Abstract
Variants of SARS-CoV-2 lineages including the most recently circulated Omicron, and previous pandemic B.1.351, B.1.1.7, which have been public concerns, contain a N501Y mutation located in the spike receptor binding domain. However, the potential interactions with host cells linking N501Y mutation to pathogenic relevance remain elusive. Recently, we and others report that kinases such as PI3K/AKT signaling are essential in SARS-CoV-2 entry. Here we analyzed the predicted potential kinases interacting with the mutation. Bioinformatics tools including structure-prediction based molecular docking analysis were applied. We found kinases such as EGFR might potentially act as new factors involving the N501Y mutation binding through possible phosphorylation at Y501 and enhanced affinity in certain variants. To our surprise, the Omicron receptor binding domain harboring N501Y mutation did not enhance binding to EGFR which might be due to the mutations of charged polar to uncharged polar side chains located on the interaction interfaces. Similarly, potent gains of phosphorylation in B.1.351 and B.1.1.7 by mutations were predicted and interaction networks were analyzed with enrichment of pathways. Given kinases might be elevated in cancer patients, the N501Y mutation containing lineages may be possibly much more infectious and additional care for cancer management might be taken into consideration by precision prevention, therapy or recovery.
Keywords: N501Y, SARS-CoV-2, EGFR, Network pharmacology
1. Introduction
Recently, Omicron (B.1.1.529) was reported or predicted to be associated with rapid increase of COVID-19 cases (https://www.who.int/news/item/28-11-2021-update-on-omicron). The new, widely reported and WHO concerned SARS-CoV-2 variant has caused many countries travel restrictions or border closures even more studies are still ongoing or urgently needed for the clinical investigations and border closure benefit [1]. The number of reported Omicron cases from early discovery dates to nowadays showed that in the short period about less than 2 weeks, the circulating of Omicron has been found in at least 65 countries and territories with more than thousands of confirmed cases and more than a thousand suspected cases (Fig. 1 ) (https://www.gisaid.org/hcov19-variants/; https://en.wikipedia.org/wiki/SARS-CoV-2_Omicron_variant).
Fig. 1.
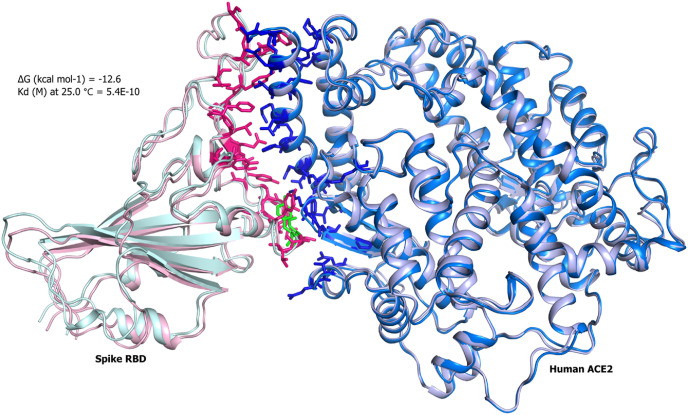
Superimposition of the spike RBD and ACE2 complexes. Docked complexes of the Spike RBD wild type (pink) and N501Y (light green) are shown in cartoon representation with hACE2 receptor. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Omicron contains many more mutations compared with other variants such as SARS-CoV-2501.V2 (B.1.351 lineage) and VOC 202012/01 (B.1.1.7 lineage) [2]. All the three variants retain N501Y mutation on spike protein receptor binding domain (RBD) which is required for binding to human receptor ACE2 (hACE2) for viral entry to host cells. The B.1.1.7 is reported to be more infectious with higher spreading rate and increased binding affinity to ACE2 [2] in particular due to N501Y, compared with other mutations such as E484K [3,4]. The N501Y mutation localizes at RBD that might help in attaining higher binding to host cells and result elevated transmission and infectious. However, not much are reported as an evidence of kinase or other signaling pathways resulting the severe complications or increased death rate by variant containing N501Y mutation has been concluded. Nevertheless, further in-depth studies on the newly emerged variant mutations induced host cell signaling is highly needed to aid the drug development and travel infection prevention.
