Abstract
U12-dependent introns containing alterations of the 3′ splice site AC dinucleotide or alterations in the spacing between the branch site and the 3′ splice site were examined for their effects on splice site selection in vivo and in vitro. Using an intron with a 5′ splice site AU dinucleotide, any nucleotide could serve as the 3′-terminal nucleotide, although a C residue was most active, while a U residue was least active. The penultimate A residue, by contrast, was essential for 3′ splice site function. A branch site-to-3′ splice site spacing of less than 10 or more than 20 nucleotides strongly activated alternative 3′ splice sites. A strong preference for a spacing of about 12 nucleotides was observed. The combined in vivo and in vitro results suggest that the branch site is recognized in the absence of an active 3′ splice site but that formation of the prespliceosomal complex A requires an active 3′ splice site. Furthermore, the U12-type spliceosome appears to be unable to scan for a distal 3′ splice site.
Two types of spliceosomal introns are known to exist in higher eukaryotic plants and animals (see reference 6 for a recent review). The major class, termed here the U2-dependent class, has been the subject of a great deal of investigation for many years. The minor or U12-dependent class has been recognized only relatively recently. Many genes contain both types of introns in an interspersed pattern, requiring cooperation between the two splicing systems to properly identify the exons. The size distributions of introns and the adjacent exons are similar for both classes of introns. This suggests that the process of splice site recognition for both classes must contend with similar problems of distinguishing correct splice sites amidst the often many tens or hundreds of thousands of nucleotides in a pre-mRNA.
The information needed to specify the sites of splicing of both classes of introns is largely located at or near the 5′ and 3′ splice sites. These regions contain conserved sequences that interact with the splicing machinery to promote the assembly of the spliceosome and to specify and activate the chemical cleavage and ligation reactions which lead to the production of spliced RNA. The 3′ splice site region contains two nucleotides that must be precisely located and activated for the two chemical steps in the splicing reaction: the branch site adenosine, with its associated 2′ hydroxyl group, which is the nucleophile in the first step of splicing, and the 3′ splice site residue, which lies immediately adjacent to the phosphodiester bond that is transesterified in the second step of splicing. The 3′ splice site is physically separate from the branch site residue and is not involved in the chemistry of the first-step reaction. Indeed, several studies have shown that the first splicing step can occur in vitro on RNA molecules in which no active 3′ splice site is present due to either truncation of the substrate RNA (1, 13, 33) or mutation of the 3′ splice site AG (15, 31, 39).
In U2-dependent introns, the branch site adenosine and the 3′ splice site are generally separated by 11 to 40 nucleotides, although cases are known where they can be over 100 nucleotides apart (18, 37). Positioned between the branch site and the 3′ splice site is a pyrimidine-rich region that appears to be a major recognition element for the 3′ end of an intron. Over the years, much effort has gone into the analysis of these sequence elements to determine the spliceosomal components with which they interact and the role that each plays in specifying the final site of splicing (See references 24 and 30 for recent reviews).
Many studies suggest that 3′ splice site selection is largely governed by a 5′-to-3′ scanning mechanism from an independently recognized branch site. Natural introns use the first AG downstream of the branch site as the 3′ splice site (25, 26). This model is also supported by evidence that 3′ splice sites can be located over 100 nucleotides from the branch site (18, 37). This implies that there is no strict limit on how far a 3′ splice site can be from the branch site. Recent in vitro studies using an artificial two-piece trans-splicing system in which the 3′ splice site is supplied on a separate piece of RNA support a model of strict scanning, with the first AG dinucleotide being selected (1, 2, 8). However, other studies have shown that potential 3′ splice sites can compete with one another, suggesting either that scanning is “leaky” or that distal sites can be more active than proximal sites due to distance or sequence preferences (10, 23, 38). A recent study of splicing in the absence of the second-step factor hSlu7 showed that 3′ splice sites both upstream and downstream of the normal 3′ splice site could be used in vitro in the second-step reaction (10). These authors suggest that the normal 3′ splice site is selected but its use is suppressed in the absence of hSlu7. A concomitant effect is that the 5′ exon is more loosely bound to the spliceosome, allowing it to react with normally cryptic 3′ splice sites.
