Abstract
Inflammatory bowel diseases (IBD), including Crohn's disease (CD) and ulcerative colitis (UC), are known as chronic gastrointestinal inflammatory disorders. The present systematic review and meta analysis was conducted to estimate the prevalence of adherent-invasive Escherichia coli (AIEC) isolates and their phylogenetic grouping among IBD patients compared with the controls. A systematic literature search was conducted among published papers by international authors until April 30, 2020 in Web of Science, Scopus, EMBASE, and PubMed databases. The pooled prevalence of AIEC isolates and their phylogenetic grouping among IBD patients as well as in controls was estimated using fixed or random effects models. Furthermore, for estimating the association of colonization by AIEC with IBD, odds ratio along with 95% confidence interval was reported. A total of 205 articles retrieved by the initial search of databases, 13 case–control studies met the eligibility criteria for inclusion in the meta analysis. There were 465 IBD cases (348 CD and 117 UC) and 307 controls. The pooled prevalence of AIEC isolates were 28% (95% CI: 18–39%), 29% (95% CI: 20–40%), 13% (95% CI: 1–30%), and 9% (95% CI: 3–19%), respectively among IBD, CD, UC, and control group, respectively. Our results revealed that the most frequent AIEC phylogroup in the IBD, CD, and control groups was B2. Fixed-effects meta analysis showed that colonization of AIEC is significantly associated with IBD (OR: 2.93; 95% CI: 1.90–4.52; P < 0.001) and CD (OR: 3.07; 95% CI: 1.99–4.74; P < 0.001), but not with UC (OR: 2.29; 95% CI: 0.81–6.51; P = 0.11). In summary, this meta analysis revealed that colonization by AIEC is more frequent in IBD and is associated with IBD (CD and UC). Our results suggested that the affects of IBD in patients colonized with the AIEC pathovar is not random, it is in fact a specific disease-related pathovar.
Keywords: adherent-invasive Escherichia coli, inflammatory bowel diseases, meta-analysis, AIEC, phylogroup
Introduction
Inflammatory bowel diseases (IBD), including Crohn's disease (CD) and ulcerative coliti (UC), are chronic incurable gastrointestinal inflammatory disorders with unknown etiology (1). Previously, IBDs were regarded as a disease prevalent in industrialized countries. However, in the 21st century, as the epidemiological trend of this disease changed, IBDs have become a global problem, and new cases in developing countries in Asia, South America, and Africa are on the rise (2).
They are probably the result of improper and continuous initiation of the intestinal mucosal immune system due to the complicated interactions of genetics, microbial, and immunological agents (3).
As shown by previous experimental and clinical research, intestinal bacteria play a role in the development of IBD and the severity of the disease (4). According to the recent molecular studies on patients with IBD, beneficial bacteria, for instance, Firmicutes and Bacteroidetes, have reduced, and pathogenic bacteria, for example, Proteobacteriae, particularly Escherichia coli, have increased (5).
Recently, the links between IBD and adherent-invasive Escherichia coli (AIEC) strains have been discussed (6–9). It is possible to classify E. coli strains into commensal or pathogenic categories based on genetic and phylogenetic characteristics. The definition of the pathogenic potential of E. coli is changing to some extent. AIEC was recently recognized as a pathotype of E. coli (late 1990s), and the variable range of AIEC was different from the six primary diarrheagenic E. coli pathotypes (Enteropathogenic E. coli (EPEC), Enteroinvasive E. coli (EIEC), Shiga toxin-producing E. coli (STEC), diffusely adherent E. coli (DAEC), Enterotoxigenic E. coli (ETEC), and Enteroaggregative E. coli (EAEC) (10). AIEC is well-known for its role in clinical and experimental epidemiological studies in IBD pathogenesis (11–14). These strains have the ability of adherence and invasion to intestinal epithelial cells (IECs) and extensive survival in macrophages by secreting high levels of tumor necrosis factor alpha (TNF-a) (15). Adherence of this pathotype through type 1 pili expression in the bacterial surface and through cell adhesion molecule 6 (CEACAM6) is attributed to the presence of carcinoembryonic antigen in the ileal epithelial cells' apical surface (16, 17). CEACAM6 is an AIEC receptor that has been indicated to have an abnormal expression in the ileal epithelial cells of adult CD patients (18).
Genetically, AIECs are very close to extraintestinal pathogenic E. coli, which includes uropathogenic E. coli as well as neonatal meningitis-related strains (5, 19). Based on the strong evidence concerning AIEC role in the promotion of gut inflammation and exacerbation of IBD pathology, most of the AIEC isolates belong to the D and B2 phylogenetic groups of E. coli, as shown by genomic studies. In addition, based on the distribution of AIEC strains from the phylogenetic perspective, the dominant force in the formation of this pathotype is convergent evolution (11, 12). Previous research has shown that AIEC encodes a large subunit of propandiol dehydratase (a fermentation product of 1,2-propandiol fucose) that is elevated in the microbiome of patients with CD and directs intestinal T cell inflammation induced by AIEC (20).
Moreover, Rath et al. reported that the main role of AIEC in Crohn's disease is to impair the mitochondrial function of epithelial cells. Additionally, different types of intestinal inflammation have shown mitochondrial dysfunction, and it has been indicated that mutations in mitochondria modulating genes are susceptible to IBD (21).
To enhance information about the epidemiology of AIEC in IBD, comprehensive study on the prevalence of AIEC in IBD patients worldwide is believed to be of great value. Therefore, the current systematic review and meta analysis aimed to investigate the prevalence of AIEC isolates and their phylogenetic in IBD patients compared with the controls and the association of colonization by AIEC with IBD, which was quantified by estimating pooled odds ratio (OR) along with 95% confidence interval for OR.
Methods
Search Strategies
The research design in the present work followed the preferred reporting items for systematic reviews and metaanalyses (PRISMA) procedures (Supplementary Table 1). The Web of Science, Scopus, EMBASE, and PubMed databases were used for a systematic literature review. The papers published by international authors up to end of April 30, 2020 were searched and reviewed. The following terms were searched as keywords of the present research: “AIEC” or “adherent-invasive Escherichia coli” AND “IBD” or “inflammatory bowel disease” OR “CD” or “Crohn's disease” OR “UC” or “ulcerative colitis” without restricting the country. We used the papers that reported the AIEC frequency/prevalence or distribution and their phylogenetic classification in patients with IBD and control group for conducting a comprehensive search. The studies in any language from any region were investigated.