It has been reported that tyrosine (Y) residue is easily phosphorylated [2]. Therefore, we hypothesized that the N501Y mutation could be more prone to the host-kinases to be modified or phosphorylated. Here we aimed to investigate the potential of the phosphorylation and predict kinases with molecular docking analysis for binding to kinases using first reported, wild type SARS-CoV-2 RBD as control and repurpose the potential therapeutic methods.
2. Methods
To test the hypothesis and predict the tyrosine kinases that might phosphorylate the mutated Spike RBD at N501Y, several bioinformatics tools are applied. Group-based Prediction System of version 5.0 web server (GPS 5.0) [5] is based on the highest probability of phosphorylation indicated by the “score”, and the “cut-off” value, which should not exceed 15% to give significant results. Another is NetPhos 3.1 web server [6,7] - DTU (www.cbs.dtu.dk › services › NetPhos) with the threshold score which is equal to 0.5.
For modeling and docking assignments, wild type hACE2, coronavirus spike RBD and kinase domain of human EGFR were retrieved from the Protein data bank (7EKG and 4I23). AlphaFold and Modeller were employed to generate mutant structures for Omicron and N501Y RBD [8,9]. In order to generate protein-protein complexes, the HDock program was utilized that samples the potent binding modes by Fast Fourier transform (FFT)–based global search optimization method and considering the available homologue information from the PDB in generating binding modes [10,11].
For the generated complexes, contact based prediction methods were used to predict and evaluate the binding-affinities(ΔG) and dissociation constant Kd (M), explained in Ref. [12]. For protein-protein interaction networks analysis, we applied a STRING database [13] and retrieved it by Cytoscape7.3 software [14]. ClusterONE plug-in of Cytoscape 7.3 was used to identify the most significant cluster (nodes as 45; density as 0.498; p-value <0.001) [15,16]. The mutation data were obtained from resources [17,18]. The most important KEGG pathways were identified by the data taken from the Kobas database. Five KEGG pathways were chosen according to the corrected p-value ≤3.40e-9 and kappa score 0.4. Analysis was also performed by an online tool [19].
3. Results and discussion
3.1. Docking analysis of a kinase with N501Y mutation in SARS-CoV-2 lineages
First we applied NetPhos 3.1 server for predicting kinases that potent potentially phosphorylate Y501, and we found the phosphorylation site was predicted whereas the gain of phosphorylation at N501 (wild type) was not found (Supplementary Fig. S1). SRC and epidermal growth factor receptor (EGFR) were potent kinases to phosphorylate Y501. Through another server GPS 5.0, EGFR and Fibroblast growth factor receptor (FGFR) were found. Interestingly, MET was predicted too even with a reduced score. It has been shown that both EGFR and MET kinases play an essential role in lung cancer [20]. On the contrary, we did not find kinases using other tools like Quokka, KinasePhos and ELM which might be explained by the limited database or different algorithm used. Thus, through the two programs prediction, EGFR potentially interacts with and phosphorylates SARS-CoV-2 spike RBD at N501Y.
