pneumoniae sepsis and outcomes are described in Supplemental Digital Content 4, http://links.lww.com/INF/E366.
FIGURE 3.
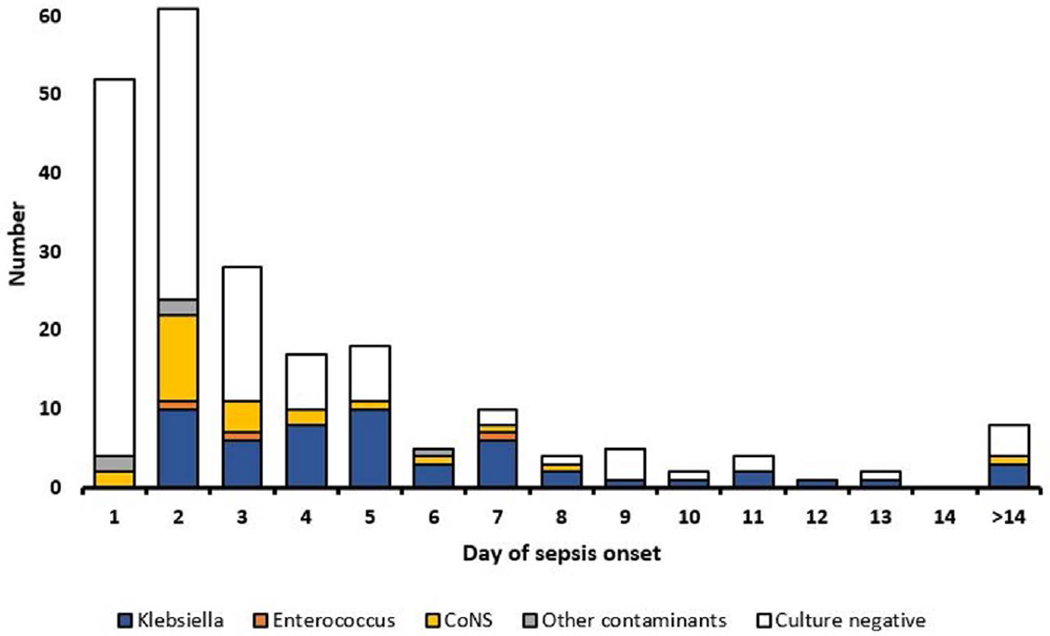
Culture results according to the day of sepsis onset from birth. Cultures were positive for K. pneumoniae starting with day two from birth. The white columns represent negative cultures. Most blood culture were collected within the first five days of life.
Testing Times
Testing times and laboratory procedures are shown in Figure 1. The median time from blood culture collection to laboratory receipt was 3 hours (IQR 2–4) and the median time to positivity for blood cultures was 18 hours (IQR 17–20). A subset of 27 (50%) InTrays were read at 5–10 hours (median 7 hours, IQR 7–7) postinoculation and the treating clinicians were notified of growth of colonies suggestive of third-generation cephalosporin-resistant K. pneumoniae at the time of reading (Fig. 4). Further 21 (39%) InTrays were read the next day, after a median of 23 hours (IQR 23–24) postinoculation. Of those, 9 were inoculated during the weekend or were positive late during the working day, and therefore, reading was not possible on the day of inoculation. For 6 InTrays, the result was delayed beyond 24 hours. Results of the early InTray reading were available at 28 hours from blood culture collection and those for the final identification and AST at 68 hours leading to a 60% reduction in the total time to result post blood culture collection.
FIGURE 4.
