Abstract
Within the last several years, the protozoan parasites Cyclospora cayetanensis, Cryptosporidium parvum, and microsporidia have become recognized as important, rapidly emerging human pathogens in immunocompromised and immunocompetent individuals. Since the early 1990s, many of the reported outbreaks of enteric illness caused by these microorganisms have been attributed to food- and water-borne contamination. Many inherent obstacles affect the success of current surveillance and detection methods used to monitor and control levels of contamination by these pathogens. Unlike methods that incorporate preenrichment for easier and unambiguous identification of bacterial pathogens, similar methods for the detection of parasitic protozoa either are not currently available or cannot be performed in a timely manner. We have developed an extraction-free, filter-based protocol to prepare DNA templates for use in PCR to identify C. cayetanensis and C. parvum oocysts and microsporidia spores. This method requires only minimal preparation to partially purify and concentrate isolates prior to filter application. DNA template preparation is rapid, efficient, and reproducible. As few as 3 to 10 parasites could be detected by PCR from direct application to the filters. In studies, as few 10 to 50 Encephalitozoon intestinalis spores could be detected when seeded in a 100-μl stool sample and 10 to 30 C. cayetanensis oocysts could be detected per 100 g of fresh raspberries. This protocol can easily be adapted to detect parasites from a wide variety of food, clinical, and environmental samples and can be used in multiplex PCR applications.
During the past decade, the parasitic protozoa Cyclospora cayetanensis and Cryptosporidium parvum and several species of microsporidia have emerged as important human pathogens (9, 17, 18). These parasites cause enteric diseases that range from acute, self-limiting diarrhea to chronic illness depending on the physical state of the infected individual. All three organisms have been identified as causative agents of AIDS-related chronic diarrhea (9, 18, 40). In several studies that have examined AIDS patients suffering from chronic diarrhea, as many as 50% were diagnosed with microsporidia. Infection with C. parvum was less common; however, it was still detected in 10 to 20% of individuals studied (18). As the number of reported cases among otherwise healthy individuals has increased within the last several years, so too has the public awareness of human susceptibility to infection by these parasites. C. cayetanensis has been found to be seasonally endemic in many developing countries (30, 31, 40) and identified as a cause of diarrhea in international travelers (21, 40); C. parvum has been linked to large community outbreaks (19, 27).
Although actual routes of transmission are unknown, the fecal-oral route appears to be the most likely. Contaminated foods and water sources resulting from deficiencies in environmental sanitation and hygienic practices are thought to be major causes in the spread of infections within groups or communities. In 1996, several major outbreaks of cyclosporiasis in North America were epidemiologically linked to the consumption of imported raspberries harvested during the spring growing season (20). Outbreaks of cyclosporiasis associated with the consumption of raspberries and other fresh produce such as basil and mesclun lettuce continued in 1997 (6, 7, 31), 1998 (8), and as recently as the summer of 1999. Unpasteurized apple cider has been cited as a source for C. parvum infections (5, 25, 29), and contaminated water sources have been suspected in illnesses involving C. parvum (19, 27), C. cayetanensis (4, 32, 37), and several species of microsporidia (15).
Increases in the number of infected cases and the growing list of potential sources of contamination warrant a greater emphasis on the development of more rapid, specific, and highly sensitive detection methods for the purposes of clinical diagnoses and environmental surveys. Classical methods using histochemical staining and microscopy are still largely used; however, proper diagnosis presents a challenge even to the most highly trained laboratory technician. Even with electron microscopy, genus and species identification may not always be conclusive or cannot be performed in a timely manner. This has become an important consideration, particularly in microsporidial infections, for which effective chemotherapy is dependent on rapid and specific diagnosis. Molecular techniques such as PCR offer many advantages over classical methods (16). The use of PCR in the detection and identification of these pathogens has been hampered by several factors that are directly related to difficulties in detecting small numbers of organisms in a complex matrix (12, 22, 36). Inefficient methods of isolating small numbers of the organism drastically reduce detection sensitivity. In many instances, the inclusion of an enrichment protocol to improve sensitivity by increasing pathogen numbers only serves to lengthen the detection time. Moreover, methods for the cultivation of many parasitic organisms in vitro are not currently available. The lack of uniformity in DNA template preparation among sample replicates is an additional concern. Protozoan parasites in particular present a challenge in achieving consistently clean and reliable DNA template preparations; this is directly related to the nature of the organism, its resistance to disruption and lysis, and the matrix in which it is presented. Matrix-derived factors that are carried through the isolation and purification procedure can significantly inhibit PCR amplification (12, 22, 36). These limit sensitivity and yield false-negative results.
In the present study, we developed a protocol that uses filter-based PCR technology to avoid the problems associated with pathogen isolation and concentration and DNA template preparation. We examined the practicality of using FTA filters, a matrix originally designed as a blood storage and processing medium, to prepare DNA templates from pure samples of C. cayetanensis and C. parvum oocysts and spores of the microsporidia species Encephalitozoon intestinalis from clinical and food samples. The filter, impregnated with denaturants, chelating agents, and free-radical traps (3), causes most cell types to lyse on contact (1) and sequesters DNA within the matrix. Cell remnants, sample debris, and other factors that may interfere with PCR are effectively removed by briefly washing the filters.
Whereas FTA filters have been used as an effective tool in PCR ribotyping methodologies for crude bacterial cultures (35), our study now extends the utility of FTA filters to include the sensitive detection of parasitic protozoa such as C. cayetanensis, C. parvum, and microsporidia. In addition, we demonstrate that this method can be effectively applied to detecting these and other pathogenic organisms in such diverse and complex matrixes as food, environmental samples, and clinical specimens.
