Supplemental Digital Content is available in the text
Keywords: distribution, genotype, hepatitis B virus, meta-analysis, phylogenetic trees, subgenotype, timetree
Abstract
Hepatitis B virus (HBV) genotypes and subgenotypes have distinct geographical distributions and influence a number of clinical disease features and responses to treatment. There are many reports on the distribution of HBV genotypes, but great differences are present between studies. What's more, a meta-analysis of HBV genotype- and subgenotype-distribution by country is lacking.
A comprehensive literature search was performed in PubMed and a systematic search of full-length HBV sequences and S gene sequences was conducted in the GenBank database. HBV genotypes were checked and subgenotypes were determined by phylogenetic comparison of full-length HBV sequences or S gene sequences. STATA 12.0 was used for the analysis for countries with multiple datasets. BEAST 2.5.2 was used for Bayesian phylogenetic analysis to infer the evolutionary time scales of HBV.
This study includes 309 datasets from 110 countries, including 188 relevant studies, 58 full-length gene datasets, and 63 S gene datasets. The meta-analysis was performed on 274 datasets from 75 countries. The distribution of genotypes is more detailed than those described by previous studies. While the overall genotype distribution is similar to that reported in previous studies, some notable aspects were different. The main genotypes present in south-eastern Africa, North Africa, and West Africa are genotypes A, D, and E, respectively. Genotypes G and H are mainly distributed in Mexico. Genotype F is mainly distributed in central and South America, but genotypes A and D are also common in Brazil, Cuba, and Haiti.
This study provides a more accurate description of the distribution of HBV genotypes and subgenotypes in different countries and suggests that the differences in genotype distribution may be related to ethnicity and human migration.
1. Introduction
Hepatitis B virus (HBV) infection is a major global public health problem and can cause both acute and chronic disease. Although there is currently a safe vaccine against HBV, hepatitis B prevalence is alarmingly high in some areas, such as Western Pacific Region and African Region. The World Health Organization has estimated that there were 257 million people suffer from chronic hepatitis B (CHB) infection globally, and nearly 900,000 people die from end-stage liver disease each year.[1–3]
HBV is a hepatotropic virus and has a partially double-stranded, relaxed circular DNA genome containing about 3.2 kilobases. Due to the lack of proofreading activity of its reverse transcriptase, the mutation rate of HBV is very high, with an estimated nucleotide replacement rate of 1.4 to 3.2 × 10−5 per site per year.[4–6] The high error rate of HBV reverse transcriptase during viral replication results in frequent nucleotide substitutions, leading to genotype, subgenotype, and quasispecies diversity.[7] In 1988, Okamoto et al[8] suggested that HBV could be divided into 4 genotypes based on genome nucleotide variation that is greater than 8%. Since then, at least 10 genotypes (A–J) have been identified. Genotypes A, B, C, D, and F have been further split into subgenotypes with 4% genome divergence.[9] In addition to the full-length HBV sequences, the S gene is also suitable for genotyping because it is more highly conserved than other genes in the HBV genome.[10]
In recent years, HBV genotypes and subgenotypes have attracted increasing attention since they influence the activity and outcome of chronic HBV infection. For example, genotype A is common in CHB patients, while genotype D is more prevalent in patients with acute hepatitis.[11] Genotype D is the most frequent genotype in patients with acute liver failure.[12] Compared to genotype B, genotype C is associated with late hepatitis B e antigen (HBeAg) seroconversion and advanced liver disease.[13] In our previous research, we have shown that the frequency of genotype C was higher than genotype B in CHB/HBV-related liver cirrhosis and hepatocellular carcinoma (HCC) patients, while genotype B was more frequent in asymptomatic carrier and liver failure patients.[14] Genotype H has a low levels of HBV DNA and HBeAg, and has been associated with occult HBV and less severe liver disease.[15] Subgenotypes A1 and D1 have been associated with lower levels of HBV DNA and early HBeAg seroconversion.[16,17] Subgenotype F1 has been associated with a higher risk of HCC compared with genotypes A, B, and D.[18] HBV genotypes are not only associated with clinical progression of disease but are also related to interferon treatment responses. In a word, genotype determination in HBV infection is important for disease progression estimations and the design of optimal antiviral treatment plans.
