Abstract
Extracellular vesicles (EVs) are lipid‐bilayer membrane structures secreted by most cell types. EVs act as messengers via the horizontal transfer of lipids, proteins, and nucleic acids, and influence various pathophysiological processes in both parent and recipient cells. Compared to EVs obtained from body fluids or cell culture supernatants, EVs isolated directly from tissues possess a number of advantages, including tissue specificity, accurate reflection of tissue microenvironment, etc., thus, attention should be paid to tissue‐derived EVs (Ti‐EVs). Ti‐EVs are present in the interstitium of tissues and play pivotal roles in intercellular communication. Moreover, Ti‐EVs provide an excellent snapshot of interactions among various cell types with a common histological background. Thus, Ti‐EVs may be used to gain insights into the development and progression of diseases. To date, extensive investigations have focused on the role of body fluid‐derived EVs or cell culture‐derived EVs; however, the number of studies on Ti‐EVs remains insufficient. Herein, we summarize the latest advances in Ti‐EVs for cancers and non‐cancer diseases. We propose the future application of Ti‐EVs in basic research and clinical practice. Workflows for Ti‐EV isolation and characterization between cancers and non‐cancer diseases are reviewed and compared. Moreover, we discuss current issues associated with Ti‐EVs and provide potential directions.
Keywords: cancer, clinical application, exosomes, extracellular vesicles, tissue‐derived extracellular vesicles
1. INTRODUCTION
Extracellular vesicles (EVs), which were firstly introduced in the 1980s (Pan et al., 1985), are heterogeneous membranous structures released from the cells through distinct biogenetic and secretory mechanisms (Fabbiano et al., 2020). In the past decades, EVs have been recognized as “divine messengers,” like Hermes (Braicu et al., 2015), which can mediate the exchange of biomolecular information between cells and their microenvironment (Costa‐Silva et al., 2015), both in prokaryotes and eukaryotes. EVs are produced constitutively or upon activation by inflammatory responses, oxidative stress, hypoxia, mechanical stress, ageing, cell death, and exposure to bacteria (Almeria et al., 2019, Bodega et al., 2019, Helmke & Vietinghoff, 2016, Kakarla et al., 2020, Shah et al., 2018, Totani et al., 2017). Growing evidence has indicated that the contents, size, and membrane components of EVs are highly heterogeneous, depending on the cell type of origin, cell state, and environment (Yáñez‐Mó et al., 2015).
Despite their significant heterogeneity, EVs can be classified into three subtypes: microvesicles (MVs), apoptotic bodies (ABs) or apoptosomes (Kerr et al., 1972), and exosomes (EXOs) based on their size distribution and biogenesis mechanism [endosomal‐ or plasma membrane (PM)‐derived] (Figure 1). MVs are larger vesicular structures shed by direct outward blebbing of the PM with a size of 50–1000 nm (Willms et al., 2018). ABs are released from cells undergoing programmed cell death through outward blebbing and fragmentation of the PM (Akers et al., 2013), with a broad size range of 100–5000 nm in diameter (Mathieu et al., 2019). EV populations unique to a disease, with specific biophysical characteristics and compositions, have also been identified (e.g., oncosomes and large oncosomes [LOs] in cancer). Oncosomes (100–400 nm) and LOs (1–10 μm) are non‐apoptotic EVs originating directly from the PM, which are unique to cancer cells (Matarredona & Pastor, 2019, Willms et al., 2018). EXOs, the smallest subgroup of EVs, are generated via an endosomal pathway that involves double invagination of the PM, the formation of intracellular multivesicular bodies (MVBs) containing intraluminal vesicles, MVBs fusion to the PM, and exocytosis (Kalluri & Lebleu, 2020), with diameters ranging from 30 to 150 nm (Kalra et al., 2016). Pioneering research has shown that exosomes may function as “waste bags” that remove redundant and non‐functional cellular contents (Abels & Breakefield, 2016, Mincheva‐Nilsson et al., 2016, Pan et al., 1985, Pegtel & Gould, 2019, Théry et al., 2002). Recently, studies have demonstrated that exosomes play indispensable roles in multiple biological processes, including angiogenesis, apoptosis, maintenance of cellular homeostasis, antigen presentation, inflammatory responses, and intercellular signal transduction (Huang & Xu, 2021, Li et al., 2020), consequently involved in pathophysiological events in a variety of diseases, such as cancer, autoimmune diseases, and infectious and neurodegenerative diseases (Gurunathan et al., 2019, Mashouri et al., 2019). The characteristics of each EV subtype are summarized in Table 1.
FIGURE 1.

Schematic representation of subtypes of extracellular vesicles (EVs) released by a cell. EVs are secreted by cells into the extracellular space, and can be classified into exosomes (EXOs), microvesicles (MVs) or ectosomes, apoptotic bodies (ABs) and large oncosomes (LOs). Exosomes are secreted by the fusion of MVBs with the plasma membrane (PM), whereas MVs and large oncosomes are released by direct outward budding of the PM. ABs are released by dying cells during the later stages of apoptosis so that cell debris can easily be eliminated by the neighbouring immune cells
TABLE 1.
Characteristics of EV subtypes
| Characteristic | Exosome | Microvesicle | Apoptotic body | Large oncosome | References |
|---|---|---|---|---|---|
| Size | 30–150 nm | 50–1000 nm | 100–5000 nm | 1–10 μm | Kalra et al. (2016); Willms et al. (2018) |
| Density | 1.13–1.19 g/ml | 1.25–1.30 g/ml | 1.16–1.28 g/ml | 1.10–1.15 g/ml | Minciacchi et al. (2017); Zhang et al. (2019) |
| Biogenesis | Multivesicular bodies fusion with plasma membrane | Direct outward budding from the plasma membrane | Direct outward budding from the plasma membrane | Direct outward budding from the plasma membrane | Akers et al. (2013); Tkach and Théry (2016); Willms et al. (2018) |
| Compositions | Nucleic acids (DNA, mRNA, miRs), lipids, proteins | Nucleic acids (DNA, mRNA, miRs), lipids, proteins | Cellular organelles, cytosolic content (DNA, mRNA, miRs), lipids, proteins | Nucleic acids (DNA, mRNA, miRs), lipids, proteins | Doyle and Wang (2019) |
| Markers | CD9, CD63, CD81 | ARF6 and VCAMP3 | Thrombospondin, C3b, annexin V | ARF6, Cav‐1, membrane‐localized cytokeratin‐18 | Akers et al. (2013); Morello et al. (2013); Muralidharan‐Chari et al. (2009); Van Engeland et al. (1998); Willms et al. (2018) |
| Functions | Maintenance of normal physiology, involvement in pathological processes | Cell‐cell communication, tissue homeostasis, tumorigenesis, diagnostics, drug delivery | Removal of dying cells, antigen presentation, antitumor immunity, autoimmunity. propagation | Propagating tumour‐promoting material and inducing transformation | Al‐Nedawi et al. (2008); Caruso and Poon (2018); Ni et al. (2020); Quesenberry et al. (2015); Ratajczak and Ratajczak (2020); Tkach and Théry (2016) |
At present, EVs can be generally classified into three subtypes: cell culture‐derived EVs, body fluid‐derived EVs, and tissue‐derived EVs (Ti‐EVs), based on the sources from which they are obtained. To date, cell culture‐derived EVs and body fluid‐derived EVs have been the focus of interest in the field of EVs. However, a number of limitations are associated with the studies described below.
Notably, caution should be exercised when working with cell culture‐derived EVs (mainly cell line‐derived EVs) which are isolated from cell culture supernatants. Cell culture‐derived EVs are reliable candidates for mechanism study due to the availability of EVs and repeatability of results. However, cultured cells may lose unique features following long‐term cell cultivation, which may lead to poor interpretation of the obtained EVs’ biological functions (Crescitelli et al., 2020, Crescitelli et al., 2021, Sahoo et al., 2021). Additionally, most cells are currently cultured in two‐dimensional (2D) environment with single‐cell type (Jensen & Teng, 2020), thus, cells lose their interactions with other cell populations that coexist in the same in vivo tumour microenvironment (TME) (Chen et al., 2020, Crescitelli et al., 2020, Crescitelli et al., 2021). Moreover, cell lines are obtained from a single individual at a specific time during disease development, therefore cell culture‐derived EVs cannot reflect dynamic progression of diseases.
Compared with cell culture‐derived EVs, growing attention has focused on EVs obtained from various biological fluids, such as blood, breast milk, cerebrospinal fluid, urine, and saliva (Belov et al., 2016, Guo et al., 2020, Leung et al., 2021, Vasconcelos et al., 2019). Body fluid‐derived EVs offer a minimally invasive way to reflect the dynamic progression of diseases in real‐time (Leung et al., 2021). However, admixtures from multiple sources are contained in body fluids, including serum proteins or a mixture of EVs from the whole body (Jingushi et al., 2018). In addition, the extent to which EVs in the circulatory system are released from specific tissues is unknown (Huang & Xu, 2021). One study reported that the location of adipocytes within the interstitial space may block the diffusion of adipocyte‐derived EVs into the circulatory system (Connolly et al., 2018). Low concentrations of EVs derived from the circulatory system have been reported (Shah et al., 2018). Moreover, it is critical to identify the tissue of origin (Shah et al., 2018). Given the limitations above, it is now the time for Ti‐EVs to make their way into history.
Ti‐EVs exist in the extracellular interstitium and are well‐established mediators of intercellular signal transduction (Figure 2). Ti‐EVs have garnered increasing attention due to several favourable features (Table 2): (I) Ti‐EVs comprise vesicles shed by most cell types within the tissue microenvironment, especially the TME, and more accurately reflect the pathophysiological characteristics and behaviours of cells as the three‐dimensional (3D) structure of tissues and cell properties are retained (Camino et al., 2020, Chen et al., 2020, Jang et al., 2019); (II) Ti‐EVs analysis enables comparison of EVs components using EVs obtained from tumour and adjacent non‐tumour tissues, which share common biological properties (Jang et al., 2019, Jingushi et al., 2018); (III) Ti‐EV samples contain minimal contaminants because of the single‐tissue source compared to body fluid‐derived EVs (Crescitelli et al., 2020, Jingushi et al., 2018); (IV) the underlying relevance of intratumoral heterogeneity (ITH) and tumour evolution has not been fully explored (Nguyen et al., 2021). Analysis of Ti‐EVs sampled at multiple sites and different stages of disease evolution may offer insights into temporal‐spatial heterogeneity within the TME. In terms of cancer, Ti‐EVs may be crucial for regulating the TME for tumour cell colonization and growth (Costa‐Silva et al., 2015, Hoshino et al., 2015).
FIGURE 2.

Biogenesis and identification of tissue‐derived EVs (Ti‐EVs). (a) Ti‐EVs are found in tissue interstitium released from various types of cells. (b) Ti‐EVs are released after double invagination and fusion of the plasma membrane with MVBs. Several proteins are implicated in EVs biogenesis and include Rab GTPases, ESCRT proteins, and SNARE family proteins. (c) Ti‐EVs have a complex composition of nucleic acids, protein, lipids, and metabolites. Some proteins are also used as markers for EVs (CD9, CD63, CD81, flotillin, TSG101, and ALIX). EVs surface proteins include cell adhesion proteins, integrins, cell‐type‐specific proteins, and so on
TABLE 2.
Comparison of characteristics of EVs from different sources
| Source of EVs | Tissue‐derived EVs | Body fluid‐derived EVs | Cell culture‐derived EVs |
|---|---|---|---|
| Advantages | Reflect pathophysiologic state accurately (Chen et al., 2020, Camino et al., 2020) | Real‐time and dynamic detection (Leung et al., 2021, Erdbrügger et al., 2021) | EVs can be collected repeatedly and are not subject to fresh sampling |
| Analyzing EVs from diseased tissues and normal tissues possessing a common histological background (Jingushi et al., 2018, Jang et al., 2019) | Minimal‐invasiveness (Leung et al., 2021, Erdbrügger et al., 2021) | Single‐cell source | |
| Relative minor contaminants with single‐tissue source (Crescitelli et al., 2020) | Large volumes (Erdbrügger et al., 2021) | Cells can be immortalized (Sahoo et al., 2021) | |
| Parental cells are grown in a three‐dimensional environment | Multi‐sources (Kaczor‐Urbanowicz et al., 2019, Siravegna et al., 2017) | ||
| Allow the analysis of temporal‐spatial heterogeneity of tissue microenvironment | |||
| Disadvantages | Mixture of EVs from multiple cell populations (Mincheva‐Nilsson et al., 2016, Jang et al., 2019) | Lower sensitivity and specificity in biomarkers identification | Cell are living in two‐dimensional environment (Jensen & Teng, 2020) |
| Invasiveness (Leung et al., 2021) | EVs admixture of other cellular and organ origin from various biological states (Crescitelli et al., 2020, Leung et al., 2021) | Loss of the interaction with the surrounding cells (Chen et al., 2020, Crescitelli et al., 2020, Crescitelli et al., 2021) | |
| Inability to reflect the dynamic progression of disease | |||
| Discrete tissue biopsies and limited sources for sampling sources (Leung et al., 2021, Jang et al., 2019, Siravegna et al., 2017) | Low concentrations of EVs in the circulatory system (Connolly et al., 2018, Shah et al., 2018) | Less representative of disease characteristics after long‐term cell culture (Domcke et al., 2013, Jang et al., 2019) | |
| Sampling bias (Siravegna et al., 2017) | Need to identify the original tissues (Shah et al., 2018, Kaczor‐Urbanowicz et al., 2019) | ||
| Applications | Mechanism study (Cheng et al., 2020) | Mechanism study (Hoshino et al., 2015) | Mechanism study (Tong et al., 2020) |
| Potential diagnostics (Hoshino et al., 2020) | Diagnostics (Sheridan, 2016, Mckiernan et al., 2018) | ||
| Potential therapeutics (Zhou et al., 2020) | Therapeutics (Usman et al., 2018) | Therapeutics (Williamson, 2018) | |
| Potential prognostics | Prognostics (Zhou et al., 2020) |
This review aims to summarize and discuss the status quo and progress of research into Ti‐EVs in cancers and non‐cancer diseases. Problems and potential solutions in the field of Ti‐EVs are proposed. Furthermore, we discuss the future directions of Ti‐EVs for clinical applications. We hope that this review will provide a comprehensive outlook on Ti‐EVs as well as information for future work to elucidate their functions for diagnostic, prognostic and therapeutic applications in a spectrum of pathological conditions.
2. TI‐EVS IN MULTIPLE CANCERS
In the past few decades, there have been remarkable developments in the study of EVs in cancers(Kalluri, 2016, Rajagopal & Harikumar, 2018). EVs are involved in all of the hallmarks of cancer (Hanahan & Weinberg, 2011, Jurj et al., 2020) (Figure 3). Tumour‐derived EVs are associated with the biogenesis and evolution of various cancer events, including TME reshaping, angiogenesis, local invasion, distant metastasis, the establishment of a pre‐metastatic niche, and the development of drug resistance (Mashouri et al., 2019, Wortzel et al., 2019). However, the exploration of Ti‐EVs in cancer development and progression has remained at a preliminary stage. In this section, Ti‐EVs in several types of cancer, including melanoma, colorectal cancer, pancreatic cancer, lung cancer, and renal cell carcinoma (Figure 4), are summarized and discussed (Table 3).
FIGURE 3.

A schematic representation of the impact of tumour‐derived EVs on the hallmarks of cancer. Ten hallmark capabilities of cancer are as follows: sustaining proliferative signalling, evading growth suppressors, evading immune destruction, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, tumour‐promoting inflammation, genome instability, and mutation, resisting cell death, and deregulating cellular energetics. Tumour‐derived EVs can contribute to all the hallmarks of cancer by transporting pro‐oncogenic molecules to recipient cells
FIGURE 4.

Schematic of tissue types involved in the research of tissue‐derived EVs (Ti‐EVs). The left part presents some types of tissue involved in non‐cancer diseases, including brain tissue (mainly Alzheimer's disease), liver tissue for a standard protocol for Ti‐EV isolation, intestinal tissue, and adipose tissue for some metabolic diseases. And the other part presents some tissues associated with cancers, including melanoma, lung cancer, pancreatic cancer, colorectal cancer, and renal cell carcinoma
TABLE 3.