MATERIALS AND METHODS
Parasites.
C. cayetanensis oocysts were obtained from M. Arrowood (Centers for Disease Control [CDC], Atlanta, Ga.). Oocysts were stored in 2.0% sodium dichromate at 4°C. C. parvum oocysts were provided by R. Fayer (U.S. Department of Agriculture, Beltsville, Md.). E. intestinalis spores were isolated from the culture medium of E6-infected mammalian cells maintained in culture as described before (39). The original E. intestinalis culture was kindly donated by G. Visvesvara (CDC). A composite fecal sample obtained from Nepalese expatriates diagnosed with cyclosporiasis and stored in 2.0% potassium dichromate was provided by John Cross (Uniformed Services University of the Health Sciences, Bethesda, Md.). All parasite counts were determined with a Petroff-Hausser counting chamber (Hausser Scientific).
Parasite spiking and washing procedure for raspberries.
The indicated number of C. cayetanensis oocysts were applied in a 10-μl volume to fresh individual raspberries and air dried at room temperature overnight. The “spiked” raspberries were then added to a 100-g berry sample and washed by the procedure detailed by Ortega et al. (31). Briefly, the berries were suspended with 100 ml of distilled water (dH2O) in a plastic bag and gently agitated for 30 min at room temperature. The wash liquid was decanted from the berries and centrifuged at 1,870 × g for 20 min at 4°C. The resulting sediment was then suspended in 5 to 10 ml of dH2O and applied to a Poly-Prep chromatography column (Bio-Rad, Hercules, Calif.) containing tightly packed glass wool presoaked with dH2O. The column was washed once with 5 ml of dH2O, and the eluent was centrifuged at 20,000 × g for 15 min at 4°C. The resulting pellet was then thoroughly suspended in 50 to 100 μl of dH2O, and 10 to 25 μl was applied to FTA filters (Fitzco, Inc., Maple Plain, Minn.).
Purification and concentration of fecal specimens.
Fecal specimens (100 to 200 μl of packed debris) were washed twice with 1 ml of dH2O and pelleted by centrifugation. The washed debris was then suspended in 1 ml of dH2O and extracted with 0.25 ml of ethyl acetate (2), vortexed for 20 to 30 s, and centrifuged. The upper, organic phase, any debris at the interface, and the lower aqueous phase were removed. The sedimented debris was then washed twice more with 1.0 ml of dH2O. The pellet was suspended in 1 ml of dH2O and passed through a glass wool column as described above.
Template preparation on FTA filter paper.
Samples were applied to FTA filters as described above, and the filters were dried on a 56°C heating block. Using an individual hole punch, 6-mm disks were punched out and placed in a 1.5-ml microcentrifuge tube. FTA disks were washed twice with 0.5 ml of FTA purification buffer (Life Technologies, Gaithersburg, Md.) for 2 min, twice with 0.5 ml of 10 mM Tris (pH 8.0) containing 0.1 mM EDTA for 2 min and again air-dried on a 56°C heating block. These washed filters were then used directly as the source of template in PCR.
PCR primers and reaction conditions.
All primers used in this study detected previously defined regions of the 18S ribosomal DNA gene in C. cayetanensis, C. parvum, and Microsporidium spp. For detection of C. cayetanensis, a slight modification to the PCR protocol as described by Relman et al. (33) was used. Primer pairs F1E-R2B and F3E-R4B were synthesized (Gibco-BRL) without the restriction site “leader” sequence. The first-round reaction was performed with the prepared FTA disk as the template in a 200-μl volume containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 2 mM MgCl2, 200 μM each dATP, dCTP, dGTP, and dTTP, and 0.2 μM each primers F1E (5′-TACCCAATGAAAACAGTTT-3′) and R2B (5′-CAGGAGAAGCCAAGGTAGG-3′) and overlaid with several drops of mineral oil. The first-round reaction mixture also contained 4 μl of a 10% powdered nonfat milk solution (13). The thermal cycling program was preceded by a host start-denaturation program of 5 min at 95°C followed by cooling to 80°C, at which time 20 μl of a Taq DNA polymerase (Promega, Madison, Wis.) stock solution (0.25 U/μl) was added. All reactions were performed in a Perkin-Elmer/Cetus DNA thermal cycler. The cycling program consisted of 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 90 s. A final extension at 72°C for 10 min followed by soaking at 4°C concluded the program. A 636-bp product will be observed when amplified with this primer pair and the C. cayetanensis DNA template (41).
The second round was conducted in a reaction volume of 100 μl, routinely using 1 to 5 μl of the first-round product as the template. Reaction component concentrations were the same as in the first-round reaction with the following exceptions: no milk solution was included in the reaction mixture; the primers used were F3E (5′-CCTTCCGCGCTTCGCTGCGT-3′) and R4B (5′-CGTCTTCAAACCCCCTACTG-3′); 10 μl of a Taq polymerase stock solution (0.25 U/μl) was added; and the annealing temperature was 60°C. This primer pair will generate a 294-bp product in the presence of the C. cayetanensis template (41).