Many studies have reported that different genotypes and subgenotypes show different geographical distributions. Genotype A is mainly found in Europe (where the main subgenotype is A2) and Africa (A1). Genotypes B and C are common in Asia. Genotype D is prevalent in Africa, Europe, the Mediterranean region, and India. Genotype E is found in West and South Africa. Genotype F is dominant in South and Central America. Genotype G has been reported in France and the United States. Genotype H is present in Mexico and Central America. Genotype I has been identified in Vietnam and Laos. The newest HBV genotype, genotype J, has been reported in the Ryukyu Islands of Japan.[19] The distribution of genotypes not only varies between countries, but also between geographical regions within countries. In China, the main genotypes are genotypes B and C, with genotype B predominating in the central region and genotype C predominating southern and northern regions.[14] Most current studies focus on single-region genotype distributions within a country, although the distribution of HBV genotypes varies greatly in different regions. As such, large variability is to be expected when using a single dataset. In addition, data regarding the distribution of subgenotypes are scarce and the sample sizes are often too small.
Genotype assessments can provide useful adjunctive information to predict the risk of liver-related morbidity and therapeutic efficacy in CHB patients. However, very few comprehensive descriptions of HBV genotype and subgenotype distributions are available. In this study, data were collected from the published literature, full-length HBV sequences, and S gene sequence data from 75 countries to be used in a meta-analysis. The aim of the meta-analysis was to obtain a comprehensive distribution of HBV genotypes and subgenotypes while providing insights regarding disease progression and therapeutic outcome predictions.
2. Methods
2.1. Search strategy
A systematic electronic search of the literature on HBV genotypes or subgenotypes in PubMed and the terms “hepatitis B virus,” “genotype or subgenotype,” and “country (specific country names)” were used. In order to improve the comprehensiveness of the search literature, this study adopts a variety of retrieval methods. Full-length HBV and S gene sequences were retrieved from original publications by searching the GenBank database using the terms: “HBV”, “genotype”, “complete genome”, “country (specific country names)” or “HBV”, “genotype”, “S-gene”, and “country (specific country names)”. In order to reduce the burden and facilitate the construction of phylogenetic trees in the future, more than 100 strains were randomly selected for 100 strains and less than 100 strains were included in this study. Literature data and gene sequence data were included if they met our stipulated inclusion and exclusion criteria. For data from China, we used meta-analysis data from our previous study, but for other countries we did not retrieve articles containing meta-analysis data. The study was approved by the Research, Ethics and Publications Committee of Anhui Medical University (LLSC2012004).
2.2. Inclusion and exclusion criteria
Studies were included from the published literature if they met the following inclusion criteria: the study was focused on the distribution of HBV genotypes or subgenotypes (the method of HBV genotyping was suitable, for example, by direct sequencing, polymerase chain reaction, restriction fragment length polymorphism, line probe assay, and enzyme-linked immunoassay); the study clearly indicated the country of HBV infection; the study contained detailed genotype or subgenotype data that could be acquired. The exclusion criteria were as follows: the country of HBV infection was not clear; no detailed genotype or subgenotype data were available; the study contained people coinfected with human immunodeficiency virus or hepatitis C virus or included hemodialysis patients (but if a country does not have other data than that, that data would be included in the study); the study contained data from previous publications; or the study contained data from unpublished sources. The inclusion criteria for gene sequences were as follows: the country was clearly indicated; the full-length HBV sequences were more than 3000 bases but less than 3400 bases; and S gene sequences were more than 450 bases but less than 800 bases. If a country had less than 7 full-length HBV sequences or S gene sequences, the data were excluded.
2.3. Gene sequence analysis and data collection
Gene sequences were aligned using the ClustalW, which is included in MEGA 7.0.26, and alignment results were further confirmed by visual inspection. Phylogenetic trees were constructed by the maximum likelihood (ML) algorithm. HBV genotypes were checked and subgenotypes were determined by the phylogenetic comparison of full-length HBV sequences or S gene sequences. Subgenotypes were identified if nucleotide sequences were phylogenetically clustered with known subgenotypes. Data from sequences and the literature were entered into Microsoft Office Excel by country, genotype, and source. HBV genotypes were classified as either genotypes A to H or genotype undetermined (limited data are available for novel genotypes I and J and they were not classified separately). All the original data will been stored in the library database of Anhui Medical University.