Inclusion and Exclusion Criteria
For determining eligibility of studies for meeting the inclusion criteria, the databases with related key terms were independently screened by two reviewers, and the titles, abstracts, and full texts were reviewed, and any inconsistencies were fixed by consensus. The inclusion criteria included: (1) the case–control research works, cohort, and retrospective studies of patients and control group with diagnosis of IBD, (2) studies reporting the AIEC prevalence in IBD patients using biopsy sample from intestine parts (colon and ileum), invasion assay, bacterial adhesion, bacterial survival, and replication in macrophages approaches for AIEC detection). We excluded clinical trials, meta analysis, review, or systematic article, case studies, editorials, letters to the editors, abstracts of meetings, congress, and non-human studies. Only biopsy samples were analyzed, and AIEC levels among the other intestinal bacteria are not mentioned.
Quality Assessment and Data Extraction
The quality of studies was evaluated independently by two authors (RD and MH) using the quality assessment tool for case–control studies developed by Joanna Briggs Institute (JBI), and disagreements were resolved by the third author (PA). Item-related title and abstract, introduction, methods, results, discussion, and other information were determined and a score was assigned to each item. Studies with a score greater than or equal to 60% were included.
Finally, detailed information on eligible studies including the first authors name, publication date, place of study, population studied (IBD patient and control group), type of sample (biopsy), sample size in both IBD and control groups, and sample size of AIEC and phylogroups analysis were extracted.
Statistical Analysis
The pooled prevalence of AIEC isolates and their phylogenetic in IBD patients and controls along with 95% confidence intervals (95% CI) was estimated by applying the “metaprop program” in STATA statistical software. In this meta analysis, confidence interval for proportion was computed by using score method. In all included studies, we evaluated the association of AIEC with IBD, and the prevalence of AIEC was compared between patients and control groups, and for quantifying the association of colonization by AIEC with IBD, the odds ratio (OR) and 95% confidence interval (95% CI) for OR was calculated as the pooled estimate of effect size using the DerSimonian and Laird method (22). Statistical heterogeneity between studies was evaluated using the Cochran Q Chi-squared test and Cochrane-I-square, and values of 25, 50, and 75% for I2 were considered as low, medium, and high levels of heterogeneity, respectively (23). When P-value <0.10 for Cochran Q Chi-squared test and the value of Cochrane-I2 was more than 50%, the heterogeneity was considered as high and a random effect approach was adopted for estimating the pooled prevalence, OR, and confidence intervals. The funnel plot, Begg's rank correlation test, and Egger's weighted regression tests were performed to evaluate possible publication bias, and any appeared asymmetry in funnel plot or P < 0.05 in used tests was considered as indication of statistically significant publication bias (24). Possible sources of heterogeneity were examined using sensitivity analysis and meta regression to evaluate the confounding role of age. Moreover, sensitivity analyses were conducted to determine the extent to which inferences (the estimated pooled prevalence and OR) might be related to a particular study. All statistical analyses were performed, using STATA Version 11 (Stata Corp., College Station, TX, USA). P-values <0.05 were considered statistically significant.
Results
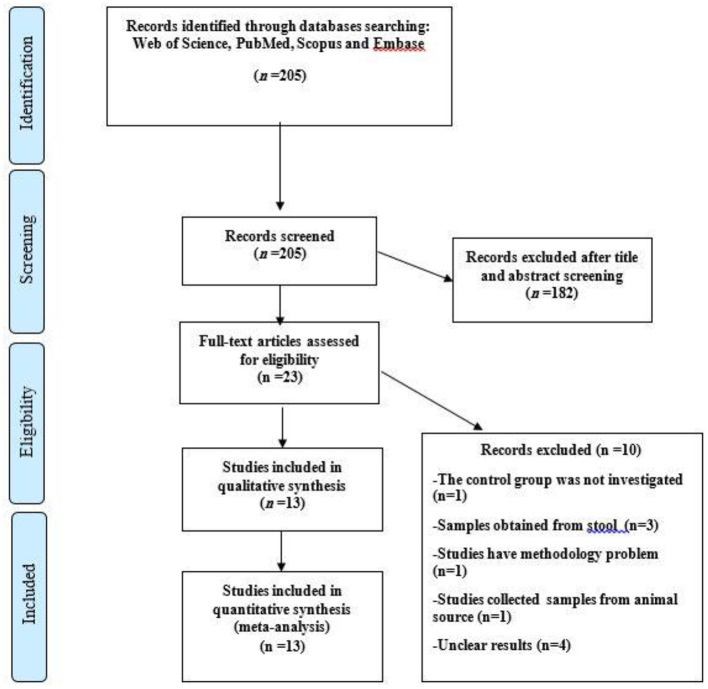
A total of 205 articles were retrieved by the initial search of databases, of which 182 were removed following selection based on titles, abstract, and index review, and 23 studies were selected for full-text analysis. After assessment of the 23 reviewed studies, three studies collected samples from stool specimen (9, 25, 26), one study had a methodology problem (27), one study collected samples from animal sources (28), two studies did not report the prevalence of AIEC isolates (12, 29), results of a study was unclear in terms of the number of patients and biopsy samples (30), one study was performed on standard isolates (6), and one study did not report the results of control group (31).
Finally, 13 case–control studies met the eligibility criteria for inclusion in the meta analysis. Figure 1 shows a flow diagram illustrating the searching procedure for the selection of eligible studies (16, 32–43). Also, the detailed features of the included articles are accessible in Tables 1, 2. All of the included studies used intestine biopsy samples, bacterial adhesion, invasion assay, bacterial survival, and replication in macrophages methods for the detection of AIEC. All of the articles were case–control studies published between 2004 and 2020.
Figure 1.
Flow chart of the study selection for inclusion in the systematic review.
Table 1.