PCR amplification for the detection of C. parvum and E. intestinalis using an FTA-based template was performed in 200-μl reaction volumes identical to those described above for C. cayetanensis. The respective primer pairs were as follows: for C. parvum, CPB-DIAGF (5′-AGCTCGTAGTTGGATTTCTG-3′) and CPB-DIAGR (5′-TAAGGTGCTGAAGGAGTAAGG-3′) (23); and for E. intestinalis, SINTF1 (5′-TTTCGAGTGTAAAGGAGTCGA-3′) and SINTR (5′-CCGTCCTCGTTCTCCTGCCCG-3′) (11). C. parvum-prepared filters were amplified using a total of 39 cycles with denaturation, annealing, and elongation temperatures and times of 94°C and 30 s, 55°C and 1 min, and 72°C and 1 min, respectively. FTA filters spotted with E. intestinalis spores were amplified using a total of 35 cycles with denaturation, annealing, and elongation temperatures and times of 94°C and 30 s, 55°C and 30 s, and 72°C and 90 s, respectively.
Multiplex PCR was carried out using the thermal cycling program described for C. cayetanensis. The following primer pairs and their expected products were use for microsporidia identification: MicroF (5′-CACCAGGTTGATTCTGCCTGA-3′) and MicroR (5′-TAATGATCCTGCTAATGGTTCTCCAAC-3′) produced a 1,300-bp product (39); Enterocytozoon bieneusi primers EBIEF1 (5′-GAAACTTGTCCACTCCTTACG-3′) and EBIER (5′-CAATGCACCACT CCTGCCATT-3′) produced a 607-bp product (14); Encephalitozoon cuniculi primers ECUNF1 (5′-ATGAGAAGTGATGTGTGTGCG-3′) and WCUNR1 (5′-TGCCATGCACTCAC AGGCATC-3′) produced a 549-bp product (38); and Encephalitozoon hellem primers EHELF1 (5′-TGAGAAGTAAGATGTTTAGCA-3′) and WHELR1 (5′-GTAAAAACACTCTCACACTCA-3′) produced a 547-bp product (38). PCR products were separated by agarose gel electrophoresis using 1.5% agarose containing ethidium bromide (0.2 μg/ml). Products were visualized on a UV transilluminator.
RESULTS
Preparation of DNA templates with FTA filters for the detection of parasitic protozoa by PCR.
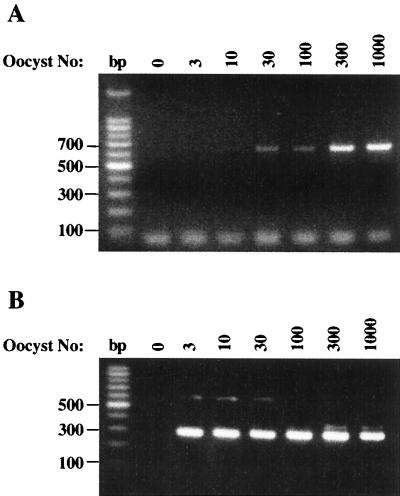
The utility of FTA filters in PCR for DNA template preparation for the detection of C. cayetanensis oocysts, C. parvum oocysts, or E. intestinalis spores was first examined by using partially purified parasite suspensions. Serial dilutions of each pathogen were made, spotted onto 6-mm FTA filters, and dried. After a brief series of washes, the filters were used directly as the source of DNA template in PCR assays. For C. cayetanensis, the two-step nested PCR method described by Relman et al. (33) was used, in which a first-round PCR will generate a product of 636 bp with the outer primer pair F1E and R2B, and the inner primer pair F3E and R4B will produce a secondary product of 294 bp. Current methods report sensitivities in the range of 10 to 50 oocysts, with a visible product only after completion of the nested reaction (22, 33, 41). With FTA filters, however, a detectible, dose-dependent DNA product of the predicted 636-bp size was obtained from filters seeded with as few as 10 to 30 oocysts after the first round of PCR (Fig. 1A). A 10-fold-greater level of sensitivity was observed from the secondary reaction, in which a strong signal at 294 bp was detected with as few as 3 oocysts (Fig. 1B).
FIG. 1.
Detection of purified Cyclospora cayetanensis oocysts by PCR using FTA filter disks as a template matrix. Serial dilutions of pure C. cayetanensis oocysts were prepared and applied to FTA filter disks in a 10-μl volume. Filters were then processed and used as template in a nested PCR analysis as described in Materials and Methods. (A) Analysis of the first-round PCR products using the primer pair F1E and R2B. The PCR product size is 636 bp. (B) Analysis of the second-round PCR products; 5 μl of first-round products was used as the template for PCR amplification with primer pair F3E and R4B in a 100-μl reaction volume as described in Materials and Methods. PCR amplification with primers F3E and R4B resulted in a 294-bp product.
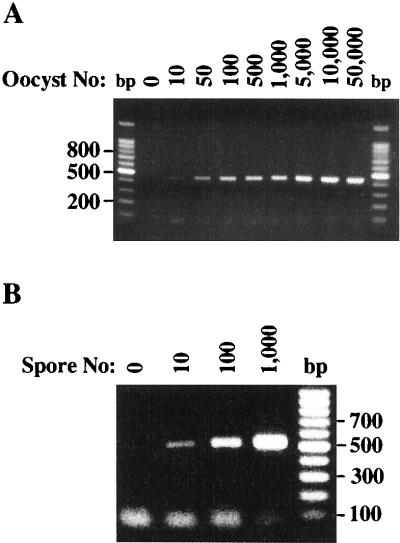
FTA filter templates were also prepared from purified C. parvum oocysts and E. intestinalis spores, and each was amplified by PCR with parasite-specific primer pairs. Similar limits of detection were observed (Fig. 2). In the presence of the C. parvum-specific primer pair CP-DIAGF and CP-DIAGR (23), the expected 435-bp product was visible from filters seeded with as few as 10 oocysts (Fig. 2A). Equally sensitive detection limits were obtained with E. intestinalis-spotted FTA filter templates amplified by PCR with primers SINTF1 and SINTR. The 520-bp product was observed in filters seeded with only 10 spores (Fig. 2B).