The many different systems and organisms used in these studies over the years make it difficult to establish a single mechanistic picture of 3′ splice site selection. It may be that more than one mechanism can be used and the rate-determining step may differ for different introns and for different organisms. For example, similar constructs containing multiple AGs show competition in Saccharomyces cerevisiae (23) but not in mammalian in vitro systems (8). At one extreme are AG-independent introns (31), in which the first splicing step can occur in the absence of a 3′ splice site. In this type of intron, a simple scanning mechanism may suffice. In AG-dependent introns, the 3′ splice site is required for spliceosome formation and the first step of splicing. Here, the 3′ splice site is recognized at least once early in the splicing pathway and then again for the chemical event of the second step. The precise AGs used for the different recognition events may not be the same (47), and likewise, the specific factors and mechanisms of recognition may also differ.
Recent results have shown that the small subunit of U2AF interacts with the 3′ splice site AG (23, 46, 48). The role of the small subunit appears to be more significant when the polypyrimidine tract, which interacts with the large subunit, is short or less pyrimidine rich (46). These also appear to be situations in which introns are AG dependent (31). In such circumstances, a strong preference for a 3′ splice site within a preferred distance of the branch site and adjacent to the polypyrimidine tract might be predicted, and indeed, there is evidence of such a constraint (46). AG-independent introns, on the other hand, may not require the small subunit of U2AF to stabilize binding of the large subunit to the polypyrimidine tract, thus allowing spliceosome formation and the first step of splicing to take place without prior selection of a 3′ splice site. In this circumstance, most readily seen in the trans-splicing assay (8), a simple scanning mechanism might then select the 3′ splice site without a strong distance constraint.
A distinctly different sequence arrangement is seen in the minor U12-dependent class of introns. These introns lack a polypyrimidine tract and have a highly conserved branch site consensus sequence that appears to be the major recognition element for the 3′ splice site (6, 16, 43). Another characteristic of this class of introns is that the distance between the branch site and the 3′ splice site is short and relatively consistent. An analysis of the information content of U12-dependent 3′ splice sites showed that the branch site consensus contained too little information to uniquely specify a 3′ splice site (6). To further constrain 3′ splice site selection, it would be useful to be able to use the existence of an appropriate 3′ splice site within a set distance of the branch site as an additional recognition element. This requires, however, that the 3′ splice site be recognized along with the branch site element during spliceosome formation. In a recent study, Frilander and Steitz showed that U12-dependent prespliceosome complex formation required the participation of both the 5′ splice site and the branch site (14). They also showed that an RNA which was truncated between the branch site and the 3′ splice site was still competent to form complexes. One interpretation of this finding is that the 3′ splice site does not play a role in the early steps of U12-dependent splicing.
In this report, we investigate the nucleotide requirements for a functional U12-dependent 3′ splice site and the consequences of altering the spacing between the branch site and the 3′ splice site both in vivo and in vitro. Our goal was to determine if this distance is mechanistically constrained to the limited range observed so far in native U12-dependent introns and to determine the consequences on splicing and spliceosome formation of violating these constraints. The results suggest that the recognition of the 3′ splice site in U12-dependent introns differs substantially from the process in U2-dependent introns.
MATERIALS AND METHODS
DNA constructs.
The four-exon, three-intron P120 intron F minigene construct and mutants derived from it have been described (11, 17, 19). Mutations used in this study were introduced by PCR methods using pairs of oligonucleotides containing the desired mutations. All mutations were confirmed by DNA sequencing.
Analysis of in vivo splicing.