The characteristics of studies included in the systematic review.
| Study | Publication year | Location | Population studied | Type of sample | IBD | CD | UC | Control | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SS of patients | SS of AIEC | SS of patients | SS of AIEC | SS of patient | SS of AIEC | SS of patient | SS of AIEC | ||||||
| 1 | Darfeuille-Michaud et al. | 2004 | France | IBD | B | 90 | 18 | 90 | 18 | - | - | 118 | 3 |
| 2 | Baumgart et al. | 2007 | USA | IBD | B | 21 | 10 | 21 | 10 | - | - | 7 | 1 |
| 3 | Medina et al. | 2009 | Spain | IBD | B | 20 | 11 | 20 | 11 | - | - | 28 | 6 |
| 4 | Raso et al. | 2011 | Italy | IBD | B | 14 | 4 | 8 | 4 | 6 | 0 | 4 | 0 |
| 5 | Negroni et al. | 2012 | Italy | IBD | B | 34 | 2 | 24 | 1 | 10 | 1 | 23 | 0 |
| 6 | Dogan et al. | 2013 | New York | IBD | B | 32 | 8 | 32 | 8 | 28 | 5 | ||
| 7 | Elliott et al. | 2013 | UK | IBD | B | 45 | 2 | 30 | 2 | 15 | 0 | 14 | 0 |
| 8 | Fuente et al. | 2014 | Chile | IBD | B | 91 | 8 | 34 | 6 | 57 | 2 | 22 | 0 |
| 9 | O'Brien et al. | 2016 | Australian | IBD | B | 19 | 5 | 14 | 3 | 5 | 2 | 21 | 5 |
| 10 | Cespedes et al. | 2017 | Spain/USA | IBD | B | 24 | 13 | 24 | 13 | 8 | 0 | ||
| 11 | Font et al. | 2019 | Spain | IBD | B | 33 | 15 | 33 | 15 | - | - | 25 | 6 |
| 12 | Lee et al. | 2019 | Korea | IBD | B | 42 | 14 | 18 | 5 | 24 | 9 | 9 | 2 |
| 13 | Abdelhalim et al. | 2020 | Turkey | IBD | B | 24 | 10 | 24 | 10 | 15 | 7 | ||
B, Biopsy; CD, Crohn's disease; UC, ulcerative colitis; IBD, Inflammatory bowel disease; SS, Sample size.
Table 2.
The details of distribution of AIEC based IBD, CD, UC, and Control.
| Study | IBD | CD | UC | Control | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SS of patients/AIEC | Location of sample AIEC (positive sample) | SS of patients/AIEC | Location of sample AIEC (positive sample) | SS of patients/AIEC | Location of sample AIEC (positive sample) | SS of patients/AIEC | Location of sample AIEC (positive sample) | |||||||||
| Darfeuille-Michaud et al. | 90/18 | Ileal | Colon | 90/18 | Ileal | Colon | - | - | 118/3 | Ileal | Colon | |||||
| 63 (17) | 27 (1) | 63 (17) | 27 (1) | 16 (1) | 102 (2) | |||||||||||
| Baumgart et al. | 21/10 | Ileal | 21/10 | Ileal | - | - | 7/1 | Ileal | ||||||||
| 21 (10) | 21 (10) | 7 (1) | ||||||||||||||
| Medina et al. | 20/11 | Ileal | Colon | Ileal + colon | 20/11 | Ileal | Colon | Ileal + colon | - | - | 28/6 | Ileal | Colon | Ileal + colon | ||
| 4 (4) | 9 (6) | 16 (1) | 4/4 | 9/6 | 7/1 | 9 (3) | 11 (3) | 8 (0) | ||||||||
| Raso et al. | 14/4 | ND | 8/4 | ND | 6/0 | ND | 4/0 | ND | ||||||||
| Negroni et al. | 34/2 | Ileal | 24/1 | Ileal | 10/1 | Colonic | 23/0 | - | ||||||||
| 34 (2) | 24 (1) | 10(1) | ||||||||||||||
| Dogan et al. | 32/8 | Ileal | 32/8 | Ileal | - | - | 28/5 | Ieal | ||||||||
| 32 (8) | 32 (8) | 28 (5) | ||||||||||||||
| Elliott et al. | 45/2 | ND | 30/2 | ND | 15/0 | ND | 14/0 | ND | ||||||||
| Fuente et al. | 91/8 | Ileal | 34/6 | Ileal | 57/2 | Ileal | 22/0 | Ileal | ||||||||
| 91 (8) | 34 (6) | 57 (2) | 22 (0) | |||||||||||||
| O'Brien et al. | 19/5 | Terminal Ileumm | 14/3 | Terminal-Ileumm | 5/2 | Terminal Ileumm | 21/5 | Terminal ileumm | ||||||||
| 19 (5) | 14 (3) | 5 (2) | 21 (5) | |||||||||||||
| Cespedes et al. | 24/13 | ND | 24/13 | ND | - | - | 8/0 | ND | ||||||||
| Font et al. | 33/15 | ND | 33/15 | ND | - | - | 25/6 | ND | ||||||||
| Lee et al. | 42/14 | Ileal | Ileocecal valve |
Colon | 18/5 | Ileal | Ileocecal valve |
Colon | 24/9 | Ileal | Ileocecal valve |
Colon | 9/2 | Ileal | Ileocecal valve |
Colon |
| 5(ND) | 10(ND) | 27(ND) | 5(ND) | 7(ND) | 6(ND) | 0(ND) | 3(ND) | 21(ND) | 0(0) | 0(0) | 9 (2) | |||||
| Abdelhalim et al. | 24/10 | Ileal | Colon | Ileocolonic | 24/10 | Ileal | Colon | Ileocolonic | - | 15/7 | Ileal | Colon | Ileocolonic | |||
| 4(ND) | 12(ND) | 8(ND) | 4(ND) | 12(ND) | 8(ND) | ND | ND | ND | ||||||||
CD, Crohn's disease; UC, ulcerative colitis; IBD, Inflammatory bowel disease; SS, Sample size; ND, No Data.
Totally, there were 465 IBD cases (348 CD and 117 UC) and 307 controls. Given that the three articles did not mention the sex of patients, the participants were almost 242 men and 239 women.