2.4. Statistical analysis
Countries with only 1 dataset were directly retained, while countries with more than 1 dataset were meta-analyzed. STATA 12.0 (Stata Corp LP, College station, TX) was used to calculate the genotype ratio by the meta-analysis of cross-sectional studies. If the genotype ratio is too large (.7 < P < 1.0) or too small (.0 < P < .3), either the confidence interval will be outside [0,1] or the variance of the study will tend toward 0, giving the study large weight. In view of this situation, we used a double arcsine data transformation.
2.5. Phylogenetic trees and divergence time estimation
Only 1 sequence was randomly selected for each genotype or major subgenotype, and a simplified dataset containing 27 sequences was constructed. For genotypes with multiple subgenotypes, 10 sequences were randomly selected from each subgenotype to construct phylogenetic trees and estimate divergence times. However, as there were less than 10 sequences of the F3 subgenotype, all were selected. Phylogenetic tree reconstruction was performed using the BEAST 2.5.2 for the Bayesian inference, and MEGA 7.0.26 for the ML method. The ML heuristic method was the Nearest-Neighbor-Interchange. The best-fitting nucleotide substitution model was determined using jModelTest 2.1.7. The Markov chains were run for 1 × 107 generations to establish convergence of all parameters. Convergence and effective sample size (effective sample size > 200) of the parameters were checked with Tracer 1.7.1. The resulting trees were summarized using maximum clade credibility topology from TreeAnnotator 2.5.2 with a burn-in of the first 10% of sampled trees. FigTree 1.4.4 was employed for the visualization of node branches.
3. Results
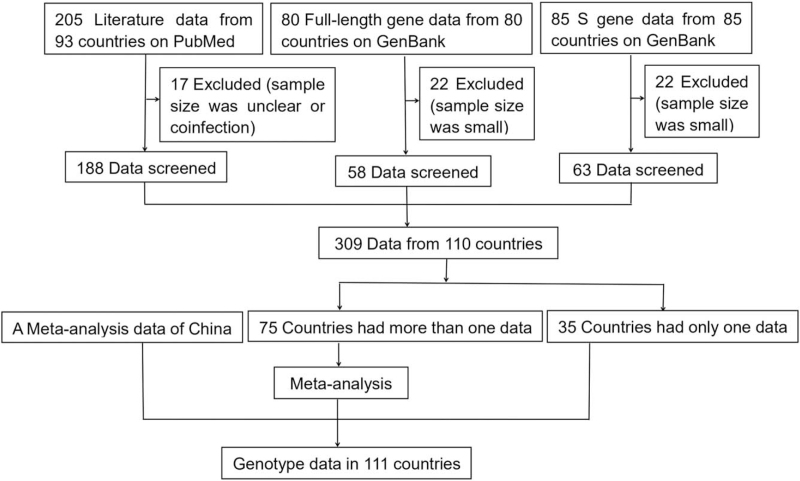
The process flow is shown in Figure 1. In total, 205 relevant studies from 93 countries from PubMed were retrieved in this study. A total of 17 studies with unclear sample size or the presence of coinfection were excluded after a preliminary review. Full-length gene data from 80 countries and S gene data from 85 countries were retrieved from the GenBank database and 44 data were excluded due to small sample size (less than 7 sequences). Our study included 309 datasets from 110 countries, of which 35 countries had only 1 dataset and the other 75 countries had more than 1 dataset. A meta-analysis was performed for multidata countries. Finally, the distribution map of HBV genotypes was obtained based on datasets that met the above criteria, including data from our previous meta-analysis (the actual ratios for each genotype are shown in Table S1, Supplemental Digital Content, http://links.lww.com/MD/G528).[14]
Figure 1.
Flowchart of the database search and study selection process. The published literature regarding HBV genotypes or subgenotypes was searched in PubMed, and the full-length HBV sequences and S gene sequences were retrieved from the GenBank database. HBV = hepatitis B virus.
