Abstract
Streptococcus mutans (S. mutans) is generally regarded as a major contributor to dental caries because of its ability to synthesize extracellular polysaccharides (EPS) that aid in the formation of plaque biofilm. The VicRKX system of S. mutans plays an important role in biofilm formation. The aim of this study was to investigate the effects of vicK gene on specific characteristics of EPS in S. mutans biofilm. We constructed single-species biofilms formed by different mutants of vicK gene. Production and distribution of EPS were detected through atomic force microscopy, scanning electron microscopy and confocal laser scanning microscopy. Microcosmic structures of EPS were analyzed by gel permeation chromatography and gas chromatography-mass spectrometry. Cariogenicity of the vicK mutant was assessed in a specific pathogen-free rat model. Transcriptional levels of cariogenicity-associated genes were confirmed by quantitative real-time polymerase chain reaction. The results showed that deletion of vicK gene suppressed biofilm formation as well as EPS production, and EPS were synthesized mostly around the cells. Molecular weight and monosaccharide components underwent evident alterations. Biofilms formed in vivo were sparse and contributed a decreased degree of caries. Moreover, expressional levels of genes related to EPS synthesis were down-regulated, except for gtfB. Our report demonstrates that vicK gene enhances biofilm formation and subsequent caries development. And this may due to its regulations on EPS metabolism, like synthesis or microcosmic features of EPS. This study suggests that vicK gene and EPS can be considered as promising targets to modulate dental caries.
Subject terms: Dental caries, Biofilms
Introduction
Dental caries is a chronic infectious disease occurring on mineralized tooth tissue, which is considered one of the most common health problems globally in 2015 posing considerable economic, social, and health-related burden.1–3 Streptococcus mutans (S. mutans) is important in caries development because of its ability to synthesize extracellular polysaccharides (EPS) matrix.4 EPS, with all the oral microorganisms embedding in,5 is essential in oral microenvironment, because it can form a well-structured biofilm on the tooth surface.1,6,7 It can also function as a sensor for environmental signals and provide a long-term dynamic protection for microbial communities.6,8 Nowadays, with the thriving development of targeted therapies, regulations on EPS metabolism and biofilm formation have gained considerable attention.9,10 Nanoparticles have been used to disrupt biofilm and a small molecule which can selectively target S. mutans biofilms has also been reported to effectively reduce dental caries in vivo without affecting the overall oral microbiota.11,12 Researchers have found that the production, distribution as well as microcosmic characteristics of EPS all take a considerable part in the pathogenicity of polymicrobial biofilm.13,14 So, an understanding of specific characteristics of EPS derived from S. mutans biofilm will provide more solid evidences for caries prevention, which have not been elucidated yet.
VicRKX signal transduction system consists of three encoding proteins: VicK, a histidine protein kinase; VicR, a global response regulator (RR); and VicX, a putative hydrolase.15–17 VicK, as an essential link between the environmental stimuli and cellular status, is supposed to sense and transmit chemical signals to downstream regulatory proteins, like VicR and GcrR.18,19 And subsequently, it can regulate transcriptional levels of biofilm associated genes, like gtfB/C, ftf and gbpB.20–22 As a result, vicK gene possesses an important position in multiple bioactivities of S. mutans, particularly biofilm formation.23,24 What’s more, an epidemiological research has reported that the frequency of a vicK C470T missense mutation was higher in the high-severity caries group than in the caries-free group.25 The mutation of C470T contributes to the alternations of PAS domain of VicK protein, and PAS domain is a major sensor to environmental stresses,26 which means vicK gene, as well as VicK protein are associated with the cariogenicity of S. mutans closely.
At present, few studies on the relationship between microcosmic structures of EPS and cariogenicity of S. mutans have been found. Although vicK gene can modulate biofilm formation and virulence of S. mutans in some degree, it is not clear whether this modulation is related to EPS metabolism, especially its structural characteristics. And the comprehensive regulation among vicK gene, transcriptional levels of downstream genes, EPS synthesis and cariogenicity of S. mutans remains unknown. Hence, this study aimed to identify the role of vicK gene in the cariogenicity of S. mutans in vitro and in vivo, as well as to elucidate effects of vicK gene on the EPS.
Results
vicK was involved in biofilm morphological changes
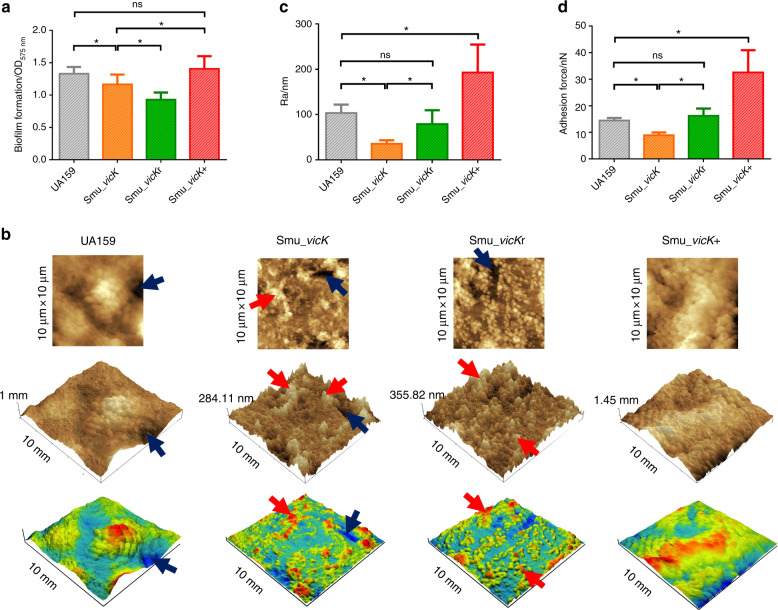
To compare the abilities of different strains to form biofilms, crystal violet (CV) assay was used and the results showed that the mean value of Smu_vicK (the vicK deficient strain) biofilms was statistically lower than that of UA159 (Fig. 1a). Atomic force microscopy (AFM) observations indicated a peak-and-valley topography in Smu_vicK and Smu_vicKr (the vicK complementary strain) biofilms (peaks: red arrows; valleys: dark blue arrows; Fig. 1b). UA159, as well as the Smu_vicK+ (the vicK overexpression strain) biofilms had smoother and more flourishing surface (Fig. 1b). Mean Ra of Smu_vicK biofilms was markedly lower than the UA159 group, while that of Smu_vicK+ was significantly higher and that of Smu_vicKr was not statistically different (Fig. 1c). Adhesion force of Smu_vicK biofilms was statistically lower, whereas Smu_vicK+ appeared an enhanced capacity (Fig. 1d).
Fig. 1. Analysis of biofilm morphologies.
