Abstract
Gene duplication is a prevalent phenomenon across the tree of life. The processes that lead to the retention of duplicated genes are not well understood. Functional genomics approaches in model organisms, such as yeast, provide useful tools to test the mechanisms underlying retention with functional redundancy and divergence of duplicated genes, including fates associated with neofunctionalization, subfunctionalization, back-up compensation and dosage amplification. Duplicated genes may also be retained as a consequence of structural and functional entanglement. Advances in human gene editing have enabled the interrogation of duplicated genes in the human genome, providing new tools to evaluate the relative contributions of each of these factors to duplicate gene retention and the evolution of genome structure.
Keywords: Gene duplication, paralogs, whole genome duplication, evolution, genetic redundancy, functional divergence
Duplication across the tree of life
Gene duplication (Glossary) events span genomes of a wide range of organisms. Gene duplication has also been shown to play an important role in the emergence of complexity during eukaryogenesis [1] and it appears to shape the natural variation and disease states in humans [2–6]. The rates of gene duplication are estimated to be as frequent as the rates of single nucleotide po4lymorphisms [7]. Other observations, such as the prevalence of duplications within large-scale copy number substitutions [8] and the rate of de novo copy number variation (CNV) associated with neurocognitive disease in the human population [9], also support the high rate of gene duplications. With the exception of rare de-novo and horizontal gene transfer events, gene repertoires in all species are contemporary snapshots of selection and drift acting on gene duplication events [10].
Mechanisms that generate gene duplicates include ‘small-scale duplication’ (SSD) events, due to tandem or segmental gene duplications reflecting error prone DNA replication [11–13], and a variety of polyploidy events that lead to the simultaneous duplication of all genomic segments, termed ‘whole-genome duplication’ (WGD) [14, 15]. Genome sequencing and mapping-based strategies have identified and classified duplicated genes resulting from these different mechanisms. For example, in yeast, ~18% of genes represent duplicates originating from WGD [14, 16], whereas ~30% of genes appear to stem from SSD [17] (Table). In humans, ~26% and ~5% of genes are duplicates that originate from ancient WGD events and various SSD events, respectively (Table) [18, 19].
Table.
Prevalence of gene duplicates across eukaryotes
| Common name | Scientific name | Total no. genes in genome | No. WGD rounds | % WGD duplicates in genome | % SSD duplicates in genome |
|---|---|---|---|---|---|
| Bacteria | Haemophilus influenzae | 1709 [129] | - | - | 17 [129] |
| Yeast | Saccharomyces cerevisiae | 6605* | 1 [14, 16] | 18 [14, 16] | 30 [17] |
| Plant | Arabidopsis thaliana | 26028 [130] | Multiple [130] | 29–59 [130] | 40 [131] |
| Worm | Caenorhabditis elegans | 20140 [132] | - | - | 33 [133] |
| Fly | Drosophila melongaster | 13601 [129] | - | - | 41 [129] |
| Human | Homo sapiens | 22980 [18] | 2 [18] | 26 [18] | 5 [19] |
YeastMine downloaded March 3, 2021 (ORF count includes genes of unknown and putative function)
Structural variation leading to differences in human haplotypes [20] is highly prevalent, such that no two people, even identical twins [21], have the same number of genes. Indeed, a survey of over 100,000 healthy individuals detected thousands of CNVs with more than half of them containing at least one gene associated with duplicate gene pair [22]. While most CNVs were recurrent and present in at least two individuals, the individual CNVs were rare since most were found at a frequency of 0.01 [22]. Because CNVs are individually rare, but collectively common, they may also contribute to the ‘missing heritability’ in human genome-wide association studies (GWAS) [22].
Most duplicated genes become non-functional by a process termed ‘nonfunctionalization’ [23]. However, those duplicates that are retained are thought to be preserved by a variety of mechanisms, including neofunctionalization [24], functional specialization by subfunctionalization [23], dosage amplification [25] and back-up compensation [26]. Interestingly, despite various modes of divergent evolution, many duplicated genes retain some level of functional redundancy [27, 28]. If genetic redundancy is inherently evolutionarily unstable due to accumulation of mutations in a functionally redundant gene, then what are the mechanisms that allow duplicated genes to persist in genomes with some level of functional overlap? A recent study using complex genetic interaction analysis supports a ‘structural and functional entanglement’ model of duplicated gene divergence [27], which suggests that the evolutionary fate of a duplicated gene is dictated by an interplay of factors that enable paralog-specific roles to evolve while simultaneously stably maintaining functional redundancy and is consistent with other studies based on simulations and literature-curation [26, 29–31]. This review will discuss functional redundancy and divergence of duplicated genes as revealed by the advances in functional genomics approaches with model systems, including yeast and human cells.
Mechanisms of gene duplication
Small-scale duplication
SSD is thought to occur through a variety of mechanisms [32] (Figure 1). Tandem duplication is observed when genes with similar sequences are detected in close proximity to each other, and this gene arrangement is often associated with the presence of repetitive sequences. Repeat sequences can generate duplicated genes by non-allelic homologous recombination due to unequal exchange between two chromatids on the same chromosome [11] or uneven crossing-over during meiosis prophase I [12]. Non-homologous recombination mechanisms involving replication accidents that lead to chromosome breakage can also give rise to duplications [13]. Horizontal (or lateral) gene transfer from one organism to another is considered another mechanism for generating duplicated genes, albeit rare [33]. Transposition events can also lead to gene duplications, and they are usually evident by the conserved terminal sequences that flank the duplicated segments [34, 35].
Figure 1.

Summary of mechanisms of gene duplication. Small scale duplication are thought to result from (a) tandem duplication, which can result from unequal exchange either between sister chromatids in mitosis or homologous chromosomes in meiosis I or non-allelic homologous recombination resulting from a misalignment of repetitive sequences; and transposition, which carries a locus from one position to another via RNA or DNA intermediates. (b) Duplication of the entire genome happens through autoploidy or alloploidy. Black arrows represent a duplication event. Grey rods represent a chromosome. Coloured blocks depict a locus.
Whole genome duplication
WGD can occur through a transient polyploid state of an organism (Figure 1). The paralogs generated by whole genome duplication are often called ‘ohnologs’ in reference to Susumo Ohno, who famously stated that gene duplication was a major evolutionary force, and proposed that at least one WGD occurred in the common ancestor of all vertebrates [24]. The ‘2R hypothesis’ is well supported and refers to evidence of a first WGD event that occurred after the divergence of invertebrates but before jawless vertebrates, followed by a second WGD which preceded the emergence of mammals [18]. A third WGD event (‘3R’) is fish-specific and occurred in the common ancestor of all teleost fish [36, 37]. The evolution of sex chromosomes in some animals (especially in mammals and birds) appears to inhibit WGD [38]. WGD disrupts sex determination in species that determine sex by the ratio of sex chromosomes to autosomes [39]. The disruptive effect of WGD is also apparent in dioecious species that developed dosage compensation mechanisms to maintain the stoichiometric expression of sex-linked genes across species [40]. Interestingly, in plants, WGD appears to provide advantages under periods of abiotic and biotic stress [41].
WGD lacks the major challenges associated with segmental duplications, as it avoids dosage imbalances for functionally-related genes, members of protein complexes, and structural genes that are duplicated along with their corresponding regulators [24]. Genome duplication can arise either through auto- or allo-polyploidization. Autopolyploidy is an intraspecies event and occurs when cytokinesis fails early during development, or if there is a fertilization event involving unreduced (i.e. diploid) gametes [42]. Although WGD lineages are rarely established, mutations that lead to WGD events are common in human embryos. A survey of ~1000 chorionic villi revealed that up to 20% of pregnancies, which result in miscarriages, exhibit trisomy, triploidy and tetraploidy [43]. Laboratory evolution experiments involving 46 haploid Saccharomyces cerevisiae populations that were evolved for over 4,000 generations reported frequent genome duplications, which often confer a fitness advantage [44]. Allopolyploidy, which is due to an interspecies cross, is another mechanism that essentially generates WGDs. Most such hybrid organisms are sterile because of the absence of pairing between similar but nonhomologous chromosomes [42]. However, tetraploidy can solve this problem of sterility by providing a true homologous chromosome for pairing [42].