Summary of key studies on Ti‐EVs
| Tissue types | Collection and pre‐processing | EV characterization | Method of analysis | Key findings | Citation | ||
|---|---|---|---|---|---|---|---|
| Pre‐EV isolation | EV‐isolation | Methods | Markers | ||||
| Freshly dissected animal or human tissue | Conditioned medium was centrifuged at 2500g for 30 min; 10,000g for 35 min. | Filter pooled supernatant through a bottle‐top or syringe‐type 0.2 μm filter; 110,000g for 2h. | NTA, WB, TEM, IEM, fluorescence staining, flow cytometry | CD9, CD63, CD8, ESCRT proteins (Alix, TSG101) | – | Separation workflow for EVs purification, characterization of components and functions. | Mincheva‐Nilsson et al. (2016) |
| Human metastatic melanoma tissue | 70 μm filter; 300g for 10 min; 2000g for 20 min; 16,500g for 20 min. | 118,000g for 2.5 h. | TEM, NTA, WB | CD9, CD63, CD81, calnexin, flotillin‐1 | NanoLC‐MS/MS analysis | A method to isolate EV subtypes; ADAM10 and mitofilin are exclusively enriched in small LD EVs and large EVs, respectively. | Crescitelli et al. (2020) |
| Melanoma tissue | 300g for 10 min; 2000g for 20 min; 16,500g for 20 min. | 118,000g for 2.5 h; iodixanol gradient 186,000g for 16 h. | TEM, WB, ExoView analysis | CD63, CD81, CD9, flotillin‐1, Calnexin | RNA profile analysis, ExoView, proteomic analysis | A protocol was provided for the isolation and characterization of Ti‐EVs from human melanoma and other potential cancer and non‐cancer tissues. | Crescitelli et al. (2021) |
| Human fresh intestine | 70‐μm strainer; 1000g for 10 min; 2000g for 20 min; 5000g for 30 min; 15,000g for 1 h; 0.22‐μm PVDF filter. | 120,000g for 130 min; Odixanol density gradient centrifugation at 288,000g for 5 h; 100,000g for 70 min. | NTA, TEM, WB | CD9, CD63, CD81, Alix, Tsg101 | RT‐qPCR | A practical method to isolate intestinal Ti‐EVs suitable for investigating the mechanism of intestinal I/R injury. | Chen et al. (2020) |
| Clear cell renal cell carcinoma tissues and adjacent normal renal tissues | Tissue‐immersed medium was centrifuged at 2000g for 30 min; 16,000g for 30 min. | 100,000g for 90 min (×2). | WB, TEM, NTA | CD9, CD63, CD81 | Quantitative LC/MS analysis | 3871 Te‐EV proteins identified; AZU1 over‐expressed in tumour‐derived Te‐EVs. | Jingushi et al. (2018) |
| Melanoma metastatic tissue | 70 μm pore size filter; 300g for 10 min; 2000g for 20 min. | 16,500g for 20 min; 110,000g for 2.5 h to collect larger and smaller vesicles, respectively. | TEM, WB, ELISA, particle measurement | CD9, CD81 | MS analysis, RNA detection | Melanoma Ti‐EVs are rich in mitochondrial membrane proteins, which are found in plasma. | Jang et al. (2019) |
| Post‐mortem brain tissue | Shaking water bath at 37°C for 20 min; 300g for 5 min; 2000g for 10 min; 10,000g for 5 min. | Triple sucrose cushion; 180,000g (average) for 3 h. | TEM, WB, IB | Calnexin | Small RNA sequencing | Disease‐associated miRNA is upregulated in brain Ti‐EVs; profiling the repertoire of miRNAs enriched in AD brain Ti‐EVs. | Cheng et al. (2020) |
| Millimetre‐sized fresh tumour and adjacent tissue | 500g for 10 min; 3000g for 20 min; 12,000g for 20 min. | 100,000g for 70 min 1 ml sucrose density cushion ultracentrifugation at 100,000g for 70 min | WB, TEM, NTA, | CD9, CD81, TSG101 | MS‐based proteomic profiling | Novel pan‐EVP markers (ACTB, MSN, RAP1B), tumour‐type‐specific EVP proteins identified. | Hoshino et al. (2020) |
| Mouse melanoma tissue | 500g for 4 min; 500g for 10 min (×2); 2000g for 15 min (×2). | 16.5K EVs: 16,500g for 24 min (×2); 100K EVs: 100,000g for 70 min (×2). | TEM, WB | Calnexin, cytochrome‐C, GM130 | LC‐MS/MS, transcriptomics analysis. | Cells from two types of melanoma phenocopy migratory behaviour through EV exchange. | Steenbeek et al. (2018) |
| Colorectal cancers and adjacent normal mucosa | 60 mm then a 40 mm mesh filter; 400g for 10 min; 2000g for 20 min; 15,000g for 40 min; 0.22 μm PES filter. | 120,000g for 4 h (×2); high‐resolution iodixanol density gradient. | IB | CD63, CD81, Syntenin‐1, Calreticulin | MS, RNA sequencing, DNA analysis, direct immunoaffinity Capture | CD9, CD63, CD81, and Annexin V are absent in non‐classical and classical exosomes, respectively; Annexin A1 and A2 are novel markers of non‐exosomal EVs. | Jeppesen et al. (2019) |
| Mouse colorectal tissue | 70 μm cell strainer; 500g for 5 min; 2000g for 10 min; 10,000g for 30 min. | 110,000g for 70 min; 134,000g for 70 min. | TEM, NTA, WB, Flow cytometry | CD9, CD81, ALIX | LC‐MS/MS, miRNA profiling, Lipidome analysis | TAM‐EVs boost inflammation and anti‐tumour immunity and support a more favourable prognosis. | Cianciaruso et al. (2019) |
| Whole brain tissues from WT and AD mice | 500g for 5 min; 2000g for 10 min; 10,000g for 30 min; 0.45 μm filter. | 100,000g for 2 h; 1.5 ml of 0.25 M sucrose buffer for gradient purification; floatation density gradient. | WB | CD63, CD81, Alix, TSG101, HSC70, Rab8a | MS, enrichment analysis | A replicable method for Ti‐EVs isolation introduced for downstream analysis and potential clinical applications. | Hurwitz et al. (2018) |
| Human brain tissues | 40 μm mesh filter; 300g for 10 min; 2000g for 10 min; 10,000g for 10 min; 0.22 μm mesh filter. | 100,000g for 70 min; 0.475 M of sucrose solution. | NTA, TEM | – | Label‐free Nano‐LC‐MS/MS analysis | Significant upregulation of Tau and Aβ1‐42; 1088 unique proteins from brain Ti‐EVs were exosomes molecules. | Muraoka et al. (2020) |
| Human brain tissue | 300g for 10 min; 1200g for 10 min (×2); 0.2 um syringe filter; 10,000g for 30 min (×2); All centrifugations were performed at 2℃. | 22,000g for 22 h. All centrifugations were performed at 2℃. | WB, TEM, Immunogold Labeling, | CD63, GAPDH, flotillin‐2 | MiRNA expression analysis, qPCR analysis | Significantly enhanced expressions of miR‐497 in SZ samples and miR‐29c in BD samples. | Banigan et al. (2013) |
| Rhesus macaque brain tissue | 40 μm mesh filter; 5 μm filter; 0.2 μm syringe filter; 300g for 10 min; 2000g for 10 min; 10,000g for 30 min. | 100,000g for 60 min (×2); 2 ml of 0.95 M sucrose solution and inserted inside a sucrose step gradient column; 200,000g for 16 h. | TEM, WB | CD9, CD63, CD81, HSP70, flotillin, TSG101 | Small RNA sequencing, qRT‐PCR | Neurotoxicity triggered by EV‐miR‐21 was not influenced by apoptosis inhibition but restricted by necrostatin‐1. | Yelamanchili et al. (2015) |
| Mouse brain tissue | 40 μm mesh filter; 0.2‐μm syringe filter; 300g for 10 min; 2000g for 10 min; 10,000g for 30 min. | 100,000g for 70 min (×2); 2 ml of 0.95 M sucrose solution centrifuged at 200,000g for 16 h. | Immunoelectron microscopy | TSG101 | Gene expression analysis, image analysis | Microglia spread tau via EVs secretion, thus contributing to the progression of tauopathy. | Asai et al. (2015) |
| Human brain tissue | 300g for 15 min; 2000g for 15 min; 0.22 μm filter; 10,000g for 30 min. | Sucrose density gradient ultracentrifugation (SDGU); 110,000g (average) for 70 min/ultrafiltration through a 10 kDa MWCO protein concentrator. | TEM, NTA, WB, NanoFCM flow analysis | CD9, CD63, CD81, Rab27, TSG101, Syntenin, Calnexin, GM130 | Small RNA sequencing, MS | Brain Ti‐EVs from three species were collected under different parameters; highlighting the importance of species specificity and technology. | Huang et al. (2020) |
| Human adipose tissue | Adipose tissue‐conditioned media was centrifuged at 800g for 10 min; 2000g for 10 min; 12,000g for 10 min; 0.22 mm sterile filter. | 100,000g for 2 h. | Fluorescence NTA, WB, EM | CD9, CD63, TSG101, Grp94 (a negative control) | MS analysis, Ingenuity pathway analysis. | Adipose Ti‐EVs mediate changes in placental functions in GDM and are involved in some pregnancy complications. | Jayabalan et al. (2019) |
| Adipose tissue | Minced tissues were cultured in conditioned α‐MEM under 100r/min for 2 days; centrifuged at 2000 × rpm for 10 min; 5000g for 30 min. | 5000g for 30 min; concentrated ATE was mixed with 0.5 volume of Total Exosome Isolation TM reagent; incubated overnight at 4°C; 10,000g for 1 h. | WB, TEM, Zetasizer Nano ZS | CD9, CD63, ALIX, TSG101 | MS analysis, Bioinformatic analysis, Real‐time PCR | 328 adipokines were identified in AT‐sEV; changes in NPM3, STEAP3, and DAD1 observed during adipogenesis. | Zhang et al. (2020) |
| Human obese white adipose tissue explant | Tissue explant was incubated in conditioned medium; 1800 rpm for 5 min; 0.22 μm filter; 10,000g for 20 min. | 100,000g for 90 min (×2). | TEM, NTA, IB | CD9, CD63, CD81, negative control GRP94 | MS DDA qualitative analysis | Obese AT release functional EVs carrying AT and obesity‐specific biomarkers depending on the original AT. | Camino et al. (2021) |
| Mouse adipose tissue | 200g for 10 min; 500 × gma× for 10 min (×2); 2000 × gma× for 15 min; at 10,000 × gma× for 30 min. | 70,000 × gma× for 60 min; 5 ml of 2.6 M sucrose at 270,000 × gma× for 16 h; Gradient fractions were centrifugated at 70,000 × gma× for 1 h. | – | – | Fluorescence activated cell sorter (FACS) analysis | Adipose Ti‐ELVs mediate crosstalk between AT and macrophages; ObELV induced TNF‐α and IL‐6 activation and insulin resistance require the TLR4/TRIF pathway. | Deng et al. (2009) |
| Mouse adipose tissue | 100 μm cell strainer; 600g for 5 min at room temperature; 1200g for 15 min; 10, 000g for 15 min; 0.22 μm syringe‐ driven filter. | 100,000g for 1.5 h (×2). | NTA, TEM, WB | CD63, Alix, TSG101 | Proteomics analysis, LC‐MS lipidomics analysis. | Adipose Ti‐EVs are involved in the response to changes in systemic nutrient conditions. | Crewe et al. (2018) |
| Mice adipose tissue | 1000g for 5 min at room temperature and re‐dissolved in medium; incubated for 30 min at 37°C, 5% CO2. | Medium exchange, tissues were incubated for 2 h at 37°C, 5% CO2. The supernatant was used for EVs isolation according to the manufacturer's instruction. | WB, TEM | CD63, Hsp70, CytoC, α‐Tubulin | MiRNA profiling | Serum concentrations of exosomal miR‐92a negatively correlate with human BAT activity. | Chen et al. (2016) |
| Mouse liver tissue | Cannulation and perfusion of tissue (0.5 h); 70 μm nylon cell strainers; 50g for 10 min; 300g for 10 min; 2000g for 20 min; 10,000g for 70 min. | 100,000g for 70 min (×2). | NTA, tunable resistive pulse sensing | – | – | An optimum and replicable procedure for the isolation of hepatic Ti‐EVs. | Ishiguro et al. (2019) |
Note: Unless stated, all centrifugations were performed at 4℃.
Abbreviations: WB, western blot; TEM, transmission electron microscopy; DLS, dynamic light scattering; IB, immunoblotting; LC‐MS/MS, liquid chromatography–mass spectrometry; NTA, nanoparticle tracking analysis; EVP, extracellular vesicle and particle; MS, mass spectrometry; EM, electron microscopy; LD, low density; AD, Alzheimer's disease; Ti‐EVs, tissue‐derived extracellular vesicles; Te‐EVs, tissue‐exudative EV; IEM, immunoelectron microscopy; sEV, small extracellular vesicles; ELISA, enzyme‐linked immunosorbent assay; TAM‐EVs, macrophage‐derived extracellular vesicles; SZ, schizophrenia; BD, bipolar disorder; qPCR, quantitative polymerase chain reaction; qRT‐PCR, quantitative real‐time polymerase chain reaction; SDGU, sucrose density gradient ultracentrifugation; GDM, gestational diabetes mellitus; AT, adipose tissue; ATE, adipose tissue extract; Ti‐ELVs, tissue exosome‐like vesicles; ObELV, obese model tissue exosome‐like vesicles; I/R injury, ischemia/reperfusion injury.
2.1. Ti‐EVs in melanoma
Melanoma is a malignancy that develops from the uncontrolled proliferation of melanocytes. Melanoma is a highly invasive malignant tumour with a high rate of metastasis and death, and prevalence has been growing (Chen et al., 2021). The development of melanoma has been attributed to multiple factors, with ultraviolet light recognized as one of the most common risk factors, as well as gene mutations (Ferguson, 2021, Lopes et al., 2021). According to Cancer Statistics, 2020, the death of melanoma accounts for over 50% of skin tumours (excluding basal and squamous) in the United States (Siegel et al., 2020). Accumulating data have shown that melanoma‐derived EVs play a multifaceted role in driving metastasis by regulating the invasion and angiogenesis of tumour cells (Gowda et al., 2020). To date, the most well‐characterized mechanisms in the formation and progression of melanoma are the (PI3K)/AKT and RAS/RAF/MEK/ERK signalling pathways (Yajima et al., 2012). Additionally, proteins such as VLA‐4, CD44, MHC I, TYRP2, Mart‐1, Her2/neu, TRP, ADAM10, MAPK4K, Annexin A2, and GP100 are overexpressed in melanoma EVs and may have potential as prognostic indicators (Mears et al., 2004, Rappa et al., 2013, Wolfers et al., 2001). Nevertheless, the function of Ti‐EVs in melanoma is yet to be defined.
A well‐developed protocol is presented in Figure 5a (Crescitelli et al., 2021). Extracellular vesicles and particles (EVPs) from melanoma primary tumour tissue and draining lymph nodes invaded by the tumour were analyzed. Researchers identified a list of tumour‐specific proteins in tissue explants (TEs) with the highest predictive values for distinguishing malignancies from normal tissues, including TNC and SRRT, which can be applied in diagnostics (Hoshino et al., 2020). Ti‐EVs from metastatic melanoma can provide more valuable information for the discovery of specific markers to separate EV subpopulations. For example, ADAM10 and EHD4 are enriched in small low‐density EVs (Crescitelli et al., 2020). Steenbeek et al. isolated cells and EVs from mouse melanoma tissue and characterized the cargo of EVs released by two types of melanoma tumours with different metastatic capacities. Different transfer efficacy was found between Ti‐EVs and cell culture‐derived EVs, emphasizing the significance of studying the contents of Ti‐EVs (Steenbeek et al., 2018). Jang et al. analyzed the surface proteomes of EVs from metastatic melanoma tissues and found that mitochondrial inner membrane proteins were enriched in melanoma Ti‐EVs, compared to non‐melanoma Ti‐EVs. Differences were identified between Ti‐EVs and cell culture‐derived EVs (Jang et al., 2019). Ti‐EVs are found to be better tools for biomarker discovery because of their high specificity and relative concentration. Investigation of Ti‐EVs in the evolution of melanoma and their potential diagnostic application may further knowledge on melanoma and help to combat the disease.
FIGURE 5.

Schematic overview of the protocol used to isolate extracellular vesicles (EVs) from multiple types of cancer tissue
2.2. Ti‐EVs in colorectal cancer
Although surgery, chemotherapy, radiotherapy, and immunotherapy have demonstrated clinical efficacy in the treatment of colorectal cancer (CRC), CRC still ranks third in terms of incidence and is the second leading cause of cancer‐related deaths worldwide (Bray et al., 2018). Growing attention has been paid to the role of EVs from cell culture supernatants or the circulatory system in CRC pathological processes (Hu et al., 2019, Takano et al., 2017, Yang et al., 2021). However, exploration of Ti‐EVs may have greater potential in the fight against CRC.
Ti‐EVs from CRC were obtained using the method shown in Figure 5b. HIST2H2AB and PYGM were found to be enriched in Ti‐EVs from patients with CRC. Ti‐EVs profiling could allow a diagnosis resulting in more targeted and efficient therapeutic strategies for patients with tumours of unknown primary focus (Hoshino et al., 2020). Using high‐resolution density gradient fractionation and direct immunoaffinity capture, Jeppesen et al. isolated EVs from colorectal tumour tissues and adjacent tissues, and identified Annexin A1 and A2 as novel markers of non‐exosomal small to large EVs. Future work will be required to ascertain the mechanisms by which heterogeneous components are released under pathological states and in the design of targeted treatment plans (Jeppesen et al., 2019). Additionally, comparative analyses of the proteomic and lipidomic profiles of EVs secreted from MC38 colorectal tumours in mice identified a panel of proteins involved in immune response, migration, metabolism, cell adhesion, and signal transduction (Cianciaruso et al., 2019). This information will be of utmost importance for the design of strategic treatment interventions to combat colorectal cancer in the near future.