Unpaired t test was used to detect the statistical significances. a Biomass was quantified by CV staining, and the optical density at 575 nm was read. (n = 6; *P < 0.05; ns no significant difference). b Surface characteristics and the 3-D reconstruction of 24 h biofilms (an area of 10 μm × 10 μm, with a bar indicating vertical deviations) with the light (red) and dark (blue) zone representing peaks and valleys. c Surface roughness (Ra) of biofilms was obtained via AFM. (n = 4; *P < 0.05; ns no significant difference). d Adhesion force data were obtained from AFM. (n = 3; *P < 0.05; ns no significant difference)
vicK changed production and distribution of EPS in biofilm
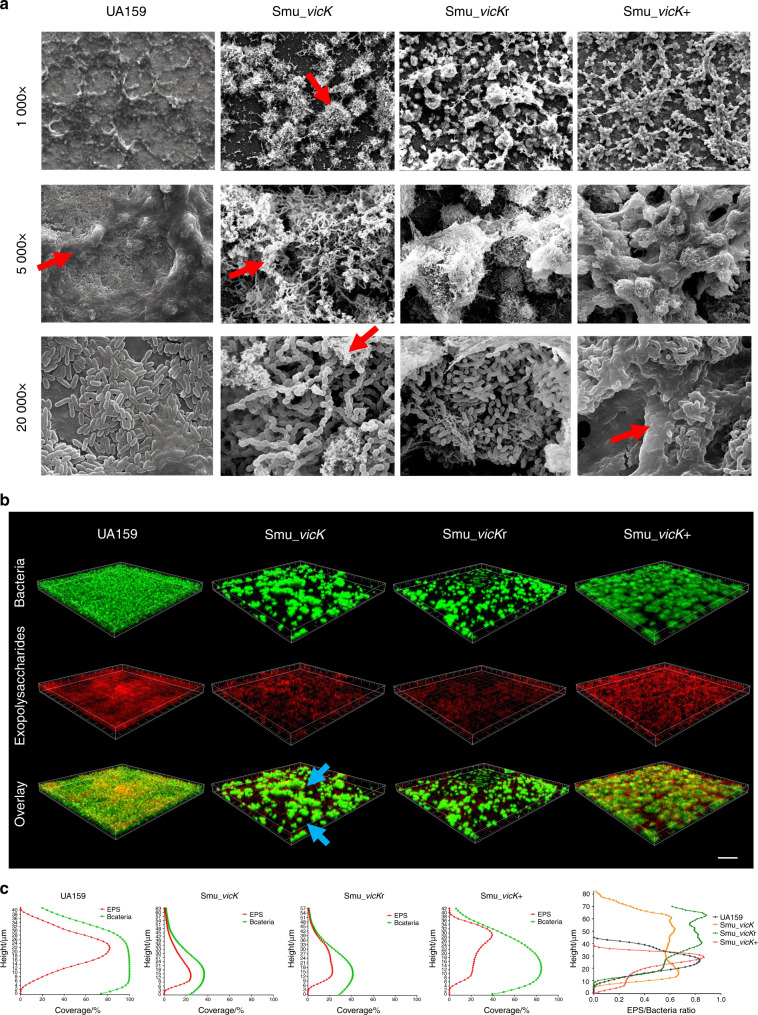
Compared with UA159, Smu_vicK biofilms demonstrated an aberrant architecture with more scattered microcolonies and thinner EPS (Fig. 2a). Evidently, Smu_vicK biofilms seemed to be devoid of EPS when scanned under a higher magnification (5 000×). On the other hand, Smu_vicK+ cells were densely encased by enriched EPS (Fig. 2a). Confocal laser scanning microscopy (CLSM) data showed that only small clusters of bacterial cells were observed in Smu_vicK and Smu_vicKr biofilms and EPS were synthesized mostly around the cells, while there were much more EPS enmeshing the bacterial aggregates and filling the space between them in UA159 and Smu_vicK+ biofilms (Fig. 2b). As shown in Fig. 2c, both EPS and bacterial cells of Smu_vicK and Smu_vicKr biofilms were less than those of UA159 and Smu_vicK+ biofilms. EPS/bacteria ratios of the biofilm formed by Smu_vicK were significantly lower than those of UA159 and Smu_vicKr.
Fig. 2. Analysis of 3-D bacteria-polysaccharides structure in mature biofilms of S. mutans strains.
a Biofilm architecture was observed by SEM, in which images were taken at 1 000×, 5 000×, and 20 000× magnifications, respectively (EPS: red arrows). b Representations of glucans distribution and biofilm reconstructions were revealed by CLSM, where EPS stained with Alexa Fluor 647 were red and total bacteria stained with SYTO9 were green. Images were taken at 20× magnification and the scale bar indicated 100 μm. Smu_vicK biofilms showed small clusters of bacterial cells without enough EPS filling the space between them (light blue arrows). c Quantification of EPS and bacteria components, as well as EPS/bacteria ratios at different heights were performed with COMSTAT
Analyses of microcosmic structures of EPS generated from biofilms
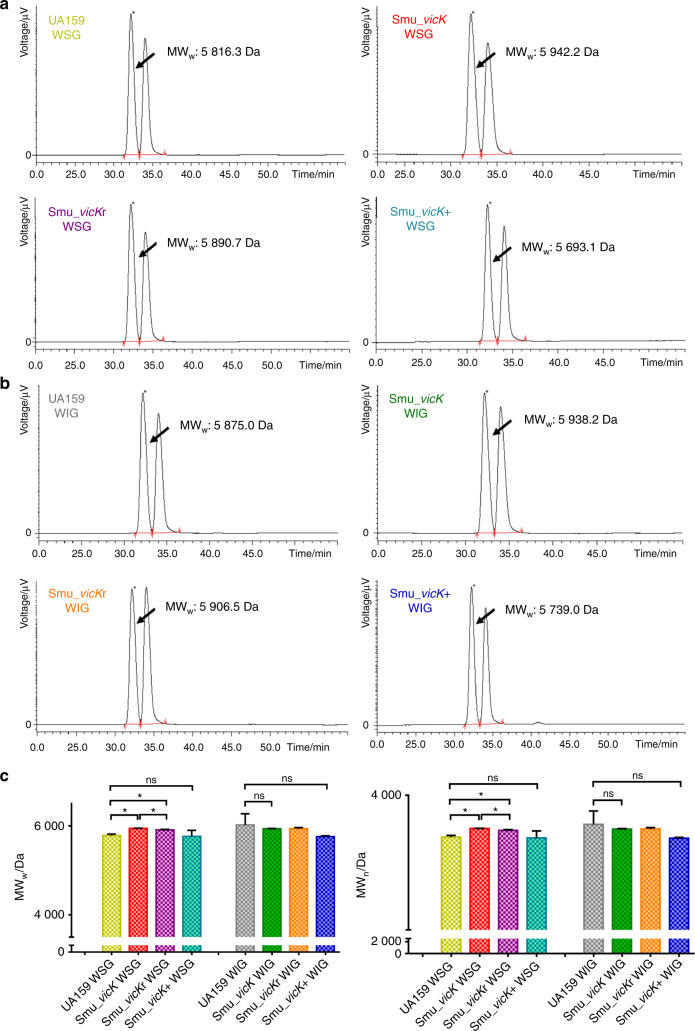
MWw (weight-average molecular weight) and MWn (number-average molecular weight) were demonstrated in Fig. 3. Water-soluble glucans (WSGs) and water-insoluble glucans (WIGs) isolated from Smu_vicK biofilms presented the highest MWw (5 942.2 Da and 5 938.2 Da, respectively), while those from Smu_vicK+ illustrated the lowest MWw when UA159 was as a reference (Fig. 3a, b). For further analysis, the mean molecular weight of WSGs from Smu_vicK biofilms were statistically higher than those from UA159 biofilms (Fig. 3c).
Fig. 3. Molecular weight distributions of EPS were estimated by GPC.
The calibration curves were constructed based on the molecular weight of standard dextran and retention time (RT), which were lgMWw = −0.285 1RT + 12.969 and lgMWn = −0.266 6RT + 12.261, with R2 = 0.994 5 and 0.993 9, respectively. a, b Typical GPC chromatograms of polysaccharides from WSGs and WIGs. a Representative MWw for WSGs. b Representative MWw for WIGs. c Distributions of mean MWw and MWn. One-way ANOVA was used to detect the statistical significances. (n = 3; *P < 0.05; ns no significant difference)
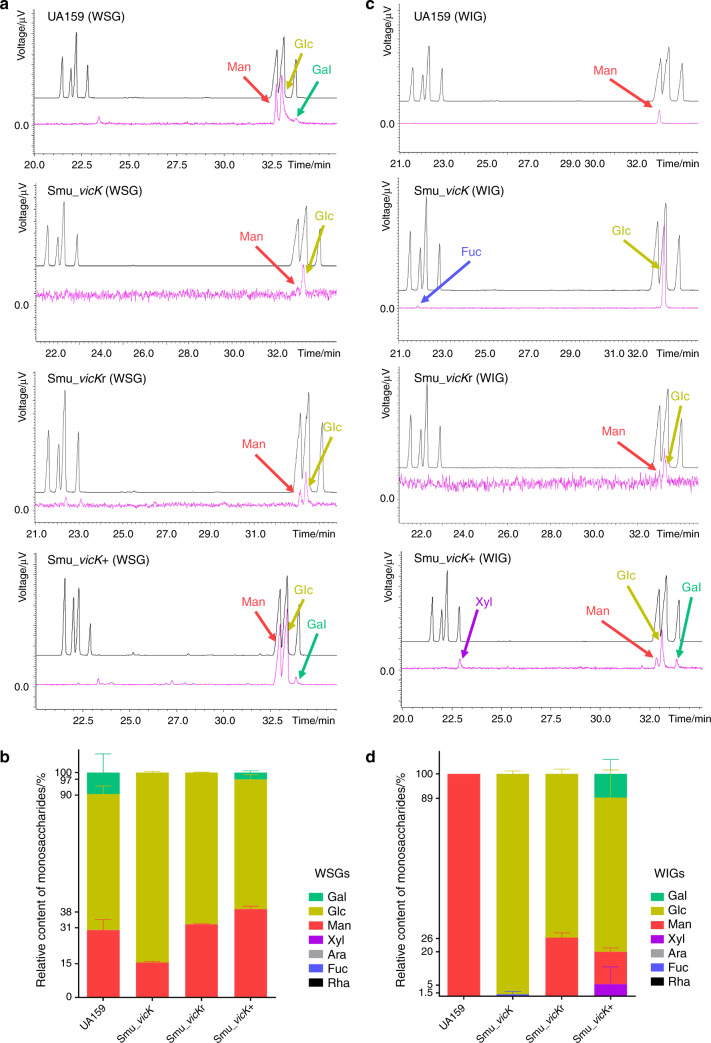
Mannose and glucose were main constituents in both WSGs and WIGs (Fig. 4). Particularly, Smu_vicK WSGs consisted of lower mannose (15.55% ± 0.36%), higher glucose (84.46% ± 0.36%) and none galactose when compared to UA159 WSGs, in which mannose was 29.96% ± 3.33%, glucose was 60.49 ± 2.53%, and galactose was 9.56% ± 5.86% (n = 2, Fig. 4b). At the same time, WSGs from Smu_vicKr biofilms were composed of mannose (32.62% ± 0.14%) and glucose (67.38% ± 0.14%); WSGs from Smu_vicK+ biofilms were more like those from UA159 (Fig. 4a, b). For WIGs (Fig. 4c, d), UA159 exhibited a 100% content of mannose and Smu_vicK showed a 99.09% ± 0.91% content of glucose. Molar ratios of Smu_vicKr WIGs were 26.41% ± 1.49% and 73.59% ± 1.49%, for mannose and glucose, respectively. And Smu_vicK+ WIGs seemed to be more complicated consisting of xylose, mannose, glucose and galactose.