Prevalence of AIEC Isolate
Thirteen studies reported the prevalence of AIEC isolates, of these the pooled prevalence of AIEC was 28% (95% CI: 18–39%) ranging from 4 to 55% among IBD patients, and among CD patients it was 29% (95% CI: 20–40%) ranging from 4 to 55%. From six studies that investigated the prevalence of AIEC isolates among UC patients, the pooled prevalence was 13% (95% CI: 1–30%) ranging from 3 to 40%. Moreover, the pooled prevalence of AIEC was 9% (95% CI: 3–19%) ranging from 0 to 47% among control subjects (Supplementary Figures 1–4).
Association of Colonization by AIEC With IBD
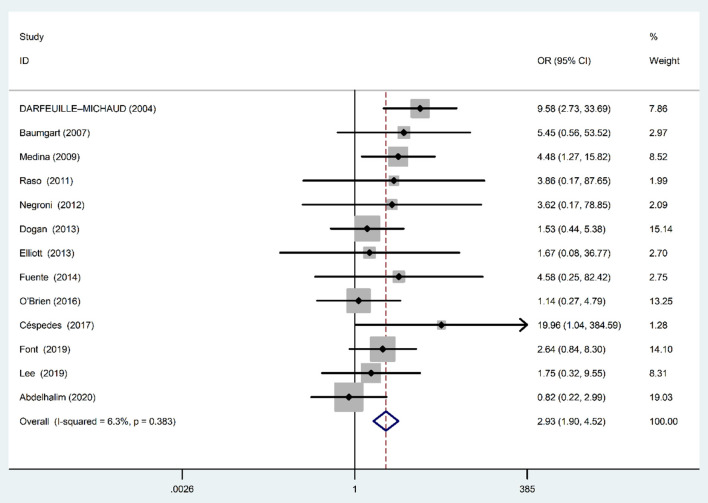
Fixed-effects meta analysis showed a significant positive association between AIEC and IBD disease (OR: 2.93; 95% CI: 1.90–4.52; P < 0.001) (Figure 2), indicating that the prevalence of AIEC is higher in IBD patients compared with controls. We found no evidence of heterogeneity among studies (χ2 = 12.81, P = 0.38; I2 = 6.3%).
Figure 2.
Forest plot of the association between AIEC rate and risk of IBD.
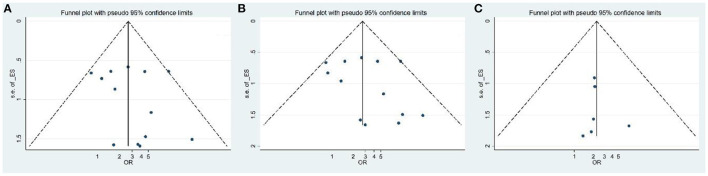
The funnel plot for publication bias did not show any evidence of asymmetry (Figure 3A). In addition, Begg's and Egger's tests were used to quantitatively evaluate the potential publication bias. According to the results of Begg's (Z = 0.31, P = 0.76) and Egger's tests (t = 0.77, P = 0.45), there was no significant publication bias.
Figure 3.
Funnel plot for evaluation of publication bias [(A) IBD; (B) CD; (C) UC patients].
Sensitivity Analysis and Meta Regression
Meta regression analysis indicated that the relationships between the association colonization by AIEC with IBD does not confound by age (OR: 1.02; 95% CI: 0.72–1.42; P = 0.84) (Supplementary Figure 5A). Additionally, the results of sensitivity analysis showed that none of the studies affects influentially the association of AIEC with effects of IBD (Supplementary Figure 6A). In this regard, each study was excluded and then the result was examined again. Then, no significant change in estimated pooled OR was obtained.
Association of Colonization by AIEC With CD
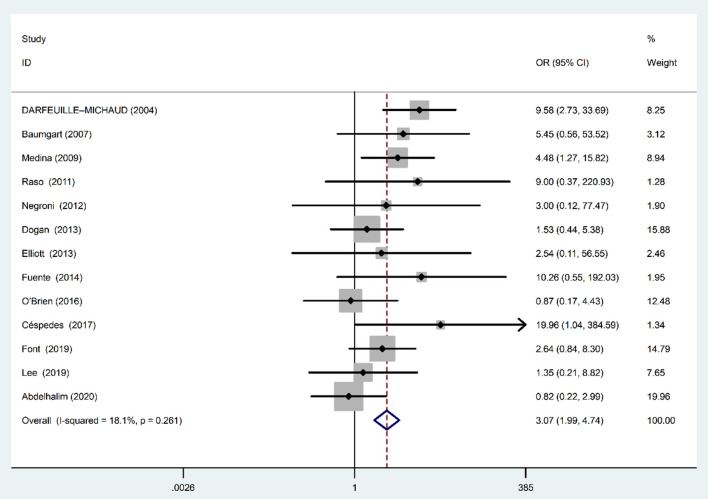
Fixed-effects meta analysis showed a significant direct association between AIEC and IBD disease (OR: 3.07; 95% CI: 1.99–4.74; P < 0.001), indicating that the AIEC prevalence is high in patients with CD compared with controls (Figure 4). We found no evidence of between-study heterogeneity (χ2 = 14.66, P = 0.26; I2 = 18.1%).
Figure 4.
Forest plot of the association between AIEC rate and risk of CD.
The funnel plot for publication bias did not show any evidence of asymmetry (Figure 3B). According to the results of Begg's (Z = 0.79, P = 0.42) and Egger's tests (t = 0.96, P = 0.35), there was no significant publication bias.
Sensitivity Analysis and Meta Regression
Meta regression analysis indicated that there is no significant relationship between age and occurrence of AIEC in patients with CD (OR: 1.07; 95% CI: 0.76–1.51; P = 0.66) (Supplementary Figure 5B). Additionally, the results of sensitivity analysis showed that none of the studies affects influentially the observed association of AIEC with UC patients (Supplementary Figure 6B). Each study was excluded and then the result was reevaluated. Accordingly, no significant change in estimated pooled OR was observed.