In yeast, WGD appears to have occurred approximately 100 Mya after the divergence of Kluyveromyces from Saccharomyces lineages [14, 15]. The timing of the duplication event is inferred from protein and nucleotide sequence alignments, which show that some regions in K. waltii correspond to two regions in the S. cerevisiae genome with blocks of conserved synteny, which are characterized by chromosomal regions where genes lie in the same order in both species [14]. A model has been proposed to explain the emergence of a novel yeast species from a polyploidy event involving fusion of two haploids from different species [45, 46]. The sterile hybrid divided mitotically until it lost a MAT locus, becoming functionally haploid, followed by mother-daughter mating to generate a diploid that resulted in a separate lineage. A recent phylogenetic analysis showed that there is evidence for an ancient interspecies hybridization that predates the expected WGD occurring before the divergence of Saccharomyces and a clade containing the genera Kluyveromyces, Lachancea and Eremothecium (Ashbya gossypii) [47]. Duplicated genes resulting from this ancient allopolypolidization event are sometimes referred to as ‘homeologs’. In S. cerevisiae there are 166 individual homeologs [47, 48] of 490 total individual paralogs that were previously reported to have originated from WGD [16]. Authors propose that autopolyploidy-driven WGD which followed the interspecies hybridization, enabled the sterile hybrid to regain fertility thereby providing an initial selective advantage.
Models of duplicated gene evolution
Neofunctionalization
The idea that gene duplication is important for the evolution of novel elements for organismal complexity first emerged around ~70 years ago [49, reviewed in 50]. In 1970 Susomo Ohno formally conceptualized this phenomenon terming it ‘neofunctionalization’ proposing that gene duplication provides a molecular landscape for functional innovation as the redundant copy can escape the constraints of natural selection and is free to acquire normally ‘forbidden mutations’, thereby allowing for the development of a novel or more specialized function [24] (Figure 2). Some of the examples of neofunctionalization include enzymes belonging to the fungal maltase family that encode α-glucosidases allowing yeast to metabolize complex carbohydrates. Maltase family of paralogs evolved a diversity of substrate specificities and may have enabled fungi to colonize new niches containing sugars provided by the emergence of angiosperms and fleshy fruits that could now be hydrolyzed by the novel Mal (Ima) enzymes [51]. Neofunctionalization can also lead to the evolution of specialization of gene expression by regulatory landscape remodeling and recruitment of novel regulatory elements. This is thought to underly morphological specialization in vertebrates as gleaned from comparative analysis of amphioxus, zebrafish, medaka and mouse [52] as well as the Atlantic salmon [53].
Figure 2.

Duplicate gene divergence by subfunctionalization or neofunctionalization. Duplicated gene divergence may proceed by subfunctionalization, which refers to the retention of partitioned complementary subfunctions of an ancestral gene (duplicates undergo ‘division of labor’) or by neofunctionalization, whereby over time one duplicate accumulates mutations and evolves a novel function which is not performed by the ancestral gene. Hypothetical different functions are illustrated in different colours.
There are numerous examples of neofunctionalized paralogs in different organisms; however, it is still unclear what is the extent of neofunctionalization among duplicates in a given species relative to paralogs retained by other mechanisms. The analysis of protein-interaction data in yeast revealed that paralogs that are annotated to different biological processes, which is indicative of a derived function, are less frequent than co-annotated pairs [54]. Another study argued that prolonged neofunctionalization follows rapid subfunctionalization, a conclusion based on the findings that the total number of protein-protein interactions and the total number of expression sites for duplicated genes is similar to two randomly chosen singletons in yeast and human cells indicating that duplicated genes act as two singletons rather than together sum up to one singleton [55]. Neofunctionalization can also result from subcellular reprogramming, which occurs when sequence changes to protein targeting regions generate new localization patterns [56]. However, the contribution of subcellular reprogramming to duplicate retention has been questioned given the similarity in frequency with which it also happens in singletons [57].
The ‘innovation-amplification-divergence’ model is an update to the neofunctionalization model [58]. It postulates that if a gene harbours a secondary nonessential weak function that becomes advantageous for survival (e.g. due to a change in the environment), then gene duplication can provide a selective advantage, first by increasing dosage of the limiting gene product and then providing an opportunity for specialization while still retaining the original parental function. This pattern was observed in experiments with Salmonella enterica. The HisA gene, which is involved in the biosynthesis of histidine, has a low level of TrpF activity, which is important for the biosynthesis of tryptophan. When grown on selective media, HisA gene can duplicate and the two paralogs specialized to either perform HisA or TrpF specific activity, a process that was observed to occur in just 3000 generations.
Subfunctionalization
Retention of duplicates may result from subfunctionalization, which is thought to occur when duplicates degenerate in function but are retained since they each provide a distinct component of the ancestral gene function, as postulated by the duplication-degeneration-complementation model (DDC) [23] (Figure 2). The concept of a distribution of multiple functions between duplicated genes that results in their specialization was first proposed by Aleksandr S. Serebrowsky, as described for scute and achaete genes that control bristle development and reside on the X chromosome in Drosophila melongaster [59]. The entire fly body is covered with bristles and these duplicated genes control distinct subsets of bristles.
A well-known example of subfunctionalization (indeed the origin of the term) involves engrailed paralogs in zebra fish that partitioned its expression, such that in the rayfinned lineage eng1 is expressed in the pectoral appendage bud and eng1b in the hindbrain/spinal cord neuron. In contrast, the most recent unduplicated ‘pro-ortholog’ [60] in chicken and mouse, En1, is expressed in the pectoral appendage bud and the hindbrain/spinal cord [23]. Floral homeotic genes in maize represent another example of duplicated genes that diverged by subfunctionalization since ZAG1 shows a high expression during maize carpel development and ZMM2 is expressed highly in maize stamens whereas the pro-ortholog in Arabidopsis and Antirrhinum are strongly expressed in both developing carpels and stamens [23].
Other examples of duplicated genes that evolved in a manner consistent with the subfunctionalization model have been reported in yeast and show partitioning of different biochemical functions. They include ORC1 and SIR3, which are involved in the origin recognition complex that is required for DNA replication and gene silencing, respectively, SNF12 and RSC6 which are involved in chromatin remodeling, RNR2 and RNR4, which are R2 subunits of ribonucleotide reductase and SKI7 and HBS1, which plays a role in RNA processing and translation [61]. In all of these examples, the loss of function mutation in both paralogs can be rescued by their pro-ortholog from, S.kluyveri, suggesting that they perform subfunctions of the single-copy ancestral gene [61]. In fact, the WGD paralog pair SKI7-HBS1 was shown to result from fixation of splice variants, which were derived from an ancestral alternatively-spliced multifunctional protein [62]. Another study characterized the evolutionary history of the components of COPII (coat protein complex II), which is important for forming membrane vesicles to transport proteins and lipids from the ER to the Golgi [63]. This phylogenetic analysis across 74 eukaryotic genomes revealed cases of subfunctionalized paralogs, such as SEC23–SEC24, which partitioned GAP activity and cargo-binding functions that may have provided a selective advantage by increasing the number or specificity of cargo proteins. Partitioning of gene expression regulatory elements has also been demonstrated in yeast [64], and human cells [65], where it leads to paralog tissue-specific expression, as well as partitioning of subcellular localization niches [56]. The specialization accomplished by subfunctionalization may contribute to modularizing of the molecular network simplifying a system [66].
Recently, a specialized version of DDC has been proposed, ‘dosage subfunctionalization’, which posits that after a duplication event paralogs are under dosage constraints and stochastic changes in gene expression are tolerated because together they sum to the pro-ortholog gene expression level contributing to the persistence of such divergently expressed paralogs [67].