Fig. 4. Monosaccharides of EPS were analyzed by GC‐MS.
a Typical chromatograms of constituent monosaccharides in WSGs. In these profiles, the black image represented standard samples in sequence: Rha, Fuc, Ara, Xyl, Man, Glc, and Gal, while the pink one represented the experimental samples. b Molar ratios of different monosaccharides in WSGs. (n = 2). c Typical chromatograms of constituent monosaccharides in WIGs. d Molar ratios of different monosaccharides in WIGs. (n = 2)
Depletion of vicK represses cariogenicity of S. mutans
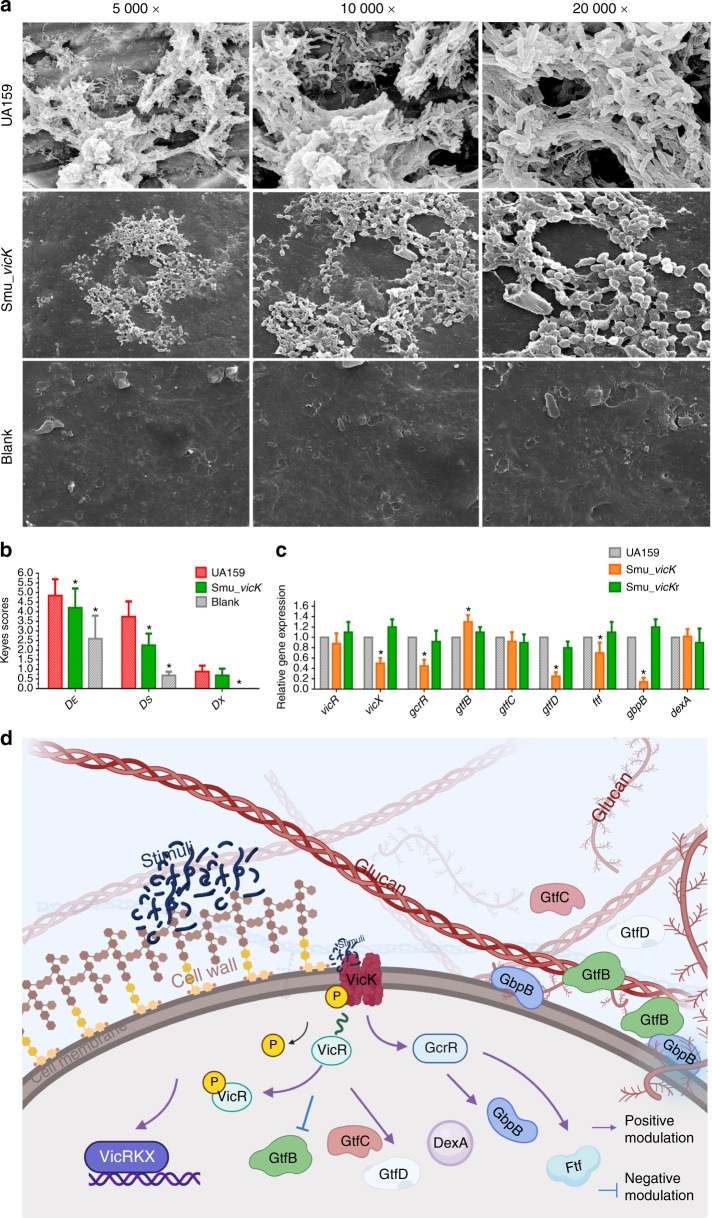
Cariogenicity of Smu_vicK in vivo was examined by a specific pathogen-free (SPF) rat model. Biofilm overview and EPS distribution were captured by scanning electron microscopy (SEM). In SEM images, Smu_vicK biofilms were sparser with thinner EPS than those formed by UA159 (Fig. 5a). According to the severity and penetration of caries, dental caries was classified into three categories: enamel only (DE), slight dentinal (DS) and extensive dentinal (DX). Smu_vicK group exhibited a significantly increased degree of caries compared to blank group and a significantly decreased degree of caries compared to UA159 group in the DE and DS categories (Fig. 5b). However, the severity of DX was unaffected by depletion of vicK gene.
Fig. 5. Influences of the deletion of vicK gene on cariogenicity and associated genes.
a Dental plaques on mandibular molars in rats were shown by SEM with pictures taken at 1 000×, 5 000×, and 20 000× magnifications. b The degrees of caries were divided into three categories by depth of penetration: DE, DS, and DX according to the modified Keyes scores. Unpaired t test was used to detect the statistical significances. (n = 10; *P < 0.05). c Expressional levels of EPS-correlated genes were measured by qRT-PCR, where UA159 was taken as the control group and the gyrA gene was used as an internal standard (not shown). One-way ANOVA was used to detect the statistical significances. (n = 3; *P < 0.05). d The schematic illustration of regulations conducted by vicK on the EPS metabolism of S. mutans. Created with BioRender.com. Adapted from “ECM (Extracellular Matrix)”, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates. After being activated by environmental stimuli, VicK has vital influences on the expression of genes correlated with EPS, whose encoding proteins promote the synthesis of EPS, subsequently enhances the biofilm virulence
vicK alters the expressional levels of virulence-related genes
To evaluate effects of vicK gene on signal transduction system and cariogenicity-related genes, the gyrA gene was used as an endogenous reference. As shown in Fig. 5c, transcriptional levels of vicX and gcrR genes were decreased in Smu_vicK when compared to UA159. Simultaneously, gtfD, ftf, and gbpB were also downregulated in Smu_vicK. On the contrary, the expressional level of the gtfB gene was significantly increased in Smu_vicK. No difference was observed in the expressions of gtfC and dexA.