FIG. 2.
Utility of FTA filters as a template matrix for the PCR detection of other parasitic protozoa. Serial dilutions were prepared from isolates of C. parvum oocysts (A) and E. intestinalis spores (B) and spotted onto FTA filter disks. FTA filters were then processed and used as the template in PCRs as described in Materials and Methods. The primer pair CPB-DIAGF and CPB-DIAGR was used for the detection of C. parvum oocysts and resulted in a 435-bp product. The primer pair SINTF1 and SINTR was used for the detection of E. intestinalis spores and resulted in a 520-bp product.
Detection of parasitic protozoa in clinical and food samples by using FTA filters for DNA template preparation.
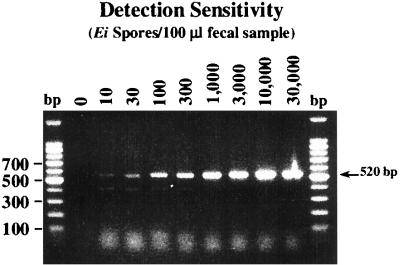
Clinical specimens (fecal, urine, and sputum), environmental samples, and foods are matrixes commonly examined for the presence of parasitic pathogens such as C. cayetanensis, C. parvum, and microsporidia. Parasite-laden fecal isolates and berries were chosen to evaluate the efficiency and sensitivity of FTA filters in the preparation of DNA templates. As was demonstrated for the detection of purified spores, the use of FTA filters allowed the detection of as few as 10 E. intestinalis spores per 100 μl of fecal material (Fig. 3). Similar results were also observed when urine and sputum isolates were tested for the presence of microsporidia (data not shown). In many instances, crude biological samples could be applied directly to individual filters without prior purification steps or any substantial loss in detection signal. Formalin fixation did not appreciably affect sensitivity.
FIG. 3.
Detection of E. intestinalis (Ei) spores in fecal samples by PCR with FTA filter disks. Aliquots (100 μl) of washed packed fecal material were spiked with the indicated number of purified spores. Samples were suspended in dH2O, passed through glass wool columns, and concentrated by centrifugation, and a portion of the resulting sediment was spotted onto FTA filters.
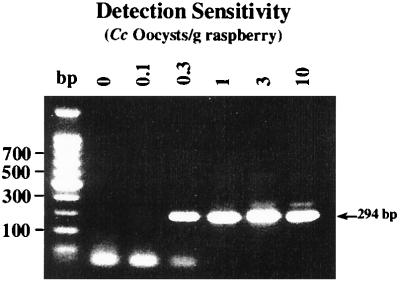
Sensitive detection of these parasitic pathogens in food matrixes is also of particular importance. Several recent outbreaks of C. cayetanensis have been linked to the contamination of produce, most notably fresh raspberries, mesclun lettuce, and basil (6, 7); cryptosporidiosis in several instances has been linked to unpasteurized apple cider (5, 25, 29). The effectiveness of FTA filters in the PCR detection of C. cayetanensis on raspberries was tested with 100-g samples of fresh raspberries that had been inoculated with decreasing levels of C. cayetanensis oocysts. As shown in Fig. 4, positive results from nested PCR analysis were seen in samples containing 0.3 to 10 oocysts per g of fruit. These results correspond to detecting as few as 30 oocysts in a 100-g sample. The observed differences in detection sensitivities between pure oocysts (Fig. 1) and those from berries were most likely attributed to the oocyst recovery and sample concentration steps prior to FTA application. In artificially contaminated apple cider, as few as 100 oocysts could easily be detected with FTA filters from direct cider sampling (data not shown), but this matrix (cider) appeared to have more noticeable effects on PCR sensitivity.
FIG. 4.
Detection of C. cayetanensis (Cc) oocysts on fresh raspberries by PCR with FTA filter disks. Individual raspberries were seeded with the indicated number of C. cayetanensis oocysts, air-dried, added to 100-g samples of fresh raspberries, and washed in water with gentle agitation for 30 min. Wash liquids were decanted from the berries and centrifuged to recover wash sediment. As detailed in Materials and Methods, wash sediments were then passed through glass wool columns, and the eluted material was concentrated by centrifugation prior to spotting onto FTA filters.
FTA filters in multiplex PCR analysis.
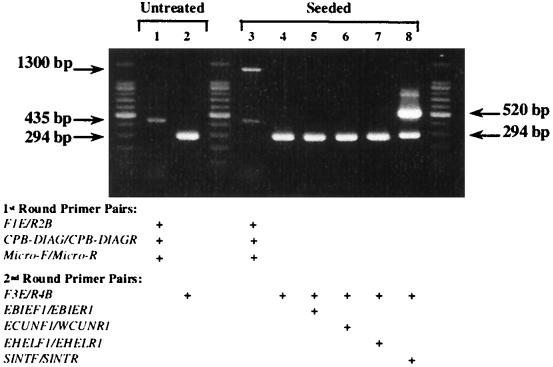
In addition to the relatively simple means of preparing DNA templates using FTA, the filter templates can also be used as the foundation for multiplex PCR analysis. We tested this capability on a composite stool sample obtained from a clinic in Nepal. Two equal samples (200 μl of a 50% suspension), one untreated and the other sample artificially contaminated with 500 E. intestinalis spores, were prepared, and a portion was spotted onto FTA filters. These filters were then used as templates for multiplex PCR amplification using three primer pairs: F1E and R2B (C. cayetanensis), CPB-DIAG and CPB-DIAGR (C. parvum), and Micro-F and Micro-R (microsporidia). A second PCR was then performed using 5 μl of each first-round product along with the nested primers for C. cayetanensis, F3E and R4B. In addition, a series of microsporidia species-specific primers were also included when first-round product from the artificially contaminated stool sample was used. When the PCR products from the first round of amplification were analyzed, a 435-bp fragment was observed, corresponding to the presence of C. parvum oocysts in the stool samples (Fig. 5, lanes 1 and 3). In addition to this product, a 1,300-bp fragment was also amplified in the “seeded” sample, confirming the presence of microsporidia spores (Fig. 5, lane 3). A subsequent series of reactions using the C. cayetanensis primer pair F3E and R4B (lanes 2 and 4 to 8) and microsporidia species-specific primers (Fig. 5, lanes 4 to 8) confirmed the presence of C. cayetanensis oocysts (Fig. 5, lanes 3 and 4 to 8) and identified the microsporidia species that had been seeded in the stool sample. The 520-bp fragment generated with the primer pair SINTF and SINTR correctly identified the inoculated microsporidia species as E. intestinalis (Fig. 5, lane 8).