‘Escape from adaptive conflict’ (EAC) is a mechanism of subfunctionalization and proposes that ‘gene sharing’ precedes gene duplication, such that the ancestral gene plays a role in more than one process or carries out more than one function and thus it is ‘shared’ between processes. Following duplication, each member of the duplicated gene pair would separately optimize those functions, which an otherwise multifunctional ancestral gene would not be able to accomplish [68]. For example, while crystallin has multiple enzymatic functions, it has also been recruited to a structural role in the lens. Its role as a structural protein does not require enzymatic activity, presenting it with an adaptive conflict, which was resolved by gene duplication and a subsequent separation of function [68]. Duplicated genes of the anthocyanin biosynthetic pathway in morning glories (Ipomoea purpurea) are also thought to have resulted from EAC, enabling them to increase the ability to metabolize different flavonoid substrates, a process that was accomplished less effectively by the ancestral gene [69].
EAC differs from the original definition of subfunctionalization, which involves neutral or deleterious mutations, by primarily involving adaptive substitutions. For example, a single base deletion can lead to a translational frameshift unveiling cryptic localization sequences [70]. Adaptive EAC allowed IDP2 and IDP3 WGD paralogs, which are NADP-dependent isocitrate dehydrogenases involved in catalyzing oxidation of isocitrate to alpha-ketoglutarate, to evolve exclusive cytosolic or peroxisomal localization compared to the ancestral gene, whose product is found in both subcellular compartments (i.e. ‘shared’) with a ‘weak trade-off’. Accordingly, reducing the levels of the cytosolic IDP by ~25% was sufficient to shift from a no-growth phenotype on petrosalinate to a growth phenotype that is only half of that observed with the ‘legitimate’ peroxisomal IDP3 protein. However, a mutation resulting in exclusive peroxisomal targeting enabled wild-type growth rates, which was conferred by duplication promoting phenotypic diversity that mediates survival in challenging environments. This finding advocates for ‘divergence before duplication’, whereby the mutations that lead to the emergence of a new function do not also undermine the original function, as postulated by neofunctionalization.
Gene duplication can also facilitate escape from adaptive conflict by resolving the conflict between conditionally responsive gene expression i.e. plasticity and expression noise i.e. stochastic cell-to-cell-variation [71, 72]. Evolutionary analysis in yeast revealed that duplicates of all ages, including SSD and WGD, are characterized by high plasticity and high noise [71]. This plasticity-noise coupling is facilitated by TATA promoters which are enriched in duplicates compared to singletons and to genes that reverted to singleton state after WGD. It was proposed that the evolution of highly responsive gene expression is limited in genes before duplication due to the detrimental consequences of noise, whereas the variation may be better tolerated after duplication as a result of functional compensation. A more specific example of this phenomenon was recently shown for Msn2-Msn4 paralog pair of transcription factors that exhibit similarity in their nuclear translocation dynamics and regulation as well as binding to the same target genes [72]. However, gene duplication enabled Msn2 to adopt a low-noise basal expression, whereas it increased the dynamic range and expression noise of Msn4 providing an opportunity for yeast to evolve a phenotypically adaptive expression tuning.
‘Minimization of paralog interference’ is another potential mechanism of subfunctionalization [73]. In this scenario the ancestral gene product participates in cooperative assemblies connected by protein-protein or protein-nucleic acid interactions. Immediately upon duplication paralogs would compete for those interactions and then partitioning of their interacting domains would resolve this interference. This model is consistent with duplicate gene divergence of MADS-box transcriptional regulator, which is found in all fungi and regulates the expression of a wide array of genes, including those involved in mating and arginine metabolism. Through degeneration of sites that mediate protein-protein interactions, Mcm1 lost its interaction with Arg81, whereas Arg80 lost its interaction with the MATα1 gene product, resulting in the divergence of the gene sets they activate while minimizing the competition for binding to each other’s partners.
Robustness against genetic or environmental perturbations
Gene duplicates may also be retained for back-up compensation (Figure 2). Although, some population genetic theories suggest that duplication is inherently genomically unstable due to accumulation of mutations [74], others show support for active selection of redundancy [26]. In general, the analysis of the yeast deletion collection showed a lower fitness cost was associated with deletion of a duplicated gene compared to a singleton gene, highlighting the important role that duplicated genes play in genetic robustness [75]. The systematic analysis of double gene deletion mutants in yeast showed that ~30% of duplicated genes display negative genetic interactions with each other, suggesting of a significant level of buffering associated with functional redundancy [76]. There have also been reports of “responsive backup circuits” that exist across various species in which a redundant gene copy is up-regulated when its paralog is subjected to an inactivating perturbation [77–79]. Detailed examination protein-protein interaction networks of 56 paralog pairs, with and without the deletion of the corresponding paralog revealed evidence of widespread compensation [28]. In total, 40% of tested pairs showed an increase of the number of detected protein interactions for the remaining paralog in response to the deletion of its sister.
Buffering capacity has also been explored under alternate growth conditions, providing evidence for duplicates contributing to adaptability to environmental insults [25, 80]. In particular, partially shared regulatory motifs predicted transcriptional patterns that provide evidence for a backup expression response, which may be important for growth across different conditions [81]. An evolution experiment of an S. cerevisiae msh2Δ strain, which was passaged over 2200 generations, revealed duplicated genes showed a greater phenotypic plasticity as measured by greater variation in gene expression when grown in various environmental stress conditions. Moreover, the paralogs tended to show stress-specific transcriptional plasticity compared to singletons that are more likely to respond more generally to all stress conditions [82, 83]. Transcriptional plasticity conferred by duplicated genes has also been demonstrated using MSN2-MSN4 paralogs, which encode stress responsive transcription factors [72]. Despite exhibiting similarity in their nuclear translocation dynamics and target gene regulation, duplication enabled Msn2 to adopt a low-noise basal expression whereas for Msn4 it enabled an increase in its dynamic range and expression noise. Thus, gene duplication led to the evolution of cooperative ‘two-factor dynamics’ resulting in a phenotypically adaptive expression tuning.
The contribution of paralogs to robustness has also been shown for duplicated genes in human cells. Paralogs protect against the deleterious effect of loss of function mutation across a panel of 455 human cell lines screened against a genome-wide CRISPR-Cas9 system for loss‐ of‐ function (LOF) mutations [84]. The CRISPR score per gene, which reflects the depletion of gRNA (guide RNA) and serves as a proxy for the relative deleteriousness of the LOF mutation on cell proliferation, is generally higher for duplicates than singletons. In another systematic study, humanized yeast cells were first generated by replacing yeast genes with their corresponding human homologs, focussing on duplicated genes, and then their compensatory ability was tested using growth rescue assays [85, 86]. By testing human–yeast ortholog pairs belonging to a variety of cellular processes, this growth assay revealed that multiple members of certain gene families are able to replace the essential roles of their yeast orthologs, thereby indicating their functional overlap. Thus, redundancy conferred by gene duplication results in back-up compensation and appears to confer fitness advantages in human cells.
Dosage amplification and stoichiometric balance
Increased gene dosage has been proposed to lead to fixation of duplicated genes by conferring a selective advantage [25, 87] (Figure 2). This concept was first proposed ~60 years ago by I. A. Rapoport who argued that duplicates are preserved directly by their impact on fitness exerted from multiple gene copies [88]. Since the yeast whole genome duplication occurred around the same time as the evolutionary emergence of large, glucose-rich fruit, fixation of duplicates involving glycolytic genes may have been favourable [89]. Increased dosage of glycolytic enzymes may have produced higher glycolytic flux, resulting in a faster growth rate, which appears to be favoured by selection despite reduced efficiency of fermentation. High enzymatic flux associated with retention of duplicated genes has also been shown in other studies [90, 91]. However, more recent work suggests that gene dosage effects of duplicates may not be sufficient to explain the fermentative capacity of yeast [92]. Interestingly, it has been suggested that since gene duplicates are significantly over-represented among vesicle trafficking genes in yeast, it is possible that their increased gene dosage was selected for enhancing the capacity of the secretory and endocytic vesicle trafficking systems [93]. Duplication of single genes has also been shown to be more likely to be associated with fitness gain and thus adaptability in yeast experimental evolution using pooled competition assays in nutrient-limiting conditions [94].