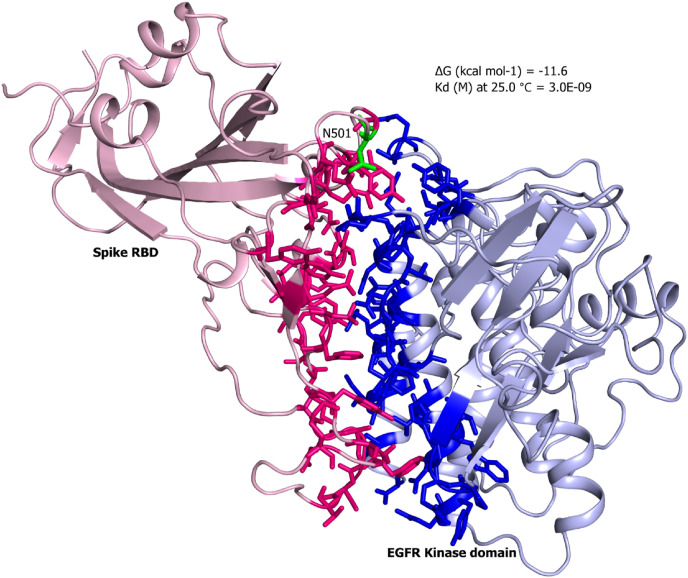
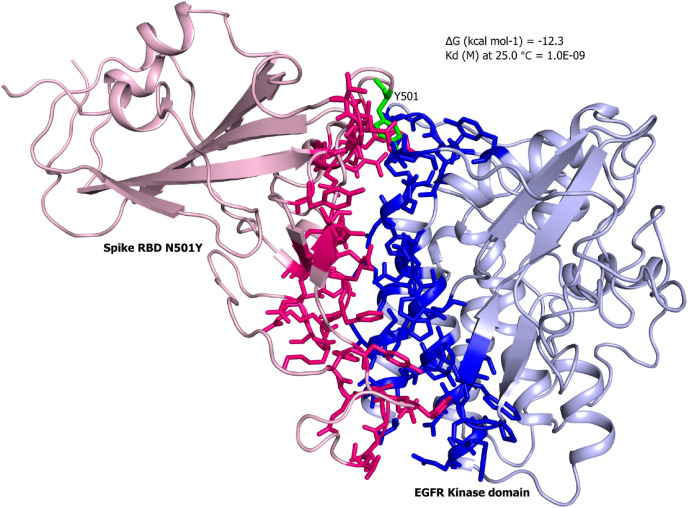
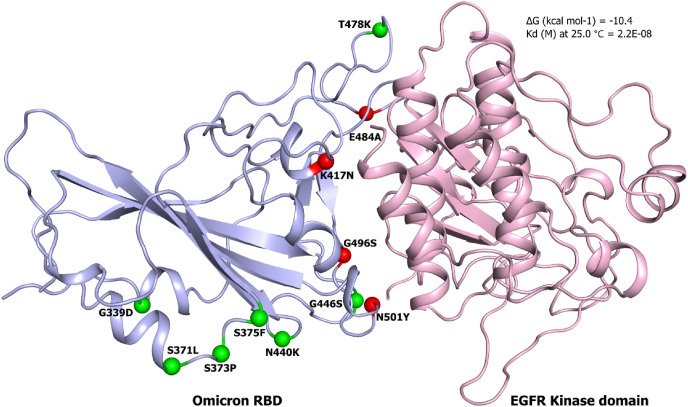
Next, we employed docking tools to generate desired complexes of the EGFR with wild and N501Y spike RBD. First, we calculated the binding affinities of the wild type PDB complex (7EKG) and dock complex of the mutant (N501Y) spike RBD against the hACE2 receptor as a control. Our results depicted that in both cases the complex retrieved same binding affinity of −12.6 kcal/mol, suggesting a negligible impact of the point mutation (Fig. 1, Table 1 ). Subsequently, dock complexes of kinase domain of EGFR-RBD wild and mutant were generated. The N501Y EGFR-RBD complex showed slightly higher affinity of −12.3 kcal/mol compared to −11.6 kcal/mol with wild type RBD (Fig. 2, Fig. 3 , Table 1). Interestingly, the Omicron RBD showed the lowest binding affinity of −10.4 kcal/mol among all the tested complexes (Fig. 4 ). Omicron RBD contains 11 mutations, where 4 of them K417 N, E484A, G496S and N501Y were found on the interface region (Fig. 4). Besides, the disappearance of the acidic and basic residues, with the interface of the Omicron RBD presented an interface region with higher uncharged polar side chains retaining amino acids.
Table 1.