2.3. Ti‐EVs in pancreatic cancer
The occurrence and mortality rate of pancreatic cancers have continued to increase, despite reductions in morbidity and mortality associated with other cancers (Vincent et al., 2011). In 2012, approximately 338,000 people were diagnosed with pancreatic cancer, making it the 11th most frequent cancer (Ilic & Ilic, 2016). Pancreatic cancers are highly malignant, easily metastatic, and immunotolerant, largely owing to the high proportion of diagnoses at advanced stages of cancer. It has been reported that 80%–85% of patients with pancreatic cancer present with terminal unresectable tumours. The metastasis and immunological tolerance of pancreatic cancers arise through communication among different types of cells. EVs are the most critical intercellular messengers (Zhao et al., 2017).
A study demonstrated that during the metastasis of pancreatic ductal adenocarcinomas (PDACs), premetastatic niche formation in the liver was highly dependent on the migration inhibitory factor (MIF) from tumour‐derived EVs. These findings suggested that EV‐MIF prepared the liver for tumour metastasis and may be a prognostic indicator for the progression of PDAC and the development of liver metastases (Costa‐Silva et al., 2015). Hoshino et al. analyzed EVP proteins obtained from the primary tumours or regional lymph nodes of patients with pancreatic cancer (Figure 5c). Proteins (e.g., THBS2, VCAN) expressed at significantly higher levels as well as those (e.g., ECM‐associated molecules, ITGs, and some proteases) exclusively expressed in pancreatic tumour tissue (TT)‐derived EVPs (PaCa EVPs) were identified, which may reflect the extent of tumour stromal invasion. Proteins associated with epithelial‐mesenchymal transformation, coagulation, and actin signalling were upregulated in PaCa EVPs compared to adjacent and distant tissue‐derived EVPs. These EVP proteins may be utilized for the detection of cancer and the determination of cancer type (Hoshino et al., 2020).
2.4. Ti‐EVs in lung cancer
As one of the most common and fatal cancers, lung cancer has a high incidence and mortality, and non‐small cell lung cancer accounts for approximately 85% of all lung cancers worldwide. Lung cancer is often diagnosed at an advanced stage when the metastatic foci have formed (Srivastava et al., 2018) owing to a poor understanding of the molecular mechanisms that drive metastasis and progression; thus, efficient diagnostic and therapeutic strategies are currently lacking (Zhang et al., 2020). To date, attention has focused on the essential roles of EVs isolated from various biological fluids and cell culture medium (Gao et al., 2018, Li et al., 2021, Liu et al., 2017, Zhang et al., 2020), making it difficult to compare EVs secreted from tumour cells to those from non‐tumour cells with common histology. In contrast, Ti‐EVs may be suitable for providing overall insights into the functions of EVs under both physical and pathological states.
Analyses of EVP proteomics in tumour and adjacent non‐tumour tissues from patients with lung cancer undergoing surgery (Figure 5b) revealed the presence of cancer‐specific EVP proteins. In addition, HIST family proteins (such as HIST2H2AB) and METTL1 were overexpressed and exclusively expressed in lung tumour tissues, respectively. Of note, proteins related to cell division, metabolism, and RNA processing events were enriched in lung cancer EVPs. These biomarkers are available in the majority of biofluids, and may therefore be used in liquid biopsies (Hoshino et al., 2020).
2.5. Ti‐EVs in renal cell carcinoma
Renal cell carcinoma (RCC) accounts for over 90% of renal malignancies, with 338,000 newly diagnosed patients every year globally. RCC is the second most frequent urogenital malignant tumour in men and the third most common in women (Barth et al., 2020, Ferlay et al., 2015, Siegel et al., 2019, Znaor et al., 2015). The morbidity of patents with RCC is increasing, and the overall prognosis remains unsatisfactory (Wang et al., 2019). The progression of RCC involves many cell types, and EVs are critical messengers that facilitate the exchange of information within the TME (Grange et al., 2019).
EVs were extracted from viable surgical specimens of tumour tissues and adjacent normal tissues from patients with clear cell renal cell carcinoma (ccRCC) (Figure 5d). Overall, 3871 tissue‐exudative EVs (Te‐EVs) proteins were identified by quantitative LC/MS analysis, among which AZU1 was found to be highly expressed, neither stage‐ nor grade‐dependent changes was found. The Jonckheere‐Terpstra test indicated that stage‐dependent upregulation of AZU1 was found in Te‐EV samples, but not in serum samples. It seems reasonable to speculate that the Te‐EV cargoes may have potential for investigations of EV functions and liquid biopsy (Jingushi et al., 2018).
3. TI‐EVS IN NON‐CANCER DISEASES
EVs are actively involved in the formation and progression of multiple cancers, and also function in the development of non‐cancer diseases, such as neurodegenerative disease, metabolic diseases, and intestinal ischemia/reperfusion injury (Figure 4). They have been found to transfer metabolites, facilitate intercellular communication, and influence the formation of aggregates of abnormally folded or unfolded proteins (Hurwitz et al., 2018, Levy, 2017, Matarredona & Pastor, 2019, Muraoka et al., 2020, Soria et al., 2017, Zhang et al., 2021). Recently, Ti‐EVs have attracted significant interest in studies on various diseases owing to their favourable properties. Herein, we focus on researches on Ti‐EVs in several non‐cancer diseases; these are summarized in Table 3.
3.1. Ti‐EVs in Alzheimer's disease
Alzheimer's disease (AD) is characterized by a decline in memory and cognitive function. In terms of incidence, it ranks first in neurodegenerative diseases, accounting for 60%–80% of dementia cases (Zhang et al., 2021), comprising most of the 50 million patients with dementia worldwide (Hurwitz et al., 2018). At present, there is no therapeutic strategy for this increasingly common disease, and current treatment plans achieve minimal effects (Levy, 2017, Soria et al., 2017). The main barrier for the diagnosis and treatment of AD is a lack of overall understanding on its pathogenesis and pathophysiology (Hurwitz et al., 2018). EVs are being increasingly considered as pivotal mediators in intercellular and cell‐microenvironment communication (Zhang et al., 2021). To date, efforts have been made to isolate brain Ti‐EVs from patients with AD (Figure 6a). Mounting evidence suggests that the characteristics of EV proteins and miRNAs make them potential diagnostic indicators for AD.
FIGURE 6.

Schematic overview of the protocol used to isolate extracellular vesicles (EVs) from multiple non‐cancer tissues
Quantitative proteomics analyses of brain Ti‐EVs from AD patients identified GPM6A, ANXA5, ACTZ, and VGF as candidate molecules for distinguishing AD patients from controls and for monitoring the progression of AD (Muraoka et al., 2020). Proteomic analyses have also identified proteins involved in neurodegenerative diseases, including tau, α‐synuclein, and SOD‐1 (Vella et al., 2017).
Recently, miRNAs carried in EVs were shown to be stable in biofluids and could be detected by high‐throughput techniques, such as microarray and RNA sequencing. In the AD brain, EVs loaded with miRNAs crossed the blood‐brain barrier and were secreted into the cerebrospinal fluid and blood circulations (Denk et al., 2015, Kumar & Reddy, 2016, Satoh et al., 2015). Some researchers have shifted their focus to miRNAs in brain Ti‐EVs. Cheng et al. reported that brain Ti‐EVs from patients with AD were enriched with disease‐associated miRNAs, and profiled the repertoire of miRNAs. This study was the first to interrogate the small RNA contents of brain Ti‐EVs and explore how they could be used to understand the early pathological changes in AD, for the development of an early diagnostic blood test (Cheng et al., 2020). Research has highlighted differences and similarities in miRNA expression from schizophrenia (SZ) and bipolar disorder (BD) tissue samples compared to control samples. RT‐PCR analysis revealed significantly enhanced expression of miR‐497 in SZ tissues and miR‐29c in BD tissues. These results suggested that Ti‐EV‐derived miRNAs are potential biomarkers of SZ and BD (Banigan et al., 2013). Profiling of miRNAs in brain Ti‐EVs showed that miR‐21‐induced neurotoxicity from EVs was not influenced by apoptosis inhibitors, but weakened by necrostatin‐1, a necroptosis inhibitor, revealing the function of this pathway in central nervous system diseases (Yelamanchili et al., 2015).
Tau, a microtubule‐associated protein, plays an essential role in AD. Asai et al. demonstrated that microglial cells and EVs were involved in the spread of tau pathology in the brain, and suppression of EVs production inhibited tau transfer both in vivo and in vitro. Evidence has shown that inhibiting EVs secretion could be a possible therapeutic target for AD (Asai et al., 2015). Muraoka et al. reported that tau and Aβ1‐42 were significantly overexpressed in brain Ti‐EVs from AD (Muraoka et al., 2020). The putative role of Ti‐EV‐tau in AD will allow the development of targeted strategies against disease progression.
3.2. Ti‐EVs in metabolic diseases
To date, multiple studies have reported different protocols to isolate and characterize adipose Ti‐EVs (Figure 6b) (Jayabalan et al., 2019) that participate in the development and progression of obesity and insulin resistance (Camino et al., 2020, Camino et al., 2021, Crewe et al., 2018, Deng et al., 2009, Thomou et al., 2017, Zhang et al., 2020), and in some obesity‐related metabolic diseases (Jayabalan et al., 2019, Koeck et al., 2014, Li et al., 2020). Adipose Ti‐EVs are critical for changes in placental function in gestational diabetes mellitus and might be involved in pregnancy complications, such as fetal overgrowth (Jayabalan et al., 2019). A complementary proteomics analysis of adipose Ti‐EVs were conducted (Camino et al., 2021, Crewe et al., 2018, Zhang et al., 2020). Proteomic analysis revealed that the expression of NPM3 and DAD1 in adipose Ti‐EVs was greatly downregulated in obese populations compared to controls. One study highlighted the function of adipokines in the regulation of biological processes within adipose tissue (Zhang et al., 2020). Neighbouring endothelial cells can transport EVs containing Cav‐1 to adipocytes, and this is influenced by fasting/refeeding and obesity, suggesting that EVs are involved in tissue responses to changes in nutritional state. Further efforts are warranted to define Ti‐EV biology and develop novel therapies against diabetes (Crewe et al., 2018). Notably, evidence has demonstrated that adipose Ti‐EVs contain more proteins than adipocyte‐conditioned culture medium, revealing the complexity of Ti‐EV profiles owing to the diversity of cell types and the vigorous interaction within the tissue microenvironment (Zhang et al., 2020). Study described striking differences in protein profile between vesicles secreted by human visceral and subcutaneous adipose TEs, paralleling the biological and endocrine behaviour of both tissues and offering more complete physiological data reflective of the real microworld (Camino et al., 2020). Research has increased the ability to decode the intricacies of intercellular cross‐talk in adipose tissue. Exploration of the Ti‐EV pathophysiology is opening new avenues to the underlying molecular mechanisms and identifying novel targets for the prevention, early diagnosis, and therapeutic management of metabolic disorders.
3.3. Ti‐EVs in intestinal ischemia/reperfusion injury
Many studies have investigated ischemia/reperfusion (I/R) injury in cardiac, cerebral (Liao et al., 2020), liver transplantation (Ye et al., 2020), as well as intestinal injury and transplantation. Among these tissues, the intestine is the most sensitive to I/R injury. Of the Toll‐like receptors (TLR), TLR2, TLR4, and myeloid differentiation primary response 88 (MyD88) are related to intestinal I/R (Nadatani et al., 2018). A rigorous workflow was recently developed to harvest small intestine Ti‐EVs from a mouse model of intestinal I/R (Figure 6c) and the expression of miRNAs of interest in these EVs of gut origin was assessed after intestinal I/R, which help the mechanism investigations (Chen et al., 2020).
4. POTENTIAL CLINICAL APPLICATION OF TI‐EVS
Over the past decades, EVs (mainly exosomes) have been characterized as emerging agents with potential clinical use owing to their ability to transfer bioactive molecules among various cells, and as ideal engineered nanocarriers for targeted drug delivery. As a subtype of EVs, Ti‐EVs have favourable characteristics as diagnostic markers, prognostic indicators, and therapeutic agents.
4.1. Ti‐EVs as diagnostic biomarkers
Ti‐EVs are ideal candidates for diagnostic markers due to their intrinsic features. Ti‐EVs are present in large numbers in the extracellular interstitium and provide a snapshot of the interplay of multiple cell types and cells with tissue microenvironment. Notably, Ti‐EVs represent biomedical tools because their molecular profile is a snapshot of parental cells in various states (Bang & Thum, 2012, Keller et al., 2011, Mathivanan et al., 2010). Thus, mapping the heterogeneous profiles of molecular components is important for defining multiple human diseases and distinguishing the pathological states from the physical states (Hoshino et al., 2020). Moreover, Ti‐EV‐harbored cargoes are not a random combination of cell fragments or garbage; however, the precise biochemical compositions are integrated by a strict sorting mechanism, which makes Ti‐EVs informative biomarkers in human disease (Yang et al., 2020). Furthermore, Ti‐EVs can protect the contents from degradation; however, the underlying mechanism has not yet been elucidated. Stable expression of the contents allows us to diagnose and detect diseases consistently and continuously (Farooqi et al., 2018). Notably, the relative concentration of Ti‐EVs outweighs that of body fluid‐derived EVs because of the diffusion of EVs from specific tissue microenvironments to the circulatory system. Moreover, EVs in the circulatory system are predominantly secreted by non‐cancer cells and may hinder the identification of biomarkers (Jang et al., 2019). While many attempts have been made to determine the functions of body fluid‐derived EVs as diagnostic tools, Ti‐EVs present higher sensitivity and specificity in diagnostics. This is because the EVs that accumulate in the tumour interstitial space are mainly derived from tumour cells (Connolly et al., 2018). Finally, the results of a recent study suggested that a certain relationship exists between the concentration of Ti‐EVs and cancer progression, indicating that the concentration of Ti‐EVs may be a reliable marker for disease evolution (Mege et al., 2016).
Taken together, these biological characteristics make Ti‐EVs attractive as promising tools for the diagnosis and detection of diseases (Chen et al., 2016) (Figure 7a). Notably, despite the advantages of Ti‐EVs, diagnosis based on Ti‐EVs cannot be performed in cases where tissue biopsy is not available, such as deep malignant tumours and tumours not suitable for biopsy (e.g., melanoma). In these cases, liquid biopsy is a valuable complementary diagnostic method, and biomarkers have been identified in mechanism studies based on Ti‐EVs.
FIGURE 7.
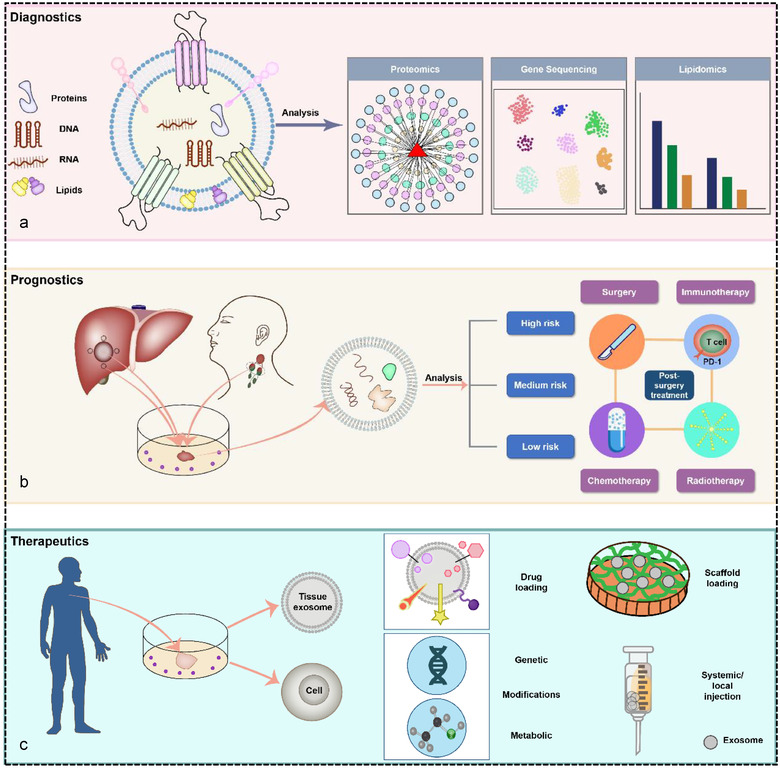
Tissue‐derived EVs (Ti‐EVs) in clinical practice. (a) Diagnostics: Ti‐EVs harbour various bioactive molecules packaged from their parent cells that can indicate pathophysiological conditions, which makes Ti‐EVs promising tools for clinical diagnosis. (b) Prognostics: Ti‐EVs isolated from tumour tissue, adjacent tissue, and draining lymph nodes can contribute to risk stratification and consequently strategic post‐surgery treatment. (c) Therapeutics: EVs can be extracted directly from tissue. Ti‐EVs are considered emerging nanocarriers for drug delivery through incorporating drugs, imaging agents, etc. Alternatively, Ti‐EVs themselves can be used as therapeutic agents employing cell modification (genetic or metabolic) in an attempt to alter the contents of Ti‐EVs for therapeutic purposes. Modified Ti‐EVs are administrated by scaffold loading, local or systemic injection
4.2. Ti‐EVs as prognostic indicators
Surgery remains the preferred treatment option for most solid tumours. While progress has been made in terms of surgical techniques, residual tumour cells in the operative margins or in the circulatory system following surgery remain a problem, leading to tumour relapse and metastasis (Baker et al., 1989, Bu et al., 2019, Gottschalk et al., 2010, Tavare et al., 2012). Local recurrence and remote metastasis during the late stage of cancer (Niibe & Hayakawa, 2010) are major challenges that need to be overcome to improve the prognosis for patients undergoing surgery. Postoperative chemoradiotherapy has been shown to improve prognosis for patients with cancer. Currently, immunotherapy, with therapeutic agents that harness the immune system to combat cancers, has been utilized as a supplementary method to traditional therapeutic regimens (Farkona et al., 2016). However, many cancer patients still die due to disease relapse, despite the use of surgery, neoadjuvant therapy, and postoperative chemoradiotherapy. Accurate diagnosis of the metastatic state is critical for the individualized design of treatment plans for patients with cancer (Li et al., 2017). Risk stratification in cancer is vital to reduce or avoid the improper treatment of patients with cancer (Krajewska et al., 2017). The stratification of patients with cancer using different factors may yield more favourable results. Therefore, efficient prognostic biomarkers are warranted for improved risk stratification, personalized postoperative adjuvant therapy, and prognostication of patients with tumours (Yu et al., 2019). It is noteworthy that molecular alterations occur ahead of changes in cell morphology and biological behaviour in patients with cancer.