Discussion
Biofilms and EPS are inseparably linked. Biofilms develop when microorganisms accumulate on tooth surfaces and secret EPS.27 At the same time, EPS initiates the adhesion for first colonizers and is critical for dental biofilms development.28 Working together, they can be virulent and cause infectious diseases, like dental caries.29 In our study, Smu_vicK showed a reduced ability in biofilm accumulation, which was consistent with previous studies that found Smu_vicK biofilm clumpy and deficient in microcolonies formation.23,30 Biofilms can act as a barrier and cause resistance against harmful factors, and thicker they are, more protective they are for microbes within.31 Thin biofilms like Smu_vicK biofilms might be less stable and more easily disrupted, which was consistent with what Senadheera had reported before.23 AFM observation of Smu_vicK biofilms demonstrated a bumpy topography with spike-like peaks, indicating that knockout of vicK gene yielded a thin biofilm with less EPS. Biofilm development is a complex process including cell to cell adhesion. Physical or molecular interactions are involved in this process.32 It also has been reported that the surface roughness does not necessarily affect bacterial adhesion.23 The limitation of this study was that we did not measure the surface roughness of a substratum surface but the surface roughness of the biofilm itself. Interestingly, Smu_vicK biofilms were thicker than those of UA159 as shown in Fig. 2c, but they seemed sparser and with less EPS (Fig. 2b). In present study, the biofilm surface roughness and adhesion force of Smu_vicK biofilms were declined, indicating biofilm surface roughness and adhesion force may be reflections of EPS production in S. mutans, which also warranted further investigations.23,33 Similar results came out in SEM and CLSM observations. It was worth mentioning that EPS of Smu_vicK biofilm did overlap with the microcolonies but it was few. Previous reports also showed that the vicK-deficient strain had a drastically reduced glycolytic rates.33
Indeed, construction of the vicK complementary strain was limited in this study, where vicK expression was slightly higher than Smu_vicK but not close to UA159 level (Fig. S1). As a result, Smu_vicKr biofilms were thin and more like Smu_vicK biofilms. One possible explanation was that the introduction of an exogenous plasmid vector might interfere with the intracellular homeostasis and growth of the mutants, which has been reported in our previous study.16 Although Smu_vicKr biofilms show less biomass than the one of Smu_vicK biofilms, the phenotypes of roughness and adhesive forces in the Smu_vicKr biofilms were partially restored when compared to those of UA159 biofilms. Smu_vicK+ biofilms did exhibit a slight increase in biomass or EPS when compared to those of UA159. We speculated that the difference was not that evident because of the introduction of the plasmid.16 Biofilms of Smu_vicK+ were not as mature as UA159 in Fig. 2, but it also showed EPS/bacteria ratios of Smu_vicK+ biofilms were higher than those of UA159. One possible explanation for Smu_vicK+ biofilms not as mature as UA159 was that Smu_vicK+ biofilms had larger microorganism clusters and more EPS.
Taken together, deletion of vicK gene not only impaired biofilm formation, but also repressed EPS production. EPS can encapsulate microbial communities and provide enough attachment sites for them to form structured biofilms, contributing to biofilm lifestyle and virulence.34 In addition to EPS, bacterial aggregation is also an essential part in biofilm formation.23 We found a rough bacterial aggregation of Smu_vicK in SEM, an evidently longer chain length and a larger colony of Smu_vicK+ (Fig. 2 and Fig. S2). Decreased EPS and aberrant biofilm might result in alteration of cariogenicity of S. mutans.
Exopolysaccharides is part of macromolecules within extracellular matrix. Both WSGs and WIGs are of great significance. WSGs is able to provide energy source; WIGs can provide protective shelters for the biofilms and modulate environmental acidification.35–37 Targeting EPS is a promising approach in biofilm control and recent studies have declared kinds of nanoparticles disrupting biofilm microenvironment, which allows disruption of the matrix.38–40 Targeted therapy for dental caries is becoming more popular,41 so a thorough understanding of the targeted EPS is a keystone. Structural features of polysaccharides like molecular weight may play an important role in various biological activities.42,43 For example, a high-molecular-weight polysaccharide fraction from Sparassis latifolia is recognized to account for a limited biological activity because it makes the fungus difficult to adhere or penetrate cell membranes.44,45 Combined with the data we got, it was speculated that the upregulation of molar mass in Smu_vicK EPS might be relevant to its impairment. Molecular weight of polysaccharides can be influenced by glycoside linkage, which is attributed to the linkage cleavage during the enzymatic degradation.46 Fructosyltransferase (Ftf, coded by ftf) converts sucrose into a predominantly β2,1-linked fructan polymer.47 Glucosyltransferases (Gtfs, encoded by gtfB/C/D genes) are also linkage-related, in which GtfB synthesizes α1,3-glycosidic linkages of WIGs and GtfD synthesizes α1,6-glycosidic linkages of WSGs.48 We assumed the altered expressional levels of ftf and gtfB/D genes in Smu_vicK (Fig. 5c) probably induced glycoside linkage alternation and subsequent molecular weight of polysaccharides. Further studies like methylation analysis will provide the information of the glycosidic linkages of EPS.49 Monosaccharides are essential substrates to exopolysaccharide chains, and different compositions of them can make different biofilm matrix.50,51 Constituent ratio of glucose experienced evident increasing in EPS of Smu_vicK biofilms, indicating a rising capability of Smu_vicK to synthesize glucose. It was speculated that polysaccharides of Smu_vicK biofilms were heteropolysaccharides containing glucose, and the side chains might be composed of mannose or fucose, which can be confirmed by connection mode analysis in further studies.52 EPS is a vital attribute for the bacterial community structuring and contributes a lot in biofilm lifecycle and subsequent caries development.53 We regarded that vicK gene and microcosmic structures of EPS were closely related, which might have a relationship with cariogenicity of S. mutans biofilms.
A dysregulation of biofilm formation and decreased cariogenicity of Smu_vicK in vivo could be seen in this study. Evidently, these results were consistent with the results in vitro. However, Senadheera had found that in low-cariogenic diet feed rats, the development of dentinal lesions was not significantly altered in Smu_vicK group compared with UA159,23 which was different from what we found when we fed the rats a normal diet with drinking water containing 2% sucrose. S. mutans can make effective use of dietary sucrose for EPS production, using GtfB/C/D.54,55 Both the diet and the nature of EPS can determine the diffusion properties of plaque and cariogenicity of S. mutans. VicK acts as a receptor of extracellular stimuli,56 and when it was deleted, the capacity of bacterial cell to get transduction signals might be impaired. On the other hand, expressions of gtfB/C/D genes could also be affected by vicK as previously reported,23 which are associated with sucrose. One possible explanation of this difference was that Smu_vicK had defects in biofilm formation while UA159 formed more compact and cariogenic biofilms under high-cariogenic circumstances. In addition, it has been found that the vicK-deficient strain produced less lactic acid,33 which might partially contribute to the low cariogenicity of the Smu_vicK biofilm. These data suggested that we should attach more importance to vicK gene and dietary sucrose.
In present study, vicK gene positively modulated vicR/X and gcrR. The vicR/K/X gene are in the same operon: the vic operon,57 so deletion of vicK gene might influence the transcriptional levels of vicR/X. GcrR (also named CovR, coded by gcrR gene) is known as an orphan response regulator in S. mutans, with no exclusively linked cognate histidine kinase.58 There is a co-regulation between GcrR and VicRK,59,60 in which, like VicR, GcrR can also directly binds to the promoter regions of some genes, but opposite to VicR, it acts like a repressor.61–63 And VicK can activate GcrR by phosphorylating it.64,65 The declined expressional level of gcrR in this study was not in coincidence with the opposite mechanism between VicR and GcrR. What’s more, we found a significant decrease of GcrR protein in Smu_vicK (Fig. S3). Although there was also a decrease in expressional level of VicR protein, it seemed relatively stable when compared with GcrR (Fig. S3). On one hand, it has been reported that deletion of vicK gene does not have a significant influence on the ratio of phosphorylated VicR to unphosphorylated VicR.66 On the other hand, it has also been authenticated that neither dimerization nor phosphorylation status of GcrR are essential for its activity.59 As a result, we speculated that VicK might also influence the expressional level of GcrR in some way to balance the regulatory system, which need further studies. Simultaneously, gtfD, ftf and gbpB were also downregulated in Smu_vicK as previously reported.23 The less-cariogenic biofilm with poorer EPS generated by Smu_vicK might be resulted from the defect of ftf and gbpB genes because Ftf and GbpB (encoded by gbpB) should have enhanced the cariogenic potential of S. mutans by extending acidification or working with Gtfs.67–69 Unexpectedly, the expressional level of gtfB gene was significantly increased and that of gtfC was unchanged in Smu_vicK, which were in contrast with a previous observation showing a 3.0 and 1.4-fold downregulation of gtfB/C in Smu_vicK at mid-log phase grown in brain heart infusion (BHI) medium.70 We considered the variation in nutrients was responsible for the differences.71,72 It was interesting that the increase of gtfB did not appear to provide a mechanically integrated and stable extracellular insoluble matrix for Smu_vicK biofilms. This phenomenon might be explained by the extreme decrease of gtfD, whose products are supposed to serve as primers for GtfB activity.73 In S. mutans, Gtfs perform a concerted action to produce glucans and affect EPS.74,75 It was once discussed that an optimal GtfB/GtfC/GtfD ratio was necessary for appropriate colonization of S. mutans in vitro.76 We speculated that the expression of vicK gene mostly influenced EPS-synthesis genes in a positive way because the expressional level of dexA gene was not changed in Smu_vicK. A schematic illustration was shown in Fig. 5d. Understanding the mechanisms involved in biofilm-associated gene expressions is crucial for us to develop new therapeutic strategies. It has been reported that expressional levels of vicR/gcrR genes, key regulators to EPS-synthesis, are maximal in S. mutans which are at mid-exponential phase.77 However, the gene expressions of planktonic cultures may not reflect the expressions of biofilm grown cells, which needs further investigations.