Statistics of docking analysis for all the generated complexes.
| Complex Name | Docking Score | Template Used | LG-score/Quality |
|---|---|---|---|
| ACE2-RBD (wild) | −253.22 | 7EKH | 5.051 |
| ACE2-RBD (N501Y) | −271.33 | 7EKH | 5.051 |
| EGFR-RBD (wild) | −267.34 | 4C02 | 5.284 |
| EGFR-RBD (N501Y) | −257.35 | 4C02 | 5.284 |
| EGFR-Omicron RBD | −266.98 | 4C02 | 5.037 |
LG-score indicates the reliability of the docking results and was calculated by ProQ program. Based on the LG-score, all the docked complexes were ranked in top hierarchy (LG-score > 5.0) [11].
Fig. 2.
Complex structure of the spike RBD against the kinase domain of EGFR. The predicted complex of spike RBD (pink) with kinase domain of EGFR (blue) are shown in cartoon representation. The stick representation are the interface residues located in the region of 5 Å. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Complex structure of the mutant spike RBD against the kinase domain of EGFR. The predicted complex of mutant spike RBD (pink) with kinase domain of EGFR (blue) are shown in cartoon representation. The residues in stick representation are of the interface located in the region of 5 Å. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
Complex structure of the Omicron spike RBD against the kinase domain of EGFR. The predicted complex with green and red spheres indicate the mutations observed in Omicron RBD and on the interface region, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Overall, these results indicate that RBD retains the capability to interact EGFR, as the wild type and mutant complex showed binding affinity almost equivalent to the control hACE2-RBD complex.
3.2. Network prediction of the mutations in SARS-CoV-2
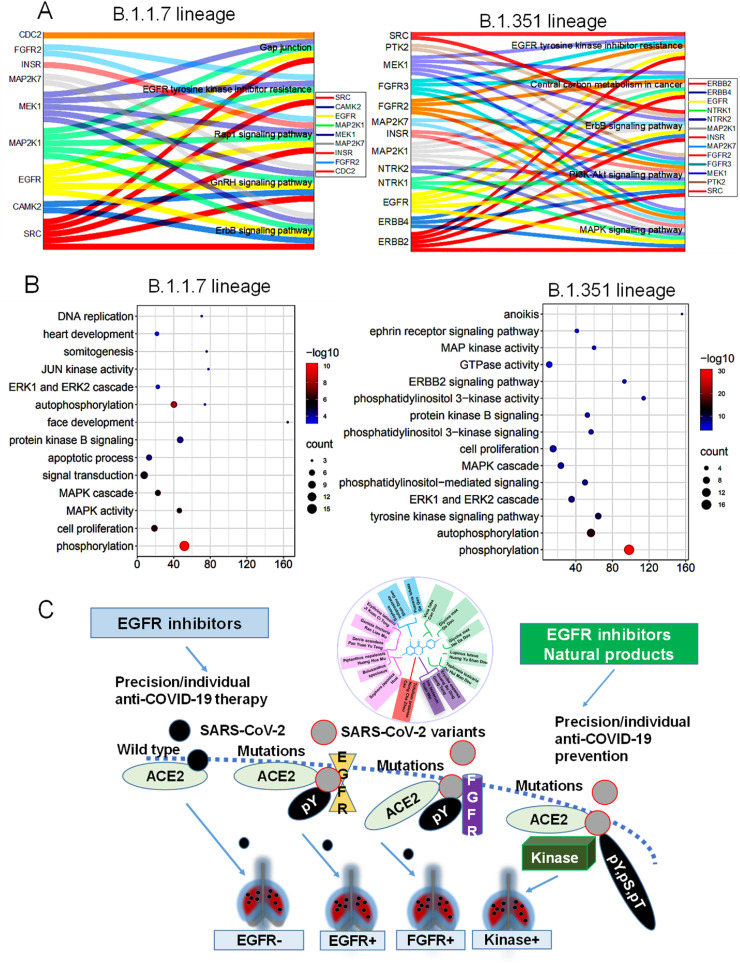
In addition to N501Y, we also performed prediction of other mutations of B.1.351 and B.1.1.7 lineages by potent phosphorylation using servers search as mentioned above. These include gain of possible phosphorylation by mutations of the two lineages with mutations of I2230T, N501Y; or H2799Y, D4527Y, N501Y to gain phosphorylation, compared with wild type sequences. All potent kinases were listed and further analyzed for gain of potent phosphorylation with protein-protein interaction maps (Supplementary Figs. S2 and S3). We found EGFR, ErbB and MAPK pathways are top ranked in the variants host cell signaling based on KEGG analysis (Fig. 5 A). Gene Ontology analysis on biological processes suggest the phosphorylation, cell proliferation, apoptosis and DNA replication pathways are involved in the two variants (Fig. 5B). Thus, the gain of potent phosphorylation by kinases might induce the possibly signaling transduction events resembling cancer induced cell death or phosphorylation events.