Association of Colonization by AIEC Isolates With UC
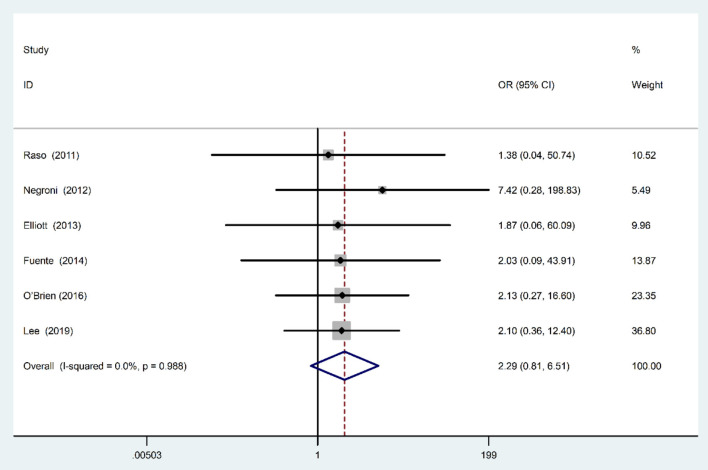
Fixed-effects meta analysis showed that although based on the estimated OR the prevalence of colonization by AIEC was higher in UC patients compared with controls; however, the observed association was not statistically significant (OR: 2.29; 95% CI: 0.81–6.51; P = 0.11) (Figure 5). We found no evidence of between-study heterogeneity (χ2 = 0.6, P = 0.98; I2 = 0%).
Figure 5.
Forest plot of the association between AIEC rate and risk of UC.
The funnel plot for publication bias did not show any evidence of asymmetry (Figure 3C). According to the results of Begg's (Z = 0.75, P = 0.4) and Egger's tests (t = 0.34, P = 0.74), there was no significant publication bias.
Sensitivity Analysis and Meta Regression
Meta regression analysis revealed a significant confounding negative effect for age in the observed association between AIEC and UC (OR: 0.83; 95% CI: 0.70–098; P = 0.041) (Supplementary Figure 5C). Additionally, the results of sensitivity analysis showed that none of the studies affects influentially the association between AIEC and UC (Supplementary Figure 6C). For follow up the sensitivity analysis, each study was excluded and then the result reexamined. No significant change in estimated pooled OR was obtained.
Prevalence of AIEC Isolate Phylogenetic Groups
Eight studies reported prevalence of different phylogroup among IBD patients (Table 3). From those studies, the most prevalent phylogroup was B2 (53%, 95% CI: 36–69%) ranging from 20 to 100%, whereas the less prevalent was B1 (1%, 95% CI: 0–6%) ranging from 0 to 20%. There was no significant heterogeneity for phylogroup among the eight studies (χ2 = 11.99, P = 0.10; I2 = 41.63). Moreover, the funnel plot for publication bias did not show any evidence of asymmetry. According to the results of Begg's and Egger's tests, there was no significant publication bias among investigated phylogroup in IBD patients. The complete results of pooled prevalence, heterogeneity, and publication bias tests of different phylogenetic groups are shown in Supplementary Table 2.
Table 3.
Phylogroups distribution among AIEC isolates.
| Authors | Type of disease (No. of AIEC) | Phylogroup | |||
|---|---|---|---|---|---|
| A | B1 | B2 | D | ||
| Baumgart et al. | IBD (10) | 3 | 2 | 2 | 3 |
| CD (10) | 3 | 2 | 2 | 3 | |
| Control (1) | 0 | 0 | 0 | 1 | |
| Medina et al. | IBD (11) | 1 | 0 | 7 | 3 |
| CD (11) | 1 | 0 | 7 | 3 | |
| Control (6) | 2 | 1 | 3 | 0 | |
| Raso et al. | IBD (4) | 0 | 0 | 3 | 1 |
| CD (4) | 0 | 0 | 3 | 1 | |
| UC (0) | 0 | 0 | 0 | 0 | |
| Control (0) | 0 | 0 | 0 | 0 | |
| Elliott et al. | IBD (2) | 0 | 0 | 2 | 0 |
| CD (2) | 0 | 0 | 2 | 0 | |
| UC (0) | 0 | 0 | 0 | 0 | |
| Control (0) | 0 | 0 | 0 | 0 | |
| Fuente et al. | IBD (8) | 1 | 0 | 3 | 4 |
| CD (6) | 1 | 0 | 2 | 3 | |
| UC (2) | 0 | 0 | 1 | 1 | |
| Control (0) | 0 | 0 | 0 | 0 | |
| Cespedes et al. | IBD (13) | 1 | 0 | 7 | 5 |
| CD (13) | 1 | 0 | 7 | 5 | |
| Control (0) | 0 | 0 | 0 | 0 | |
| Font et al. | IBD (15) | 2 | 0 | 11 | 2 |
| CD (15) | 2 | 0 | 11 | 2 | |
| Control (6) | 2 | 1 | 3 | 0 | |
| Lee et al. | IBD (14) | 4 | 1 | 5 | 4 |
| CD (5) | 1 | 0 | 3 | 1 | |
| UC (9) | 3 | 1 | 2 | 3 | |
| Control (2) | 0 | 1 | 0 | 1 | |
AIEC, adherent–invasive Escherichia coli; CD, Crohn's disease; UC, ulcerative colitis; IBD, Inflammatory bowel disease; SS, Sample size.
Moreover, eight studies investigated prevalence of phylogroup among CD patients. From those studies, the most frequent phylogroup was B2 (57%, 95% CI: 40–73%) ranging from 20 to 100%, while the less frequent was B1 (0%, 95% CI: 0–5%) ranging from 0 to 20%. There was no significant heterogeneity for phylogroup among the eight studies. Moreover, the funnel plot for publication bias did not show any evidence of asymmetry. According to the results of Begg's and Egger's tests, there was no significant publication bias among investigated phylogroup in CD patients (Supplementary Table 2).
In addition, among four studies that investigated prevalence of different phylogroup among control group, the highest phylogroup was B2 (36%, 95% CI: 8–68%) ranging from 0 to 50%, whereas the lowest was D (11%, 95% CI: 0–62%) ranging from 0 to 100%. There was no significant heterogeneity against phylogroup among the four studies. Moreover, the funnel plot for publication bias did not show any evidence of asymmetry. According to the results of Begg's and Egger's tests, except phylogroup D, there was significant publication bias among investigated phylogroup in the control group (Supplementary Table 2).