En masse duplication from WGD events would ensure proper stoichiometry of key protein complexes [95, 96]. Maintaining a stoichiometrically balance in gene dosage is especially apparent for genes encoding subunits of the ribosome, the majority of which are duplicated [30]. Dosage amplification is also illustrated by yeast genes encoding histones, which are identical in coding sequence, expressed at high and similar copy number. Indeed, yeast histone genes show many shared negative genetic interactions consistent with their inability to fully buffer each other due to being selected for dosage amplification [97]. Genetic tug-of-war (gTOW) experiments in yeast demonstrated that protein complex subunits (including duplicated genes) respond to each other’s change in copy number by post-transcriptional regulation [98] rather than protein synthesis feedback regulation as evident by ribosome profiling [99]. For example, if a subunit of a protein complex is reduced, then another complex member would exhibit a decrease in protein abundance due to enhanced rate of protein degradation by the proteasome and vice versa. Additionally, a systematic analysis of protein complexes in yeast revealed that a reduction in the activity of a protein complex is caused by a deletion rather than an overexpression of one of the subunits [100]. It was proposed that the tolerance of protein complexes to subunit overexpression may facilitate evolution of novel complexes that are able to duplicate with no adverse phenotypic effect. Another study showed that a set of haploinsufficient genes tend to also result in a growth defect when they are tested in a haploid organism which gained an extra gene copy from a centromeric plasmid under the regulation of its native promoter [101]. Duplicated genes were also suggested to buffer against fluctuations in gene expression thereby escaping haploinsufficiency. In human cells, the combined analysis of mRNA expression across 374 cell lines and protein expression across 49 cell lines revealed that paralogs often show symmetric expression in heteromers, a finding that differs from that of nonheteromeric paralogs, indicating that paralogs that form heteromeric complexes are more dosage balanced than nonheteromeric paralogs [84], a finding that suggests heteromeric paralogs are under selection to maintain stoichiometric balance.
Structural and functional entanglement
Systematic analysis of complex genetic interactions in yeast has recently been used to illuminate evolutionary trajectories of duplicated genes [27]. A complex genetic interaction occurs when a perturbation in three or more genes results in an unexpected effect on fitness, given the effects of lower order combinations of genetic perturbations [102]. A systematic analysis of trigenic interactions involving LOF mutations of three genes revealed insight into the key role of complex genetic interactions in the genotype-to-phenotype relation and genome evolution [103]. Trigenic interaction profiles of 240 double mutants and their corresponding single mutants, involving pairs of dispensable WGD gene duplicates, generated ~550,000 double and ~260,000 triple mutants, which revealed ~4700 negative and ~2500 positive digenic interactions and ~2500 negative and ~2100 positive trigenic interactions [27]. Functional specificity of paralogs was captured by negative digenic interactions, whereas the core functionality shared by paralogs was captured by negative trigenic interactions (Figure 3). The extent of the functional overlap between ohnologs was gauged by their ‘trigenic interaction fraction’, which quantified the fraction of their negative trigenic interactions relative to the total negative, trigenic plus digenic, interactions. A bimodal distribution of the trigenic interaction fraction revealed two basic paralog classes, a functionally redundant class and another more divergent one (Figure 3). Using correlation of position-specific evolutionary rate patterns between paralog proteins in relation to that of the pre-WGD homolog, as specified in the Yeast Gene Order Browser [16], showed that the incomplete subfunctionalization of functionally overlapping paralogs was due to structural constraints acting on the protein sequence. This finding was also supported by in silico modeling that demonstrated that paralogs which began their trajectory with an increasing extent of overlapping functions (i.e. ‘entangled’) evolved asymmetrically (partitioned domains unequally to each paralog). These paralogs also reached steady state with a higher range of functional overlap, which appears to be associated with constrained domains at steady state. These findings suggested a ‘structural and functional entanglement’ model of evolution of paralogs, in which highly entangled duplicates reverted to a singleton state; those that were minimally entangled and unconstrained diverged; and those with intermediate level of entanglement that were somewhat constrained diversified and evolved paralog specific functions, while retaining functional overlap at steady-state (Figure 3).
Figure 3.
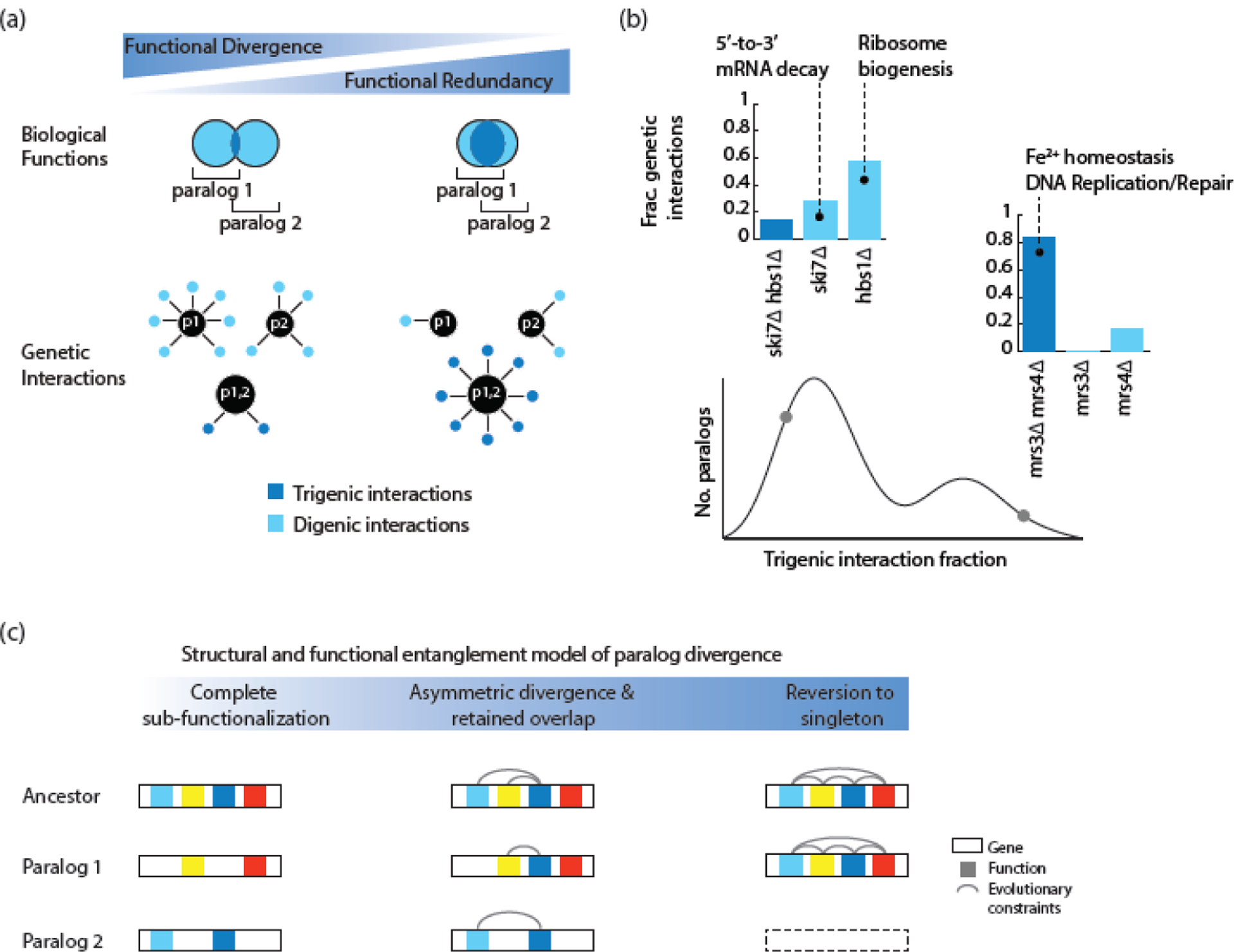
The structural and functional entanglement model of paralog divergence. (a) Digenic and trigenic interactions reveal paralog-specific and redundant functions as denoted by light blue and dark blue colours, respectively. (b) Distribution of negative trigenic interaction fraction obtained from screening 240 double mutants and 480 single mutants involving dispensable duplicated genes for digenic and trigenic interactions [27]. Examples of functionally divergent (SKI7-HBS1) and redundant (MRS3-MRS4) paralog pairs are depicted. Their respective trigenic interaction fractions are shown using a grey circle. (c) Members of a duplicated gene pair will diverge by subfunctionalization if their structure and function are modular and are composed of partitionable functions (left). A duplicated gene pair that is highly structurally and functionally entangled will tend to revert to a singleton state because one of its paralogs will rapidly degenerate by accumulating intrinsically deleterious mutations (right). Duplicated genes that are characterized by an intermediate level of structural entanglement at the time of duplication will tend to partition some and retain some overlapping functions, allowing for both specialization and retention of a common activity (center). This figure was adapted from a previous publication [27].