Fig. 5.
Signaling analysis of potent network kinases and suggested precision therapy or prevention. KEGG pathways (A) and Gene Ontology (GO) biological process (B) were shown. The top five KEGG pathways were identified by Kobas database and conducted by OriginLab 2020b software and bioinformatics tools online. C. Suggested precision therapy, and prevention medicine based on the role of EGFR/FGFR as an example of targeting potent virus-host cell interactions to disrupt potent membrane fusion for precision prevention infection especially in kinase elevated cancer patients.
3.3. Potent kinase inhibitors in treatment and prevention of the N501Y mutations induced gain of phosphorylation
The EGFR, a receptor tyrosine kinase, is known to be frequently mutated and upregulated in some cancers in response to their internal and external stresses to survive and resist against different types of therapies. EGFR has also been reported acting as a receptor in viral entry for influenza, and hepatitis C viruses [21]. According to Ref. [22] SARS-CoV-2 infection causes activation of aberrant STAT pathway which results in an acute lung injury upregulating EGFR that leads to constitutive STAT3 activation. Moreover, EGFR signaling can disrupt antiviral events induced by INF-α, thus promoting severity of viral infection [23]. A recent study by proteomics found that the SARS-CoV-2 proteins are mostly phosphorylated and network analysis identified EGFR as a central hint of the proteomic interactome [21]. The study also reported that sorafenib, a RAF inhibitor and RO5126766, a RAF/MEK dual inhibitor can disrupt viral entry and infection which is promising in anti-COVID-19 [21]. For mechanistic analysis, EGFR likely plays essential roles in cell fusion during infection of respiratory syncytial virus predicted by the other virus studies.
Given the specificity of over-expression of EGFR or SRC kinases in some cancers and its predicted role in COVID-19, it suggests an additional consideration with the treatment of cancer patients, as cancer patients with elevated EGFR or SRC might be susceptible to the variants of SARS-CoV-2 as predicted above. Our data also suggest that the mutated N501Y and other sites of the RBD has the potential to be phosphorylated by different kinases including EGFR. Thus kinase inhibitors could possibly be considered and studied as an agent for precision treatment of cancer patients with SARS-CoV-2 infection after experimental validation. For example, erlotinib, as EGFR inhibitor, could be recommended because its combination with INF-α showed antiviral activity [24].
Moreover, we may apply some herbal food which contains EGFR inhibitors compound of natural products for preventing the new variants induced COVID-19 (Fig. 5C). For example, we may select Genistein rich food to prevent the infection, including different species of soybean, black soybean, and other herbs or herbal food analyzed by a database of TCMSP Version 2.3 [[25], [26], [27]]. While processing of natural products of herbal food or traditional Chinese Medicine, possibly generates nanozyme of phosphatase which can block the kinases activity by combination, as we reported, the herbal food resources of kinase inhibitors may benefit the prevention of the variants. For example, Huangjing, which is already recommended as a component of anti-COVID-19 herbal food by clinical agencies [28,29], could be recommended for prevention of infection of variant. Moreover, integration with the traditional Chinese Medicine Huangjing, and EGFR inhibitor for treatment by applying the phosphatase activity of nanocale Huangjing of nanozyme, as we recently proposed in cancer cells [27], would also be promising. Thus, cancer patients may benefit the integration method if undergoing cancer precision therapy, with the target inhibitors as repurposed or rehabilitation medicine during Omicron circulation [30].