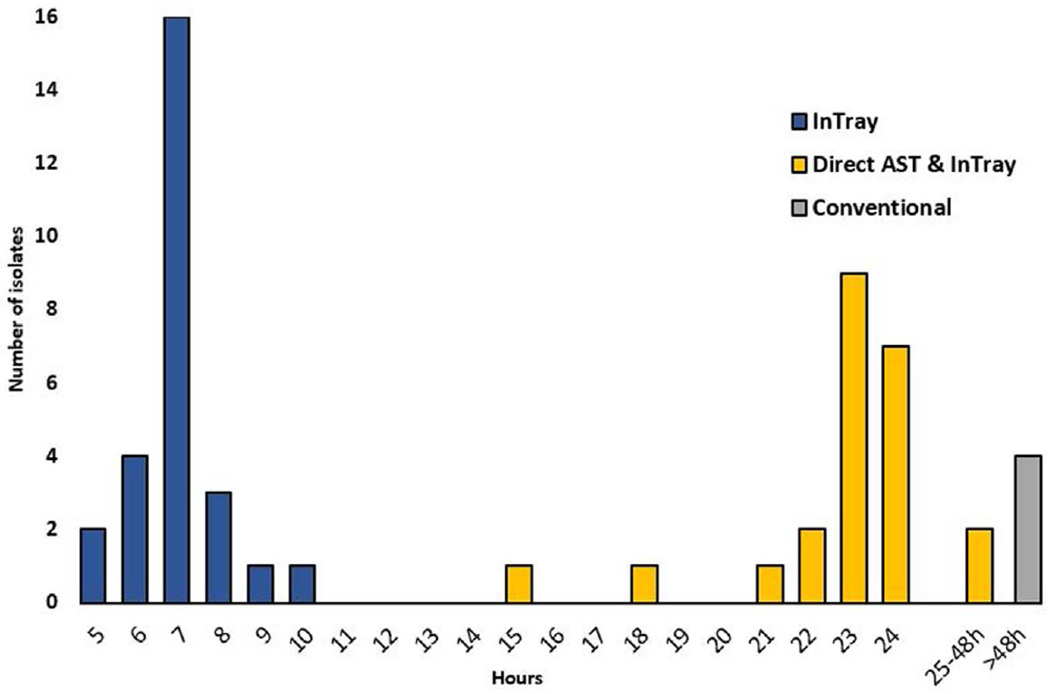
Time from blood culture positivity to presumptive identification and determination of third-generation cephalosporin resistance using the different methods for blood cultures positive for K. pneumoniae. InTray method: growth of blue colonies on the InTray and ESBL plate was interpreted as presumptive K. pneumoniae with third-generation cephalosporin resistance; Direct AST and InTray: AST was performed directly from the blood culture bottle and read after ~24 hours. The horizontal axis shows the time in hours from blood culture positivity. AST indicates antimicrobial susceptibility testing; ESBL, extended-spectrum beta-lactamase.
Outcomes of Neonates with ESBL-K. pneumoniae Sepsis
The median duration of hospital stay was 11 days (IQR 6–21) in neonates with a positive blood culture for K. pneumoniae compared with 7 days (IQR 4–12, P = 0.004) in babies with either a negative or contaminated blood culture. In-hospital mortality was 25/54 (46%) in neonates with K. pneumoniae sepsis and 27/162 (17%, P < 0.001, Supplemental Digital Content 5, http://links.lww.com/INF/E366) in neonates without a positive blood culture for K. pneumoniae. Of the 30 deaths which occurred within 72 hours of the blood culture collection, 15 (50%) were in neonates with confirmed Klebsiella sepsis. Follow-up calls were successful for 162/164 neonates; one baby died after discharge.
DISCUSSION
This study found that one in four neonates with suspected sepsis had a positive blood culture with K. pneumoniae and isolates were almost universally resistant to first- and second-line antibiotics as recommended by the WHO guidelines for low- and middle-income countries ( LMICs).14 The use of a novel culture system and feeding-back laboratory results in real-time led to a time to preliminary pathogen identification and detection of cephalosporin resistance of as little as 28 hours from blood culture collection (7 hours from blood culture positivity) as compared with 3 days when using conventional laboratory methods and 5 days for the HCH laboratory.
The predominance of K. pneumoniae among the isolates is not surprising and reflects findings from across sub-Saharan Africa.7 The high burden of Klebsiella sepsis in neonates is likely related to suboptimal infection prevention and control procedures that enable nosocomial transmission, although the early onset of infection in many neonates also raises the possibility of perinatal transmission.7 Nosocomial transmission is facilitated by the long survival of the pathogen on dry surfaces,16 unit overcrowding and limited adherence to aseptic techniques and hand hygiene.17
Mortality in this study was very high with every second neonate with a positive culture for K. pneumoniae dying during the course of admission. The study was conducted in a referral unit admitting babies following high-risk pregnancies, with hypoxic ischemic encephalopathy, congenital abnormalities or who were born prematurely. These babies require a high level of care and frequent handling by healthcare staff increasing the risk of colonization and infection, particularly in the absence of stringent infection prevention and control procedures and hand hygiene.