It was shown that one of the members of duplicated gene pairs tend to become nonfunctional within a few million years, and thus those that have been retained for over 100 million years post-WGD represent stably-retained duplicated genes in the genome [7]. Structurally and functionally overlapping ancestral domains have been proposed as constraints that prevent the complete divergence of duplicated genes, resulting in incomplete subfunctionalization [30]. Other studies that conducted in silico simulations, which were based on mutation rates of genes and the varying contribution of their functions to overall fitness, have also revealed instances of indefinite retention of paralogs with functional redundancy [26, 29, 31]. Simulation of duplicated gene evolution within a protein complex has been tested using a simulation platform for protein evolution [104, 105]. The results of these simulations, based on a ubiquitin-like protein and its binding peptide, showed that a duplicated gene encoding a ubiquitin-like protein can diverge through dosage imbalance [106]. As a duplicated gene diverges, the binding partner of its product will acquire substitutions to enhance their protein-protein interactions, thereby revealing that the protein interface constrains the evolution of the proteins within the context of a complex. These findings are consistent with another study that analyzed protein-protein interaction data based on PCA and crystal structures, showing that functional divergence of duplicated genes is impacted by the binding interface required for maintenance of heteromeric complexes involving duplicates [48]. These findings support the potential for paralogs to evolve divergently within the context of structural constraints, thereby maintaining core functionality while also developing a functional specificity.
Concluding remarks
To understand the forces that shape genomes, it is imperative to understand the evolution of duplicated genes. While functional genomics tools have certainly enabled us to explore the mechanisms that lead to the functional divergence of duplicated genes more deeply, it is important to further refine our experimental techniques to learn about the functional relationship between paralogs, especially since the current limiting factor is the sparsity of data associated with some methodologies. More recent developments in complex genetic interaction mapping and phenomic experimental approaches and the associated machine learning analytics will undoubtedly be useful to dissect the precise roles of duplicated genes.
The large majority of studies have been conducted in standard growth conditions and the few studies that investigated conditional activation of duplicated genes have been limited to the assessment of their transcriptional plasticity under environmental stress conditions. Using other functional readouts, such as genetic interaction profiling, protein interaction profiling, as well as phenomic analyses, is key to gain insight into the role that adaptation to environmental changes impacted duplicate gene retention (see Outstanding Questions).
Outstanding Questions.
-
What specific structural domains constrain the evolution of duplicated genes and to what extent?
Use in-depth structural and mutational analyses to understand which structural domains can diverge versus which ones are intolerant to mutations to remain functional.
-
How important or prevalent is conditional functional redundancy for duplicated gene evolution?
Use functional genomic approaches such as complex genetic interaction profiling, protein interaction profiling, and phenomic analyses to understand how the adaptation to environmental changes impacted duplicate gene retention.
-
Which models of duplicated gene evolution hold in humans and to what extent?
Draw on functional genomic approaches that have been used to interrogate duplicated gene evolution in model organisms, such as yeast, to characterize fates of duplicated genes in human cell models.
-
What is the extent of duplicated gene buffering in the human genome?
Use CRISPR-Cas methodology to conduct systematic dual perturbation screens involving duplicated genes to identify synthetic sick/lethal pairs in human cell models as well as whole-organism mammalian models, such as mice. Screen across cell lines originating from multiple tissues to understand how does tissue of origin modify this buffering relationship.
Systematic perturbation studies using CRISPR-Cas9 methodology have started to interrogate duplicated genes in human cells. Indeed, paralogous genes are less likely to be essential than singleton genes and show a fitness defect using pooled whole-genome CRISPR-Cas9 LOF fitness screens, suggesting that paralogs provide back-up compensation in human cells [107]. This effect was even more pronounced for genes with multiple paralogs and paralogs with a high sequence similarity [107] and in cases that did not exhibit heteromerization [84]. Dual-gene loss of function studies using Cas9, Cas12a and a Cas9-Cas12a hybrid system uncovered negative genetic interactions within duplicated gene pairs, which is consistent with a direct buffering relationship between paralogs [108–110]. However, since screening different cancer cell lines revealed a rate of negative genetic interactions within the duplicated gene pairs between ~ 6 – 17%, it remains unclear to what extent there is buffering among human paralogs, and, if there is in fact paralog buffering, how dependent this is on a particular tissue or cell type [108, 109] (see Outstanding Questions). As we continue to functionally characterize the human genome, a deeper insight into the functional divergence of duplicated genes will be key, and systems biology approaches, similar to those employed in model organisms, such as yeast, will undoubtedly be highly useful in this endeavor (Box).
Box: Functional genomic approaches for interrogating duplicates in yeast.
A range of experimental approaches have been utilized in yeast to answer the question of why duplicated genes have been maintained during evolution. Analysis of genome sequences allows estimates of rates of divergence of both coding and regulatory regions and protein domain architecture [14, 16, 111] and identified gene families with accelerated and decelerated evolution which has helped to explain compensatory ability of duplicates. Gene ontology (GO) semantic distance has also been utilized to study duplicates by computing the semantic similarity of their annotated GO terms [54]. Metabolic flux balance analysis has also provided insight into evolutionary constraints on gene duplicate retention by examining buffering accomplished by metabolic network structure and function as well as flux reorganization [90, 91].
The yeast deletion mutant collection has been used to generate a wealth of functional genomics data. Single mutant fitness of duplicated genes in nutrient-rich and alternative growth conditions has been used to examine dispensability of duplicates relative to singletons [75, 112–115]. Analysis of duplicate gene expression levels. Gene expression profiles have also been used to probe gene function and understand transcriptional circuitry associated with duplicated genes [55, 64, 66, 81, 113, 116–119].
Analyses of similarity of protein-protein interactions within networks [48, 54, 55, 95, 116, 120, 121], changes in protein abundance [78] or protein-interactions [28] of one paralog upon perturbation of another combine to provide functional readouts for estimating functional redundancy of duplicated genes. However, analysis of protein interaction networks has been limited by the sparsity of the data given that TAP-MS experiments, even when gathered from multiple sources [122, 123], reveal at least one shared protein for only 8% of all possible duplicate pairs. The yeast GFP collection was used to survey subcellular localizations and then reconstruct phylogenetic relationship of proteins in a protein family to infer cases of sub- and neofunctionalization [56, 57].
Systematic analysis of digenic interactions within the duplicate gene pair offered a means to capture the extent of buffering relationship of paralogs [76, 80, 97, 124]. Integration of digenic and trigenic interactions offers a riche functional read-out for interrogating duplicated genes by capturing paralog-specific as well as overlapping functions [27, 125–128]. Developing experimental assays that can deeply probe the duplicate gene functional divergence is importance for understanding the functional relationship of duplicated genes and their evolutionary trajectories.
Highlights.
Gene duplication events are major factors in shaping eukaryotic genomes.