3.1. The distribution of HBV genotypes
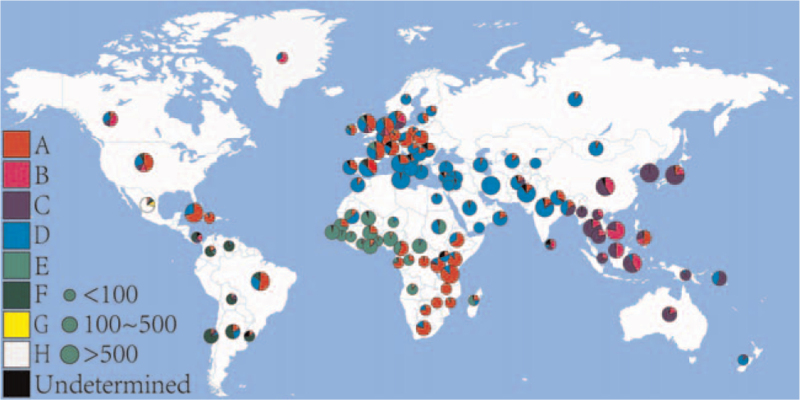
The geographical distributions of HBV genotypes are distinct among the 5 continents (Fig. 2). Genotype A is mainly prevalent in south-eastern Africa, north-western Europe, and some countries in America. Genotypes B and C are prevalent in Southeast Asia, China, and Japan. Genotype C is almost exclusively present in South Korea. Genotype D has been found worldwide but is predominant in the Mediterranean basin and some parts of Asia, such as Western, Central, and South Asia. In West Africa, genotype E is prevalent and its distribution appears to also stretch into parts of central Africa. Genotype F is found in South and Central America and is thought to be the most common genotype in most of these countries, although genotypes A and D are also present in Brazil, Cuba, and Haiti. Genotypes G and H are found in Mexico.
Figure 2.
The geographical distribution of HBV genotypes. The small pie chart indicates the sample size was less than 100, the medium pie chart indicates the sample size was between 100 and 500, and the large pie chart indicates the sample size was more than 500. A indicates HBV genotype A (red); B indicates HBV genotype B (pink); C indicates HBV genotype C (dark blue); D indicates HBV genotype D (light blue); E indicates HBV genotype E (light green); F indicates HBV genotype F (dark green); G indicates HBV genotype G (yellow); H indicates HBV genotype H (white); Undetermined indicates the genotype is unknown (black). HBV = hepatitis B virus.
3.2. The distribution of HBV subgenotypes
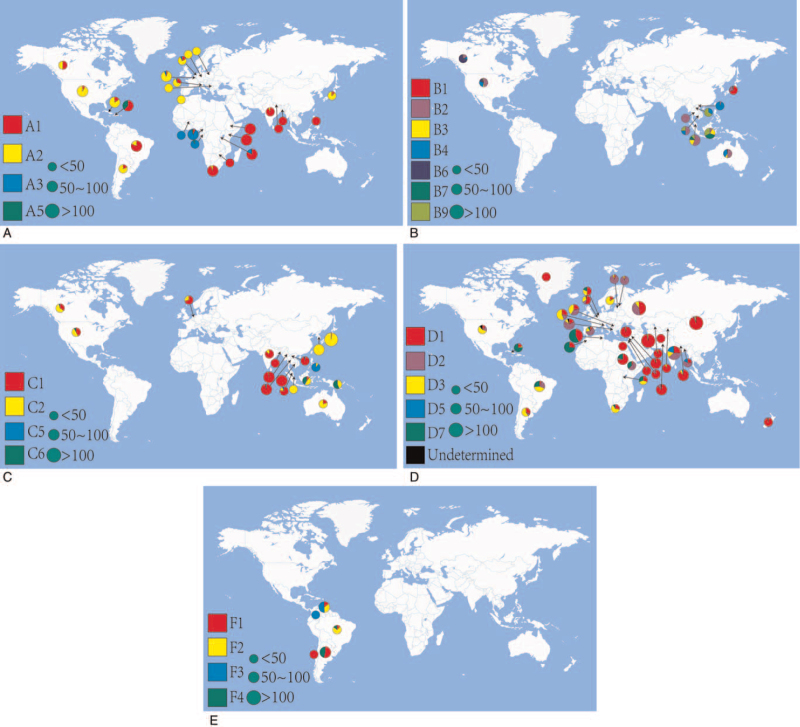
We analyzed the HBV major subgenotype in each country. If the sample size of full-length genes or S genes from the genotype was less than 10 gene sequences, it was discarded. Genotype A included 1109 sequences from 26 countries. Subgenotype A1 is predominant in Asia and south-eastern Africa, while A2 prevails in Europe, America, and Japan. In central and western Africa, subgenotype A3 is prevalent, and subgenotype A5 is found in Haiti (Fig. 3A). Genotype B includes 297 sequences from 10 countries (excluding China), and the subgenotype distribution appears disordered due to its large number subgenotypes. However, subgenotype B1 is prevalent in Japan, B2 can be seen in most countries, and B6 is mainly found in Canada. Subgenotypes B3, B7, and B9 are restricted almost entirely to Southeast Asia, especially Indonesia and Malaysia (Fig. 3B). Genotype C includes 688 sequences from 17 countries. Subgenotype C1 is prevalent in South Asia and Southeast Asia, C2 is most common in East Asia, North America, and Oceania. Subgenotype C5 prevails in the Philippines and its neighboring countries, while C6 is mainly found in Indonesia and Papua New Guinea (Fig. 3C). Genotype D includes 1932 sequences from 36 countries, and its subgenotype distribution