Blood cultures are essential for the identification of pathogens causing sepsis and for determining effective treatment. Furthermore, in an outbreak setting, early detection and implementation of infection prevention and control measures are crucial in preventing neonatal deaths. In many LMICs, however, laboratory testing capacity is limited by the unavailability of laboratory consumables and staff.18 Conventional bacterial identification and AST require frequent media preparation, as well as the presence of reliable supply chains to avoid stock-outs. Use of InTrays may bypass some of these challenges because they are ready-to-use, easy to inoculate and have a relatively long shelf life of 6–12 months. Their use may result in important reductions in TAT for presumptive pathogen identification and detection of cephalosporin resistance with an excellent sensitivity and specificity. This is essential given that cephalosporin resistance is often associated with resistance to gentamicin,7 and thus, resistant pathogens cannot be effectively treated with the recommended empiric antimicrobials for neonatal sepsis.14 Other strategies to reduce TAT such as dehydrated chromogenic media for ESBL detection19 and rapid AST20 are available, but they may be more difficult to implement in LMICs requiring higher-skilled technicians. In addition, dehydrated media has a relatively short shelf life once prepared and requires more stringent quality control.
Prolonged TATs are commonly reported as a challenge in LMICs21,22 and rapid communication of preliminary results such as Gram-stains are often insufficient.23 For blood cultures, a study in Ethiopia reported TATs of seven days although the authors did not differentiate between TATs for positive and negative cultures.24 The study cited repeated stock-outs, high work-loads and insufficient staffing as the main reasons for delays in TATs for a wide range of laboratory tests.24 An earlier study conducted in the same neonatal unit in Zimbabwe before the implementation of this study’s testing and notification algorithm reported TATs of five days for positive cultures. The long TAT was partly due to delays in preanalytical and postanalytical times and only 4% of culture results arrived early enough to impact on clinical management.13The current study showed that TAT from blood culture collection to presumptive pathogen identification and AST could be shortened to as little as 28 hours, or 5–8 hours from blood culture positivity. InTrays were easy to use and required minimal training which also contributed to the short TATs. Presumptive pathogen identification using colony color and the potential to reduce the requirements for subcultures can be valuable especially in settings where highly trained laboratory staff may not be always available.
The study is limited by its enrollment of babies from a single facility and by its recruitment restricted to working hours and weekdays due to logistics and research laboratory operating times. As a result, blood cultures becoming positive overnight were only investigated the next morning. Similarly, blood cultures that became positive late during the working day or during weekends were not read at 6–8 hours. Had the study been done in a routine laboratory shift system, TATs could have further been decreased. The samples were processed in a well-functioning research bacteriology laboratory and testing was supervised by trained clinical microbiologists. Heightened efforts were made to minimize laboratory TAT to provide timely results to clinicians and optimize patient care. Thus, these results may not be generalizable and replicated in routine hospital laboratories in LMICs operating under resource and staff constraints. This study used an automated blood culture system with continuous monitoring for growth. These systems are often not available in LMICs despite the added advantage of higher yields and shorted TATs compared with manually-read blood cultures prepared in house.25 We were only able to evaluate the InTrays for a single Gram-negative pathogen however ESBL-Klebsiella spp. is particularly problematic on neonatal units in LMICs. This study was not designed and powered to show an impact of InTray use and rapid result notification on neonatal mortality.
This is the first report evaluating the use of InTrays in reducing TATs for blood cultures in LMICs. These tests have several advantages: prolonged shelf life and easy inoculation, reading and interpretation requiring less-skilled laboratory staff.
This study highlights the burden of Klebsiella sepsis in a tertiary neonatal care unit in sub-Saharan Africa. The implementation of a novel culture method paired with rapid communication of results was feasible, led to simplified laboratory procedures, and reduced TAT by 60%. Further research is needed to demonstrate cost-effectiveness of these methods and their impact on patient outcomes.
Supplementary Material
Acknowledgments
This work was supported by the Foundation for Innovative New Diagnostics (FIND). Research reported in this publication was supported by the Fogarty International Center of the National Institutes of Health under Award Number D43 TW009539. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. FIND was supported by the UK aid from the British people for the acquisition of the InTrays which were used in the study. F.F. is supported by the Academy of Medical Sciences, the funders of the Starter Grant for Clinical Lecturers scheme and UCL Great Ormond Street NIHR Biomedical Research Centre.
Footnotes
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.pidj.com)
REFERENCES
- 1.UNICEF. Levels & trends in child mortality—report 2020. 2020. Available at: https://www.unicef.org/reports/levels-and-trends-child-mortality-report-2020. Accessed February 20, 2021.