Transient transfection of the P120 minigene plasmids into cultured CHO cells was done as described (17, 19). For these experiments, 1 μg of P120 plasmid and 9 μg of pUC19 carrier DNA were added to 106 cells. Total RNA was isolated from cells 48 h after transfection, treated with DNase I, reverse transcribed using a vector-specific primer, and amplified by PCR using P120 exon 6- and 7-specific primers as described (11, 19). For high-resolution mapping, the exon 7 primer was 5′-end labeled with 32P, and the PCR products were analyzed by electrophoresis in 8% polyacrylamide gels containing 7 M urea and 40% formamide. To verify the sites of splicing used in the various mutants, PCR products were separated on agarose or nondenaturing polyacrylamide gels. Bands were excised, reamplified, purified by gel electrophoresis, and sequenced. For Fig. 5, the exon 7 primer (TCAGACAGAGGGAAGAGGTCCATGAG) was located at the 3′ end of the exon. The PCR products were analyzed by agarose gel electrophoresis and visualized using ethidium bromide.
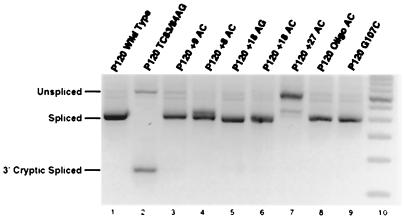
FIG. 5.
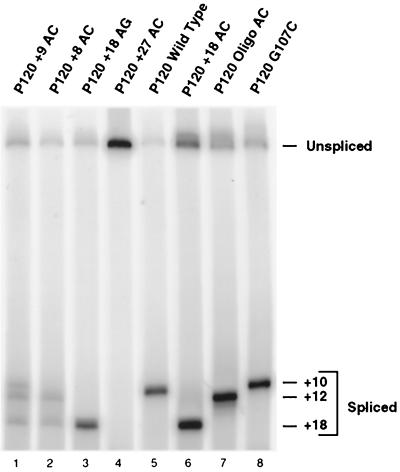
Cryptic U12-dependent 3′ splice site usage in branch and 3′ splice site mutants. The indicated minigene constructs were transfected and assayed as in Fig. 3 except that a primer at the 3′ end of exon 7 was used in the PCR amplification and the products were seperated on an agarose gel. Lane 2 shows the activation of the cryptic U12-dependent 3′ splice site at position 124 of exon 7 when the branch site is mutated from the consensus sequence UCUUAAC to AGUUAAC. Lane 10 is 50-bp ladder molecular size markers (Life Technologies).
In vitro splicing and spliceosome formation.
DNA templates for in vitro transcription were prepared by PCR amplification using the minigene constructs with 3′ splice site sequences shown in Fig. 2 as described (11). RNA was synthesized from these templates using T7 RNA polymerase and gel purified, and equal amounts of each RNA were spliced in vitro for 3 h in the presence of an antisense 2′-O-methyl oligonucleotide directed against U2 snRNA as described (11). An antisense 2′-O-methyl oligonucleotide against U12 snRNA was included where indicated. RNA from splicing reactions was separated on 8% polyacrylamide gels under denaturing conditions and detected with a Molecular Dynamics PhosphorImager.
FIG. 2.
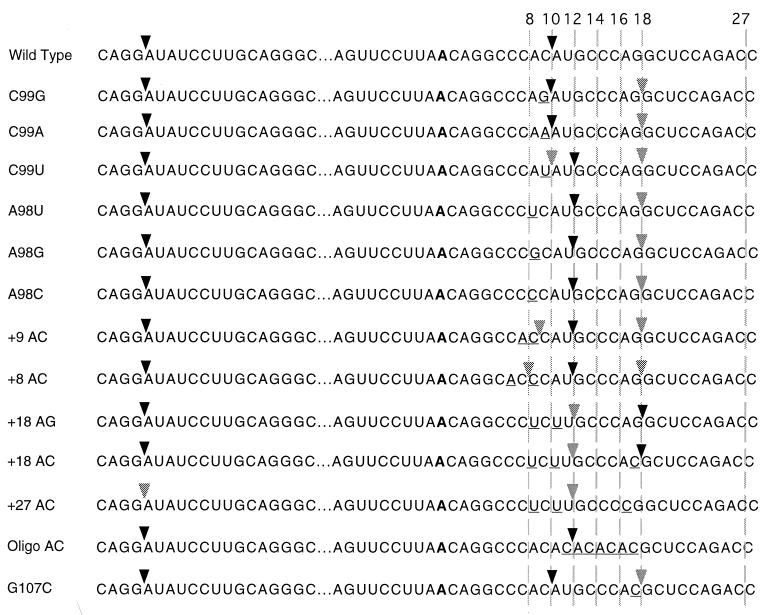
Sequences of the 3′ splice site mutants used in this study. The common 5′ splice site is shown at the left. The 3′ splice sites are shown at the right. The common branch site is shown in bold, and the mutated positions are underlined. The sites of in vivo splicing for each construct are indicated by the arrows. Major sites are indicated by dark arrowheads, and minor sites are indicated by light arrowheads. The numbers at the top refer to the distance in nucleotides between the branch site adenosine and the dashed lines.