is wide-ranging. Subgenotype D1 is widely distributed in all continents, but is highly prevalent in Asia, and D2 is prevalent in Europe, Bangladesh, and India. Subgenotype D3 is common and appears to be present worldwide, while D5 is only found in some Asian countries. Subgenotype D7 is common in Africa, Brazil, and Cuba (Fig. 3D). Genotype F includes 221 sequences from 5 countries, and little information is available for this genotype because it is restricted almost entirely to South America (Fig. 3E).
Figure 3.
The geographical distribution of HBV subgenotypes. (A) The geographical distribution of HBV genotype A. (B) The geographical distribution of HBV genotype B. (C) The geographical distribution of HBV genotype C. (D) The geographical distribution of HBV genotype D. (E) The geographical distribution of HBV genotype F. The small pie chart indicates the sample size was less than 50, the medium pie chart indicates the sample size was between 50 and 100, and the large pie chart indicates the sample size was more than 100. HBV = hepatitis B virus.
3.3. Phylogenetic trees and relative divergence times
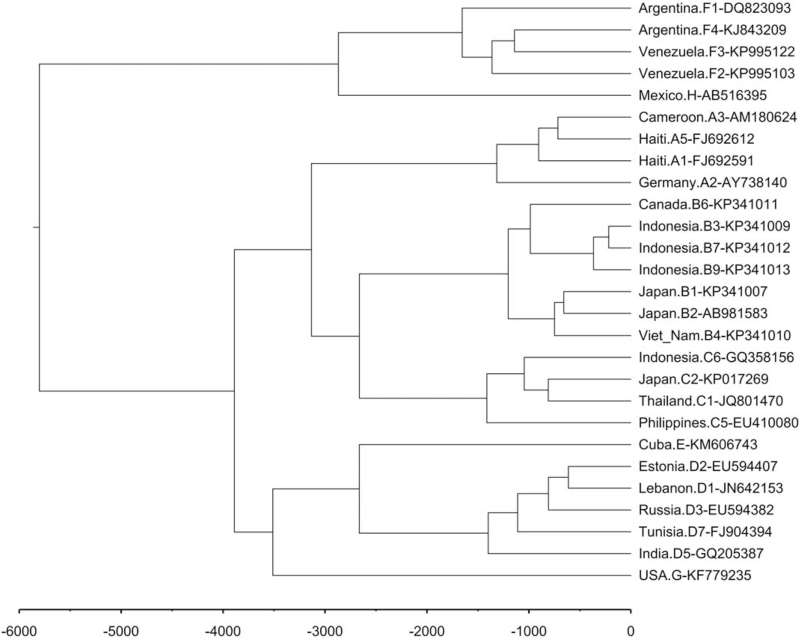
Based on the nucleotide substitution rate of 1.4 to 3.2 × 10−5 per site per year for the full-length of the viral genome, the relative evolution time of the HBV genotypes and subgenotypes was estimated. The genotypes of HBV likely originated from a common ancestor approximately 5800 years ago (Fig. 4). Genotype A originated before the year 1300; subgenotype A1 is predominant in Africa and in Central and South America, and we can see that Haiti and South Africa are closer in the phylogenetic tree. Subgenotype A2 prevails in Europe and the USA (Figure S1, Supplemental Digital Content, http://links.lww.com/MD/G527). Genotype B appeared about 1200 years ago, and subgenotypes B3, B7, and B9 are closer in phylogenetic tree and are mainly from Indonesia (Figure S2, Supplemental Digital Content, http://links.lww.com/MD/G527). Genotype C evolved about 1400 years ago, and subgenotypes C5 and C6 are more ancient than C1 and C2, with C5 and C6 found almost exclusively in Southeast Asia (Figure S3, Supplemental Digital Content, http://links.lww.com/MD/G527). Genotype D arose about 1400 years ago, and given the worldwide distribution of subgenotypes D1, D2, and D3 and the presence of multiple sequences and limited selected sequences, there is no obvious distribution from the phylogenetic tree. Subgenotype D7 is commonly found in Tunisia and Cuba, while D5 is mainly distributed in India (Figure S4, Supplemental Digital Content, http://links.lww.com/MD/G527). Genotype F existed about 1600 years ago, the differentiation time of subgenotype F1 was earlier and the distribution range was wider, while the distribution of other subgenotypes was more limited (Figure S5, Supplemental Digital Content, http://links.lww.com/MD/G527).