Carbohydrate metabolism is necessary to viability and virulence of S. mutans, in which production of lactic acid, acid tolerance and production of EPS are all essential.78 S. mutans is able to produce lactic acid as a metabolic product, which induces lower pH value in environment, as well as the enamel erosion and dental caries. The acid tolerance response becomes a key survival mechanism for the acid stress adaption.79 Senadheera et al. had studied the effects of S. mutans vicK gene on acidogenicity and aciduricity in the presence of glucose. They found vicK deletion mutant produced less lactic acid with its acid tolerance enhanced.33 However, the effects of S. mutans vicK gene on EPS production in the presence of sucrose were not fully elucidated. So, in current study, we provided evidences proving the modulation of vicK gene on EPS of S. mutans biofilm. The vicK gene regulated biofilm characteristics, including synthesis as well as component and structural modifications of EPS. These data illustrated a significant situation where vicK was a promising governor, being involved in cariogenicity of S. mutans through EPS metabolism. We hypothesized vicK gene enhanced transcriptional levels of EPS-induced genes in a signal transduction cascade and it could increase the production of EPS, change the microcosmic features of EPS as well as strengthen the virulence of S. mutans.
Materials and methods
Strains and bacterial culture conditions
S. mutans standard strain,36 UA159 (Table 1) was provided by State Key Laboratory of Oral Diseases, West China School of Stomatology, Sichuan University. An erythromycin resistance cassette was utilized to knock out the vicK gene in UA159 chromosome by polymerase chain reaction (PCR) ligation mutagenesis to get Smu_vicK.23,80 Recombinant pDL278 plasmids which contained the vicK gene coding region were introduced into Smu_vicK and UA159 cells respectively to construct the vicK complementary strain (Smu_vicKr) and the vicK overexpression strain (Smu_vicK+).81 The expressions of vicK gene in the mutants were verified by quantitative real-time PCR (qRT-PCR) (Fig. S1). Conventionally, UA159, Smu_vicK, Smu_vicKr, and Smu_vicK+ were cultured into mid-exponential phase in BHI medium (Becton, Dickinson and Company, Sparks, MD 21152 USA) at 37 °C, anaerobically, with appropriate antibiotics when necessary. The achieved final concentrations of antibiotics were 10 μg·mL−1 erythromycin for Smu_vicK, 1 000 μg·mL−1 spectinomycin for Smu_vicK+, 10 μg·mL−1 erythromycin and 1 000 μg·mL−1 spectinomycin for Smu_vicKr.
Table 1.
Bacterial strains and plasmid used in this study
| Strain or plasmid | Description | Source of reference |
|---|---|---|
| UA159 | S. mutans UA159 | ATCC 700610 |
| Smu_vicK | S. mutans UA159 with in-frame replacement with an erythromycin cassette | This study |
| Smu_vicKr | Smu_vicK transformed with pDL278 encoding vicK gene | This study |
| Smu_vicK + | S. mutans UA159 transformed with pDL278 encoding vicK gene | This study |
| pDL278 | Escherichia coli-Streptococcus shuttle vector and expression plasmid (spectinomycin) | Gifted by Dr. Huichun Dong, Institute of Microbiology, Chinese Academy of Sciences |
Biofilm formation
Strains were grown to mid-exponential phase at an OD600 nm of 0.5 and then were diluted 1:100 into BHI medium containing 1% (w/v) sucrose (BHIS). Biofilms were formed at 37 °C, anaerobically for 24 h.
CV assay
Biofilm mass was quantified by CV assay as previously described.82 Briefly, biofilms formed on a polystyrene surface in a 96-well plate were washed by sterilized deionized distilled water three times to remove planktonic bacteria cells, then were stained with 0.1% (w/v) CV for 15 min. The stained biofilms were gently rinsed again and air dried, followed by an addition of 33% (v/v) glacial acetic acid to solubilize the dye. At last, the optical density was read at 575 nm (SpectraMax® iD5, Molecular Devices, San Jose, California, USA).
Atomic force microscopy
Surface topography and adhesion force of the biofilms were analyzed by AFM as conducted previously.83,84 Biofilms were incubated in a 24-well plate for 24 h with round, sterile glass slides, then the slides were washed by sterilized deionized distilled water and dried for two minutes in air. Shimadzu SPM-9700 system (SHIMADZU, Kyoto, Japan) was conducted in the contact mode using a specialized probe (HYDRA-ALL-G-20, APPNANO, USA) for the measurements.
Scanning electron microscopy
Architecture of the biofilms was measured by SEM (Inspect F, FEI, Eindhoven, Holland) using round, sterile glass slides.85 The biofilms on slides were washed by sterilized deionized distilled water then fixed with 2.5% (v/v) glutaraldehyde overnight. Next day, they experienced a sequential dehydration in ethanol solutions and then were prepared for imaging.86 The specimens were examined at 1 000×, 5 000×, and 20 000× magnifications, and representative pictures are shown.
Confocal laser scanning microscopy
Three-dimensional (3-D) structure and EPS distribution of the biofilms were observed by CLSM (Olympus FV1000, Japan).87 1 μmol·L−1 Alexa Fluor 647 (Invitrogen, Eugene, Oregon, USA) were added into 2 mL BHIS with 20 µL bacterial cultures before biofilm incubation to label dextran conjugate. Biofilms were incubated in a 24-well plate for 24 h in dark, with round, sterile glass slides. After biofilm formation, nucleic acid of S. mutans cells were labeled with 2.5 μmol·L−1 SYTO9 (Invitrogen, Eugene, Oregon, USA). Microscopic observations were performed with a 20× objective lens, and 3-D images were reconstructed by Imaris 7.0.0 software (Bitplane, Zürich, Switzerland). Calculation of EPS/bacteria biomass were performed with COMSTAT.