Discussion
The current study was a comprehensive systematic review, and meta analysis was conducted to investigate the association of colonization by AIEC with IBD, IBD UC, and DC types. Based on previous studies, the mucosa-associated E. coli may be important in the pathogenesis of IBD, UC, and CD.
Additionally, AIEC has the ability to invade Peyer's patches and the lamina propria through M cells (44). AIEC could be adopted into macrophages, replicate, and survive within them because of the host autophagy defect. It then triggers the secretion of TNF-α through activating infected macrophages and increasing proinflammatory cytokine expression (45). Based on the studies reviewed, the incidence of IBD between colonized individuals with AIEC was different. These differences may be explained by the distribution and composition of different intestinal microbiota depending on the involvement of host and/or environmental factors (43).
In recent years, several studies have shown the role of intestinal microbiota in the development of IBD. It has been shown that in patients with IBD, the balance of intestinal bacteria is disturbed and the number of beneficial bacteria such as Bifidobacteria, Lactobacilli, and Firmicutes is reduced and the number of possible pathogenic bacteria such as Bacteroides and E. coli is increased (46). Studies have shown that an increase in Bacteroides and E. coli and a change in intestinal microbiota composition due to a high-fat/high-sugar diet increases the sensitivity to AIEC and intestinal inflammation in CEABAC10 transgenic mice (34, 45).
Majority of included studies in our systematic review and meta analysis showed significant association between IBD and the presence of AIEC; however, there were few studies with insignificant results. The non-significant results in these studies may be attributed to the low sample size and high type 2 statistical error rate. Totally, the estimated pooled OR from all included studies in our meta analysis resulted in significant relationship between AIEC rate and IBD, in which the strength of association was 2.93 and 3.07 for IBD (irrespective of disease type) and IBD CD type, respectively.
Based on literature review, individuals with high levels of CEACAM6 and CHI3L1 receptors, which are overexpressed in inflammation, promote AIEC adhesion and invasion to IECs located in the ileum via the type-1 pili's FimH adhesion (44, 47) or colonic IECs via the chitinase ChiA (48), and subsequently, the colonization of AIEC strains promote the secretion of IFN-γ and TNF-α by macrophages, which are likely to stimulate granuloma formation and is a common histological feature of CD (45). Overall, these findings suggest that AIEC strains in CD patients can promote their own colonization and the ensuing inflammatory amplification cycle (47).
Meta regression analysis indicated the association of AIEC with IBD and CD does not significantly confound by age.
Our results suggested that the affect by IBD in patients colonized with the AIEC is not random, it is a specific disease-related pathovar. That is because this pathovar has the ability to invade epithelial cells and attach to receptors of CEACAM with specificity for the oncofetal carbohydrate antigens that are overexpressed by mucosal glycoconjugates in the inflammation condition. AIEC is pertinent in IBD due to the contribution of genetic mutations associated with defects in handling intracellular microbes in the disease pathogenesis and to the intestinal injury. Thus, the mucosal environment of an individual susceptible to IBD could be exploited by these bacteria; otherwise, their proliferation might be an outcome of a normal flora depletion (49).
However, in patients undergoing AIEC colonization, the use of antibiotics may be effective in certain conditions. Antibiotics such as ciprofloxacin and rifaximin are safer alternatives for patients with CD concomitant AIEC because they have fewer side effects than immunosuppressive drugs (50). On the other hand, heptilmenoside derivatives have been shown to have strong antiadhesion effects and in vivo/in vitro protective effect against colitis, which means that they can be useful compounds for the treatment of patients with AIEC colonized patients (51). Moreover, according to previous report, the effect of diet on intestinal homeostasis and AIEC severity suggests that combined dietary use can be used to alter the availability of luminal nutrients, along with drug therapies, to limit AIEC growth and implantation (17).
In the present work, E. coli strains included A, B1, B2, and D phylogroups based on the availability of chuA, yjaA, and TspE4.C2 genes (52). These were described on the basis of their multilocus enzyme electrophoresis patterns (MLEE). Subsequently, they were grouped by DNA-based multilocus sequence typing (MLST), and whole genome sequences confirmed it (53). B2 and D groups were the main parts of extraintestinal pathogenic strains of E. coli. However, A and B1 groups were the lowest pathogenic E. coli strains and are defined as non-human enteropathogenic strains (54).
Researchers have recently compared non-AIEC and AIEC strains of the same phylogroup and identified three genomic regions in all the B2 phylogroup AIEC strains, which are absent in AIEC strains of other phylogroups and commensal strains with any phylogenetic origin (e.g., B2) (12). Nevertheless, it is not known if these regions are only specifically present in B2-AIEC strains or are also available in other pathogenic groups with the same phylogenetic origin, like B2 ExPEC strains (12). Our results implied that most frequent AIEC phylogroup in the IBD, CD, and control groups was B2 and the least frequent phylogroup in the IBD and CD was B1, which was D in the control group. Therefore, it could be concluded that AIEC strains belonging to phylogenetic groups B2 might have the ability to colonize and survive in epithelial cells and macrophages in patients, particularly those with chuA gene.
Conclusions
In summary, this meta analysis revealed that colonization by AIEC is more prevalent in IBD. Our meta-analysis results indicated that there is a significant association between colonization by AIEC with IBD totally and IBD CD type. In addition, the most prevalent AIEC phylogroup among the IBD patients was B2 and the least prevalent one was B1. Meanwhile, the most frequent AIEC phylogroup among the control group was B2 and the least frequent one was D. Our results suggested that the affect by IBD in patients colonized with the AIEC pathovar is not random, it is in fact a specific disease-related pathovar. Based on our findings, AIEC is neither commensal nor a real pathogenic strain, but it is a pathobiont that expands rapidly in the host and can apply special pathogenic influences. Although, our study showed a significant association between colonization by AIEC and IBD, these findings are based on case–control studies therefor the cause-and-effect relationship as well as directional dependency cannot be inferred. Longitudinal prospective studies will provide more reliable evidence about the directional association. However, these findings provide evidence on the importance of this strain in the treatment of IBD patients.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
This study used publicly available data. The studies involving human participants were reviewed and approved at the time of original publications (please see references and data availability statement). Written informed consent was not necessary to obtain for this study.