Recently, we reported that PI3K/AKT signal pathway is essential in COVID-19 and capivasertib as an inhibitor restricts SARS-CoV-2 entry into host cells [31,32]. In addition, based on our hypothesis mentioned above, to further investigate the potential of combination therapy by EGFR and AKT inhibitors would be promising in anti-variants. Thus the crosstalk pathways would also be deserved to clarify.
Given kinases are usually elevated in cancer patients, the potent kinase associated new variant mutation binding may induce more cancer signaling and sever infection, even there is no clear evidence that the SARS-CoV-2 can promote tumorigenesis. In addition, cancer patients should be noticed that the potent kinase associated infection by spike RBD binding to cell membrane receptor kinases. Thus the potent risk may exist that cancer patient might be more vulnerable for infection due to elevated kinase expression levels.
Most recently, while we are revising the manuscript, a study was published on which N501Y mutation's superiority among different mutations tested for transmission [33]. The tested mutations include spike mutants of T716I, N501Y, A570D, P681H, D1118H, 982A, deletion 69–70, deletion 145, with lineage B.1.1.7 variant spike [33]. The experimental data by infected hamsters suggest that in addition to deletion 69–70, N501Y showed much more advantages of fitness compared with wild type in replication efficiency in the upper airway, and shedding in nasal cavity or secretions which are related to air-borne transmission [33]. It has been shown that N501Y alone can exert the function as a powerful driver for elevated transmission but not inducing immune disruption suggesting the vaccine escape is minimum [[33], [34]].
4. Conclusion
In summary, though our prediction and hypothesis are limited which needs experimental investigation, we provided potential new mutations mediated interactions and its predicted phosphorylation landscape with host cell kinases based on current evidence from published reports in database. Our results depict that EGFR receptor is one of the potential binary partner of the spike RBD that binds almost with equal affinity as RBD-hACE2 complex. Keeping the fact of EGFR kinase activity, this report suggests the possibility of phosphorylating N501Y mutation that is located on the interface region to further potentiate its importance. We also suggest a prevention approach, and applicable solutions to fight against the new variants in the current pandemic era. Cancer patients might consider the kinase elevation induced binding to spike protein for increased risk of infection and applying kinase inhibitors or natural products for both precision cancer therapy and preventive or rehabilitative medicine. Potent host cell kinases elevation as biomarker for prevention of viral infection might be considered for cancer patients during Omicron circulation.
Data and code availability
Not applicable.
Ethics declarations
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Funding
This research is not supported specifically by any funding related to bioinformatics and COVID-19. However, the authors received a cancer research funding during the work period: Faculty Development Competitive Research Grants of Nazarbayev University (16797152).
Author information
All authors agreed to the current form.
Contributions
Bexultan Kazybay: results of supplementary Figures of network and old version draft of structure prediction, pathways, and draft writing; Ashfaq Ahmad: bioinformatics analysis, new docking data including omicron docking and methods, results writing with editing; Chenglin Mu: bioinformatics analysis, and part of methods writing; Diana Mengdesh: data gathering of kinase prediction and results with part of draft writing; Yingqiu Xie: designing as supervisor, data analysis and draft writing.
Declaration of competing interest
There is no conflict of interest to claim.