Figure 4.
Evolutionary timescales of HBV genotypes. Estimates of divergence times for major clades are listed near nodes. HBV = hepatitis B virus.
4. Discussion
The distribution of HBV genotypes varies from region to region, even within 1 country, HBV genotypes may present considerable spatial and temporal variation. However, existing studies are either a general prevalence of a genotype or genotypes in a country or region. For the latter, the results vary widely from person to person or from different parts of the country. As such, we searched all published articles on genotype distributions, collected gene sequences, and conducted a meta-analysis on countries with multiple datasets to obtain detailed information regarding the distribution of HBV genotypes and subgenotypes. This study is comprehensive analysis of the distribution of HBV genotypes and subgenotypes, including hundreds of data from 110 countries, and presents clear genotype and subgenotype distributions for each country considered.
The distribution of HBV genotypes and subgenotypes may be influenced by socio-demographic, ethnic, or migratory factors. In Australia, the only genotype in northern aboriginal populations was genotype C,[20] while in the Australian immigrants from Africa, genotypes A, D, and E comprised 32.5%, 32.5%, and 35%, respectively.[21] Another study showed that in Australian immigrants from Myanmar, B and C comprised 10.5% and 89.5%, respectively, of all HBV genotypes in the population, with C1 being the predominant subgenotype.[22] In this study, the prevalence of genotypes A, B, C, and D in the Australian population was 2.78%, 17.40%, 76.34%, and 3.47%, respectively, reflecting the multicultural nature of Australian society and different immigrant groups.
In most South and Central American countries, genotype F prevails. However, in Brazil, Cuba, and Haiti, genotype A is very common, accounting for 53.55%, 70.71% and 92.06%, respectively, of total HBV genotypes. In addition, in Cuba, subgenotype A2 was dominant (84.42%). Subgenotype A1 was dominant in Brazil (82.05%) and Haiti (62.07%). As can be seen in Figure 3A and Supplemental Figure S1a, Supplemental Digital Content, http://links.lww.com/MD/G527, A1 was predominant in south-eastern Africa, A2 prevails in Europe, and Haiti and South Africa are closer in phylogenetic tree. These results suggest that there may be some connection between these regions, which can be explained by human migration[23]: during the 15th and 19th centuries, the Atlantic slave trade brought millions of slaves to the New World. During this period, Brazil and Haiti were the main recipients of African slaves. Similarly, subgenotype A2 prevails in Europe and Cuba, and may be explained given that Cuba was a colony of Spain from the 15th century until the Spanish American War of 1898.
There are significant differences in HBV infection regarding clinical manifestation, disease progression, and response to antiviral therapy. These differences are affected by several factors, including HBV genotypes and subgenotypes. Genotype C is associated with later HBeAg seroconversion and advanced liver disease, such as liver cirrhosis and HCC. Subgenotype C2 has a higher risk of HCC than C1.[24] Antiviral therapy reduces the risk of liver disease and the development of HCC and may even contribute to the reversion of liver fibrosis in some patients with CHB.[25–28] The treatment response of interferon is closely related to HBV genotypes. In general, the response probability to interferon is ranked in descending order from genotype A to D for HBeAg-positive patients. Limited data are currently available for genotypes E, F, and H with regard to patient responses to interferon therapy, although these genotypes appear to respond more favorably than genotype G, which has shown poor responsiveness.[29]
HBV genotyping (genotypes and subgenotypes) has guiding significance for clinical results and future treatments. For example, in China, genotypes B and C account for the majority, while genotype A is only about 0.26%, so guidelines recommend entecavir, tenofovir disoproxil fumarate, or interferon for treatment-naïve patients with HBeAg-positive CHB.[30] In the UK, approximately 39.31% of people have genotype A, which has a higher response to interferon therapy. As such, NICE guidelines recommend interferon as first-line of treatment for HBV.