Author Contributions
HF and AF conceived the study. AF, RK, MH, and PA wrote the study protocol and data analysis plan. RK and MH did the systematic review, requested individual participant data, and did the study quality assessments. AF did the statistical analysis. AF, HF, MH, RK, and PA interpreted the data. MH, RK, HF, and AF wrote the first draft of the manuscript. All authors reviewed and approved the final manuscript and had full access to all the data in the study and final responsibility for the decision to submit for publication.
Funding
This study was supervised by AF and supported in part by a grant from Isfahan University of Medical Sciences [Grant no. 199279, Ethics Code: IR.MUI.RESEARCH.REC.1399.287].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.730243/full#supplementary-material
References
- 1.Wark G, Samocha-Bonet D, Ghaly S, Danta M. The role of diet in the pathogenesis and management of inflammatory bowel disease: a review. Nutrients. (2021) 13:135. 10.3390/nu13010135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park J, Cheon JH. Incidence and Prevalence of Inflammatory Bowel Disease across Asia. Yonsei Med J. (2021) 62:99. 10.3349/ymj.2021.62.2.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ananthakrishnan AN, Kaplan GG, Ng SC. Changing global epidemiology of inflammatory bowel diseases: sustaining health care delivery into the 21st century. Clin Gastroenterol Hepatol. (2020) 18:1252–60. 10.1016/j.cgh.2020.01.028 [DOI] [PubMed] [Google Scholar]
- 4.Caruso R, Lo BC, Núñez G. Host–microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. (2020) 20:411–26. 10.1038/s41577-019-0268-7 [DOI] [PubMed] [Google Scholar]
- 5.Vanderploeg R, Panaccione R, Ghosh S, Rioux K. Influences of intestinal bacteria in human inflammatory bowel disease. Infect Dis Clin. (2010) 24:977–93. 10.1016/j.idc.2010.07.008 [DOI] [PubMed] [Google Scholar]
- 6.Barnich N, Denizot J, Darfeuille-Michaud A. E. coli-mediated gut inflammation in genetically predisposed Crohn's disease patients. Pathol Biol. (2013) 61:e65–9. 10.1016/j.patbio.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 7.Prorok-Hamon M, Friswell MK, Alswied A, Roberts CL, Song F, Flanagan PK, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. (2014) 63:761–70. 10.1136/gutjnl-2013-304739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aygun H, Karamese M, Ozic C, Uyar F. The effects of mucosal media on some pathogenic traits of Crohn's disease-associated Escherichia coli LF82. Future Microbiol. (2018) 13:141–9. 10.2217/fmb-2017-0133 [DOI] [PubMed] [Google Scholar]
- 9.Barrios-Villa E, de la Peña CFM, Lozano-Zaraín P, Cevallos MA, Torres C, Torres AG, et al. Comparative genomics of a subset of adherent/invasive Escherichia coli strains isolated from individuals without inflammatory bowel disease. Genomics. (2020) 112:1813–20. 10.1016/j.ygeno.2019.10.013 [DOI] [PubMed] [Google Scholar]
- 10.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. (2013) 26:822–80. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL, et al. Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm Bowel Dis. (2014) 20:1919–32. 10.1097/MIB.0000000000000183 [DOI] [PubMed] [Google Scholar]
- 12.Desilets M, Deng X, Rao C, Ensminger AW, Krause DO, Sherman PM, et al. Genome-based definition of an inflammatory bowel disease-associated adherent-invasive Escherichia coli pathovar. Inflamm Bowel Dis. (2016) 22:1–12. 10.1097/MIB.0000000000000574 [DOI] [PubMed] [Google Scholar]
- 13.O'Brien EJ, Utrilla J, Palsson BO. Quantification and classification of E. coli proteome utilization and unused protein costs across environments. PLoS Comput Biol. (2016) 12:e1004998. 10.1371/journal.pcbi.1004998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camprubí-Font C, Lopez-Siles M, Ferrer-Guixeras M, Niubó-Carulla L, Abellà-Ametller C, Garcia-Gil LJ, et al. Comparative genomics reveals new single-nucleotide polymorphisms that can assist in identification of adherent-invasive Escherichia coli. Sci Rep. (2018) 8:1–11. 10.1038/s41598-018-20843-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glasser A-L, Boudeau J, Barnich N, Perruchot M-H, Colombel J-F, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect Immun. (2001) 69:5529–37. 10.1128/IAI.69.9.5529-5537.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelhalim KA, Uzel A, Ünal NG. Virulence determinants and genetic diversity of adherent-invasive Escherichia coli (AIEC) strains isolated from patients with Crohn's disease. Microb Pathog. (2020) 145:104233. 10.1016/j.micpath.2020.104233 [DOI] [PubMed] [Google Scholar]
- 17.Chervy M, Barnich N, Denizot J. Adherent-Invasive E. coli: Update on the lifestyle of a troublemaker in Crohn's disease. Int J Mol Sci. (2020) 21:3734. 10.3390/ijms21103734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boudeau J, Glasser A-L, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun. (1999) 67:4499–509. 10.1128/IAI.67.9.4499-4509.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, et al. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics. (2010) 11:667. 10.1186/1471-2164-11-667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viladomiu M, Metz ML, Lima SF, Jin W-B, Chou L, Bank JLC, et al. Adherent-invasive E. E. coli metabolism of propanediol in Crohn's disease regulates phagocytes to drive intestinal inflammation. Cell Host Microbe. (2021) 29:607–19. e608. 10.1016/j.chom.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rath E, Moschetta A, Haller D. Mitochondrial function—gatekeeper of intestinal epithelial cell homeostasis. Nat Rev Gastroenterol Hepatol. (2018) 15:497–516. 10.1038/s41575-018-0021-x [DOI] [PubMed] [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. (1986) 7:177–88. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 24.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. (1997) 315:629–34. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol. (2009) 9:1–7. 10.1186/1471-2180-9-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ormsby MJ, Logan M, Johnson SA, McIntosh A, Fallata G, Papadopoulou R, et al. Inflammation associated ethanolamine facilitates infection by Crohn's disease-linked adherent-invasive Escherichia coli. EBioMedicine. (2019) 43:325–32. 10.1016/j.ebiom.2019.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asiabar AS, Aghdaei HA, Sabokbar A, Zali MR, Feizabadi MM. Investigation of adherent-invasive E. coli in patients with Crohn's disease. Med J Islam Repub Iran. (2018) 32:11. 10.14196/mjiri.32.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberc AM, Fiebig-Comyn AA, Tsai CN, Elhenawy W, Coombes BK. Antibiotics potentiate adherent-invasive E. coli infection and expansion. Inflamm Bowel Dis. (2019) 25:711–21. 10.1093/ibd/izy361 [DOI] [PubMed] [Google Scholar]