Systematic analysis of complex genetic interactions of duplicated genes revealed that their functional redundancy is evolutionary stable and can co-evolve with acquisition of functional specialization due to structural and functional entanglement factors.
Structural constraints that lead to maintenance of functionally overlapping duplicated genes include protein-protein interactions that maintain heteromers between sister duplicates.
Glossary
- Allopolyploidy
A type of polyploidy in which the sets of chromosomes originate from the different but related species
- Autopolyploidy
A type of polyploidy in which all the sets of chromosomes originate from the same species
- Back-up compensation
The ability of a copy of a gene to functionally replace a gene when the latter is perturbed by mutational or environmental processes
- Copy-number variation
Variation in the number of copies of a genomic region (> 50 bp) among individuals of a population within the same species and can include duplications and deletions of coding and noncoding regions. The term copy-number polymorphism is used interchangeably with copy-number variant.
- Complex genetic interaction
A genetic interaction, involving more than two genes, which results in an unexpected phenotype from a combination of mutations which cannot be predicted from individual mutations and lower-order combinations. It is also referred to as a higher-order genetic interaction.
- Dosage duplicates
Gene duplicates which were selected in evolution for increased number of copies their gene resulting in an elevated level of their gene products
- Duplication
Event that leads to doubling of a genomic region
- Escape from adaptive conflict
A special case of subfunctionalization which postulates that gene duplication selects for separate optimization of function in each duplicate
- Homeolog
Pairs of genes in the same species that originate from an interspecies hybridization event. Spelling variations such as ‘homoelog’ or ‘homoeolog’ are also occasionally used. This term should not be confused with ‘homolog’.
- Homolog
A homolog or a homologous gene refers to a gene in two different species which arose in a common ancestor. Homologs includes orthologs and paralogs.
- Minimization of paralog interference
A special case of subfunctionalization in which interacting domains in duplicates genes are divided to prevent competitive interference in cooperative assemblies
- Neofunctionalization
A model of duplicate gene divergence in which one of the duplicates evolves a novel or more specialized function due to the redundant copy escaping the constraints of natural selection and acquiring mutations
- Nonfunctionalization
A process during which a gene acquires a null mutation rendering it nonfunctional
- Ohnolog
Genes that originate from a whole genome duplication event
- Ortholog
Genes in two different species that originated from the same common ancestor of those species
- Paralog
Genes that originate from a duplication event in a single species
- Polyploidy
State of the genome characterized by three or more complete sets of chromosomes
- Pro-ortholog
A single copy gene that is an ancestral ortholog to paralogs of interest
- Redundancy
Robustness originating from at least two entities, such as genes, performing similar functions, such that they can compensate for each other’s loss
- Robustness
The property of a system to generate a stable output when faced with a perturbation
- Singe nucleotide polymorphism
Variation at a single nucleotide (A-adenine, T-thymine, G-guanine, C-cytosine) in a DNA sequence among individuals of the same species or between paired chromosomes in an individual
- Small-scale duplication
Type of a duplication event that leads to tandem or segmental gene duplication of a locus
- Stoichiometric balance
Maintaining a specific ratio of members in a protein complex
- Structural and functional entanglement
A model of duplicate divergence which postulates that the evolutionary fate of a duplicated gene is ruled by an interplay of structural and functional entanglement factors leading to evolution of paralog specific roles in the cell while maintaining functional redundancy at an evolutionary steady state.
- Structural variation
A broad type of genomic variation which includes inversions, translocations and copy number variants
- Subfunctionalization
A model of duplicated gene divergence, which leads to partitioning of subfunctions of an ancestral gene
- Whole-genome duplication
Type of a duplication event that leads to simultaneous duplication of all genomic segments
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vosseberg J, van Hooff JJE, Marcet-Houben M, van Vlimmeren A, van Wijk LM, Gabaldon T, et al. Timing the origin of eukaryotic cellular complexity with ancient duplications. Nat Ecol Evol. 2021;5(1):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho CM, Zhang F, Lupski JR. Evolution in health and medicine Sackler colloquium: Genomic disorders: a window into human gene and genome evolution. Proc Natl Acad Sci U S A. 2010;107 Suppl 1:1765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. [DOI] [PubMed] [Google Scholar]
- 4.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 2010;107(20):9270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saitou M, Gokcumen O. An Evolutionary Perspective on the Impact of Genomic Copy Number Variation on Human Health. J Mol Evol. 2020;88(1):104–19. [DOI] [PubMed] [Google Scholar]
- 6.van Ommen GJ. Frequency of new copy number variation in humans. Nat Genet. 2005;37(4):333–4. [DOI] [PubMed] [Google Scholar]
- 7.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. [DOI] [PubMed] [Google Scholar]
- 8.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–8. [DOI] [PubMed] [Google Scholar]
- 9.Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20(11):1469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wacholder A, Carvunis AR. New genes from borrowed parts. Science. 2021;371(6531):779–80. [DOI] [PubMed] [Google Scholar]
- 11.Taylor JH, Woods PS, Hughes WL. The Organization and Duplication of Chromosomes as Revealed by Autoradiographic Studies Using Tritium-Labeled Thymidinee. Proc Natl Acad Sci U S A. 1957;43(1):122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smithies O Chromosomal Rearrangements and Protein Structure. Cold Spring Harb Symp Quant Biol. 1964;29:309–19. [DOI] [PubMed] [Google Scholar]
- 13.Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23(1):234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428(6983):617–24. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387(6634):708–13. [DOI] [PubMed] [Google Scholar]
- 16.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15(10):1456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175(2):933–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dehal P, Boore JL. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005;3(10):e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17(11):661–9. [DOI] [PubMed] [Google Scholar]
- 20.Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, et al. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell. 2019;176(3):663–75 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdellaoui A, Ehli EA, Hottenga JJ, Weber Z, Mbarek H, Willemsen G, et al. CNV Concordance in 1,097 MZ Twin Pairs. Twin Res Hum Genet. 2015;18(1):1–12. [DOI] [PubMed] [Google Scholar]
- 22.Li YR, Glessner JT, Coe BP, Li J, Mohebnasab M, Chang X, et al. Rare copy number variants in over 100,000 European ancestry subjects reveal multiple disease associations. Nat Commun. 2020;11(1):255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno S Why Gene Duplication? Evolution by Gene Duplication. New York: Springer-Verlag New York Inc.; 1970. [Google Scholar]
- 25.Kondrashov FA, Kondrashov AS. Role of selection in fixation of gene duplications. J Theor Biol. 2006;239(2):141–51. [DOI] [PubMed] [Google Scholar]
- 26.Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388(6638):167–71. [DOI] [PubMed] [Google Scholar]
- 27.Kuzmin E, VanderSluis B, Nguyen Ba AN, Wang W, Koch EN, Usaj M, et al. Exploring whole-genome duplicate gene retention with complex genetic interaction analysis. Science. 2020;368(6498):1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diss G, Gagnon-Arsenault I, Dion-Cote AM, Vignaud H, Ascencio DI, Berger CM, et al. Gene duplication can impart fragility, not robustness, in the yeast protein interaction network. Science. 2017;355(6325):630–4. [DOI] [PubMed] [Google Scholar]
- 29.Vavouri T, Semple JI, Lehner B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008;24(10):485–8. [DOI] [PubMed] [Google Scholar]
- 30.Dean EJ, Davis JC, Davis RW, Petrov DA. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008;4(7):e1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner A The role of population size, pleiotropy and fitness effects of mutations in the evolution of overlapping gene functions. Genetics. 2000;154(3):1389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durand D, Hoberman R. Diagnosing duplications--can it be done? Trends Genet. 2006;22(3):156–64. [DOI] [PubMed] [Google Scholar]