FIG. 5.
Multiplex PCR analysis for the detection of C. cayetanensis, C. parvum, and microsporidia in a stool sample with FTA filters. Two 100-μl composite stool samples obtained from a Nepali clinic were examined; one untreated sample and one sample artificially contaminated with 500 E. intestinalis spores were prepared and applied to FTA filters as described in Materials and Methods. The filters were then used as the initial template in a two-step multiplex PCR. Primer pairs F1E and R2B, CP-DIAGF and CP-DIAGR, and Micro-F and Micro-R were used during the first amplification. From 1 to 5 μl of the resulting product was then used in a second reaction with the primer pair F3E and R4B and microsporidia species-specific primers was then carried out (see Materials and Methods). Thermal cycling parameters for both sets of reactions were identical to those used for amplifying C. cayetanensis DNA.
DISCUSSION
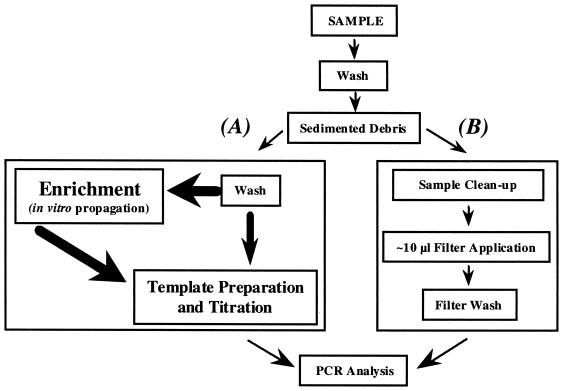
Conventional methods currently used to detect pathogenic microorganisms by PCR often require multiple steps to achieve suitable DNA template preparations (Fig. 6). In addition, selective enrichment or concentration steps may also be required to obtain detectable levels of the targeted pathogen. As a consequence, analysis times are increased considerably. Another complication also exists for the detection of many parasitic protozoa: culture methods are either not available or cannot be performed in a timely manner.
FIG. 6.
General flow diagram for the isolation, detection, and identification of pathogenic organisms. Method A is representative of current protocols in DNA template preparation for PCR analysis; method B represents a preparative method using the FTA filter format. Bold arrows and text denote the need for multiple, intervening processes.
Detection of protozoan parasites such as C. cayetanensis, C. parvum, and microsporidia by PCR is highly dependent on the method used to extract DNA (12). Current methods to prepare DNA templates can be inefficient and labor-intensive and yield nonuniform results. Sonication, freeze-thawing, and glass bead disruption are three frequently employed methods (12, 33, 41); additional purification steps are often necessary. DNA binding in the presence of chaotropic agents is another method favored by some laboratories and has proven to be effective (12). With smaller sample sizes, however, these methods can result in significant losses and yield inconsistent results. Whereas current DNA template preparation and PCR detection methods using purified parasite isolates may yield satisfactory results, detection sensitivities can be greatly affected by substances derived from the sample matrix and its processing. A high percentage of false-negative results may occur. With regard to the detection of C. cayetanensis, C. parvum, and microsporidia, these problems are most notable in the preparation of DNA templates from clinical specimens, foods, and environmental samples (12, 22, 34).
In our study, PCR analysis using the FTA filter format for DNA template preparation was routinely unaffected by the matrix from which the sample was derived while still maintaining an extremely high level of detection sensitivity. Similar sensitivities were demonstrated with both purified organisms and isolates from clinical or food samples (Table 1). With the increasing recognition that these enteric human parasites cause debilitating diarrheal disease, the development of a rapid and sensitive PCR method not susceptible to matrix-derived inhibitors was paramount. The importance of this is exemplified by the statistics that indicate that a growing percentage of AIDS patients suffering from chronic diarrhea are infected with either microsporidia or C. parvum (18). Current PCR methods are greatly affected by fecal components and lack of uniformity. Urine and sputum samples also contain many endogenous substances that will inhibit PCR and yield false-negative results. The use of FTA filters, however, appeared to limit or negate the effects of endogenous substances. In many instances, particularly with microsporidia, we found that sputum, urine, and stool suspensions could be spotted directly onto filters without any additional preparative steps prior to application. Formalin-fixed specimens could also be used directly with the FTA filter format. Our results suggested that even with minimal preparation, the detection of E. intestinalis spores from a particulate matrix was quite efficient and sensitive.
TABLE 1.