Ti‐EVs provide a “snapshot” of the cells of origin owing to their lineage. Thus, it is tempting to speculate that Ti‐EVs may represent tools for risk stratification for personalized postoperative therapy (Figure 7b). For example, Ti‐EVs can be used to determine whether the scope of surgery is adequate to eliminate tumour cells or whether the sentinel lymph nodes have been invaded by analysis of Ti‐EVs obtained from the surgical margin and draining lymph nodes, respectively. Based on the results of such analyses, a comprehensive post‐surgery management strategy (two‐stage operation, chemotherapy, radiotherapy, immunotherapy, or a combination of these methods) can be developed, which could improve the overall survival rate for patients with cancer.
4.3. Ti‐EVs as therapeutic agents
Mesenchymal stromal cells (MSCs) have been widely reported therapeutic efficacy in many pre‐clinical trials of various diseases and safety in human patients. Growing and convincing evidence have shown that MSCs exert many, if not most, of paracrine effects through the secretion of EVs (Witwer et al., 2019). EVs from cell culture supernatants of MSCs have been demonstrated to be therapeutically effective in multiple pre‐clinical models, therefore, promising therapeutic agents that are proposed to test in clinical trials (Börger et al., 2017, Lener et al., 2015, Reiner et al., 2017). MSCs can be isolated in multiple types of tissues, such as bone marrow (Luo et al., 2019), fat tissue (Lou et al., 2015), the umbilical cord (Yaghoubi et al., 2019), the placenta (Bier et al., 2018), the amniotic membrane (Farhadihosseinabadi et al., 2018), gingiva, and dental pulp (Zhang et al., 2018). Hopefully, MSC‐EVs can be extracted directly from tissues through multiplex bead‐based platform (Koliha et al., 2016), microfluidic EVs isolation method, etc. However, challenges remain in Ti‐EVs for therapeutic application, such as heterogeneity of Ti‐EVs, low yield, limited tissue sources, difficult preserving, and accurate determination of sampling sites, among others. In terms of therapeutics, cell culture‐derived EVs may be better source for disease treatment due to larger yield, certainty of cellular origin of EVs, simple and economical production procedures, etc. At present, therapeutic application of EVs is still in its infancy. There is still a long way to go in the future.
5. PROBLEMS AND SOLUTIONS
5.1. Optimized methodology for the isolation of Ti‐EVs
In 2017, Vella et al. presented an efficient protocol for separating Ti‐EVs from brain tissue (Vella et al., 2017). Since then, a multitude of research has been performed. To date, considerable attention has focused on the potential clinical applications of Ti‐EVs. However, several technical hindrances have restricted basic and applied research on Ti‐EVs. The first involves optimization of the Ti‐EV isolation procedure (Lötvall et al., 2014, Yang et al., 2020).
While progress has been made in elucidating the role of Ti‐EVs in recent decades, challenges in the effective enrichment of Ti‐EVs still need to be addressed (Yang et al., 2020). This is mainly attributed to the complexities of Ti‐EVs with preserved biochemical properties from the intact extracellular milieu and similarities in biochemical and physicochemical properties among Ti‐EVs, viruses, lipoproteins, as well as the heterogeneity of Ti‐EVs themselves (Chen et al., 2019, Sahoo et al., 2021). Multiple commonly used isolation methods impact EV purity, integrity, and active state, as well as yield (Xu et al., 2016). Huang et al. showed that different parameters and techniques can influence the recovery and contents of Ti‐EVs, and developed a protocol for the isolation of Ti‐EVs from brain tissue (Huang et al., 2020). As there is no optimal method for the isolation and characterization of Ti‐EVs that fulfils experimental and clinical needs, selecting a suitable method for use in studies on Ti‐EVs may be challenging. Several factors need to be considered: the type of EVs‐containing matrix and the expected Ti‐EVs yield, purity, integrity, and concentration, which are dictated by the downstream analysis and scientific questions to be addressed (Witwer et al., 2017).
Methods vary depending on the variety of tissue samples. For example, a well‐established method for the isolation of Ti‐EVs has never been performed on bone tissue (Crescitelli et al., 2021), which should be optimized, such as the use of enzymes, including the type, concentration, and incubation period (Allen et al., 2016, Hurwitz et al., 2019, Jeppesen et al., 2019, Steenbeek et al., 2018, Yang et al., 2020).
Methods vary depending on the downstream application(s). The type of study—discovery, clinical, or preparatory research—mandates a different yield, purity, and concentration (Sahoo et al., 2021). These factors are mutually restrictive. For example, “the purity” of Ti‐EVs is technically sensitive, which likely affects the interpretation of results, and accounts for the poor concordance among the results of individual studies (Xing et al., 2020). Conversely, in some studies on biomarker discovery, there may not be strict requirements for purity if certain biomarkers can be detected repeatedly in EV solutions. In some cases, the combination of several methods or specific modifications to existing methods may improve outcomes when addressing specific scientific questions (Sahoo et al., 2021). Therefore, the choice of separation and concentration methods is subject to factors that vary between studies and the heterogeneity of tissue samples, such that there is no one‐size‐fits‐all approach (Théry et al., 2018).
Notably, some conclusions can be drawn in terms of Ti‐EV isolation. Firstly, for tissues rich in blood, such as liver and heart tissue, a pre‐rinse or perfusion is critical before Ti‐EV isolation (Figure 6d), which can reduce contamination from serum‐EVs and other particles. In addition, differences exist among non‐tumour tissues, benign tumours, malignant tumours, solid tumours, and non‐solid tumours in terms of texture, interstitial components, and blood abundance. These factors mandate the optimization of isolation methods, such as the dissociation method, type of digestive enzymes, filter pore size, centrifugation time and force.
Moreover, it is important to confirm that separated EVs derive from the interstitial space, and not from the intracellular space or soluble nanoparticles secreted as a result of EVs‐containing matrix harvest, processing, and storage (Jayabalan et al., 2019, Sahoo et al., 2021). For example, Ti‐EV isolation protocols should be performed on fresh tissue samples to avoid excessive cell death (Huang et al., 2020, Hyatt & Wilber, 1959, Mincheva‐Nilsson et al., 2016, Vella et al., 2017) or on TEs, for which a short culture time is suitable (Mincheva‐Nilsson et al., 2016). Moreover, it is recommended that gentle dissociation be performed to reduce cell damage. For example, homogenization and shear stress can induce the release of intracellular vesicles (Mincheva‐Nilsson et al., 2016, Vella et al., 2017). In addition, the correct duration for enzymatic digestion, enzyme type, and gentle dissociation are necessary to be determined to reduce cell damage. It was reported that enzymatic treatment of tissues digests the extracellular matrix, possibly compromising cell secretory functions. Evidence has shown that the enzymatic methods may be too aggressive and destroy Ti‐EVs during processing (García‐Contreras et al., 2012). A key point for Ti‐EV studies lies in the proportion of inactive cells when Ti‐EVs are extracted, which should be specified. Even a small number of dead cells can release more membrane structures than the Ti‐EVs of interest (Théry et al., 2018). The absence of negative biomarkers for Ti‐EV, such as calnexin, makes Ti‐EV preparations more convincing (Vella et al., 2017).
While differences exist among individual protocols for the isolation of Ti‐EVs, some shared methods can be used to extract Ti‐EVs from multiple types of tissues (Figure 8).
FIGURE 8.

Common methods can be used to extract tissue‐derived EVs (Ti‐EVs) from multiple types of tissue. Tissue samples were processed immediately after dissection by gentle mechanical forces or enzymes and 1 h has been considered a favourable duration; centrifugation at a speed of ∼500–3000g at 4°C can be performed to remove apoptotic bodies and large cellular debris/intact cells; centrifugation at a speed of 10–14,000g at 4°C, and then passing through a 0.22 μm filter can be to used obtain large EVs after the removal of any contaminating microvesicles and remaining cellular debris; ultracentrifugation speeds exceeding 100,000g at 4°C are used to pellet small EVs, and this procedure should take more than 1 h; finally, the extracted Ti‐EVs can be used immediately or stored at −80°C for several years
1) Tissue samples were processed immediately after dissection, and 1 h has been considered a favourable duration (Crescitelli et al., 2021). For TEs cultured in conditioned medium, culture time should be given special attention (preferably 24–48 h; at most 72 h, if possible) (Mincheva‐Nilsson et al., 2016). In addition, it is recommended that tissues be dissociated gently by mechanical forces or enzymes. Great caution should be exercised when selecting enzymes. For example, collagenase D (Cianciaruso et al., 2019, Jang et al., 2019) is preferred when it is necessary to maintain the functionality and integrity of cell‐surface biomolecules. Collagenase D has enabled the isolation of cells from multiple tissue samples, including epithelial, liver, fat, muscle, pancreatic islet, and endothelial cells (Crescitelli et al., 2021, Angyal et al., 2010, Fan et al., 2012, 2014, Benck et al., 2016, Uchea et al., 2015). Finally, the extracted Ti‐EVs can be used immediately or stored at −80°C for several years (Crescitelli et al., 2021). Different tissue sources and isolation procedures mandate optimum temperature conditions to ensure the stability of Ti‐EVs during long‐term storage (Zhang et al., 2020). The results showed that compared to freshly isolated Ti‐EVs, the morphology and certain biological characteristics of Ti‐EVs change during storage at ‐80°C for 4 days and 4 weeks, respectively (Lőrincz et al., 2014, Maroto et al., 2017). Lyophilization is a candidate for long‐term storage; however, its influence on Ti‐EVs structure during recovery varies depending on the use of cryoprotectants. Currently, storage at −80°C remain a common option, despite the challenging and less economical characteristics (Herrmann et al., 2021).
Notably, collected Ti‐EVs may be from multiple cell populations (Jang et al., 2019), even from single tissue. For an in‐depth study on Ti‐EVs, and for use in clinical applications, the purification of specific cell type‐EVs is required. Tissues comprise of multiple cell types, including tumour cells, mesenchymal cells, endothelial cells, fibroblasts, and immune cells, from which Ti‐EVs inherit cell‐specific markers during their biogenesis (Koliha et al., 2016). Cell populations responsible for the release of metastatic EVs can be identified based on surface markers unique to certain cells in combination with classical exosome markers (e.g., CD9, CD63, and CD81 (Mathieu et al., 2021). In addition to cell markers, potent technologies are crucial for the isolation of cell subpopulation‐EVs from tissues, such as single particle interferometric reflectance imaging sensing (SP‐IRIS) with fluorescence (Arab et al., 2021), microfluidic resistive pulse sensing (MRPS) (Ahn et al., 2021), nanoflow cytometry measurement (NFCM) (Tian et al., 2020), immuno‐magnetic activated cell sorting (IMACS) (Sutermaster & Darling, 2019), immuno‐magnetic nanoparticles (IMNPs) (Zhang et al., 2017), etc. However, EVs derived from the same cell population can exhibit distinct characteristics (Sahai et al., 2020). For example, MSCs are highly heterogeneous cells, wide variability exists in MSC‐sEV preparations that permeates the whole process from the starting materials through the production and purification to the end preparations (Witwer et al., 2019). Thus, Ti‐EVs derived from specific cell types with definite properties are needed to be identified. In addition, according to MISEV 2018 (Minimal Information for Studies of Extracellular Vesicles), EV subtypes can be defined based on their physical characteristics. For example, “small EVs” (sEVs) and “medium/large EVs” medium/large EVs (m/lEVs) refer to EVs with diameters of <100 nm or <200 nm (small), or >200 nm (large and/or medium), respectively. Alternatively, density can be used for classification, such as low‐, medium‐, and high‐density EVs (Crescitelli et al., 2020, Crescitelli et al., 2021). Precise purification of Ti‐EVs is critical for elucidating the roles of Ti‐EVs under various pathophysiological conditions.
5.2. Standardized protocol for the characterization of Ti‐EVs
It is important to determine whether something of interest has been obtained, regardless of the tissue source or research type. Several techniques are necessary to assess the concentration, yield, purity, and subtypes of separated Ti‐EVs. Herein, we recommend complying with the MISEV2018 guidelines (Théry et al., 2018). However, it may be difficult to conduct all characterization assays as the MISEV guidelines suggest when the EV yield is low, especially for in vivo—derived specimens. The main approaches for characterizing isolated Ti‐EVs involve the following: (i) determining the concentration and size distribution of EVs (with nanoparticle tracking analysis [NTA] being the most common method) and normalization of the Ti‐EV concentration to the weight of the tissues; (ii) visualization of EVs using TEM; (iii) characterizing the presence of EV‐specific molecules using techniques such as western blotting, flow cytometric analyses, ELISA, mass spectrometry, ExoView, and other candidates (Bachurski et al., 2019, Crescitelli et al., 2021, Hoshino et al., 2020, Théry et al., 2018, Welsh et al., 2020). Recently, a panel of novel pan‐EVP markers were identified, including ACTB, MSN, and RAP1B, which can be applied for the purification and detection of EVPs (Hoshino et al., 2020). In the past decades, promising techniques for the characterization of Ti‐EVs have been emerging, including cryo‐electron microscopy (Brisson et al., 2017), liquid‐mode atomic force microscopy with peak force tapping (Hardij et al., 2013), electron tomography (van Niel et al., 2011), immunogold labelling (Brisson et al., 2017), flow cytometry (Van Der Pol et al., 2014), novel scatter‐ and fluorescence‐based flow cytometry (Pospichalova et al., 2015, Van Der Vlist et al., 2012), and resistive pulse sensing (Besse et al., 2016, Fraikin et al., 2011).
In addition, the definition of the bioactive substance(s) will remain a critical precondition before the fundamental study and clinical application of Ti‐EVs (Willis et al., 2017). Evaluation of Ti‐EVs’ potency would be an excellent procedure in addressing the challenges of the inconsistencies in preparations and batch‐to‐batch variations (Gupta et al., 2021). Thus, a fit‐for‐purpose exosome potency unit (EPU) could be applied to purify bioactive EVs and standardize dosage among different EV preparations (Willis et al., 2017). Potency may be evaluated utilizing biological in vivo and in vitro assays, non‐biological assays or a combinatorial assay matrix (Nguyen et al., 2020).
In summary, there is a critical point to consider when characterizing isolated Ti‐EVs: different combinations should be conducted to define Ti‐EVs based on four dimensions, size, shape, molecular markers, and specific functional properties. The criteria above may be updated with advances in the “state of the art.” Standardization of characterization methods for Ti‐EV preparations could advance research outcomes towards powerful clinical applications.
5.3. Sampling problems and heterogeneity exploration of Ti‐EVs
Compared with EVs from other sources, sampling is a hurdle unique to Ti‐EV research. Firstly, the availability of Ti‐EVs is limited. The clinical utilization of Ti‐EVs requires the detection of a minimum amount of Ti‐EVs in order to exert the expected results. In terms of diagnostics, this can be addressed by improvements of detection sensitivity and specificity, while for potential therapeutics, in vitro amplification of Ti‐EVs may represent a potent and practical solution. Second, an invasive sampling process cannot be avoided (Leung et al., 2021). Moreover, sampling for Ti‐EVs may be unavailable in some instances, including deep malignant tumours and tumours that are not suitable for biopsy, such as arterial aneurysms. In these cases, liquid biopsy may be a more qualified candidate. Finally, Ti‐EV‐based analysis is subject to sampling bias (Siravegna et al., 2017). Multi‐site sampling provides an understanding of tumour heterogeneity; however, this can mislead diagnostics, such as the collection of specimens at the centre of the tumour, adjacent tumour tissue, invasive margin, and bulky metastases. Multi‐region analysis revealed spatial heterogeneity in esophageal squamous cell carcinoma (Yan et al., 2019). Similarly, variations exist between different cancer stages, from carcinoma initiation to tumour progression or metastasis. Conceivably, Ti‐EVs, which provide a vivid snapshot of parental cells and are mediators of various biological events, demonstrate significant heterogeneity. Thus, analysis of EVs from tissues sampled at different disease stages and from multiple regions may have potential to guide more personalized clinical management. Therefore, the immediate next challenge will be to explore the spatial‐temporal heterogeneity of Ti‐EVs. Feasible strategy can be to conduct large‐scale clinical trials, sample at multiple sites, and analyze the yield and molecular expression differences to determine the best sampling sites, and develop standard operating procedure (SOP) to guide future researches and clinical practice.