- 33.Hilario E, Gogarten JP. Horizontal transfer of ATPase genes--the tree of life becomes a net of life. Biosystems. 1993;31(2–3):111–9. [DOI] [PubMed] [Google Scholar]
- 34.Hughes AL, Friedman R, Ekollu V, Rose JR. Non-random association of transposable elements with duplicated genomic blocks in Arabidopsis thaliana. Mol Phylogenet Evol. 2003;29(3):410–6. [DOI] [PubMed] [Google Scholar]
- 35.Zdobnov EM, Campillos M, Harrington ED, Torrents D, Bork P. Protein coding potential of retroviruses and other transposable elements in vertebrate genomes. Nucleic Acids Res. 2005;33(3):946–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. 2005;27(9):937–45. [DOI] [PubMed] [Google Scholar]
- 37.Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13(3):382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spoelhof JP, Keeffe R, McDaniel SF. Does reproductive assurance explain the incidence of polyploidy in plants and animals? New Phytol. 2020;227(1):14–21. [DOI] [PubMed] [Google Scholar]
- 39.Muller HJ. Why Polyploidy is Rarer in Animals Than in Plants. The American Naturalist. 1925;59(663):346–53. [Google Scholar]
- 40.Orr HA. “Why Polyploidy is Rarer in Animals Than in Plants” Revisited. The American Naturalist. 1990;136(6):759–70. [Google Scholar]
- 41.Van de Peer Y, Ashman T, Soltis PS, Soltis DE. Polyploidy: an evolutionary and ecological force in stressful times. The Plant Cell. 2021:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spring J Vertebrate evolution by interspecific hybridisation--are we polyploid? FEBS Lett. 1997;400(1):2–8. [DOI] [PubMed] [Google Scholar]
- 43.Soler A, Morales C, Mademont-Soler I, Margarit E, Borrell A, Borobio V, et al. Overview of Chromosome Abnormalities in First Trimester Miscarriages: A Series of 1,011 Consecutive Chorionic Villi Sample Karyotypes. Cytogenet Genome Res. 2017;152(2):81–9. [DOI] [PubMed] [Google Scholar]
- 44.Fisher KJ, Buskirk SW, Vignogna RC, Marad DA, Lang GI. Adaptive genome duplication affects patterns of molecular evolution in Saccharomyces cerevisiae. PLoS Genet. 2018;14(5):e1007396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440(7082):341–5. [DOI] [PubMed] [Google Scholar]
- 46.Conant GC. Comparative genomics as a time machine: How relative gene dosage and metabolic requirements shaped the time-dependent resolution of yeast polyploidy. Mol Biol Evol. 2014. [DOI] [PubMed] [Google Scholar]
- 47.Marcet-Houben M, Gabaldon T. Beyond the Whole-Genome Duplication: Phylogenetic Evidence for an Ancient Interspecies Hybridization in the Baker’s Yeast Lineage. PLoS Biol. 2015;13(8):e1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchant A, Cisneros AF, Dube AK, Gagnon-Arsenault I, Ascencio D, Jain H, et al. The role of structural pleiotropy and regulatory evolution in the retention of heteromers of paralogs. eLife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metz CW. Duplication of chromosome parts as a factor in evolution. Am Nat. 1947;81(797):81–103. [DOI] [PubMed] [Google Scholar]
- 50.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–43. [DOI] [PubMed] [Google Scholar]
- 51.Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, Maere S, et al. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012;10(12):e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marlétaz F, Firbas PN, Maeso I, Tena JJ, Bogdanovic O, Perry M, et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature. 2018;564(7734):64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533(7602):200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baudot A, Jacq B, Brun C. A scale of functional divergence for yeast duplicated genes revealed from analysis of the protein-protein interaction network. Genome Biol. 2004;5(10):R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He XL, Zhang JZ. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169(2):1157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marques AC, Vinckenbosch N, Brawand D, Kaessmann H. Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol. 2008;9(3):R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian W, Zhang J. Protein subcellular relocalization in the evolution of yeast singleton and duplicate genes. Genome Biol Evol. 2009;1:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nasvall J, Sun L, Roth JR, Andersson DI. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338(6105):384–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Serebrovsky SA. Genes scute and achaete in Drosophila melanogaster and a hypothesis of gene divergency. C R Acad SciURSS. 1938;19:77–81. [Google Scholar]
- 60.Sharman AC. Some new terms for duplicated genes. Seminars in cell & developmental biology. 1999;10(5):561–3. [DOI] [PubMed] [Google Scholar]
- 61.van Hoof A Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171(4):1455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marshall AN, Montealegre MC, Jiménez-López C, Lorenz MC, van Hoof A. Alternative Splicing and Subfunctionalization Generates Functional Diversity in Fungal Proteomes. PLoS Genet. 2013;9(3):e1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlacht A, Dacks JB. Unexpected ancient paralogs and an evolutionary model for the COPII coat complex. Genome Biol Evol. 2015;7(4):1098–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conant GC, Wolfe KH. Functional Partitioning of Yeast Co-Expression Networks after Genome Duplication. PLoS Biol. 2006;4(4):e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154(1):459–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449(7158):54–U36. [DOI] [PubMed] [Google Scholar]
- 67.Gout JF, Lynch M. Maintenance and Loss of Duplicated Genes by Dosage Subfunctionalization. Mol Biol Evol. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piatigorsky J, Wistow G. The recruitment of crystallins: new functions precede gene duplication. Science. 1991;252(5009):1078–9. [DOI] [PubMed] [Google Scholar]
- 69.Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454(7205):762–5. [DOI] [PubMed] [Google Scholar]
- 70.Yanagida H, Gispan A, Kadouri N, Rozen S, Sharon M, Barkai N, et al. The Evolutionary Potential of Phenotypic Mutations. PLoS Genet. 2015;11(8):e1005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lehner B Conflict between noise and plasticity in yeast. PLoS Genet. 2010;6(11):e1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chapal M, Mintzer S, Brodsky S, Carmi M, Barkai N. Resolving noise-control conflict by gene duplication. PLoS Biol. 2019;17(11):e3000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baker CR, Hanson-Smith V, Johnson AD. Following Gene Duplication, Paralog Interference Constrains Transcriptional Circuit Evolution. Science. 2013;342(6154):104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brookfield J Can genes be truly redundant? Curr Biol. 1992;2(10):553–4. [DOI] [PubMed] [Google Scholar]
- 75.Gu ZL, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421(6918):63–6. [DOI] [PubMed] [Google Scholar]
- 76.VanderSluis B, Bellay J, Musso G, Costanzo M, Papp B, Vizeacoumar FJ, et al. Genetic interactions reveal the evolutionary trajectories of duplicate genes. Mol Syst Biol. 2010;6:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A. 2006;103(31):11653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeLuna A, Springer M, Kirschner MW, Kishony R. Need-Based Up-Regulation of Protein Levels in Response to Deletion of Their Duplicate Genes. PLoS Biol. 2010;8(3):e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480(7376):250–3. [DOI] [PubMed] [Google Scholar]
- 80.Musso G, Costanzo M, Huangfu M, Smith AM, Paw J, San Luis BJ, et al. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res. 2008;18(7):1092–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kafri R, Bar-Even A, Pilpel Y. Transcription control reprogramming in genetic backup circuits. Nature Genetics. 2005;37(3):295–9. [DOI] [PubMed] [Google Scholar]
- 82.Keane OM, Toft C, Carretero-Paulet L, Jones GW, Fares MA. Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae. Genome Res. 2014;24(11):1830–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mattenberger F, Sabater-Munoz B, Toft C, Fares MA. The Phenotypic Plasticity of Duplicated Genes in Saccharomyces cerevisiae and the Origin of Adaptations. G3 (Bethesda). 2017;7(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dandage R, Landry CR. Paralog dependency indirectly affects the robustness of human cells. Mol Syst Biol. 2019;15(9):e8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garge RK, Laurent JM, Kachroo AH, Marcotte EM. Systematic Humanization of the Yeast Cytoskeleton Discerns Functionally Replaceable from Divergent Human Genes. Genetics. 2020;215(4):1153–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laurent JM, Garge RK, Teufel AI, Wilke CO, Kachroo AH, Marcotte EM. Humanization of yeast genes with multiple human orthologs reveals functional divergence between paralogs. PLoS Biol. 2020;18(5):e3000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3(2):RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rapoport IA. Mnogokratnye linejnye povtoreniya uchastkov khromosom i ikh evolyucionnoe znachenie. [Multiple linear repeats of chromosome segments and their evolutionary significance]. Zhurnal Obshchej Biologii. 1940;1:235–70. [Google Scholar]