Summary of PCR detection sensitivities for parasitic protozoa using FTA filters
| Parasite | Primer pair | Product size (bp) | No. of oocysts/organisms detecteda
|
|
|---|---|---|---|---|
| Purified isolate | Matrixes | |||
| C. cayetanensis | F1E and R2B | 636 | 30 | — |
| F1E and R2B, F3E and R4B | 294 | 3 | Stool; ND | |
| Raspberries; 30b | ||||
| Pasta salad; ND | ||||
| C. parvum | CPB-DIAGF and CPB-DIAGR | 435 | 10 | Stool; ND |
| Apple cider; <100 | ||||
| E. intestinalis | SINTF1 and SINTR | 520 | 10 | Stool; 10b |
| Sputum; ND | ||||
| Urine; ND | ||||
| E. bieneusi | EBIEF1 and EBIER | 607 | — | Stool; ND |
| E. cuniculi | ECUNF1 and WCUNR1 | 549 | — | Sputum; ND |
| E. hellem | EHELF1 and WHELR1 | 547 | — | Urine; ND |
| Microsporidia | Micro-F and Micro-R | 1,300 | ND | Bile-NDc |
| Tissue culture | ||||
Parasites were detected using the FTA filter application, although detection limits were not established. ND, number not determined; —, not done.
Detection of C. cayetanensis from 100 g of fresh raspberries; C. cayetanensis, C. parvum, and microsporidia were detected in 100 μl of packed fecal material.
E. bieneusi.
The need for a rapid and sensitive method of detecting these parasites from matrixes such as food and water samples has become as important as the current need in clinical diagnoses. Illnesses arising from the contamination of fresh produce and water are well documented. Unlike clinical specimens, however, foods such as produce and fruit hamper PCR analysis for pathogens due to additional factors (13). For raspberries and strawberries in particular, many difficulties have been reported in obtaining useful preparations of nucleic acids (24, 28). Acidity, particularly from fruit extracts, and other plant-derived factors such as polyphenolics and polysaccharides isolated during DNA template preparation have all been shown to significantly inhibit PCR (24, 28) and are leading causes in the failure to detect small numbers of pathogenic organisms. These factors have significantly influenced the ability to reliably detect C. cayetanensis on raspberries. Steps to abrogate their effects have relied heavily on methods either to adsorb inhibitory substances from DNA extracts with polyvinyl polypyrrolidone (12, 24), to bind DNA to a silica matrix in the presence of chaotropic reagents (12, 26), or to dilute template preparations (22). While these steps, alone or in combination, have reduced PCR inhibition, a concomitant loss in detection signal has also been observed, and success has been marginal. Whereas the sensitivity of detecting C. cayetanensis has been suggested to be 10 to 50 pure oocysts using other current protocols (22, 33, 41), detection from matrixes such as raspberries, basil, and stool specimens has been inconsistent and relatively unreliable (22). As shown here, however, the FTA filter format allowed the detection of as few as 30 oocysts from pure parasite preparations after the primary reaction and from as few as 3 when the nested reaction was completed. The use of FTA filters in DNA template preparation from raspberries, though not as sensitive as pure isolates, was nonetheless able to detect as few as 30 oocysts per 100 g of berries after completion of the nested reaction. It is important to note, however, that while FTA is capable of detecting extremely small numbers of microorganisms, the sample preparation, washing, and recovery efficiencies of any method employed in conjunction with FTA template preparation will still influence detection limits independent of FTA's potential.
As shown in the present study, we expanded the utility of FTA-impregnated filters to include the detection of parasitic protozoa by PCR. The inherent properties of FTA-impregnated filters caused oocysts and spores to lyse on contact and sequestered DNA within the paper matrix. FTA filters eliminated time-consuming and inefficient methods usually necessary for reliable pathogen detection. No extensive purification and enrichment steps were required for those parasitic microorganisms examined in this study. The FTA filter format provided an extraction-free means of preparing DNA templates without the need for laborious and often cumbersome isolation and purification schemes. Preparation of DNA templates from FTA filters was therefore rapid, uniform, and reproducible. DNA losses were avoided, as additional purification steps were not needed. Likewise, these filters preserved DNA integrity and eliminated potential sources of target DNA losses through degradative processes normally associated with conventional methods.
The FTA format has been shown in this study to be a useful tool in the PCR detection of parasites from various sources. The use of these filters has been shown to alleviate many of the current difficulties inherent in DNA template preparation and in achieving sensitive detection of pathogens from diverse sources. Moreover, these filters are time- and cost-effective. It is reasonable to expect that protocols employing the FTA filter format can be easily adapted to detect diverse parasitic and other pathogenic microorganisms from a wide variety of clinical, food, and environmental sources. This format's potential for multiplex PCR protocols in diagnostic screening is currently being investigated.
REFERENCES
- 1.Belgrader P, Del Rio S A, Turner K A, Marino M A, Weaver K R, Williams P E. Automated DNA purification and amplification from blood-stained cards using a robotic workstation. Biotechniques. 1995;19:426–432. [PubMed] [Google Scholar]
- 2.Bukhari Z, Smith H V. Effects of three concentration techniques on viability of Cryptosporidium parvum oocysts recovered from bovine feces. J Clin Microbiol. 1995;33:2592–2595. doi: 10.1128/jcm.33.10.2592-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgoyne, L. A. March 1996. Solid medium and method for DNA storage. U.S. patent 5,496,562.