Acknowledgements
We would like to thank Assel Ibadulla, for kind help on outline draft, and Wen Li, for docking analysis by different tools. We also thank the Nazarbayev University Faculty Development Competitive Research Grants (16797152). We thank Guldan Nazarbek for the herbal food search and screening and Zhonghao Chen for public health data analysis even though we did not use Mr. Chen's results.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2021.102242.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Timur L., Xie Y. Is border closure effective in containing COVID-19? Trav Med Infect Dis. 2021;44 doi: 10.1016/j.tmaid.2021.102137. Nov-Dec. Epub 2021 Jul 9. PMID: 34252572; PMCID: PMC8268676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang T.J., Yu P.Y., Chang Y.C., Liang K.H., Tso H.C., Ho M.R., Chen W.Y., Lin H.T., Wu H.C., Hsu S.D. Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat Struct Mol Biol. 2021 Aug:1–9. doi: 10.1038/s41594-021-00652-z. [DOI] [PubMed] [Google Scholar]
- 3.Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J Mol Biol. 2021 Jul 23;433(15) doi: 10.1016/j.jmb.2021.167058. Epub 2021 May 21. PMID: 34023401; PMCID: PMC8139174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villoutreix B.O., Calvez V., Marcelin A.G., Khatib A.M. In silico investigation of the new UK (B.1.1.7) and South African (501Y.V2) SARS-CoV-2 variants with a focus at the ACE2-spike RBD interface. Int J Mol Sci. 2021 Feb 8;22(4):1695. doi: 10.3390/ijms22041695. PMID: 33567580; PMCID: PMC7915722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C., Xu H., Lin S., Deng W., Zhou J., Zhang Y., Shi Y., Peng D., Xue Y. GPS 5.0: an update on the prediction of kinase-specific phosphorylation sites in proteins. GPB. 2020;18(1):72–80. doi: 10.1016/j.gpb.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 7.Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics: Jun. 2004;4(6):1633–1649. doi: 10.1002/pmic.200300771. submitted for publication. [DOI] [PubMed] [Google Scholar]
- 8.Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webb B., Sali A. John Wiley & Sons, Inc.; 2016. Comparative protein structure modeling using modeller. Current protocols in bioinformatics 54; pp. 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Y., Tao H., He J., Huang S.-Y. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020 doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 11.Larsson P., Skwark M.J., Wallner B., Elofsson A. Assessment of global and local model quality in CASP8 using Pcons and ProQ. Proteins. 2009;77(Suppl 9):167–172. doi: 10.1002/prot.22476. [DOI] [PubMed] [Google Scholar]
- 12.Vangone A., Bonvin A.M. Contacts-based prediction of binding affinity in protein-protein complexes. Elife. 2015 Jul 20;4 doi: 10.7554/eLife.07454. PMID: 26193119; PMCID: PMC4523921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C.Y., Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Origin (Pro) OriginLab Corporation USA; Northampton (MA): 2021. Release 2021. Accompanied by:1 user manual. [Google Scholar]
- 17.Wilton T., Bujaki E., Klapsa D., Majumdar M., Zambon M., Fritzsche M., Mate R., Martin J. Rapid increase of SARS-CoV-2 variant B.1.1.7 detected in sewage samples from England between October 2020 and January 2021. mSystems. 2021;6(3) doi: 10.1128/mSystems.00353-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gravagnuolo A.M., Faqih L., Cronshaw C., Wynn J., Klapper P., Wigglesworth M. High throughput diagnostics and dynamic risk assessment of SARS-CoV-2 variants of concern. EBioMedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WeiShengxin . 2013. An online platform for data analysis and visualization.http://www.bioinformatics.com.cn [Google Scholar]