Several limitations exist in this study. In the literature, some studies may not have been retrieved by the search terms, and data from some countries may not be available due to language issues. Furthermore, the interaction between virus, host, and environmental factors is complex, and the understanding of the relationship between HBV genotypes and subgenotypes on the outcomes of HBV infection in patients is limited. However, the number of studies on HBV genotypes and subgenotypes is increasing rapidly, and the relationship between HBV genotypes/subgenotypes and disease progression and long-term prognosis of HBV infection will be more clear, which will certainly contribute to the control and treatment of HBV infection worldwide.
In summary, this study obtained a relatively accurate description of the distributions of HBV genotypes and subgenotypes and found that HBV distribution is related to ethnicity and human migration.
Author contributions
Conceptualization: Zhongping Liu, Zhenhua Zhang.
Data curation: Zhongping Liu, Yafei Zhang, Mengyuan Xu.
Formal analysis: Zhongping Liu, Yafei Zhang, Mengyuan Xu.
Funding acquisition: Xu Li, Zhenhua Zhang.
Methodology: Zhongping Liu, Yafei Zhang.
Project administration: Zhenhua Zhang.
Statistical analysis: Zhenhua Zhang.
Supervision: Zhenhua Zhang.
Writing – original draft: Zhongping Liu.
Writing – review & editing: Xu Li, Zhenhua Zhang.
Footnotes
Abbreviations: CHB = chronic hepatitis B, HBeAg = hepatitis B e antigen, HBV = hepatitis B virus, HCC = hepatocellular carcinoma, ML = maximum likelihood.
How to cite this article: Liu Z, Zhang Y, Xu M, Li X, Zhang Z. Distribution of hepatitis B virus genotypes and subgenotypes: a meta-analysis. Medicine. 2021;100:50(e27941).
The study was supported by Anhui Provincial Natural Science Foundation (2108085MH298), Scientific research project of the Second Hospital of Anhui Medical University (2019GMFY02), and the National Science Foundation of China (81273142). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
References
- [1].Tian T, Song C, Jiang L, et al. Hepatitis B virus infection and the risk of cancer among the Chinese population. Int J Cancer 2020;147:3075–84. [DOI] [PubMed] [Google Scholar]
- [2].Yang H, Yeh M, Wong G, et al. Real-world effectiveness from the Asia Pacific rim liver consortium for HBV risk score for the prediction of hepatocellular carcinoma in chronic hepatitis B patients treated with oral antiviral therapy. J Infect Dis 2020;221:389–99. [DOI] [PubMed] [Google Scholar]
- [3].World Health Organization (WHO). Hepatitis B. Available at: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. Accessed July 24, 2020. [Google Scholar]
- [4].Girones R, Miller RH. Mutation rate of the hepadnavirus genome. Virology 1989;170:595–7. [DOI] [PubMed] [Google Scholar]
- [5].Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci U S A 1996;93:4398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chotiyaputta W, Lok AS. Hepatitis B virus variants. Nat Rev Gastroenterol Hepatol 2009;6:453–62. [DOI] [PubMed] [Google Scholar]
- [7].Zhang ZH, Wu CC, Chen XW, Li X, Li J, Lu MJ. Genetic variation of hepatitis B virus and its significance for pathogenesis. World J Gastroenterol 2016;22:126–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Okamoto H, Tsuda F, Sakugawa H, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol 1988;69:2575–83. [DOI] [PubMed] [Google Scholar]
- [9].Norder H, Couroucé AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 1994;198:489–503. [DOI] [PubMed] [Google Scholar]
- [10].Norder H, Hammas B, Löfdahl S, Couroucé AM, Magnius LO. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J Gen Virol 1992;73:1201–8. [DOI] [PubMed] [Google Scholar]