- 29.Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, et al. Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. (2013) 9:e1003141. 10.1371/journal.ppat.1003141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conte MP, Longhi C, Marazzato M, Conte AL, Aleandri M, Lepanto MS, et al. Adherent-invasive Escherichia coli (AIEC) in pediatric Crohn's disease patients: phenotypic and genetic pathogenic features. BMC Res Notes. (2014) 7:1–12. 10.1186/1756-0500-7-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. (2004) 127:80–93. 10.1053/j.gastro.2004.03.054 [DOI] [PubMed] [Google Scholar]
- 32.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. (2004) 127:412–21. 10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- 33.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. (2007) 1:403–18. 10.1038/ismej.2007.52 [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. (2009) 15:872–82. 10.1002/ibd.20860 [DOI] [PubMed] [Google Scholar]
- 35.Raso T, Crivellaro S, Chirillo MG, Pais P, Gaia E, Savoia D. Analysis of Escherichia coli isolated from patients affected by Crohn's disease. Curr Microbiol. (2011) 63:131–7. 10.1007/s00284-011-9947-8 [DOI] [PubMed] [Google Scholar]
- 36.Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, et al. Multidrug resistance is common in Escherichia coli associated with ileal Crohn's disease. Inflamm Bowel Dis. (2012) 19:141–50. 10.1002/ibd.22971 [DOI] [PubMed] [Google Scholar]
- 37.Negroni A, Costanzo M, Vitali R, Superti F, Bertuccini L, Tinari A, et al. Characterization of adherent-invasive Escherichia coli isolated from pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. (2012) 18:913–24. 10.1002/ibd.21899 [DOI] [PubMed] [Google Scholar]
- 38.Elliott TR, Hudspith BN, Wu G, Cooley M, Parkes G, Quiñones B, et al. Quantification and characterization of mucosa-associated and intracellular Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. (2013) 19:2326–38. 10.1097/MIB.0b013e3182a38a92 [DOI] [PubMed] [Google Scholar]
- 39.De la Fuente M, Franchi L, Araya D, Díaz-Jiménez D, Olivares M, Álvarez-Lobos M, et al. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int J Med Microbiol. (2014) 304:384–92. 10.1016/j.ijmm.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Céspedes S, Saitz W, Del Canto F, De la Fuente M, Quera R, Hermoso M, et al. Genetic diversity and virulence determinants of Escherichia coli strains isolated from patients with Crohn's disease in Spain and Chile. Front. Microbiol. (2017) 8:639. 10.3389/fmicb.2017.00639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Brien CL, Bringer M-A, Holt KE, Gordon DM, Dubois AL, Barnich N, et al. Comparative genomics of Crohn's disease-associated adherent-invasive Escherichia coli. Gut. (2017) 66:1382–9. 10.1136/gutjnl-2015-311059 [DOI] [PubMed] [Google Scholar]
- 42.Camprubí-Font C, Ewers C, Lopez-Siles M, Martinez-Medina M. Genetic and phenotypic features to screen for putative adherent-invasive Escherichia coli. Front Microbiol. (2019) 10:108. 10.3389/fmicb.2019.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JG, Han DS, Jo SV, Lee AR, Park CH, Eun CS, et al. Characteristics and pathogenic role of adherent-invasive Escherichia coli in inflammatory bowel disease: Potential impact on clinical outcomes. PLoS ONE. (2019) 14:e0216165. 10.1371/journal.pone.0216165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, et al. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. (2009) 206:2179–89. 10.1084/jem.20090741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agus A, Massier S, Darfeuille-Michaud A, Billard E, Barnich N. Understanding host-adherent-invasive Escherichia coli interaction in Crohn's disease: opening up new therapeutic strategies. BioMed Res Int. (2014) 2014:567929. 10.1155/2014/567929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrière J, Darfeuille-Michaud A, Nguyen HTT. Infectious etiopathogenesis of Crohn's disease. World J Gastroenterol. (2014) 20:12102–17. 10.3748/wjg.v20.i34.12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnich N, Carvalho FA, Glasser A-L, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Investig. (2007) 117:1566–74. 10.1172/JCI30504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Low D, Tran HT, Lee IA, Dreux N, Kamba A, Reinecker HC, et al. Chitin-binding domains of Escherichia coli ChiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology. (2013) 145:602–12.e609. 10.1053/j.gastro.2013.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amoroso C, Perillo F, Strati F, Fantini M, Caprioli F, Facciotti F. The role of gut microbiota biomodulators on mucosal immunity and intestinal inflammation. Cells. (2020) 9:1234. 10.3390/cells9051234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. (2018) 67:574–87. 10.1136/gutjnl-2017-314903 [DOI] [PubMed] [Google Scholar]
- 51.Sivignon A, Yan X, Alvarez Dorta D, Bonnet R, Bouckaert J, Fleury E, et al. Development of Heptylmannoside-Based Glycoconjugate Antiadhesive Compounds against Adherent-Invasive Escherichia coli Bacteria Associated with Crohn's Disease. MBio. (2015) 6:e01298–15. 10.1128/mBio.01298-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordon DM, Clermont O, Tolley H, Denamur E. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol. (2008) 10:2484–96. 10.1111/j.1462-2920.2008.01669.x [DOI] [PubMed] [Google Scholar]
- 53.Chaudhuri RR, Henderson IR. The evolution of the Escherichia coli phylogeny. Infect Genet Evol. (2012) 12:214–26. 10.1016/j.meegid.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 54.Wassenaar TM. E. coli and colorectal cancer: complex relationship that deserves a critical mindset. Crit Rev Microbiol. (2018) 44:619–32. 10.1080/1040841X.2018.1481013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.