- 2.Ranjeva SL, Warf BC, Schiff SJ. Economic burden of neonatal sepsis in sub-Saharan Africa. BMJ Glob Health. 2018;3:e000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaidi AK, Huskins WC, Thaver D, et al. Hospital-acquired neonatal infections in developing countries. Lancet. 2005;365:1175–1188. [DOI] [PubMed] [Google Scholar]
- 4.Mazzucchelli I, Garofoli F, Angelini M, et al. Rapid detection of bacteria in bloodstream infections using a molecular method: a pilot study with a neonatal diagnostic kit. Mol Biol Rep. 2020;47:363–368. [DOI] [PubMed] [Google Scholar]
- 5.Iroh Tam PY, Bendel CM. Diagnostics for neonatal sepsis: current approaches and future directions. Pediatr Res. 2017;82:574–583. [DOI] [PubMed] [Google Scholar]
- 6.National Institute for Healthcare Excellence (NICE). Neonatal infection (early onset): antibiotics for prevention and treatment. Clinical guideline (August 2012). NICE. Available at: https://www.nice.org.uk/guidance/cg149. Accessed February 20, 2021. [Google Scholar]
- 7.Okomo U, Akpalu ENK, Le Doare K, et al. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect Dis. 2019;19:1219–1234. [DOI] [PubMed] [Google Scholar]
- 8.Guidelines Review Committee. WHO Recommendations on Newborn Health: Guidelines Approved by the WHO. World Health Organization; 2017. [Google Scholar]
- 9.Iregbu KC, Anwaal U. Extended spectrum Beta-Lactamase-producing Klebsiella pneumoniae septicaemia outbreak in the Neonatal Intensive Care Unit of a tertiary hospital in Nigeria. Afr J Med Med Sci. 2007;36:225–228. [PubMed] [Google Scholar]
- 10.Kayange N, Kamugisha E, Mwizamholya DL, et al. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr. 2010;10:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sehgal R, Gaind R, Chellani H, et al. Extended-spectrum beta lactamase-producing gram-negative bacteria: clinical profile and outcome in a neonatal intensive care unit. Ann Trop Paediatr. 2007;27:45–54. [DOI] [PubMed] [Google Scholar]
- 12.Blomberg B, Jureen R, Manji KP, et al. High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar es Salaam, Tanzania. J Clin Microbiol. 2005;43:745–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chimhini G, Chimhuyaa S, Madzudzoa L, et al. Auditing use of antibiotics in Zimbabwean neonates. Infection PreventionPrac. 2020;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Pocket Book of Hospital Care for Children: Guidelines for the Management of Common Childhood Illnesses. 2nd ed. WHO; 2013. Available at: https://www.who.int/maternal_child_adolescent/documents/child_hospital_care/en/. Accessed February 20, 2021. [PubMed] [Google Scholar]
- 15.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0, 2020. Available at: http://www.eucast.org. Accessed February 20, 2021.
- 16.Okomo U, Senghore M, Darboe S, et al. Investigation of sequential outbreaks of Burkholderia cepacia and multidrug-resistant extended spectrum β-lactamase producing Klebsiella species in a West African tertiary hospital neonatal unit: a retrospective genomic analysis. Lancet Microbe. 2020;1:e119–e129. [DOI] [PubMed] [Google Scholar]
- 17.Essel V, Tshabalala K, Ntshoe G, et al. A multisectoral investigation of a neonatal unit outbreak of Klebsiella pneumoniae bacteraemia at a regional hospital in Gauteng Province, South Africa. S Afr Med J. 2020;110:783–790. [DOI] [PubMed] [Google Scholar]
- 18.Sayed S, Cherniak W, Lawler M, et al. Improving pathology and laboratory medicine in low-income and middle-income countries: roadmap to solutions. Lancet. 2018;391:1939–1952. [DOI] [PubMed] [Google Scholar]
- 19.HiMedia. HiCrome™ ESBL Agar Base. Product information sheet. Available at https://www.himedialabs.com/. Accessed February 20, 2021.
- 20.Åkerlund A, Jonasson E, Matuschek E, et al. ; RAST Study Group. EUCAST rapid antimicrobial susceptibility testing (RAST) in blood cultures: validation in 55 European laboratories. J Antimicrob Chemother. 2020;75:3230–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuijn CJ, Msoka E, Mushi DL, et al. The interface between clinicians and laboratory staff: a field study in northern Tanzania. Afr J Lab Med. 2014;3:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moyo K, Porter C, Chilima B, et al. Use of laboratory test results in patient management by clinicians in Malawi. Afr J Lab Med. 2015;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dailey PJ, Osborn J, Ashley EA, et al. Defining system requirements for simplified blood culture to enable widespread use in resource-limited settings. Diagnostics (Basel). 2019;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiferaw MB, Yismaw G. Magnitude of delayed turnaround time of laboratory results in Amhara Public Health Institute, Bahir Dar, Ethiopia. BMC Health Serv Res. 2019;19:240.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ombelet S, Barbé B, Affolabi D, et al. Best practices of blood cultures in low- and middle-income countries. Front Med (Lausanne). 2019;6:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