- 4.Centers for Disease Control. Outbreaks of diarrheal illness associated with cyanobacteria (blue-green algae)-like bodies—Chicago and Nepal, 1989 and 1990. Morbid Mortal Wkly Rep. 1991;40:325–327. [PubMed] [Google Scholar]
- 5.Centers for Disease Control. Outbreaks of Escherichia coli O175:H7 infection and cryptosporidiosis associated with drinking unpasteurized apple cider—Connecticut and New York, October 1996. Morbid Mortal Wkly Rep. 1997;46:4–8. [PubMed] [Google Scholar]
- 6.Centers for Disease Control. Update: outbreaks of cyclosporiasis—United States and Canada. Morbid Mortal Wkly Rep. 1997;46:521–523. [PubMed] [Google Scholar]
- 7.Centers for Disease Control. Outbreak of cyclosporiasis—Northern Virginia-Washington, D.C.-Baltimore, Maryland, metropolitan area, 1997. Morbid Mortal Wkly Rep. 1997;46:689–691. [PubMed] [Google Scholar]
- 8.Centers for Disease Control. Update: outbreak of cyclosporiasis—Ontario, Canada, May 1998. Morbid Mortal Wkly Rep. 1998;47:806–809. [PubMed] [Google Scholar]
- 9.Curry A, Smith H V. Emerging pathogens: Isospora, Cyclospora, and Microsporidia. Parasitology. 1998;117(Suppl):S143–S159. doi: 10.1017/s0031182099004904. [DOI] [PubMed] [Google Scholar]
- 10.da Silva A J, Schwartz D A, Visvesvara G S, De Moura H, Slemenda S B, Pieniazek N J. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (Microsporidia) using primers based on the region coding for small-subunit rRNA. J Clin Microbiol. 1996;34:986–987. doi: 10.1128/jcm.34.4.986-987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva A J, Slemenda S B, Visvesvara G S, Schwartz D A, Wilcox C M, Wallace S, Pieniazek N J. Detection of Septata intestinalis (microsporidia) Cali et al. 1993 using polymerase chain reaction primers targeting the small subunit ribosomal RNA coding region. Mol Diagn. 1997;2:47–52. doi: 10.1054/MODI00200047. [DOI] [PubMed] [Google Scholar]
- 12.da Silva A J, Bornay-Llinares F J, Moura I N S, Slemenda S B, Tuttle J L, Pieniazek N J. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol Diagn. 1999;4:57–64. doi: 10.1016/s1084-8592(99)80050-2. [DOI] [PubMed] [Google Scholar]
- 13.De Boer S H, Ward L J, Li X, Chittaranjan S. Attenuation of PCR inhibition in the presence of plant compounds by addition of BLOTTO. Nucleic Acids Res. 1995;23:2567–2568. doi: 10.1093/nar/23.13.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Aguila C, Lopez-Velez R, Fenoy S, Turrientes C, Cobo J, Navajas R, Visvesvara G S, Croppo G P, da Silva A J, Pieniazek N J. Identification of Enterocytozoon bieneusi spores in respiratory samples from an AIDS patient with a 2-year history of intestinal microsporidiosis. J Clin Microbiol. 1997;35:1862–1866. doi: 10.1128/jcm.35.7.1862-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowd S E, Gerba C P, Pepper I L. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl Environ Microbiol. 1998;64:3332–3335. doi: 10.1128/aem.64.9.3332-3335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedorko D P, Hijazi Y M. Application of molecular techniques to the diagnosis of microsporidial infection. Emerg Infect Dis. 1996;2:183–190. doi: 10.3201/eid0203.960304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flynn P M. Emerging diarrheal pathogens: Cryptosporidium parvum, Isospora belli, Cyclospora species, and Microsporidia. Pediatr Ann. 1996;25:480–481. doi: 10.3928/0090-4481-19960901-04. , 485–487. [DOI] [PubMed] [Google Scholar]
- 18.Goodgame R W. Understanding intestinal spore-forming protozoa: Cryptosporidia, Microsporidia, Isospora, and Cyclospora. Ann Intern Med. 1996;124:429–441. doi: 10.7326/0003-4819-124-4-199602150-00008. [DOI] [PubMed] [Google Scholar]
- 19.Hayes E B, Matte T D, O'Brien T R, McKinley T W, Logsdon G S, Rose J B, Ungar B L P, Word D M, Pinskey P F, Cummings M L, Wilson M A, Long E G, Hurwitz E S, Juranek D D. Large community outbreak of cryptosporidiosis due to contamination of a filtered public water supply. N Engl J Med. 1989;320:1372–1376. doi: 10.1056/NEJM198905253202103. [DOI] [PubMed] [Google Scholar]
- 20.Herwaldt B L, Ackers M-L the Cyclospora Working Group. An outbreak in 1996 of cyclosporiasis associated with imported raspberries. N Engl J Med. 1997;336:1548–1556. doi: 10.1056/NEJM199705293362202. [DOI] [PubMed] [Google Scholar]
- 21.Hoge C W, Shlim D R, Ghimire M, et al. Placebo controlled trial of co-trimoxazole for Cyclospora infection among travelers and foreign residents in Nepal. Lancet. 1995;345:6991–6993. doi: 10.1016/s0140-6736(95)90868-4. [DOI] [PubMed] [Google Scholar]
- 22.Jinneman K C, Wetherington J H, Hill W E, Adams A M, Johnson J M, Tenge B J, Dang N-L, Manger R L, Wekell M M. Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. and Eimeria spp. oocysts directly from raspberries. J Food Prot. 1998;61:1497–1503. doi: 10.4315/0362-028x-61.11.1497. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for the sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones C S, Iannetta P P M, Woodhead M, Davies H V, McNicol R J, Taylor M A. The isolation of RNA from raspberry (Rubus idaeus) fruit. Mol Biotech. 1997;8:219–221. doi: 10.1007/BF02760775. [DOI] [PubMed] [Google Scholar]