5.4. Discovery of biomarkers based on single Ti‐EVs
Notably, given the heterogeneity of Ti‐EVs, it is of particular importance to exploit their diagnostic and therapeutic potential at both the population and single EVs levels. At present, the majority of diagnostic applications for EVs are at the level of “bulk measurements,” which require 105‐106 EVs for each biomarker to detect proteins of interest, or 102‐103 EVs for the more sensitive approaches (Xiong et al., 2021). Thus, the results are indicative of average levels for the Ti‐EVs population. High‐speed centrifugation induces the formation of EV aggregates (Linares et al., 2015). Therefore, proteins trapped in the centre of aggregates may be omitted if they are present at a low level. Profiling of single Ti‐EV can avoid erroneous interpretation and enable more accurate detection data to be obtained. Novel technologies for single EV analysis have been emerging, and include transmission electron microscopy (TEM) coupled with immunogold labelling (Gärtner et al., 2019), atomic force microscopy (AFM) (Matsumura et al., 2019), SP‐IRIS (Arab et al., 2021), MRPS (Ahn et al., 2021), NFCM with fluorescence (Tian et al., 2020), and laser tweezers Raman spectroscopy (Smith et al., 2015), among other methods. In addition, large‐scale clinical trials based on single‐EV level by sampling among various types of cancer are required to potentially identify common and specific Ti‐EV‐specific markers, which will benefit further standardization and quality control of Ti‐EVs.
5.5. Prospects and challenges for the therapeutic application of Ti‐EVs
EVs isolated from some special tissues can be potentially utilized for clinical treatment, such as bone marrow, fat, the umbilical cord, the placenta, the amniotic membrane, gingiva, and dental pulp, and so on. Of concern, there are still some challenges to which we need to pay attention. Firstly, the exploration of the biodistribution of Ti‐EVs as therapeutic agents after administrated. Several strategies have been developed to maintain a high concentration of EVs in target tissues, such as enhancing their ability to cross biological barriers, prolonging their half‐life, improving targeting (by cloaking and surface display), and reducing clearance (Antes et al., 2018, De Jong et al., 2020, EL Andaloussi et al., 2013, Fais et al., 2016). Secondly, the achievement of clinical scale yield of Ti‐EVs, possible solutions can be reconstitution of 3D tissue‐like structures (organoids), bioreactor‐based culture systems, or the improvement of treatment efficiency. In addition to these obstacles, heterogeneity of Ti‐EVs, storage problem, biocompatibility, etc., which need to be addressed urgently. Hopefully, future studies on Ti‐EVs will focus on these issues. Nonetheless, to harmonize research practice into Ti‐EVs, and to ensure an acceptable level of data comparability, detailed technical and experimental information should be provided in scientific articles, and characterizations should include a minimal set of evidence for the EVs reported (Lötvall et al., 2014).
6. CONCLUSIONS AND PERSPECTIVES
Although EVs from body fluids provide a source for liquid biopsies, and cell culture‐derived EVs can be harvested for tissue regeneration, understanding the reality of the tissue microenvironment, especially the TME, remains challenging. Ti‐EVs, with additional traits, have been shown to be promising agents for basic research and clinical applications. There are opportunities for practitioners, scientists, and pharmacologists to work together to achieve this goal. Nonetheless, recent developments in Ti‐EV research should not mask the significant endeavours that are still need to be made to increase our knowledge of Ti‐EVs and translate this into clinical practice.
(1) Optimization of the methods for Ti‐EV isolation should be prioritized. Of note, the isolation of Ti‐EVs from specific cell populations is necessary for in‐depth mechanism studies and applications in clinical setting. (2) Standardized protocol for the characterization of Ti‐EVs and potency assays are critical to confirm bioactive Ti‐EVs and establish the standards for clinical applications. (3) Exploration of the spatial‐temporal heterogeneity of Ti‐EV is required. (4) Further investigations on the detection and identification of new biomarkers for clinical diagnosis at single Ti‐EV levels are warranted. (5) There are also several other problems to be solved, such as preclinical studies to determine the characteristics of Ti‐EVs, animal studies to elucidate the biodistribution of administrated Ti‐EVs. Based on these considerations, an authorized guideline and emerging technologies for Ti‐EV research are desired to improve the consistency and repeatability of results, and to optimize clinical applications.
Despite the above challenges, the clinical application of Ti‐EVs will be promising. Globally, at least 204 clinical trials focusing on EVs had been reported by August 2021. Further knowledge on the intrinsic properties of Ti‐EVs, as well as standardization and quality control of Ti‐EV researches, will greatly accelerate the clinical application of Ti‐EVs (Xu et al., 2016). It is foreseeable that with further efforts, these obstacles will be gradually removed and Ti‐EVs may have potential clinical applications in the future.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China [81922038 and 81870361 to G. Chen]; National Natural Science Foundation of China [81702703 to L.L. Bu]; National Natural Science Foundation of China [82001066 to Q.W. Man]; National Natural Science Foundation of China [81872203 to B. Liu]; China Postdoctoral Science Foundation [2018M630883 and 2019T120688 to L.L. Bu]; Applied Basic Research Project of Wuhan Municipal Science and Technology Bureau [2020020601012249 to G. Chen], Hubei Natural Science Foundation Outstanding Young Talents Project [2020CFA068 to G. Chen].
Li, S.‐R. , Man, Q.‐W. , Gao, X. , Lin, H. , Wang, J. , Su, F.‐C. , Wang, H.‐Q. , Bu, L.‐L. , Liu, B. , & Chen, G. (2021). Tissue‐Derived Extracellular Vesicles in Cancers and Non‐cancer Diseases: Present and Future. Journal of Extracellular Vesicles, 10, e12175. 10.1002/jev2.12175.
Su‐Ran Li and Qi‐Wen Man contributed equally to this work
Contributor Information
Lin‐Lin Bu, Email: lin-lin.bu@whu.edu.cn.
Bing Liu, Email: liubing9909@whu.edu.cn.
Gang Chen, Email: geraldchan@whu.edu.cn.
REFERENCES
- Abels, E. R. , & Breakefield, X. O. (2016). Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cellular and Molecular Neurobiology. 36(3), 301‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, J. Y. , Datta, S. , Bandeira, E. , Cano, M. , Mallick, E. , Rai, U. , Powell, B. , Tian, J. , Witwer, K. W. , Handa, J. T. , & Paulaitis, M. E. (2021). Release of extracellular vesicle miR‐494‐3p by ARPE‐19 cells with impaired mitochondria. Biochimica et Biophysica Acta. General subjects. 1865(4), 129598. [DOI] [PubMed] [Google Scholar]
- Akers, J. C. , Gonda, D. , Kim, R. , Carter, B. S. , & Chen, C. C. (2013). Biogenesis of extracellular vesicles (EV), exosomes, microvesicles, retrovirus‐like vesicles, and apoptotic bodies. Journal of Neuro‐Oncology. 113(1), 1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M. , Bjerke, M. , Edlund, H. , Nelander, S. , & Westermark, B. (2016). Origin of the U87MG glioma cell line: Good news and bad news. Science Translational Medicine. 8(354), 354re353. [DOI] [PubMed] [Google Scholar]
- Almeria, C. , Weiss, R. , Roy, M. , Tripisciano, C. , Kasper, C. , Weber, V. , & Egger, D. (2019). Hypoxia conditioned mesenchymal stem cell‐derived extracellular vesicles induce increased vascular tube formation in vitro. Frontiers in Bioengineering and Biotechnology. 7, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Nedawi, K. , Meehan, B. , Micallef, J. , Lhotak, V. , May, L. , Guha, A. , & Rak, J. (2008). Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature Cell Biology. 10(5), 619‐624. [DOI] [PubMed] [Google Scholar]
- Angyal, A. , Szekeres, Z. , Balogh, P. , Neer, Z. , Szarka, E. , Virag, V. , Medgyesi, D. , Prechl, J. , & Sarmay, G. (2010). CD16/32‐specific biotinylated 2.4G2 single‐chain Fv complexed with avidin‐FITC enhances FITC‐specific humoral immune response in vivo in a CD16‐dependent manner. International Immunology. 22(2), 71‐80. [DOI] [PubMed] [Google Scholar]
- Antes, T. J. , Middleton, R. C. , Luther, K. M. , Ijichi, T. , Peck, K. A. , Liu, W. J. , Valle, J. , Echavez, A. K. , & Marbán, E. (2018). Targeting extracellular vesicles to injured tissue using membrane cloaking and surface display. Journal of Nanobiotechnology. 16(1), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab, T. , Mallick, E. R. , Huang, Y. , Dong, L. , Liao, Z. , Zhao, Z. , Gololobova, O. , Smith, B. , Haughey, N. J. , Pienta, K. J. , Slusher, B. S. , Tarwater, P. M. , Tosar, J. P. , Zivkovic, A. M. , Vreeland, W. N. , Paulaitis, M. E. , & Witwer, K. W. (2021). Characterization of extracellular vesicles and synthetic nanoparticles with four orthogonal single‐particle analysis platforms. Journal of Extracellular Vesicles. 10(6), e12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, H. , Ikezu, S. , Tsunoda, S. , Medalla, M. , Luebke, J. , Haydar, T. , Wolozin, B. , Butovsky, O. , Kügler, S. , & Ikezu, T. (2015). Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nature Neuroscience. 18(11), 1584‐1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autengruber, A. , Gereke, M. , Hansen, G. , Hennig, C. , & Bruder, D. (2012). Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. European Journal of Microbiology & Immunology. 2(2), 112‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski, D. , Schuldner, M. , Nguyen, P. ‐. H. , Malz, A. , Reiners, K. S. , Grenzi, P. C. , Babatz, F. , Schauss, A. C. , Hansen, H. P. , Hallek, M. , & Pogge Von Strandmann, E. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis—An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. Journal of Extracellular Vesicles. 8(1), 1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D. , Masterson, T. , Pace, R. , Constable, W. C. , & Wanebo, H. (1989). The influence of the surgical wound on local tumor recurrence. Surgery. 106(3), 525‐532. [PubMed] [Google Scholar]
- Bang, C. , & Thum, T. (2012). Exosomes: New players in cell‐cell communication. The International Journal of Biochemistry & Cell Biology. 44(11), 2060‐2064. [DOI] [PubMed] [Google Scholar]
- Banigan, M. , Kao, P. , Kozubek, J. , Winslow, A. R., Medina, J. , Costa, J. , Schmitt, A. , Schneider, A. , Cabral, H. , Cagsal‐Getkin, O. , Vanderburg, C. R. , & Delalle, I. . (2013). Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS ONE. 8(1), e48814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, D. A. , Drula, R. , Ott, L. , Fabris, L. , Slaby, O. , Calin, G. A. , & Pichler, M. (2020). Circulating non‐coding RNAs in renal cell carcinoma‐pathogenesis and potential implications as clinical biomarkers. Frontiers in Cell and Developmental Biology. 8, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov, L. , Matic, K. J. , Hallal, S. , Best, O. G. , Mulligan, S. P. , & Christopherson, R. I. (2016). Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. Journal of Extracellular Vesicles. 5, 25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benck, C. J. , Martinov, T. , Fife, B. T. , & Chatterjea, D. (2016). Isolation of infiltrating leukocytes from mouse skin using enzymatic digest and gradient separation. Journal of Visualized Experiments: JoVE. (107), e53638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse, B. , Charrier, M. , Lapierre, V. , Dansin, E. , Lantz, O. , Planchard, D. , Le Chevalier, T. , Livartoski, A. , Barlesi, F. , Laplanche, A. , Ploix, S. , Vimond, N. , Peguillet, I. , Théry, C. , Lacroix, L. , Zoernig, I. , Dhodapkar, K. , Dhodapkar, M. , Viaud, S. , Soria, J. ‐. C. , … Chaput, N. (2016). Dendritic cell‐derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. 5(4), e1071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier, A. , Berenstein, P. , Kronfeld, N. , Morgoulis, D. , Ziv‐Av, A. , Goldstein, H. , Kazimirsky, G. , Cazacu, S. , Meir, R. , Popovtzer, R. , Dori, A. , & Brodie, C. (2018). Placenta‐derived mesenchymal stromal cells and their exosomes exert therapeutic effects in Duchenne muscular dystrophy. Biomaterials. 174, 67‐78. [DOI] [PubMed] [Google Scholar]
- Bodega, G. , Alique, M. , Puebla, L. , Carracedo, J. , & Ramírez, R. M. (2019). Microvesicles: ROS scavengers and ROS producers. Journal of Extracellular Vesicles. 8(1), 1626654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börger, V. , Bremer, M. , Ferrer‐Tur, R. , Gockeln, L. , Stambouli, O. , Becic, A. , & Giebel, B. (2017). Mesenchymal stem/stromal cell‐derived extracellular vesicles and their potential as novel immunomodulatory therapeutic agents. International Journal of Molecular Sciences. 18(7), 1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braicu, C. , Tomuleasa, C. , Monroig, P. , Cucuianu, A. , Berindan‐Neagoe, I. , & Calin, G. A. (2015). Exosomes as divine messengers: Are they the Hermes of modern molecular oncology? Cell Death and Differentiation. 22(1), 34‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 68(6), 394‐424. [DOI] [PubMed] [Google Scholar]
- Brisson, A. R. , Tan, S. , Linares, R. , Gounou, C. , & Arraud, N. (2017). Extracellular vesicles from activated platelets: A semiquantitative cryo‐electron microscopy and immuno‐gold labeling study. Platelets. 28(3), 263‐271. [DOI] [PubMed] [Google Scholar]
- Bu, L. ‐. L. , Yan, J. , Wang, Z. , Ruan, H. , Chen, Q. , Gunadhi, V. , Bell, R. B. , & Gu, Z. (2019). Advances in drug delivery for post‐surgical cancer treatment. Biomaterials. 219, 119182. [DOI] [PubMed] [Google Scholar]
- Camino, T. , Lago‐Baameiro, N. , Bravo, S. , Molares‐Vila, A. , Sueiro, A. , Couto, I. , Baltar, J. , Casanueva, E. F. , & Pardo, M. (2021). Human obese white adipose tissue sheds depot‐specific extracellular vesicles and reveals candidate biomarkers for monitoring obesity and its comorbidities. Translational Research: the Journal of Laboratory and Clinical Medicine. S1931‐5244(21)00006‐2. [DOI] [PubMed] [Google Scholar]
- Camino, T. , Lago‐Baameiro, N. , Martis‐Sueiro, A. , Couto, I. , Santos, F. , Baltar, J. , & Pardo, M. (2020). Deciphering adipose tissue extracellular vesicles protein cargo and its role in obesity. International Journal of Molecular Sciences. 21(24), 9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, S. , & Poon, I. K. H. (2018). Apoptotic cell‐derived extracellular vesicles: More than just debris. Frontiers in Immunology. 9, 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. , Sun, D. , Qin, X. , & Gao, X.‐H. (2021). Screening and identification of potential biomarkers and therapeutic drugs in melanoma via integrated bioinformatics analysis. Investigational New Drugs. 39(4):928–948. [DOI] [PubMed] [Google Scholar]
- Chen, Bo‐Y , Sung, C. W. ‐. H. , Chen, C. , Cheng, C. ‐. M. , Lin, D. P. ‐. C. , Huang, C‐Te , & Hsu, M. ‐. Y. (2019). Advances in exosomes technology. Clinica Chimica Acta; International Journal of Clinical Chemistry. 493, 14‐19. [DOI] [PubMed] [Google Scholar]