Exopolysaccharides assessment
Molecular weights of EPS were analyzed by gel permeation chromatography (GPC) and monosaccharide composites were analyzed by gas chromatography–mass spectrometry (GC–MS). Both of WSGs and WIGs derived from 24 h biofilms were isolated and purified for GPC and GC–MS analysis as reported before.16 In short, biofilms suspended by sterilized deionized distilled water were centrifuged to separate WSGs solution and other insoluble precipitates containing WIGs. The insoluble precipitates were dissolved in 1 mol·L−1 NaOH solution and then filtered through a 0.22 μm filter (Merck Millipore Ltd. Tullagreen, Carrigtwohill, Co. Cork, IRL) to get WIGs solution. Then, 20% (w/v) trichloroacetic acid was added into the solution mentioned above for protein precipitations. After that, solutions were dialyzed (cellulose membrane with a molecular weight cut-off of 500–1 000 Da) for further purification and were lyophilized afterwards. The lyophilized powder was used for GPC and GC-MS.88 For GPC analysis, sample polysaccharides in a final concentration of 5 mg mL−1 and standard solutions of dextran in different molecular weights (MW; MW 1 152, 5 200, 11 600, 148 000, 273 000, 410 000 Da; Waters, Massachusetts, USA) were loaded in a high performance liquid chromatograph (LC-10A, SHIMADZU, Japan).51 For GC–MS analysis (QP 2010 Plus, SHIMADZU, Japan), samples were further acetylated and the compositions of EPS were identified with standard monosaccharide substances (Sigma, San Francisco, CA, USA), which included rhamnose (Rha), fucose (Fuc), arabinose (Ara), xylose (Xyl), mannose (Man), glucose (Glc), galactose (Gal).89
Cariogenicity ability in vivo
Effects of vicK gene on cariogenicity of S. mutans in vivo were assessed in a SPF rat model.90 This study was approved by the Ethics Committee of West China Hospital of Stomatology, Sichuan University (NO: WCHSIRB-D-2019-144), and conducted in IVC Experimental Animal Center of Public Health, Sichuan University (Chengdu, China). Thirty caries-susceptible, Osborne–Mendel rats (15 male rats and 15 female rats), aged 14 day old (Dashuo Company, Chengdu, China) were included and randomly assigned into 3 groups: Blank (the negative control), UA159 (the positive control), and Smu_vicK (the experimental group). Posterior to endogenous Streptococci test,91 on days 23–30, rats were infected orally, daily for one week, using 200 μL bacterial suspensions that comprised UA159 or Smu_vicK, respectively. Bacterial suspensions were collected when strains grew to mid-exponential phase at the same OD600nm 0.4. All rats received drinking water containing 2% sucrose as well as a normal diet, and animals were sacrificed on day 50. Then, their lower jaws were excised for plaque observation using SEM and caries level determination using modified Keyes score.92
Gene expression assay
S. mutans cells, which included UA159, Smu_vicK and Smu_vicKr, were collected at mid-exponential phase in BHIS by centrifugation at 4 000 r·min−1 for 15 min (Thermo Fisher SCIENTIFIC, Germany). Gene expression was analyzed by qRT-PCR. Total RNAs of S. mutans cells were isolated and purified using MasterPureTM Complete DNA and RNA Purification kit (Lucigen Corporation, Wisconsin, USA) according to the instructions. Concentration and purity of total RNAs were assessed by NanoDrop OneC Microvolume UV–-Vis Spectrophotometer (Thermo Scientific, Waltham, MA, USA). After that, RNAs were reverse transcribed into cDNA using PrimeScriptTM RT reagent kit with gDNA eraser (TAKARA BIO INC. Kusatsu, Shiga, Japan). Optimal sample or primer concentrations were followed by the instruction of TB GreenTM Premix Ex TaqTM II kit (TAKARA BIO INC. Kusatsu, Shiga, Japan). In brief, each 20 μL reaction mixture included 10 μL of TB GreenTM Premix Ex TaqTM II, 0.8 μL of 10 μmol·L−1 PCR Forward Primer, 0.8 μL of 10 μmol·L−1 PCR Reverse Primer, 2.0 μL of template cDNA and 6.4 μL of deionized water. And qRT-PCR was conducted in a LightCycler® 480 System (Roche, Basel, Switzerland) according to the instruction above.93 The expressional levels of gyrA, vicR/X, gcrR, gtfB/C/D, ftf, gbpB and dexA genes were evaluated. All primers for qRT-PCR were designed based on the sequence of the S. mutans UA159 genome according to previous studies and obtained commercially (Sangon Biotech, Shanghai, China).23,88 They were listed in Table 2. Gene expressions were calculated by the 2−ΔΔCT method in which P < 0.05 were thought to be significantly different.94
Table 2.
List of the primers used in this study
| Primer | Nucleotide sequence | Annealing temperature/°C | Size/bp |
|---|---|---|---|
| PCR-ligation mutagenesis | |||
| erm-PF | 5′ GGCGCGCCCCGGGCCCAAAATTTGTTTGAT 3′ | 52.3 | 876 |
| erm-PR | 5′ GGCCGGCCAGTCGGCAGCGACTCATAGAAT 3′ | ||
| vicK-P1 | 5′ TGGTAAAGCAGTATCTGGCGAGG 3′ | 53.1 | 886 |
| vicK-P2 | 5′ GGCGCGCCATAGTGAGGAAGGCGAAGGGTC 3′ | ||
| vicK-P3 | 5′ GGCCGGCCCCAGGGACTTGATTCAAACACATTAG 3′ | 54.1 | 666 |
| vicK-P4 | 5′ GGCTAAGGAAGGTTATGACACG 3′ | ||
| primers for qRT-PCR | |||
| gyrA-F | 5′ ATTGTTGCTCGGGCTCTTCCAG 3′ | 62 | 105 |
| gyrA-R | 5′ ATGCGGCTTGTCAGGAGTAACC 3′ | ||
| vicR-F | 5′ CGCAGTGGCTGAGGAAAATG 3′ | 53 | 157 |
| vicR-R | 5′ ACCTGTGTGTGTCGCTAAGTGATG 3′ | ||
| vicK-F | 5’ CACTTTACGCATTCGTTTTGCC 3′ | 52 | 102 |
| vicK-R | 5′ CGTTCTTCTTTTTCCTGTTCGGTC 3′ | ||
| vicX-F | 5′ TGCTCAACCACAGTTTTACCG 3′ | 51.4 | 127 |
| vicX-R | 5′ GGACTCAATCAGATAACCATCAGC 3′ | ||
| gcrR-F | 5′ ACCAGAGATGGACGGGTATG 3′ | 55 | 120 |
| gcrR-R | 5′ CACGATAGGTAGTGTCATTTTTAG 3′ | ||
| gtfB-F | 5′ ACACTTTCGGGTGGCTTG 3′ | 49 | 127 |
| gtfB-R | 5’ GCTTAGATGTCACTTCGGTTG 3′ | ||
| gtfC-F | 5′ CCAAAATGGTATTATGGCTGTCG 3′ | 50.5 | 136 |
| gtfC-R | 5′ TGAGTCTCTATCAAAGTAACGCAG 3′ | ||
| gtfD-F | 5′ AATGAAATTCGCAGCGGACTTGAG 3′ | 55 | 245 |
| gtfD-R | 5′ TTAGCCTGACGCATGTCTTCATTGTA 3′ | ||
| ftf-F | 5′ ATTGGCGAACGGCGACTTACTC 3′ | 52.8 | 103 |
| ftf-R | 5’ CCTGCGACTTCATTACGATTGGTC 3′ | ||
| gbpB-F | 5′ AGCAACAGAAGCACAACCATCAG 3′ | 55 | 150 |
| gbpB-R | 5′ CCACCATTACCCCAGTAGTTTCC 3′ | ||
| dexA-F | 5′ AGGGCTGACTGCTTCTGGAGT 3′ | 55 | 142 |
| dexA-R | 5′ AGTGCCAAGACTGACGCTTTG 3′ | ||
Statistical analysis
Statistical analysis was executed by SPSS 16.0 (SPSS Inc, Chicago, IL, USA). Unpaired t test and one-way ANOVA were utilized to detect the statistical significance. Generally, the differences between the means of study data were statistically significant if P < 0.05.
Supplementary information
Acknowledgements
This study was conducted at State Key Laboratory of Oral Diseases, West China School of Stomatology, Sichuan University and IVC Experimental Animal Center of Public Health, Sichuan University. This work was supported by grants from the National Natural Science Foundation of China (grant Nos. 81771068, 81670980, and 81800964), and Sichuan International Science and Technology Innovation Cooperation (Grant No. 2020YFH0010).
Author contributions
Tao Hu and Lei Lei designed the experiments, analyzed the data, and wrote this paper; Yalan Deng and Yingming Yang performed the experiments, analyzed the data, and wrote this paper; Bin Zhang, Hong Chen, Yangyu Lu and Shirui Ren contributed to data acquisition and analysis. All authors gave final approval and agreed to be accountable for all aspects of the work.
Competing interests
The authors declare no competing interests.
Statement of ethics
Animal study in this paper conformed to the ethical guidelines of the IVC Experimental Animal Center of Public Health, Sichuan University (Chengdu, China), and was approved by the Ethics Committee of West China Hospital of Stomatology, Sichuan University (NO: WCHSIRB-D-2019-144).
Footnotes
These authors contributed equally: Yalan Deng, Yingming Yang
Contributor Information
Lei Lei, Email: leilei@scu.edu.cn.
Tao Hu, Email: hutao@scu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41368-021-00149-x.