- 25.Laberge I, Griffths M W, Griffiths M W. Prevalence, detection, and control of Cryptosporidium parvum in food. Int J Food Microbiol. 1996;32:1–26. doi: 10.1016/0168-1605(96)00977-4. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz H, Jager C, Willems H, Baljer G. PCR detection of Coxiella burnetti from different clinical specimens, especially bovine milk, on the basis of DNA preparation with a silica matrix. Appl Environ Microbiol. 1998;64:4234–4237. doi: 10.1128/aem.64.11.4234-4237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacKenzie W R, Hoxie N J, Proctor M E, Gradus M S, Blair K A, Peterson D E, Kazmierczak J J, Adiss D S, Fox K R, Rose J B, Davis J P. A massive outbreak in Milwaukee of Cryptosporidium infection through the public water supply. N Engl J Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 28.Manning K. Isolation of nucleic acids from plants by differential solvent precipitation. Anal Biochem. 1995;195:45–50. doi: 10.1016/0003-2697(91)90292-2. [DOI] [PubMed] [Google Scholar]
- 29.Millard P S, Gensheimer K F, Addiss D G, Sosin D M, Beckett G A, Houck-Jankoski A, Hudson A. An outbreak of cryptosporidiosis from fresh-pressed apple cider. J Am Med Assoc. 1994;272:1592–1596. [PubMed] [Google Scholar]
- 30.Ortega Y R, Sterling C R, Gilman R H, Cama V A, Diaz F. Cyclospora species—a new protozoan pathogen of humans. N Engl J Med. 1993;328:1308–1312. doi: 10.1056/NEJM199305063281804. [DOI] [PubMed] [Google Scholar]
- 31.Ortega Y R, Roxas C, Gilman R, Miller N, Cabera L, Taquiri C, Sterling C. Isolation of Cryptosporidium parvum and Cyclospora cayetanensis from vegetables collected in markets of an endemic region of Peru. Am J Trop Med Hyg. 1997;57:683–686. doi: 10.4269/ajtmh.1997.57.683. [DOI] [PubMed] [Google Scholar]
- 32.Rabold J G, Hoge C W, Shlim D R, Kefford C, Rajah R, Echeverria P. Cyclospora outbreak associated with chlorinated drinking water. Lancet. 1994;344:1360–1361. doi: 10.1016/s0140-6736(94)90716-1. [DOI] [PubMed] [Google Scholar]
- 33.Relman D A, Schmidt T M, Gajadhar A, Sogin M, Cross J, Yoder K, Sethabutr O, Echeverria P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J Infect Dis. 1996;173:440–445. doi: 10.1093/infdis/173.2.440. [DOI] [PubMed] [Google Scholar]
- 34.Rinder H, Janitschke K, Aspock H, da Silva A J, Deplazes P, Fedorko D P, Franzen C, Futh U, Hunger F, Lehmacher A, Meyer C G, Molina J M, Sandfort J, Weber R, Loscher T. Blinded, externally controlled multicenter evaluation of light microscopy and PCR for the detection of microsporidia in stool specimens: the Diagnostic Multicenter Study Group on Microsporidia. J Clin Microbiol. 1998;36:1814–1818. doi: 10.1128/jcm.36.6.1814-1818.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers C, Burgoyne L. Bacterial typing: storage and processing of stabilized reference bacteria for polymerase chain reaction without preparing DNA—an example of an automatable procedure. Anal Biochem. 1997;247:223–227. doi: 10.1006/abio.1997.2031. [DOI] [PubMed] [Google Scholar]
- 36.Sluter S D, Tzipori S, Widmer G. Parameters affecting polymerase chain reaction detection of waterborne Cryptosporidium parvum oocysts. Appl Microbiol Biotechnol. 1997;48:325–330. doi: 10.1007/s002530051057. [DOI] [PubMed] [Google Scholar]
- 37.Sturbaum G D, Ortega Y R, Gilman R H, Sterling C R, Cabrera L, Klein D A. Detection of Cyclospora cayetanensis in waste water. Appl Environ Microbiol. 1998;64:2284–2286. doi: 10.1128/aem.64.6.2284-2286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visvesvara G S, Leitch G J, da Silva A J, Croppo G P, de Moura H, Wallace S, Slemenda S B, Schwartz D A, Moss D, Bryan R T, Pieniazek N J. Polyclonal and monoclonal antibody and PCR-amplified small-subunit rRNA identification of a microsporidian, Encephalitozoon hellem, isolated from an AIDS patient with disseminated infection. J Clin Microbiol. 1994;32:2760–2768. doi: 10.1128/jcm.32.11.2760-2768.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visvesvara G S, da Silva A J, Croppo G P, Pieniazek N J, Leitch G J, Ferguson D, de Moura H, Wallace S, Slemenda S B, Tyrrell I, Moore D F, Meador J. In vitro culture and serologic and molecular identification of Septata intestinalis isolated from urine of a patient with AIDS. J Clin Microbiol. 1995;33:930–936. doi: 10.1128/jcm.33.4.930-936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wurtz R. Cyclospora: a newly identified intestinal pathogen of humans. Clin Infect Dis. 1994;18:620–623. doi: 10.1093/clinids/18.4.620. [DOI] [PubMed] [Google Scholar]
- 41.Yoder K E, Sethabutr O, Relman D A. PCR-based detection of the intestinal pathogen Cyclospora. In: Persing D H, editor. PCR protocols for emerging infectious diseases, a supplement to diagnostic molecular microbiology: principles and applications. Washington, D.C.: ASM Press; 1996. pp. 169–176. [Google Scholar]