- Chen, X. ‐. D. , Zhao, J. , Yan, Z. , Zhou, Bo‐W , Huang, W. ‐. F. , Liu, W.‐. F. , Li, C. , & Liu, Ke‐X (2020). Isolation of extracellular vesicles from intestinal tissue in a mouse model of intestinal ischemia/reperfusion injury. BioTechniques. 68(5), 257‐262. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Buyel, J. J. , Hanssen, M. J. W. , Siegel, F. , Pan, R. , Naumann, J. , Schell, M. , Van Der Lans, A. , Schlein, C. , Froehlich, H. , Heeren, J. , Virtanen, K. A. , Van Marken Lichtenbelt, W. , & Pfeifer, A. (2016). Exosomal microRNA miR‐92a concentration in serum reflects human brown fat activity. Nature Communications. 7, 11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Vella, L. J. , Barnham, K. J. , Mclean, C. , Masters, C. L. , & Hill, A. F. (2020). Small RNA fingerprinting of Alzheimer's disease frontal cortex extracellular vesicles and their comparison with peripheral extracellular vesicles. Journal of Extracellular Vesicles. 9(1), 1766822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciaruso, C. , Beltraminelli, T. , Duval, F. , Nassiri, S. , Hamelin, R. , Mozes, A. , Gallart‐Ayala, H. , Ceada Torres, G. , Torchia, B. , Ries, C. H. , Ivanisevic, J. , & De Palma, M. (2019). Molecular profiling and functional analysis of macrophage‐derived tumor extracellular vesicles. Cell Reports. 27(10), 3062‐3080.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, K. D. , Wadey, R. M. , Mathew, D. , Johnson, E. , Rees, D. A. , & James, P. E. (2018). Evidence for adipocyte‐derived extracellular vesicles in the human circulation. Endocrinology. 159(9), 3259‐3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa‐Silva, B. , Aiello, N. M. , Ocean, A. J. , Singh, S. , Zhang, H. , Thakur, B. K. , Becker, A. , Hoshino, A. , Mark, M. T. , Molina, H. , Xiang, J. , Zhang, T. , Theilen, T. ‐. M. , García‐Santos, G. , Williams, C. , Ararso, Y. , Huang, Y. , Rodrigues, G. , Shen, T. ‐. L. , Labori, K. J. , …, Lyden, D. (2015). Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nature Cell Biology. 17(6), 816‐826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli, R. , Lässer, C. , Jang, SuC , Cvjetkovic, A. , Malmhäll, C. , Karimi, N. , Höög, J. L. , Johansson, I. , Fuchs, J. , Thorsell, A. , Gho, Y. S. , Olofsson Bagge, R. , & Lötvall, J. (2020). Subpopulations of extracellular vesicles from human metastatic melanoma tissue identified by quantitative proteomics after optimized isolation. Journal of Extracellular Vesicles. 9(1), 1722433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli, R. , LäSser, C. , & LöTvall, J. (2021). Isolation and characterization of extracellular vesicle subpopulations from tissues. Nature Protocols. 16(3), 1548‐1580. [DOI] [PubMed] [Google Scholar]
- Crewe, C. , Joffin, N. , Rutkowski, J. M. , Kim, M. , Zhang, F. , Towler, D. A. , Gordillo, R. , & Scherer, P. E. (2018). An endothelial‐to‐adipocyte extracellular vesicle axis governed by metabolic state. Cell. 175(3), 695‐708.e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, B. , Barros, E. R. , Hoenderop, J. G. J. , & Rigalli, J. P. (2020). Recent advances in extracellular vesicles as drug delivery systems and their potential in precision medicine. Pharmaceutics. 12(11), 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Z. , Poliakov, A. , Hardy, R. , Clements, R. , Liu, C. , Liu, Y. , Wang, J. , Xiang, X. , Zhang, S. , Zhuang, X. , Shah, S. V. , Sun, D. , Michalek, S. , Grizzle, W. E. , Garvey, T. , Mobley, J. , & Zhang, H.‐G. (2009). Adipose tissue exosome‐like vesicles mediate activation of macrophage‐induced insulin resistance. Diabetes. 58(11), 2498‐2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk, J. , Boelmans, K. , Siegismund, C. , Lassner, D. , Arlt, S. , & Jahn, H. (2015). MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer`s disease. PLoS ONE. 10(5), e0126423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke, S. , Sinha, R. , Levine, D. A. , Sander, C. , & Schultz, N. (2013). Evaluating cell lines as tumour models by comparison of genomic profiles. Nature Communications. 4, 2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, L. , & Wang, M. (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 8(7). 727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EL Andaloussi, S. , Mäger, I. , Breakefield, X. , & Wood, M. J. A. (2013). Extracellular vesicles: Biology and emerging therapeutic opportunities. Nature Reviews. Drug Discovery. 12(5), 347‐357. [DOI] [PubMed] [Google Scholar]
- Erdbrügger, U. , Blijdorp, C. J. , Bijnsdorp, I. V. , Borràs, F. E. , Burger, D. , Bussolati, B. , Byrd, J. B. , Clayton, A. , Dear, J. W. , Falcón‐Pérez, J. M. , Grange, C. , Hill, A. F. , Holthöfer, H. , Hoorn, E. J. , Jenster, G. , Jimenez, C. R. , Junker, K. , Klein, J. , Knepper, M. A. , Koritzinsky, E. H. , …, Martens‐Uzunova, E. S. (2021). Urinary extracellular vesicles: A position paper by the urine task force of the international society for extracellular vesicles. Journal of Extracellular Vesicles. 10(7), e12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbiano, F. , Corsi, J. , Gurrieri, E. , Trevisan, C. , Notarangelo, M. , & D'agostino, V. G. (2020). RNA packaging into extracellular vesicles: An orchestra of RNA‐binding proteins?. Journal of Extracellular Vesicles. 10(2), e12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais, S. , O'driscoll, L. , Borras, F. E. , Buzas, E. , Camussi, G. , Cappello, F. , Carvalho, J. , Cordeiro Da Silva, A. , Del Portillo, H. , El Andaloussi, S. , Ficko Trček, T. , Furlan, R. , Hendrix, An , Gursel, I. , Kralj‐Iglic, V. , Kaeffer, B. , Kosanovic, M. , Lekka, M. E. , Lipps, G. , Logozzi, M. , Marcilla, A. , Sammar, M. , … Giebel, B. (2016). Evidence‐based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano. 10(4), 3886‐3899. [DOI] [PubMed] [Google Scholar]
- Fan, Y. , Hu, S. , Liu, J. , Xiao, F. , Li, X. , Yu, W. , Cui, Y. , Sun, M. , Lv, T. , He, Q. , & Jin, J. (2014). Low intraprostatic DHT promotes the infiltration of CD8+ T cells in BPH tissues via modulation of CCL5 secretion. Mediators of Inflammation. 2014, 397815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhadihosseinabadi, B. , Farahani, M. , Tayebi, T. , Jafari, A. , Biniazan, F. , Modaresifar, K. , Moravvej, H. , Bahrami, S. , Redl, H. , Tayebi, L. , & Niknejad, H. (2018). Amniotic membrane and its epithelial and mesenchymal stem cells as an appropriate source for skin tissue engineering and regenerative medicine. Artificial Cells, Nanomedicine, and Biotechnology. 46(sup2), 431‐440. [DOI] [PubMed] [Google Scholar]
- Farkona, S. , Diamandis, E. P. , & Blasutig, I. M. (2016). Cancer immunotherapy: The beginning of the end of cancer?. BMC Medicine. 14, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi, A. A. , Desai, N. N. , Qureshi, M. Z. , Librelotto, D. R. N. , Gasparri, M. L. , Bishayee, A. , Nabavi, S. M. , Curti, V. , & Daglia, M. (2018). Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnology Advances. 36(1), 328‐334. [DOI] [PubMed] [Google Scholar]
- Ferguson, N. N. (2021). Challenges and controversy in determining UV exposure as a risk factor for cutaneous melanoma in skin of color. JAMA Dermatology. 157(2), 151‐153. [DOI] [PubMed] [Google Scholar]
- Ferlay, J. , Soerjomataram, I. , Dikshit, R. , Eser, S. , Mathers, C. , Rebelo, M. , Parkin, D. M. , Forman, D. , & Bray, F. (2015). Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. International Journal of Cancer. 136(5), E359‐E386. [DOI] [PubMed] [Google Scholar]
- Fraikin, J. ‐. L. , Teesalu, T. , Mckenney, C. M. , Ruoslahti, E. , & Cleland, A. N. (2011). A high‐throughput label‐free nanoparticle analyser. Nature Nanotechnology. 6(5), 308‐313. [DOI] [PubMed] [Google Scholar]
- Gao, J. , Qiu, X. , Li, X. , Fan, H. , Zhang, F. , Lv, T. , & Song, Y. (2018). Expression profiles and clinical value of plasma exosomal Tim‐3 and Galectin‐9 in non‐small cell lung cancer. Biochemical and Biophysical Research Communications. 498(3), 409‐415. [DOI] [PubMed] [Google Scholar]
- García‐Contreras, M. , Messaggio, F. , Jimenez, O., & Mendez, A. (2012). Differences in exosome content of human adipose tissue processed by non‐enzymatic and enzymatic methods. CellR4 2015, 3(1), e1423. [Google Scholar]
- Gärtner, K. , Luckner, M. , Wanner, G. , & Zeidler, R. (2019). Engineering extracellular vesicles as novel treatment options: Exploiting herpesviral immunity in CLL. Journal of Extracellular Vesicles. 8(1), 1573051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk, A. , Sharma, S. , Ford, J. , Durieux, M. E. , & Tiouririne, M. (2010). Review article: the role of the perioperative period in recurrence after cancer surgery. Anesthesia and Analgesia. 110(6), 1636‐1643. [DOI] [PubMed] [Google Scholar]
- Gowda, R. , Robertson, B. , Iyer, S. , Barry, J. , Dinavahi, S. S. , & Robertson, G. P. (2020). The role of exosomes in metastasis and progression of melanoma. Cancer Treatment Reviews. 85, 101975. [DOI] [PubMed] [Google Scholar]
- Grange, C. , Brossa, A. , & Bussolati, B. (2019). Extracellular vesicles and carried miRNAs in the progression of renal cell carcinoma. International Journal of Molecular Sciences. 20(8), 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Wang, J. , Zhao, Y. , Feng, Y. , Han, S. , Dong, Q. , Cui, M. , & Tieu, K. (2020). Microglial exosomes facilitate α‐synuclein transmission in Parkinson's disease. Brain: A Journal of Neurology. 143(5), 1476‐1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, D. , Zickler, A. M. , & El Andaloussi, S. (2021). Dosing extracellular vesicles. Advanced Drug Delivery Reviews. 178, 113961. [DOI] [PubMed] [Google Scholar]
- Gurunathan, S. , Kang, M. ‐. H. , Jeyaraj, M. , Qasim, M. , & Kim, J. ‐. H. (2019). Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells. 8(4), 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. , & Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell. 144(5), 646‐674. [DOI] [PubMed] [Google Scholar]
- Hardij, J. , Cecchet, F. , Berquand, A. , et al. (2013) Characterisation of tissue factor‐bearing extracellular vesicles with AFM: comparison of air‐tapping‐mode AFM and liquid Peak Force AFM. Journal of extracellular vesicles. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmke, A. , & Vietinghoff, S. V. (2016). Extracellular vesicles as mediators of vascular inflammation in kidney disease. World journal of nephrology. 5(2), 125‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, I. K. , Wood, M. J. A. , & Fuhrmann, G. (2021). Extracellular vesicles as a next‐generation drug delivery platform. Nature nanotechnology. 748‐759. [DOI] [PubMed] [Google Scholar]
- Hoshino, A. , Costa‐Silva, B. , Shen, T. ‐. L. , Rodrigues, G. , Hashimoto, A. , Tesic Mark, M. , Molina, H. , Kohsaka, S. , Di Giannatale, A. , Ceder, S. , Singh, S. , Williams, C. , Soplop, N. , Uryu, K. , Pharmer, L. , King, T. , Bojmar, L. , Davies, A. E. , Ararso, Y. , Zhang, T. , & Lyden, D. (2015). Tumour exosome integrins determine organotropic metastasis. Nature. 527(7578), 329‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Kim, H. S. , Bojmar, L. , Gyan, K. E. , Cioffi, M. , Hernandez, J. , Zambirinis, C. P. , Rodrigues, G. , Molina, H. , Heissel, S. , Mark, M. T. , Steiner, L. , Benito‐Martin, A. , Lucotti, S. , Di Giannatale, A. , Offer, K. , Nakajima, M. , Williams, C. , Nogués, L. , Pelissier Vatter, F. A. , Lyden, D. (2020). Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell. 182(4), 1044‐1061.e18.e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. L. , Wang, W. , Lan, X. L. , Zeng, Z. C. , Liang, Y. S. , Yan, Y. R. , Song, F. Y. , Wang, F. F. , Zhu, X. H. , Liao, W. J. , Liao, W. T. , Ding, Y. Q. , & Liang, L. (2019). CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial‐mesenchymal transition in colorectal cancer. Molecular cancer. 18(1), 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Cheng, L. , Turchinovich, A. , Mahairaki, V. , Troncoso, J. C. , Pletniková, O. , Haughey, N. J. , Vella, L. J. , Hill, A. F. , Zheng, L. , & Witwer, K. W. (2020). Influence of species and processing parameters on recovery and content of brain tissue‐derived extracellular vesicles. Journal of extracellular vesicles, 9(1), 1785746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z. , & Xu, A. (2021). Adipose Extracellular Vesicles in Intercellular and Inter‐Organ Crosstalk in Metabolic Health and Diseases. Frontiers in immunology. 12, 608680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, S N. , Sun, Li , Cole, K Y. , Ford, C R. , Olcese, J M. , & Meckes, D G. (2018). An optimized method for enrichment of whole brain‐derived extracellular vesicles reveals insight into neurodegenerative processes in a mouse model of Alzheimer's disease. Journal of neuroscience methods. 307, 210‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, S. N. , Olcese, J. M. , & Meckes, D. G. (2019). Extraction of Extracellular Vesicles from Whole Tissue. Journal of visualized experiments : JoVE. (144), e59143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt, G. W. , & Wilber, M. C. (1959). The storage of human tissues for surgical use. Postgraduate medical journal. 35(404), 338‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, M. , & Ilic, I. (2016). Epidemiology of pancreatic cancer. World journal of gastroenterology. 22(44), 9694‐9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro, K. , Yan, I. K. , & Patel, T. (2019). Isolation of Tissue Extracellular Vesicles from the Liver. Journal of visualized experiments : JoVE. (150), e58649. [DOI] [PubMed] [Google Scholar]
- Jang, SuC , Crescitelli, R. , Cvjetkovic, A. , Belgrano, V. , Olofsson Bagge, R. , Sundfeldt, K. , Ochiya, T. , Kalluri, R. , & Lötvall, J. (2019). Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. Journal of extracellular vesicles. 8(1), 1635420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan, N. , Lai, A. , Ormazabal, V. , Adam, S. , Guanzon, D. , Palma, C. , Scholz‐Romero, K. , Lim, R. , Jansson, T. , McIntyre, H. D. , Lappas, M. , & Salomon, C. . (2019). Adipose Tissue Exosomal Proteomic Profile Reveals a Role on Placenta Glucose Metabolism in Gestational Diabetes Mellitus. The Journal of clinical endocrinology and metabolism. 104(5), 1735‐1752. [DOI] [PubMed] [Google Scholar]
- Jensen, C. , & Teng, Y. (2020). Is It Time to Start Transitioning From 2D to 3D Cell Culture?. Frontiers in molecular biosciences. 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Qi , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of Exosome Composition. Cell. 177(2), 428‐445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingushi, K. , Uemura, M. , Ohnishi, N. , Nakata, W. , Fujita, K. , Naito, T. , Fujii, R. , Saichi, N. , Nonomura, N. , Tsujikawa, K. , & Ueda, K. (2018). Extracellular vesicles isolated from human renal cell carcinoma tissues disrupt vascular endothelial cell morphology via azurocidin. International journal of cancer. 142(3), 607‐617. [DOI] [PubMed] [Google Scholar]
- Jurj, A. , Pop‐Bica, C. , Slaby, O. , Ştefan, C. , Cho, W. C. , Korban, S. , & Berindan‐Neagoe, I. (2020). Tiny Actors in the Big Cellular World: Extracellular Vesicles Playing Critical Roles in Cancer. International journal of molecular sciences. 21(20), 7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczor‐Urbanowicz, KElż , Wei, F. , Rao, S. L. , Kim, J. , Shin, H. , Cheng, J. , Tu, M. , Wong, D. T. W. , & Kim, Y. (2019). Clinical validity of saliva and novel technology for cancer detection. Biochimica et biophysica acta. Reviews on cancer. 1872(1), 49‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarla, R. , Hur, J. , Kim, Y. , Kim, J. & Chwae, Y. ‐. J. (2020). Apoptotic cell‐derived exosomes: messages from dying cells. Experimental & molecular medicine. 52(1), 1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri, R. (2016). The biology and function of exosomes in cancer. The Journal of clinical investigation. 126(4), 1208‐1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri, R. , & Lebleu, V. S. (2020). The biology function and biomedical applications of exosomes. Science (New York, N.Y.) 367(6478). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra, H. , Drummen, G. , & Mathivanan, S. (2016). Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. International journal of molecular sciences. 17(2), 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, S. , Ridinger, J. , Rupp, A. ‐. K. , Janssen, JWg , & Altevogt, P. (2011). Body fluid derived exosomes as a novel template for clinical diagnostics. Journal of translational medicine. 9, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, J. F. R. , Wyllie, A. H. , & Currie, A. R. (1972). Apoptosis: a basic biological phenomenon with wide‐ranging implications in tissue kinetics. British journal of cancer. 26(4), 239‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck, E. , Iordanskaia, T. , Sevilla, S. , et al. (2014). Adipocyte exosomes induce transforming growth factor beta pathway dysregulation in hepatocytes: a novel paradigm for obesity‐related liver disease. The Journal of surgical research. 192(2), 268‐275. [DOI] [PubMed] [Google Scholar]