References
- 1.Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26:229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin W, et al. Oral health status in Sichuan Province: findings from the oral health survey of Sichuan, 2015–2016. Int. J. Oral. Sci. 2017;9:10–15. doi: 10.1038/ijos.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peres MA, et al. Oral diseases: a global public health challenge. Lancet. 2019;394:249–260. doi: 10.1016/S0140-6736(19)31146-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, et al. Evaluating Streptococcus mutans strain dependent characteristics in a polymicrobial biofilm community. Front. Microbiol. 2018;9:1498. doi: 10.3389/fmicb.2018.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao J, Fiscella KA, Gill SR. Oral microbiome: possible harbinger for children’s health. Int. J. Oral. Sci. 2020;12:12. doi: 10.1038/s41368-020-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 2015;39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 8.Cugini C, Shanmugam M, Landge N, Ramasubbu N. The role of exopolysaccharides in oral biofilms. J. Dent. Res. 2019;98:739–745. doi: 10.1177/0022034519845001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Ren Z, Hwang G, Koo H. Therapeutic strategies targeting cariogenic biofilm microenvironment. Adv. Dent. Res. 2018;29:86–92. doi: 10.1177/0022034517736497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao Y, Tay FR, Niu LN, Chen JH. Advancing antimicrobial strategies for managing oral biofilm infections. Int J. Oral. Sci. 2019;11:28. doi: 10.1038/s41368-019-0062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia SS, et al. Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. J. Dent. Res. 2017;96:807–814. doi: 10.1177/0022034517698096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naha PC, et al. Dextran-coated iron oxide nanoparticles as biomimetic catalysts for localized and pH-activated biofilm disruption. ACS Nano. 2019;13:4960–4971. doi: 10.1021/acsnano.8b08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon-Soro A, Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23:76–82. doi: 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Mao MY, et al. The rnc gene promotes exopolysaccharide synthesis and represses the vicRKX gene expressions via microRNA-size small RNAs in Streptococcus mutans. Front. Microbiol. 2016;7:687. doi: 10.3389/fmicb.2016.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner C, et al. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 2002;70:6121–6128. doi: 10.1128/IAI.70.11.6121-6128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei L, et al. Modulation of biofilm exopolysaccharides by the Streptococcus mutans vicX gene. Front. Microbiol. 2015;6:1432. doi: 10.3389/fmicb.2015.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei, L. et al. Activity of Streptococcus mutans VicR is modulated by antisense RNA. J. Dent. Res. 10.1177/0022034518781765 (2018). [DOI] [PMC free article] [PubMed]
- 18.Lei L, et al. Carbohydrate metabolism regulated by antisense vicR RNA in cariogenicity. J. Dent. Res. 2020;99:204–213. doi: 10.1177/0022034519890570. [DOI] [PubMed] [Google Scholar]
- 19.Zschiedrich CP, Keidel V, Szurmant H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016;428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng DM, Liu MJ, ten Cate JM, Crielaard W. The VicRK system of Streptococcus mutans responds to oxidative stress. J. Dent. Res. 2007;86:606–610. doi: 10.1177/154405910708600705. [DOI] [PubMed] [Google Scholar]
- 21.Senadheera DB, et al. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J. Bacteriol. 2012;194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayala E, et al. A biochemical characterization of the DNA binding activity of the response regulator VicR from Streptococcus mutans. PLoS ONE. 2014;9:e108027. doi: 10.1371/journal.pone.0108027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senadheera MD, et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 2005;187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alves LA, et al. The two-component system VicRK regulates functions associated with Streptococcus mutans resistance to complement immunity. Mol. Oral. Microbiol. 2017;32:419–431. doi: 10.1111/omi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang PL, et al. Relationship between the genetic polymorphisms of vicR and vicK Streptococcus mutans genes and early childhood caries in two-year-old children. BMC Oral Health. 2018;18:39. doi: 10.1186/s12903-018-0501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, et al. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 2013;11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horev B, et al. pH-activated nanoparticles for controlled topical delivery of farnesol to disrupt oral biofilm virulence. ACS Nano. 2015;9:2390–2404. doi: 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flemming HC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 29.Davies D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 30.Duque C, et al. Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans. Infect. Immun. 2011;79:786–796. doi: 10.1128/IAI.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schilcher K, Horswill AR. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 2020;84:e00026–19. doi: 10.1128/MMBR.00026-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senadheera D, et al. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 2009;191:6415–6424. doi: 10.1128/JB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karygianni L, Ren Z, Koo H, Thurnheer T. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Marsh PD, Zaura E. Dental biofilm: ecological interactions in health and disease. J. Clin. Periodontol. 2017;44:S12–s22. doi: 10.1111/jcpe.12679. [DOI] [PubMed] [Google Scholar]
- 36.Hwang G, et al. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017;13:e1006407. doi: 10.1371/journal.ppat.1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang G, et al. Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure. Sci. Rep. 2016;6:32841. doi: 10.1038/srep32841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y, et al. Size and charge adaptive clustered nanoparticles targeting the biofilm microenvironment for chronic lung infection management. ACS Nano. 2020;14:5686–5699. doi: 10.1021/acsnano.0c00269. [DOI] [PubMed] [Google Scholar]
- 39.Sims KR, Jr, et al. Dual antibacterial drug-loaded nanoparticles synergistically improve treatment of Streptococcus mutans biofilms. Acta Biomater. 2020;115:418–431. doi: 10.1016/j.actbio.2020.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, et al. Oral biofilm elimination by combining iron-based nanozymes and hydrogen peroxide-producing bacteria. Biomater. Sci. 2020;8:2447–2458. doi: 10.1039/c9bm01889a. [DOI] [PubMed] [Google Scholar]
- 41.Fulaz S, Vitale S, Quinn L, Casey E. Nanoparticle-biofilm interactions: the role of the EPS matrix. Trends Microbiol. 2019;27:915–926. doi: 10.1016/j.tim.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Qi J, Kim SM. Effects of the molecular weight and protein and sulfate content of Chlorella ellipsoidea polysaccharides on their immunomodulatory activity. Int. J. Biol. Macromol. 2018;107:70–77. doi: 10.1016/j.ijbiomac.2017.08.144. [DOI] [PubMed] [Google Scholar]
- 43.Hou N, et al. Polysaccharides and their depolymerized fragments from Costaria costata: molecular weight and sulfation-dependent anticoagulant and FGF/FGFR signal activating activities. Int. J. Biol. Macromol. 2017;105:1511–1518. doi: 10.1016/j.ijbiomac.2017.06.042. [DOI] [PubMed] [Google Scholar]
- 44.Sun L, Wang C, Shi Q, Ma C. Preparation of different molecular weight polysaccharides from Porphyridium cruentum and their antioxidant activities. Int. J. Biol. Macromol. 2009;45:42–47. doi: 10.1016/j.ijbiomac.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Liu W, et al. Characterization and antioxidant activity of two low-molecular-weight polysaccharides purified from the fruiting bodies of Ganoderma lucidum. Int. J. Biol. Macromol. 2010;46:451–457. doi: 10.1016/j.ijbiomac.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Xu X, et al. Gastric protective activities of sea cucumber fucoidans with different molecular weight and chain conformations: a structure-activity relationship investigation. J. Agric. Food Chem. 2018;66:8615–8622. doi: 10.1021/acs.jafc.8b01497. [DOI] [PubMed] [Google Scholar]
- 47.Zeng L, Burne RA. Comprehensive mutational analysis of sucrose-metabolizing pathways in Streptococcus mutans reveals novel roles for the sucrose phosphotransferase system permease. J. Bacteriol. 2013;195:833–843. doi: 10.1128/JB.02042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castillo Pedraza, M. C. et al. Modulation of lipoteichoic acids and exopolysaccharides prevents streptococcus mutans biofilm accumulation. Molecules.25, 2232 (2020). [DOI] [PMC free article] [PubMed]
- 49.Kan L, Chai Y, Li X, Zhao M. Structural analysis and potential anti-tumor activity of Sporisorium reilianum (Fries) polysaccharide. Int. J. Biol. Macromol. 2020;153:986–994. doi: 10.1016/j.ijbiomac.2019.10.228. [DOI] [PubMed] [Google Scholar]
- 50.Xu Z, et al. Characteristics and bioactivities of different molecular weight polysaccharides from camellia seed cake. Int. J. Biol. Macromol. 2016;91:1025–1032. doi: 10.1016/j.ijbiomac.2016.06.067. [DOI] [PubMed] [Google Scholar]