- 20.Robinson K.W., Sandler A.B. The role of MET receptor tyrosine kinase in non-small cell lung cancer and clinical development of targeted anti-MET agents. Oncol. 2013;18(2):115–122. doi: 10.1634/theoncologist.2012-0262. Epub 2013 Jan 23. PMID: 23345546; PMCID: PMC3579594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klann K., Bojkova D., Tascher G., Ciesek S., Münch C., Cinatl J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol Cell. 2020;80(1):164–174. doi: 10.1016/j.molcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama T., Kubli S.P., Yoshinaga S.K., Pfeffer K., Mak T.W. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020;27(12):3209–3225. doi: 10.1038/s41418-020-00633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupberger J., Duong F.H., Fofana I., Zona L., Xiao F., Thumann C., Durand S.C., Pessaux P., Zeisel M.B., Heim M.H., et al. Epidermal growth factor receptor signaling impairs the antiviral activity of interferon-alpha. Hepatology. 2013;58(4):1225–1235. doi: 10.1002/hep.26404. [DOI] [PubMed] [Google Scholar]
- 24.Currier M.G., Lee S., Stobart C.C., Hotard A.L., Villenave R., Meng J., Pretto C.D., Shields M.D., Nguyen M.T., Todd S.O., et al. EGFR interacts with the fusion protein of respiratory syncytial virus strain 2-20 and mediates infection and mucin expression. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanjak P., Thiantanawat A., Watcharasit P., Satayavivad J. Genistein reduces the activation of AKT and EGFR, and the production of IL6 in cholangiocarcinoma cells involving estrogen and estrogen receptors. Int J Oncol. 2018;53(1):177–188. doi: 10.3892/ijo.2018.4375. [DOI] [PubMed] [Google Scholar]
- 26.Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y., Mu C., Kazybay B., Sun Q., Kutzhanova A., Nazarbek G., Xu N., Nurtay L., Wang Q., Amin A., Li X. Network pharmacology and experimental investigation of Rhizoma polygonati extract targeted kinase with herbzyme activity for potent drug delivery. Drug Deliv. 2021 Dec;28(1):2187–2197. doi: 10.1080/10717544.2021.1977422. PMID: 34662244; PMCID: PMC8526013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu C., Sheng Y., Wang Q., Amin A., Li X., Xie Y. Potential compound from herbal food of Rhizoma Polygonati for treatment of COVID-19 analyzed by network pharmacology: viral and cancer signaling mechanisms. J Funct Foods. 2021;77 doi: 10.1016/j.jff.2020.104149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benassi E., Fan H., Sun Q., Dukenbayev K., Wang Q., Shaimoldina A., Assanbiyeva A., Nurtay L., Nurkesh A., Kutzhanova A., et al. Herbal food processing generation of particle assembly mimicking enzymatic activity: the case of Rhizoma polygonati and other natural ingredients of traditional Chinese medicine. Nanoscale Adv. 2021;3:2222–2235. doi: 10.1039/d0na00958j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nurtay Lazzat, Sun Qinglei, Mu Chenglin, Cao Zhongshan, Wang Qian4, Liang Zongsuo, Ma Cuiping, Li Xugang, Amin Amr, Xie Yingqiu. Rhizoma polygonati from Mount Tai: nutritional value and usefulness as a traditional Chinese medicine, source of herbzyme, and potential remediating agent for COVID-19 and chronic and hidden hunger. Acupuncture and Herbal Medicine. September 2021;1(1):31–38. doi: 10.1097/HM9.0000000000000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santamaria S. Targeting the PI3K/AKT pathway: a potential new weapon in the global fight against SARS-CoV-2? Int J Biol Sci. 2021 Jul 5;17(11):2770–2771. doi: 10.7150/ijbs.63969. PMID: 34345206; PMCID: PMC8326129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun F., Mu C., Kwok H.F., Xu J., Wu Y., Liu W., Sabatier J.M., Annweiler C., Li X., Cao Z., Xie Y. Capivasertib restricts SARS-CoV-2 cellular entry: a potential clinical application for COVID-19. Int J Biol Sci. 2021;17(9):2348–2355. doi: 10.7150/ijbs.57810. https://www.ijbs.com/v17p2348.htm Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., Widen S.G., Menachery V.D., Shi P.Y., Weaver S.C. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2021 Nov 24 doi: 10.1038/s41586-021-04245-0. Epub ahead of print. PMID: 34818667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., Wang J., Cong Q., Kinch L.N., Schaeffer R.D., Millán C. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373(6557):871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.