- Koliha, N. , Wiencek, Y. , Heider, U. , Jüngst, C. , Kladt, N. , Krauthäuser, S. , Johnston, I. C. D. , Bosio, A. , Schauss, A. , & Wild, S. (2016). A novel multiplex bead‐based platform highlights the diversity of extracellular vesicles. Journal of extracellular vesicles. 5, 29975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska, J. , Chmielik, E. , & Jarząb, B. (2017). Dynamic risk stratification in the follow‐up of thyroid cancer: what is still to be discovered in 2017?. Endocrine‐related cancer. 24(11), R387‐R402. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , & Reddy, P. (2016). Are circulating microRNAs peripheral biomarkers for Alzheimer's disease?. Biochimica et biophysica acta. 1862(9), 1617‐1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lener, T. , Gimona, M. , Aigner, L. , Börger, V. , Buzas, E. , Camussi, G. , Chaput, N. , Chatterjee, D. , Court, F. A. , Portillo, H. A. D. , O'driscoll, L. , Fais, S. , Falcon‐Perez, J. M. , Felderhoff‐Mueser, U. , Fraile, L. , Gho, Y. S. , Görgens, A. , Gupta, R. C. , Hendrix, An , Hermann, D. M. , Hill, A. F. , … Giebel, B. (2015). Applying extracellular vesicles based therapeutics in clinical trials ‐ an ISEV position paper. Journal of extracellular vesicles. 4, 30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, L. L. , Riaz, M. K. , Qu, X. , Chan, J. , & Meehan, K. (2021). Profiling of extracellular vesicles in oral cancer, from transcriptomics to proteomics. Seminars in cancer biology. 74, 3‐23. [DOI] [PubMed] [Google Scholar]
- Levy, E. (2017). In vivoExosomes in the Diseased Brain: First Insights from Studies. Frontiers in neuroscience. 11, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. ‐. J. , Fang, Q. ‐. H. , Liu, M.‐. L. , & Lin, J‐Na (2020). Current understanding of the role of Adipose‐derived Extracellular Vesicles in Metabolic Homeostasis and Diseases: Communication from the distance between cells/tissues. Theranostics. 10(16), 7422‐7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. ‐. Y. , Liu, Li‐Z , & Dong, M. (2021). Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Molecular cancer. 20(1), 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Hong, G. , Cheng, J. , Li, J. , Cai, H. , Li, X. , Guan, Q. , Tong, M. , Li, H. , & Guo, Z. (2017). Identifying Reproducible Molecular Biomarkers for Gastric Cancer Metastasis with the Aid of Recurrence Information. Scientific reports. 2016(6), 24869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Cheng, J. , Kong, X. , Li, X. , Li, X. , Zhang, M. , Zhang, H. , Yang, T. , Dong, Y. , Li, J. , Xu, Y. , & Yuan, Z. (2020). HDAC3 inhibition ameliorates ischemia/reperfusion‐induced brain injury by regulating the microglial cGAS‐STING pathway. Theranostics. 10(21), 9644‐9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares, R. , Tan, S. , Gounou, C. , Arraud, N. , & Brisson, A. R. (2015). High‐speed centrifugation induces aggregation of extracellular vesicles. Journal of Extracellular Vesicles. 4, 29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Yu, Z. , Yuan, S. , Xie, W. , Li, C. , Hu, Z. , Xiang, Y. , Wu, Na , Wu, L. , Bai, Li , & Li, Y. (2017). Circulating exosomal microRNAs as prognostic biomarkers for non‐small‐cell lung cancer. Oncotarget. 8(8), 13048‐13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, F. , Sleiman, M. , Sebastian, K. , Bogucka, R. , Jacobs, E. A. , & Adamson, A. S. (2021). UV Exposure and the Risk of Cutaneous Melanoma in Skin of Color: A Systematic Review. JAMA dermatology. 157(2), 213‐219. [DOI] [PubMed] [Google Scholar]
- Lőrincz, Á. M. , Timár, C. I. , Marosvári, K. A. , Veres, D. S. , Otrokocsi, L. , Kittel, Á. , & Ligeti, E. (2014). Effect of storage on physical and functional properties of extracellular vesicles derived from neutrophilic granulocytes. Journal of extracellular vesicles. 3, 25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall, J. , Hill, A. F. , Hochberg, F. , Buzás, E. I. , Di Vizio, D. , Gardiner, C. , Gho, Y. S. , Kurochkin, I. V. , Mathivanan, S. , Quesenberry, P. , Sahoo, S. , Tahara, H. , Wauben, M. H. , Witwer, K. W. , & Théry, C. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of extracellular vesicles. 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, G. , Song, X. , Yang, F. , et al. (2015). Exosomes derived from miR‐122‐modified adipose tissue‐derived MSCs increase chemosensitivity of hepatocellular carcinoma. Journal of hematology & oncology. 8, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Z. ‐. W. , Li, Fu‐X‐Zi , Liu, Yi‐W , Rao, S. ‐. S. , Yin, H. , Huang, J. , Chen, C. ‐. Y. , Hu, Y. , Zhang, Y. , Tan, Yi‐J , Yuan, L. ‐. Q. , Chen, T. ‐. H. , Liu, H. ‐. M. , Cao, J. , Liu, Z. ‐. Z. , Wang, Z. ‐. X. , & Xie, H. (2019). Aptamer‐functionalized exosomes from bone marrow stromal cells target bone to promote bone regeneration. Nanoscale. 11(43), 20884‐20892. [DOI] [PubMed] [Google Scholar]
- Maroto, R. , Zhao, Y. , Jamaluddin, M. , Popov, V. L. , Wang, H. , Kalubowilage, M. , Zhang, Y. , Luisi, J. , Sun, H. , Culbertson, C. T. , Bossmann, S. H. , Motamedi, M. , & Brasier, A. R. (2017). Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. Journal of extracellular vesicles. 6(1), 1359478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashouri, L. , Yousefi, H. , Aref, A. R. , Ahadi, A. M. , Molaei, F. , & Alahari, S. K. (2019). Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Molecular cancer. 18(1), 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarredona, E. R. , & Pastor, A. M. (2019). Extracellular Vesicle‐Mediated Communication between the Glioblastoma and Its Microenvironment. Cells. 9(1), 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, M. , Martin‐Jaular, L. , Lavieu, G. , & Théry, C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell‐to‐cell communication. Nature cell biology. 21(1), 9‐17. [DOI] [PubMed] [Google Scholar]
- Mathieu, M. , Névo, N. , Jouve, M. , Valenzuela, J. I. , Maurin, M. , Verweij, F. J. , Palmulli, R. , Lankar, D. , Dingli, F. , Loew, D. , Rubinstein, E. , Boncompain, G. , Perez, F. , & Théry, C. (2021). Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nature communications. 12(1), 4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan, S. , Ji, H. , & Simpson, R. J. (2010). Exosomes: extracellular organelles important in intercellular communication. Journal of proteomics. 73(10), 1907‐1920. [DOI] [PubMed] [Google Scholar]
- Matsumura, S. , Minamisawa, T. , Suga, K. , Kishita, H. , Akagi, T. , Ichiki, T. , Ichikawa, Y. , & Shiba, K. (2019). Subtypes of tumour cell‐derived small extracellular vesicles having differently externalized phosphatidylserine. Journal of extracellular vesicles. 8(1), 1579541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckiernan, J. , Donovan, M. J. , Margolis, E. , Partin, A. , Carter, B. , Brown, G. , Torkler, P. , Noerholm, M. , Skog, J. , Shore, N. , Andriole, G. , Thompson, I. , & Carroll, P. (2018). A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High‐grade Prostate Cancer in Patients with Prostate‐specific Antigen 2–10ng/ml at Initial Biopsy. European urology. 74(6), 731‐738. [DOI] [PubMed] [Google Scholar]
- Mears, R. , Craven, R. A. , Hanrahan, S. , Totty, N. , Upton, C. , Young, S. L. , Patel, P. , Selby, P. J. , & Banks, R. E. (2004). Proteomic analysis of melanoma‐derived exosomes by two‐dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 4(12), 4019‐4031. [DOI] [PubMed] [Google Scholar]
- Mege, D. , Panicot‐Dubois, L. , Ouaissi, M. , Robert, S. , Sielezneff, I. , Sastre, B. , Dignat‐George, F. , & Dubois, C. (2016). The origin and concentration of circulating microparticles differ according to cancer type and evolution: A prospective single‐center study. International journal of cancer. 138(4), 939‐948. [DOI] [PubMed] [Google Scholar]
- Mincheva‐Nilsson, L. , Baranov, V. , Nagaeva, O. , & Dehlin, E. (2016). Isolation and Characterization of Exosomes from Cultures of Tissue Explants and Cell Lines. Current protocols in immunology. 115, 14.42.11‐14.42.21. [DOI] [PubMed] [Google Scholar]
- Minciacchi, V. R. , Spinelli, C. , Reis‐Sobreiro, M. , Cavallini, L. , You, S. , Zandian, M. , Li, X. , Mishra, R. , Chiarugi, P. , Adam, R. M. , Posadas, E. M. , Viglietto, G. , Freeman, M. R. , Cocucci, E. , Bhowmick, N. A. , & Di Vizio, D. (2017). MYC Mediates Large Oncosome‐Induced Fibroblast Reprogramming in Prostate Cancer. Cancer research. 77(9), 2306‐2317. [DOI] [PubMed] [Google Scholar]
- Morello, M. , Minciacchi, V. , De Candia, P. , Yang, J. , Posadas, E. , Kim, H. , Griffiths, D. , Bhowmick, N. , Chung, L. , Gandellini, P. , Freeman, M. , Demichelis, F. , & Divizio, D. (2013). Large oncosomes mediate intercellular transfer of functional microRNA. Cell cycle (Georgetown, Tex. 12(22), 3526‐3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan‐Chari, V. , Clancy, J. , Plou, C. , Romao, M. , Chavrier, P. , Raposo, G. , & D'souza‐Schorey, C. (2009). ARF6‐regulated shedding of tumor cell‐derived plasma membrane microvesicles. Current biology : CB. 19(22), 1875‐1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka, S. , DeLeo, A. , Sethi, M. , et al. (2020). Proteomic and biological profiling of extracellular vesicles from Alzheimer's disease human brain tissues. Alzheimer's & dementia : the journal of the Alzheimer's Association. 16(6):896‐907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadatani, Y. , Watanabe, T. , Shimada, S. , Otani, K. , Tanigawa, T. , & Fujiwara, Y. (2018). Microbiome and intestinal ischemia/reperfusion injury. Journal of clinical biochemistry and nutrition. 63(1), 26‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, P. H. D. , Ma, S. , Phua, C. Z. J. , Kaya, N. A. , Lai, H. L. H. , Lim, C. J. , Lim, JQi , Wasser, M. , Lai, L. , Tam, W. L. , Lim, T. K. H. , Wan, W. K. , Loh, T. , Leow, W. Q. , Pang, Y. H. , Chan, C. Y. , Lee, S. Y. , Cheow, P. C. , Toh, H. C. , Ginhoux, F. , Iyer, S. , Kow, A. W. C. , Young Dan, Y. , Chung, A. , Bonney, G. K. , Goh, B. K. P. , Albani, S. , Chow, P. K. H. , Zhai, W. , & Chew, V. (2021). Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. Nature communications. 12(1), 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, V. V. T. , Witwer, K. W. , Verhaar, M. C. , Strunk, D. , & Balkom, B. W. M. (2020). Functional assays to assess the therapeutic potential of extracellular vesicles. Journal of Extracellular Vesicles. 10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z. , Zhou, S. , Li, S. , Kuang, L. , Chen, H. , Luo, X. , Ouyang, J. , He, M. , Du, X. , & Chen, L. (2020). Exosomes: roles and therapeutic potential in osteoarthritis. Bone Research. 8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niibe, Y. , & Hayakawa, K. Oligometastases and oligo‐recurrence: the new era of cancer therapy. Japanese journal of clinical oncology. 40(2), 107‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, B. T. , Teng, K. , Wu, C. , Adam, M. , & Johnstone, R. M. (1985). Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. The Journal of cell biology. 101(3), 942‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, B. T. , Teng, K. , Wu, C. , Adam, M. , & Johnstone, R. M. (1985). Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. The Journal of cell biology. 101(3), 942‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel, D. M. , & Gould, S. J. (2019). Exosomes. Annual review of biochemistry. 88, 487‐514. [DOI] [PubMed] [Google Scholar]
- Pospichalova, V. , Svoboda, J. , Dave, Z. , Kotrbova, A. , Kaiser, K. , Klemova, D. , Ilkovics, L. , Hampl, A. , Crha, I. , Jandakova, E. , Minar, L. , Weinberger, V. , & Bryja, V. (2015). Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. Journal of extracellular vesicles. 4, 25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesenberry, P. J. , Aliotta, J. , Deregibus, M. C. , et al. (2015). Role of extracellular RNA‐carrying vesicles in cell differentiation and reprogramming. Stem cell research & therapy. 6, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal, C. , & Harikumar, K. B. (2018). The Origin and Functions of Exosomes in Cancer. Frontiers in oncology. 8, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappa, G. , Mercapide, J. , Anzanello, F. , Pope, R. M. , & Lorico, A. (2013). Biochemical and biological characterization of exosomes containing prominin‐1/CD133. Molecular cancer. 12, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak, M. Z. , & Ratajczak, J. (2020). Extracellular microvesicles/exosomes: discovery, disbelief, acceptance, and the future?. Leukemia. 34(12), 3126‐3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, A. T. , Witwer, K. W. , Van Balkom, B. W. M. , De Beer, J. , Brodie, C. , Corteling, R. L. , Gabrielsson, S. , Gimona, M. , Ibrahim, A. G. , De Kleijn, D. , Lai, C. P. , Lötvall, J. , Del Portillo, H. A. , Reischl, I. G. , Riazifar, M. , Salomon, C. , Tahara, H. , Toh, W. S. , Wauben, M. H. M. , Yang, V. K. , … Lim, S. K. (2017). Concise Review: Developing Best‐Practice Models for the Therapeutic Use of Extracellular Vesicles. Stem cells translational medicine. 6(8), 1730‐1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, E. , Astsaturov, I. , Cukierman, E. , Denardo, D. G. , Egeblad, M. , Evans, R. M. , Fearon, D. , Greten, F. R. , Hingorani, S. R. , Hunter, T. , Hynes, R. O. , Jain, R. K. , Janowitz, T. , Jorgensen, C. , Kimmelman, A. C. , Kolonin, M. G. , Maki, R. G. , Powers, R. S. , Puré, E. , Ramirez, D. C. , …, Werb, Z. (2020). A framework for advancing our understanding of cancer‐associated fibroblasts. Nat Rev Cancer. 20(3), 174‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo, S. , Adamiak, M. , Mathiyalagan, P. , Kenneweg, F. , Kafert‐Kasting, S. , & Thum, T. (2021). Therapeutic and Diagnostic Translation of Extracellular Vesicles in Cardiovascular Diseases: Roadmap to the Clinic. Circulation. 143(14), 1426‐1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, J. , Kino, Y. , & Niida, S. (2015). MicroRNA‐Seq Data Analysis Pipeline to Identify Blood Biomarkers for Alzheimer's Disease from Public Data. Biomarker insights. 10, 21‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, R. , Patel, T. , & Freedman, J. E. (2018). Circulating Extracellular Vesicles in Human Disease. The New England journal of medicine. 379(10), 958‐966. [DOI] [PubMed] [Google Scholar]
- Sheridan, C. (2016). Exosome cancer diagnostic reaches market. Nature biotechnology. 34(4), 359‐360. [DOI] [PubMed] [Google Scholar]
- Siegel, R. , Miller, K. , & Jemal, A. (2019). Cancer statistics, 2019. CA: a cancer journal for clinicians. 69(1), 7‐34. [DOI] [PubMed] [Google Scholar]
- Siegel, R. , Miller, K. , & Jemal, A. (2020). Cancer statistics, 2020. CA: a cancer journal for clinicians. 70(1), 7‐30. [DOI] [PubMed] [Google Scholar]
- Siravegna, G. , Marsoni, S. , Siena, S. , & Bardelli, A. (2017). Integrating liquid biopsies into the management of cancer. Nature reviews. Clinical oncology. 14(9), 531‐548. [DOI] [PubMed] [Google Scholar]