For the spliceosome formation assay, splicing reactions were assembled under conditions identical to those of the splicing assays and included the antisense 2′-O-methyl oligonucleotide directed against U2 snRNA. After 30 and 90 min of incubation at 30°C, aliquots were removed and heparin was added to a concentration of 250 μg/ml (40, 41). Following 15 min on ice, the samples were loaded onto 4% polyacrylamide (80:1, acrylamide-bisacrylamide)–Tris–glycine native gels (20). The gels were run for 3 h at 5 W and dried, and the RNA was detected with a Molecular Dynamics PhosphorImager. For the adenovirus major late intron precursor control reaction, the anti-U2 oligonucleotide was omitted and incubation was for 15 min.
RESULTS
Natural distribution of branch site to 3′ splice site distances.
At the time that we first described the conserved branch site element in U12-dependent introns, we noted that the distance between this element and the 3′ splice site was short compared to U2-dependent introns and varied in our limited sample of introns over only a few nucleotides (16). Subsequently, we have greatly expanded the list of putative U12-dependent introns by searches through the genomic databases (5).
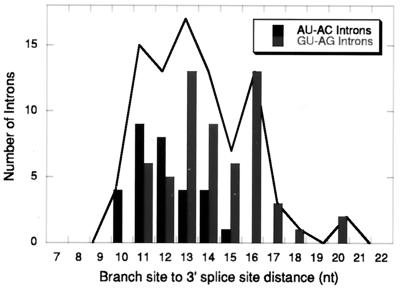
The observed distribution of distances from an all-phylum collection is shown in Fig. 1. The distribution shows clear limits of 10 nucleotides at the minimum and 20 nucleotides at the maximum. Within this collection, only the plant and mammal phylogenetic subgroups contained enough introns to compare, and these two subgroups showed no significant differences in mean distance or limits. The other subgroup comparison we made was between U12-dependent introns that have AU and AC terminal dinucleotides and those that have GU and AG termini. The AU-AC subgroup contained introns with distances from 10 to 16, with a peak at 12, while the GU-AG subgroup had distances between 11 and 20 nucleotides, with a broad peak between 12 and 16. It is not clear if this difference is functionally significant. In this report we have used the human nucleolar protein P120 intron F, which is a member of the AU-AC subgroup with a wild-type spacing of 10 nucleotides.
FIG. 1.
Distribution of branch site-to-3′ splice site distances in putative U12-dependent introns. The data are from the compilation of putative U12-dependent introns in Burge et al. (5). The solid line shows the distribution for all introns, while the black and grey bars show the distribution of distances in the AU-AC and GU-AG subclasses of U12-dependent introns, respectively. The branch site is assumed to be the position of the second adenosine in the branch site consensus sequence UCCUUAAC. All branch sites could be aligned with the consensus without ambiguity. In a few cases, the presumed branch site was a G residue.
Nucleotide requirements for 3′ splice site function in vivo.