- 51.Chen QL, et al. Laser microdissection hyphenated with high performance gel permeation chromatography-charged aerosol detector and ultra performance liquid chromatography-triple quadrupole mass spectrometry for histochemical analysis of polysaccharides in herbal medicine: Ginseng, a case study. Int. J. Biol. Macromol. 2018;107:332–342. doi: 10.1016/j.ijbiomac.2017.08.162. [DOI] [PubMed] [Google Scholar]
- 52.Duan GL, Yu XB. Isolation, purification, characterization, and antioxidant activity of low-molecular-weight polysaccharides from Sparassis latifolia. Int. J. Biol. Macromol. 2019;137:1112–1120. doi: 10.1016/j.ijbiomac.2019.06.177. [DOI] [PubMed] [Google Scholar]
- 53.Paula AJ, Hwang G, Koo H. Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization. Nat. Commun. 2020;11:1354. doi: 10.1038/s41467-020-15165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemos, J. A. et al. The biology of Streptococcus mutans. Microbiol. Spectr. 7, 10.1128/microbiolspec.GPP3-0051-2018 (2019). [DOI] [PMC free article] [PubMed]
- 55.Florez Salamanca EJ, Klein MI. Extracellular matrix influence in Streptococcus mutans gene expression in a cariogenic biofilm. Mol. Oral. Microbiol. 2018;33:181–193. doi: 10.1111/omi.12212. [DOI] [PubMed] [Google Scholar]
- 56.Gushchin, I. et al. Mechanism of transmembrane signaling by sensor histidine kinases. Science.356, eaah6345 (2017). [DOI] [PubMed]
- 57.Senadheera MD, et al. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J. Bacteriol. 2007;189:1451–1458. doi: 10.1128/JB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biswas I, Mohapatra SS. CovR alleviates transcriptional silencing by a nucleoid-associated histone-like protein in Streptococcus mutans. J. Bacteriol. 2012;194:2050–2061. doi: 10.1128/JB.06812-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alves LA, et al. CovR and VicRKX regulate transcription of the collagen binding protein Cnm of Streptococcus mutans. J. Bacteriol. 2018;200:e00141–18. doi: 10.1128/JB.00141-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negrini TC, et al. Influence of VicRK and CovR on the interactions of Streptococcus mutans with phagocytes. Oral Dis. 2012;18:485–493. doi: 10.1111/j.1601-0825.2011.01896.x. [DOI] [PubMed] [Google Scholar]
- 61.Biswas S, Biswas I. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans. J. Bacteriol. 2006;188:988–998. doi: 10.1128/JB.188.3.988-998.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Idone V, et al. Effect of an orphan response regulator on Streptococcus mutans sucrose-dependent adherence and cariogenesis. Infect. Immun. 2003;71:4351–4360. doi: 10.1128/IAI.71.8.4351-4360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SF, Delaney GD, Elkhateeb M. A two-component covRS regulatory system regulates expression of fructosyltransferase and a novel extracellular carbohydrate in Streptococcus mutans. Infect. Immun. 2004;72:3968–3973. doi: 10.1128/IAI.72.7.3968-3973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chong P, Drake L, Biswas I. Modulation of covR expression in Streptococcus mutans UA159. J. Bacteriol. 2008;190:4478–4488. doi: 10.1128/JB.01961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Downey JS, et al. In vitro manganese-dependent cross-talk between Streptococcus mutans VicK and GcrR: implications for overlapping stress response pathways. PLoS ONE. 2014;9:e115975. doi: 10.1371/journal.pone.0115975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S, et al. Dissecting the role of VicK phosphatase in aggregation and biofilm formation of Streptococcus mutans. J. Dent. Res. 2021;100:631–638. doi: 10.1177/0022034520979798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burne RA, et al. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J. Dent. Res. 1996;75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- 68.Alves LA, et al. CovR regulates Streptococcus mutans susceptibility to complement immunity and survival in blood. Infect. Immun. 2016;84:3206–3219. doi: 10.1128/IAI.00406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koo H, Falsetta ML, Klein MI. The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J. Dent. Res. 2013;92:1065–1073. doi: 10.1177/0022034513504218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stipp RN, et al. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS ONE. 2013;8:e58271. doi: 10.1371/journal.pone.0058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao W, et al. Effect of sucrose concentration on sucrose-dependent adhesion and glucosyltransferase expression of S. mutans in children with severe early-childhood caries (S-ECC) Nutrients. 2014;6:3572–3586. doi: 10.3390/nu6093572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Decker EM, Klein C, Schwindt D, von Ohle C. Metabolic activity of Streptococcus mutans biofilms and gene expression during exposure to xylitol and sucrose. Int J. Oral. Sci. 2014;6:195–204. doi: 10.1038/ijos.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanada N, Kuramitsu HK. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect. Immun. 1989;57:2079–2085. doi: 10.1128/iai.57.7.2079-2085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klein MI, et al. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front. Cell Infect. Microbiol. 2015;5:10. doi: 10.3389/fcimb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu F, Zhang H, Wu H. Glycosyltransferase-mediated sweet modification in oral streptococci. J. Dent. Res. 2015;94:659–665. doi: 10.1177/0022034515574865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ooshima T, et al. Contributions of three glycosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J. Dent. Res. 2001;80:1672–1677. doi: 10.1177/00220345010800071401. [DOI] [PubMed] [Google Scholar]
- 77.Stipp RN, et al. Transcriptional analysis of gtfB, gtfC, and gbpB and their putative response regulators in several isolates of Streptococcus mutans. Oral. Microbiol. Immunol. 2008;23:466–473. doi: 10.1111/j.1399-302X.2008.00451.x. [DOI] [PubMed] [Google Scholar]
- 78.Abranches, J. et al. Biology of oral streptococci. Microbiol. Spectr. 6, 10.1128/microbiolspec.GPP3-0042-2018 (2018). [DOI] [PMC free article] [PubMed]
- 79.McNeill K, Hamilton IR. Acid tolerance response of biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 2003;221:25–30. doi: 10.1016/S0378-1097(03)00164-2. [DOI] [PubMed] [Google Scholar]
- 80.Lau PC, et al. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 81.Weidmann S, et al. Production of the small heat shock protein Lo18 from Oenococcus oeni in Lactococcus lactis improves its stress tolerance. Int. J. Food Microbiol. 2017;247:18–23. doi: 10.1016/j.ijfoodmicro.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 82.Fong SA, et al. Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front. Cell Infect. Microbiol. 2017;7:418. doi: 10.3389/fcimb.2017.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marka S, Anand S. Feed substrates influence biofilm formation on reverse osmosis membranes and their cleaning efficiency. J. Dairy Sci. 2018;101:84–95. doi: 10.3168/jds.2017-13249. [DOI] [PubMed] [Google Scholar]
- 84.Kannan A, et al. Nanoscale investigation on Pseudomonas aeruginosa biofilm formed on porous silicon using atomic force microscopy. Scanning. 2014;36:551–553. doi: 10.1002/sca.21148. [DOI] [PubMed] [Google Scholar]
- 85.Neu TR, Lawrence JR. Innovative techniques, sensors, and approaches for imaging biofilms at different scales. Trends Microbiol. 2015;23:233–242. doi: 10.1016/j.tim.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 86.da Silva BR, et al. Antibacterial activity of a novel antimicrobial peptide [W7]KR12-KAEK derived from KR-12 against Streptococcus mutans planktonic cells and biofilms. Biofouling. 2017;33:835–846. doi: 10.1080/08927014.2017.1374378. [DOI] [PubMed] [Google Scholar]
- 87.Zhang K, et al. Effect of antibacterial dental adhesive on multispecies biofilms formation. J. Dent. Res. 2015;94:622–629. doi: 10.1177/0022034515571416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mao MY, et al. The regulator gene rnc is closely involved in biofilm formation in Streptococcus mutans. Caries Res. 2018;52:347–358. doi: 10.1159/000486431. [DOI] [PubMed] [Google Scholar]
- 89.Blanco Y, et al. Environmental parameters, and not phylogeny, determine the composition of extracellular polymeric substances in microbial mats from extreme environments. Sci. Total Environ. 2019;650:384–393. doi: 10.1016/j.scitotenv.2018.08.440. [DOI] [PubMed] [Google Scholar]
- 90.Bowen WH. Rodent model in caries research. Odontology. 2013;101:9–14. doi: 10.1007/s10266-012-0091-0. [DOI] [PubMed] [Google Scholar]
- 91.Yucesoy, D. T. et al. Early caries in an in vivo model: structural and nanomechanical characterization. J. Dent. Res. 10.1177/0022034518789898 (2018). [DOI] [PubMed]
- 92.Keyes PH. Dental caries in the molar teeth of rats. I. Distribution of lesions induced by high-carbohydrate low-fat diets. J. Dent. Res. 1958;37:1077–1087. doi: 10.1177/00220345580370060801. [DOI] [PubMed] [Google Scholar]
- 93.Monette P, et al. Autoregulation of the S. mutans SloR metalloregulator is constitutive and driven by an independent promoter. J. Bacteriol. 2018;200:e00214–18. doi: 10.1128/JB.00214-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.