- Smith, Z. J. , Lee, C. , Rojalin, T. , Carney, R. P. , Hazari, S. , Knudson, A. , Lam, K. , Saari, H. , Ibañez, E. L. , Viitala, T. , Laaksonen, T. , Yliperttula, M. , & Wachsmann‐Hogiu, S. (2015). Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. Journal of extracellular vesicles. 4, 28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria, F N. , Pampliega, O. , Bourdenx, M. , Meissner, W G. , Bezard, E. , & Dehay, B. (2017). Exosomes, an Unmasked Culprit in Neurodegenerative Diseases. Frontiers in neuroscience. 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, A. , Amreddy, N. , Razaq, M. , et al. (2018). Exosomes as Theranostics for Lung Cancer. Advances in cancer research. 139, 1‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbeek, S. C. , Pham, T. V. , Ligt, J. , Zomer, A. , Knol, J. C. , Piersma, S. R. , Schelfhorst, T. , Huisjes, R. , Schiffelers, R. M. , Cuppen, E. , Jimenez, C. R. , & Rheenen, J. (2018). Cancer cells copy migratory behavior and exchange signaling networks via extracellular vesicles. The EMBO journal. 37(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutermaster, B. A. , & Darling, E. (2019). M. Considerations for high‐yield, high‐throughput cell enrichment: fluorescence versus magnetic sorting. Scientific reports. 9(1), 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, Y. , Masuda, T. , Iinuma, H. , Yamaguchi, R. , Sato, K. , Tobo, T. , Hirata, H. , Kuroda, Y. , Nambara, S. , Hayashi, N. , Iguchi, T. , Ito, S. , Eguchi, H. , Ochiya, T. , Yanaga, K. , Miyano, S. , & Mimori, K. (2017). Circulating exosomal microRNA‐203 is associated with metastasis possibly via inducing tumor‐associated macrophages in colorectal cancer. Oncotarget. 8(45), 78598‐78613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavare, A. N. , Perry, N. J. S. , Benzonana, L. L. , Takata, M. , & Ma, D. (2012). Cancer recurrence after surgery: direct and indirect effects of anesthetic agents. International journal of cancer. 130(6), 1237‐1250. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. ‐. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , Beckham, C. , Bedina Zavec, A. , …, Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018), a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of extracellular vesicles. 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Zitvogel, L. , & Amigorena, S. (2002). Exosomes: composition, biogenesis and function. Nature reviews. Immunology. 2(8), 569‐579. [DOI] [PubMed] [Google Scholar]
- Thomou, T. , Mori, M. , Dreyfuss, J. , et al. (2017). Adipose‐derived circulating miRNAs regulate gene expression in other tissues. Nature. 542(7642), 450‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Ye , Gong, M. , Hu, Y. , Liu, H. , Zhang, W. , Zhang, M. , Hu, X. , Aubert, D. , Zhu, S. , Wu, L. , & Yan, X. (2020). Quality and efficiency assessment of six extracellular vesicle isolation methods by nano‐flow cytometry. Journal of extracellular vesicles. 9(1), 1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach, M. , & Théry, C. (2016). Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 164(6), 1226‐1232. [DOI] [PubMed] [Google Scholar]
- Tong, F. , Mao, X. , Zhang, S. , Xie, H. , Yan, B. , Wang, B. , Sun, Ji , & Wei, L. (2020). HPV + HNSCC‐derived exosomal miR‐9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer letters. 478, 34‐44. [DOI] [PubMed] [Google Scholar]
- Totani, L. , Plebani, R. , Piccoli, A. , Di Silvestre, S. , Lanuti, P. , Recchiuti, A. , Cianci, E. , Dell'elba, G. , Sacchetti, S. , Patruno, S. , Guarnieri, S. , Mariggiò, M. A. , Mari, V. C. , Anile, M. , Venuta, F. , Del Porto, P. , Moretti, P. , Prioletta, M. , Mucilli, F. , Marchisio, M. , Pandolfi, A. , Evangelista, V. , & Romano, M. (2017). Mechanisms of endothelial cell dysfunction in cystic fibrosis. Biochimica et biophysica acta. Molecular basis of disease. 1863(12), 3243‐3253. [DOI] [PubMed] [Google Scholar]
- Uchea, C. , Owen, S. F. , & Chipman, J. (2015). K. Functional xenobiotic metabolism and efflux transporters in trout hepatocyte spheroid cultures. Toxicology research. 4(2), 494‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman, W. M. , Pham, T. C. , Kwok, Y. Y. , Vu, L. T. , Ma, V. , Peng, B. , Chan, Y. S. , Wei, L. , Chin, S. M. , Azad, A. , He, A. B. ‐. L. , Leung, A. Y. H. , Yang, M. , Shyh‐Chang, Ng , Cho, W. C. , Shi, J. , & Le, M. T. N. (2018). Efficient RNA drug delivery using red blood cell extracellular vesicles. Nature communications. 9(1), 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Pol, E. , Coumans, F. A. W. , Grootemaat, A. E. , Gardiner, C. , Sargent, I. L. , Harrison, P. , Sturk, A. , Van Leeuwen, T. G. , & Nieuwland, R. (2014). Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. Journal of thrombosis and haemostasis : JTH. 12(7), 1182‐1192. [DOI] [PubMed] [Google Scholar]
- Van Der Vlist, E. J. , Nolte‐'T Hoen, E. N. M. , Stoorvogel, W. , Arkesteijn, G. J. A. , & Wauben, M. H. M. (2012). Fluorescent labeling of nano‐sized vesicles released by cells and subsequent quantitative and qualitative analysis by high‐resolution flow cytometry. Nature protocols. 7(7), 1311‐1326. [DOI] [PubMed] [Google Scholar]
- Van Engeland, M. , Nieland, L. J. W. , Ramaekers, F. C. S. , Schutte, B. , & Reutelingsperger, C. P. M. (1998). Annexin V‐affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 31(1), 1‐9. [DOI] [PubMed] [Google Scholar]
- van Niel, G. , Charrin, S. , Simoes, S. , et al. (2011). The tetraspanin CD63 regulates ESCRT‐independent and ‐dependent endosomal sorting during melanogenesis. Developmental cell. 21(4), 708‐721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos, M. H. , Caires, H. R. , Ābols, A. , Xavier, C. P. R. , & Linē, A. (2019). Extracellular vesicles as a novel source of biomarkers in liquid biopsies for monitoring cancer progression and drug resistance. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 47, 100647. [DOI] [PubMed] [Google Scholar]
- Vella, L J. , Scicluna, B J. , Cheng, L. , Bawden, E G. , Masters, C L. , Ang, C. ‐. S. , Willamson, N. , Mclean, C. , Barnham, K J. , & Hill, A F. (2017). A rigorous method to enrich for exosomes from brain tissue. Journal of extracellular vesicles. 6(1), 1348885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, A. , Herman, J. , Schulick, R. , Hruban, R. H. , & Goggins, M. (2011). Pancreatic cancer. Lancet (London, England). 378(9791), 607‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Yang, G. , Zhao, D. , Wang, J. , Bai, Y. , Peng, Q. , Wang, H. , Fang, R. , Chen, G. , Wang, Z. , Wang, K. , Li, G. , Yang, Y. , Wang, Z. , Guo, P. , Peng, L. , Hou, D. , & Xu, W. (2019). CD103‐positive CSC exosome promotes EMT of clear cell renal cell carcinoma: role of remote MiR‐19b‐3p. Molecular cancer. 18(1), 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh, J. A. , Pol, E. , Bettin, B. A. , Carter, D. R. F. , Hendrix, An , Lenassi, M. , Langlois, M. ‐. A. , Llorente, A. , Nes, A. S. , Nieuwland, R. , Tang, V. , Wang, L. , Witwer, K. W. , & Jones, J. C. (2020). Towards defining reference materials for measuring extracellular vesicle refractive index, epitope abundance, size and concentration. Journal of extracellular vesicles. 9(1), 1816641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, B. (2018). Aegle Therapeutics Corp. Receives IND Clearance from FDA for Stem Cell‐Derived Extracellular Vesicle Therapy for Burns (prweb.com). 5, 11. [Google Scholar]
- Willis, G. R. , Kourembanas, S. , & Mitsialis, S. A. (2017). Toward Exosome‐Based Therapeutics: Isolation, Heterogeneity, and Fit‐for‐Purpose Potency. Frontiers in cardiovascular medicine. 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms, E. , Cabañas, C. , Mäger, I. , Wood, M. J. A. , & Vader, P. (2018). Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Frontiers in immunology. 9, 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Soekmadji, C. , Hill, A. F. , Wauben, M. H. , Buzás, E. I. , Di Vizio, D. , Falcon‐Perez, J. M. , Gardiner, C. , Hochberg, F. , Kurochkin, I. V. , Lötvall, J. , Mathivanan, S. , Nieuwland, R. , Sahoo, S. , Tahara, H. , Torrecilhas, A. C. , Weaver, A. M. , Yin, H. , Zheng, L. , Gho, Y. S. , Quesenberry, P. , & Théry, C. (2017). Updating the MISEV minimal requirements for extracellular vesicle studies: building bridges to reproducibility. Journal of extracellular vesicles. 6(1), 1396823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Van Balkom, B. W. M. , Bruno, S. , Choo, A. , Dominici, M. , Gimona, M. , Hill, A. F. , De Kleijn, D. , Koh, M. , Lai, R. C. , Mitsialis, S. A. , Ortiz, L. A. , Rohde, E. , Asada, T. , Toh, W. S. , Weiss, D. J. , Zheng, L. , Giebel, B. , & Lim, S. K. (2019). Defining mesenchymal stromal cell (MSC)‐derived small extracellular vesicles for therapeutic applications. Journal of extracellular vesicles. 8(1), 1609206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfers, J. , Lozier, A. , Raposo, G. , Regnault, A. , Théry, C. , Masurier, C. , Flament, C. , Pouzieux, S. , Faure, F. , Tursz, T. , Angevin, E. , Amigorena, S. , & Zitvogel, L. (2001). Tumor‐derived exosomes are a source of shared tumor rejection antigens for CTL cross‐priming. Nature medicine. 7(3), 297‐303. [DOI] [PubMed] [Google Scholar]
- Wortzel, I. , Dror, S. , Kenific, C. M. , & Lyden, D. (2019). Exosome‐Mediated Metastasis: Communication from a Distance. Developmental cell. 49(3), 347‐360. [DOI] [PubMed] [Google Scholar]
- Xing, X. , Han, S. , Li, Z. , & Li, Z. (2020). Emerging role of exosomes in craniofacial and dental applications. Theranostics. 10(19), 8648‐8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, H. , Huang, Z. , Yang, Z. , Lin, Q. , Yang, B. , Fang, X. , Liu, B. , Chen, H. , & Kong, J. (2021). Recent Progress in Detection and Profiling of Cancer Cell‐Derived Exosomes. Small. 17(35), e2007971. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Greening, D. W. , Zhu, H. ‐. J. , Takahashi, N. , & Simpson, R. J. (2016). Extracellular vesicle isolation and characterization: toward clinical application. The Journal of clinical investigation. 126(4), 1152‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghoubi, Y. , Movassaghpour, A. , Zamani, M. , Talebi, M. , Mehdizadeh, A. , & Yousefi, M. (2019). Human umbilical cord mesenchymal stem cells derived‐exosomes in diseases treatment. Life sciences. 233, 116733. [DOI] [PubMed] [Google Scholar]
- Yajima, I. , Kumasaka, M. Y. , Thang, N. D. , Goto, Y. , Takeda, K. , Yamanoshita, O. , Iida, M. , Ohgami, N. , Tamura, H. , Kawamoto, Y. , & Kato, M. (2012). RAS/RAF/MEK/ERK and PI3K/PTEN/AKT Signaling in Malignant Melanoma Progression and Therapy. Dermatology research and practice. 2012, 354191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, T. , Cui, H. , Zhou, Y. , Yang, B. , Kong, P. , Zhang, Y. , Liu, Y. , Wang, B. , Cheng, Y. , Li, J. , Guo, S. , Xu, E. , Liu, H. , Cheng, C. , Zhang, L. , Chen, L. , Zhuang, X. , Qian, Yu , Yang, J. , … Cui, Y. (2019). Multi‐region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma. Nature communications. 10(1), 1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R. ‐. M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , & De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of extracellular vesicles. 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. , Dou, R. , Wei, C. , Liu, K. , Shi, D. , Zhang, C. , Liu, Q. , Wang, S. , & Xiong, B. (2021). Tumor‐derived exosomal microRNA‐106b‐5p activates EMT‐cancer cell and M2‐subtype TAM interaction to facilitate CRC metastasis. Molecular therapy: the journal of the American Society of Gene Therapy. 29(6), 2088‐2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D. , Zhang, W. , Zhang, H. , Zhang, F. , Chen, L. , Ma, L. , Larcher, L. M. , Chen, S. , Liu, N. , Zhao, Q. , Tran, P. H. L. , Chen, C. , Veedu, R. N. , & Wang, T. (2020). Progress, opportunity, and perspective on exosome isolation ‐ efforts for efficient exosome‐based theranostics. Theranostics. 10(8), 3684‐3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D. , Zhang, W. , Zhang, H. , Zhang, F. , Chen, L. , Ma, L. , Larcher, L. M. , Chen, S. , Liu, N. , Zhao, Q. , Tran, P. H. L. , Chen, C. , Veedu, R. N. , & Wang, T. (2020). Progress, opportunity, and perspective on exosome isolation ‐ efforts for efficient exosome‐based theranostics. Theranostics. 10(8), 3684‐3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L. , He, S. , Mao, X. , Cai, Y. , & Li, S. . (2020). Effect of Hepatic Macrophage Polarization and Apoptosis on Liver Ischemia and Reperfusion Injury During Liver Transplantation. Frontiers in immunology. 11, 1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamanchili, S. , Lamberty, B. , Rennard, D. , Morsey, B.M. , Hochfelder, C.G. , Meays, B. M. , Levy, E. , & Fox, H. S. , (2015). MiR‐21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease. PLoS pathogens. 11(7), e1005032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Quan, F. , Xu, J. , Zhang, Y. , Xie, Yi , Zhang, J. , Lan, Y. , Yuan, H. , Zhang, H. , Cheng, S. , Xiao, Y. , & Li, X. (2019). Breast cancer prognosis signature: linking risk stratification to disease subtypes. Briefings in bioinformatics. 20(6), 2130‐2140. [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Vu, L. , Ismail, N. , Minh T N Le, & Grimson, A. (2021). Landscape of extracellular vesicles in the tumour microenvironment: Interactions with stromal cells and with non‐cell components, and impacts on metabolic reprogramming, horizontal transfer of neoplastic traits, and the emergence of therapeutic resistance. Seminars in cancer biology. 74, 24‐44. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Nan, A. , Chen, L. , Li, X. , Jia, Y. , Qiu, M. , Dai, X. , Zhou, H. , Zhu, J. , Zhang, H. , & Jiang, Y. (2020). Circular RNA circSATB2 promotes progression of non‐small cell lung cancer cells. Molecular cancer. 19(1), 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Yin, T. , Xu, R. , Gao, W. , Zhao, H. , Shapter, J. G. , Wang, K. , Shen, Y. , Huang, P. , Gao, G. , Wu, Y. , & Cui, D. (2017). Large‐scale immuno‐magnetic cell sorting of T cells based on a self‐designed high‐throughput system for potential clinical application. Nanoscale. 9(36), 13592‐13599. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Ma, S. , Lv, J. , et al. (2021). The emerging role of exosomes in Alzheimer's disease. Ageing research reviews. 68, 101321. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Huang, F. , Li, W. , Dang, J. ‐. L. , Yuan, J. , Wang, J. , Zeng, D. ‐. L. , Sun, C. ‐. X. , Liu, Y. ‐. Y. , Ao, Q. , Tan, H. , Su, W. , Qian, X. , Olsen, N. , & Zheng, S. G. (2018). Human Gingiva‐Derived Mesenchymal Stem Cells Modulate Monocytes/Macrophages and Alleviate Atherosclerosis. Frontiers in immunology. 9, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, Y. , Liu, H. , & Tang, W. H. (2019). Exosomes: biogenesis, biologic function and clinical potential. Cell & bioscience. 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Yu, M. , Dong, J. , Wu, Y. , & Tian, W. , (2020). Identification of Novel Adipokines through Proteomic Profiling of Small Extracellular Vesicles Derived from Adipose Tissue. Journal of proteome research. 19(8), 3130‐3142. [DOI] [PubMed] [Google Scholar]
- Zhang, Yi , Bi, J. , Huang, J. , Tang, Y. , Du, S. , & Li, P. (2020). Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. International journal of nanomedicine. 15, 6917‐6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Gao, F. , Weng, S. , & Liu, Q. (2017). Pancreatic cancer and associated exosomes. Cancer biomarkers : section A of Disease markers. 20(4), 357‐367. [DOI] [PubMed] [Google Scholar]
- Zhou, B. , Xu, K. , Zheng, Xi , Chen, T. , Wang, J. , Song, Y. , Shao, Y. , & Zheng, S. (2020). Application of exosomes as liquid biopsy in clinical diagnosis. Signal transduction and targeted therapy. 5(1), 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Li, Z. , Qi, M. , Zhao, P. , Duan, Y. , Yang, G. , & Yuan, L. (2020). Brown adipose tissue‐derived exosomes mitigate the metabolic syndrome in high fat diet mice. Theranostics. 10(18), 8197‐8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znaor, A. , Lortet‐Tieulent, J. , Laversanne, M. , Jemal, A. , & Bray, F. (2015). International variations and trends in renal cell carcinoma incidence and mortality. European urology. 67(3), 519‐530. [DOI] [PubMed] [Google Scholar]