- [11].Mayerat C, Mantegani A, Frei PC. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J Viral Hepat 1999;6:299–304. [DOI] [PubMed] [Google Scholar]
- [12].Anastasiou OE, Widera M, Westhaus S, et al. Clinical outcome and viral genome variability of hepatitis B virus-induced acute liver failure. Hepatology 2019;69:993–1003. [DOI] [PubMed] [Google Scholar]
- [13].Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol 2004;72:363–9. [DOI] [PubMed] [Google Scholar]
- [14].Wu X, Wei SF, Xu N, Su Q, Li X, Zhang ZH. Meta-analysis on distribution of hepatitis B virus genotypes and related clinical outcomes in China [in Chinese]. J Clin Hepatol 2017;271–5. [Google Scholar]
- [15].Sozzi V, Shen F, Chen J, et al. In vitro studies identify a low replication phenotype for hepatitis B virus genotype H generally associated with occult HBV and less severe liver disease. Virology 2018;519:190–6. [DOI] [PubMed] [Google Scholar]
- [16].Tanaka Y, Hasegawa I, Kato T, et al. A case-control study for differences among hepatitis B virus infections of genotypes A (subtypes Aa and Ae) and D. Hepatology 2004;40:747–55. [DOI] [PubMed] [Google Scholar]
- [17].Ozaras R, Inanc Balkan I, Yemisen M, Tabak F. Epidemiology of HBV subgenotypes D. Clin Res Hepatol Gastroenterol 2015;39:28–37. [DOI] [PubMed] [Google Scholar]
- [18].McMahon BJ. The influence of hepatitis B virus genotype and subgenotype on the natural history of chronic hepatitis B. Hepatol Int 2009;3:334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin C, Kao J. Natural history of acute and chronic hepatitis B: the role of HBV genotypes and mutants. Best Pract Res Clin Gastroenterol 2017;31:249–55. [DOI] [PubMed] [Google Scholar]
- [20].Davies J, Littlejohn M, Locarnini SA, et al. The molecular epidemiology of hepatitis B in the Indigenous people of northern Australia. J Gastroenterol Hepatol 2013;28:1234–41. [DOI] [PubMed] [Google Scholar]
- [21].Thurnheer MC, Edwards R, Schulz TR, et al. Genotypic profiles of hepatitis B in African immigrants and their clinical relevance. J Med Virol 2017;89:1000–7. [DOI] [PubMed] [Google Scholar]
- [22].Schulz TR, Edwards R, Thurnheer MC, et al. Hepatitis B among immigrants from Myanmar: genotypes and their clinical relevance. J Med Virol 2018;90:271–6. [DOI] [PubMed] [Google Scholar]
- [23].Kramvis A, Paraskevis D. Subgenotype A1 of HBV--tracing human migrations in and out of Africa. Antivir Ther 2013;18:513–21. [DOI] [PubMed] [Google Scholar]
- [24].Chan HL, Tse CH, Mo F, et al. High viral load and hepatitis B virus subgenotype ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol 2008;26:177–82. [DOI] [PubMed] [Google Scholar]
- [25].Gordon SC, Lamerato LE, Rupp LB, et al. Antiviral therapy for chronic hepatitis B virus infection and development of hepatocellular carcinoma in a US population. Clin Gastroenterol Hepatol 2014;12:885–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lai CL, Yuen MF. Prevention of hepatitis B virus-related hepatocellular carcinoma with antiviral therapy. Hepatology 2013;57:399–408. [DOI] [PubMed] [Google Scholar]
- [27].Papatheodoridis GV, Lampertico P, Manolakopoulos S, Lok A. Incidence of hepatocellular carcinoma in chronic hepatitis B patients receiving nucleos(t)ide therapy: a systematic review. J Hepatol 2010;53:348–56. [DOI] [PubMed] [Google Scholar]
- [28].Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018;67:1560–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Erhardt A, Göbel T, Ludwig A, et al. Response to antiviral treatment in patients infected with hepatitis B virus genotypes E-H. J Med Virol 2009;81:1716–20. [DOI] [PubMed] [Google Scholar]
- [30].Hou J, Wang G, Wang F, et al. Guideline of prevention and treatment for chronic hepatitis B (2015 update). J Clin Transl Hepatol 2017;5:297–318. [DOI] [PMC free article] [PubMed] [Google Scholar]