- 89.Conant GC, Wolfe KH. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. 2007;3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Papp B, Pal C, Hurst LD. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429(6992):661–4. [DOI] [PubMed] [Google Scholar]
- 91.Vitkup D, Kharchenko P, Wagner A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 2006;7(5):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Solis-Escalante D, Kuijpers NG, Barrajon-Simancas N, van den Broek M, Pronk JT, Daran JM, et al. A Minimal Set of Glycolytic Genes Reveals Strong Redundancies in Saccharomyces cerevisiae Central Metabolism. Eukaryotic cell. 2015;14(8):804–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Purkanti R, Thattai M. Paralogous gene modules derived from ancient hybridization drive vesicle traffic evolution in yeast. bioRxiv. 2021. [Google Scholar]
- 94.Payen C, Sunshine AB, Ong GT, Pogachar JL, Zhao W, Dunham MJ. High-Throughput Identification of Adaptive Mutations in Experimentally Evolved Yeast Populations. PLoS Genet. 2016;12(10):e1006339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hakes L, Pinney JW, Lovell SC, Oliver SG, Robertson DL. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 2007;8(10):R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Presser A, Elowitz MB, Kellis M, Kishony R. The evolutionary dynamics of the Saccharomyces cerevisiae protein interaction network after duplication. Proc Natl Acad Sci U S A. 2008;105(3):950–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ihmels J, Collins SR, Schuldiner M, Krogan NJ, Weissman JS. Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol. 2007;3:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ishikawa K, Makanae K, Iwasaki S, Ingolia NT, Moriya H. Post-Translational Dosage Compensation Buffers Genetic Perturbations to Stoichiometry of Protein Complexes. PLoS Genet. 2017;13(1):e1006554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taggart JC, Li GW. Production of Protein-Complex Components Is Stoichiometric and Lacks General Feedback Regulation in Eukaryotes. Cell Syst. 2018;7(6):580–9 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Semple JI, Vavouri T, Lehner B. A simple principle concerning the robustness of protein complex activity to changes in gene expression. BMC Systems Biology. 2008;2(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morrill SA, Amon A. Why haploinsufficiency persists. Proc Natl Acad Sci U S A. 2019;116(24):11866–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Costanzo M, Kuzmin E, van Leeuwen J, Mair B, Moffat J, Boone C, et al. Global Genetic Networks and the Genotype-to-Phenotype Relationship. Cell. 2019;177:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuzmin E, VanderSluis B, Wang W, Tan G, Deshpande R, Chen Y, et al. Systematic analysis of complex genetic interactions. Science. 2018;360(6386). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, Marcotte EM. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348(6237):921–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Teufel AI, Wilke CO. Accelerated simulation of evolutionary trajectories in origin-fixation models. J R Soc Interface. 2017;14(127). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Teufel AI, Johnson MM, Laurent JM, Kachroo AH, Marcotte EM, Wilke CO. The many nuanced evolutionary consequences of duplicated genes. Mol Biol Evol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.De Kegel B, Ryan CJ. Paralog buffering contributes to the variable essentiality of genes in cancer cell lines. PLoS Genet. 2019;15(10):e1008466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gonatopoulos-Pournatzis T, Aregger M, Brown KR, Farhangmehr S, Braunschweig U, Ward HN, et al. Genetic interaction mapping and exon-resolution functional genomics with a hybrid Cas9-Cas12a platform. Nat Biotechnol. 2020;38(5):638–48. [DOI] [PubMed] [Google Scholar]
- 109.Dede M, McLaughlin M, Kim E, Hart T. Multiplex enCas12a screens detect functional buffering among paralogs otherwise masked in monogenic Cas9 knockout screens. Genome Biol. 2020;21(1):262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thompson NA, Ranzani M, van der Weyden L, Iyer V, Offord V, Droop A, et al. Combinatorial CRISPR screen identifies fitness effects of gene paralogues. Nature Communications. 2021;12(1):1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grassi L, Fusco D, Sellerio A, Cora D, Bassetti B, Caselle M, et al. Identity and divergence of protein domain architectures after the yeast whole-genome duplication event. Mol Biosyst. 2010;6(11):2305–15. [DOI] [PubMed] [Google Scholar]
- 112.He XL, Zhang JZ. Higher duplicability of less important genes in yeast genomes. Mol Biol Evol. 2006;23(1):144–51. [DOI] [PubMed] [Google Scholar]
- 113.Wagner A Decoupled evolution of coding region and mRNA expression patterns after gene duplication: Implications for the neutralist-selectionist debate. Proceedings of the National Academy of Sciences. 2000;97(12):6579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.DeLuna A, Vetsigian K, Shoresh N, Hegreness M, Colon-Gonzalez M, Chao S, et al. Exposing the fitness contribution of duplicated genes. Nature Genetics. 2008;40(5):676–81. [DOI] [PubMed] [Google Scholar]
- 115.Plata G, Vitkup D. Genetic robustness and functional evolution of gene duplicates. Nucleic Acids Res. 2014;42(4):2405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wagner A Asymmetric Functional Divergence of Duplicate Genes in Yeast. Mol Biol Evol. 2002;19(10):1760–8. [DOI] [PubMed] [Google Scholar]
- 117.Gu X, Zhang Z, Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc Natl Acad Sci U S A. 2005;102(3):707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8(4):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wapinski I, Pfiffner J, French C, Socha A, Thompson DA, Regev A. Gene duplication and the evolution of ribosomal protein gene regulation in yeast. Proc Natl Acad Sci U S A. 2010;107(12):5505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Makino T, Suzuki Y, Gojobori T. Differential evolutionary rates of duplicated genes in protein interaction network. Gene. 2006;385:57–63. [DOI] [PubMed] [Google Scholar]
- 121.Musso G, Zhang Z, Emili A. Retention of protein complex membership by ancient duplicated gene products in budding yeast. Trends Genet. 2007;23(6):266–9. [DOI] [PubMed] [Google Scholar]
- 122.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–6. [DOI] [PubMed] [Google Scholar]
- 123.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–43. [DOI] [PubMed] [Google Scholar]
- 124.Li J, Yuan Z, Zhang Z. The Cellular Robustness by Genetic Redundancy in Budding Yeast. PLoS Genet. 2010;6(11):e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haber JE, Braberg H, Wu Q, Alexander R, Haase J, Ryan C, et al. Systematic triple-mutant analysis uncovers functional connectivity between pathways involved in chromosome regulation. Cell Rep. 2013;3(6):2168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zou J, Friesen H, Larson J, Huang D, Cox M, Tatchell K, et al. Regulation of cell polarity through phosphorylation of Bni4 by Pho85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20(14):3239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moir RD, Gross DA, Silver DL, Willis IM. SCS3 and YFT2 link transcription of phospholipid biosynthetic genes to ER stress and the UPR. PLoS Genet. 2012;8(8):e1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lai X, Beilharz T, Au WC, Hammet A, Preiss T, Basrai MA, et al. Yeast hEST1A/B (SMG5/6)-like proteins contribute to environment-sensing adaptive gene expression responses. G3 (Bethesda). 2013;3(10):1649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, et al. Comparative genomics of the eukaryotes. Science. 2000;287(5461):2204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422(6930):433–8. [DOI] [PubMed] [Google Scholar]
- 131.Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 2005;102(15):5454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thomas JH. Genome evolution in Caenorhabditis. Briefings in Functional Genomics. 2008;7(3):211–6. [DOI] [PubMed] [Google Scholar]
- 133.Cavalcanti AR, Ferreira R, Gu Z, Li WH. Patterns of gene duplication in Saccharomyces cerevisiae and Caenorhabditis elegans. J Mol Evol. 2003;56(1):28–37. [DOI] [PubMed] [Google Scholar]