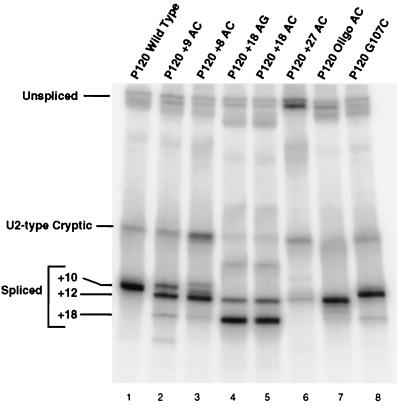
In order to manipulate the location of the 3′ splice site of the P120 intron F, we needed to determine the sequence requirements for efficient 3′ splice site function in vivo. While most U12-dependent introns that begin with AU end in AC, examples of AU-AG and AU-AA introns have been found (5, 11, 43). To clarify the roles of the last and penultimate nucleotides of this intron, we tested single mutations of these positions in vivo. All mutations were made in the four-exon, three-intron minigene construct described previously (17, 19). The constructs were transfected into CHO cells, and RNA was harvested after 48 h. The splicing pattern of the transfected P120 intron F was determined by reverse transcription (RT) of the RNA using a minigene-specific primer and PCR amplification of the products using primers in the flanking exons 6 and 7. Figure 2 shows the sequences of the mutants and indicates the sites of in vivo splicing used in each case.
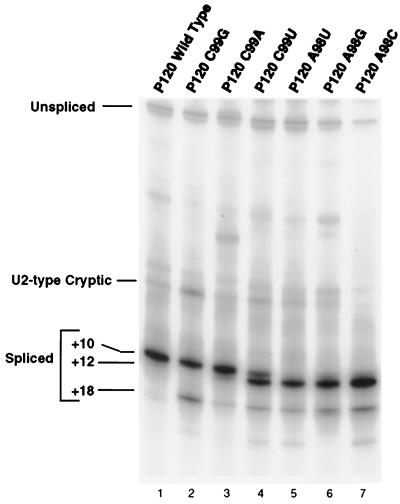
The RT-PCR results for mutations in the 3′ splice site terminal AC dinucleotide (residues A98 and C99) are shown in Fig. 3. The results for mutations of C99 show that any nucleotide can serve as the terminal residue, but there are clear differences in the efficiency with which the different nucleotides are used. Note that in each case, an AU dinucleotide is present immediately following the normal 3′ splice site at position +12 and an AG dinucleotide is present six nucleotides further downstream at +18. With the wild-type AC, only the +10 site is used to a detectable level. In the C99G mutant, in which the +10 splice site is AG, the AU at +12 is not used but the AG at +18 is used to a small extent. In addition, there is activation of a previously observed internal cryptic U2-type intron with a 3′ splice site at −6 (11, 40, 41). In the C99A mutant, the majority of splicing is to the AA at +10, with slight activation of the +12 AU and +18 AG. In the C99U mutant, the AUs at both +10 and +12 are used, with the +12 position predominating. Some use of the +18 AG is also apparent. This result suggests that the +12 position is favored over the +10 position. This is confirmed by the results presented below.
FIG. 3.
Pattern of in vivo splicing of 3′ splice site dinucleotide mutants. The indicated minigene constructs were transfected into CHO cells, and total RNA was prepared after 48 h. The splicing pattern of the P120 intron F was determined by RT-PCR amplification and seperation of the products on a denaturing gel. The positions of spliced and unspliced PCR products are shown, as well as the position of a U2-dependent cryptic splice that is activated in some constructs. The numbers at the left indicate the distance in nucleotides between the branch site and the 3′ splice site in each product.
To address the importance of the penultimate A residue at the 3′ splice site, A98 was mutated to U, G, or C and the mutants were tested in vivo. Figure 3 (lanes 5 to 7) shows the splicing patterns of these mutants. In all cases, the mutants showed no or very minimal splicing to the +10 site. Instead, splicing occurred at the +12 AU and, to a small extent, at the +18 AG site. From these results, we concluded that a U12-dependent 3′ splice could be inactivated by mutation of the penultimate A residue. For the experiments below, we used A to U or A to C mutations to achieve inactivation of the wild-type or potential alternative 3′ splice sites.
Spacing requirements for 3′ splice site function in vivo.
To investigate the functional constraints on the distance between the branch site and 3′ splice site in vivo, we varied this distance in the P120 intron between 8 and 27 nucleotides. The natural sequence around the wild-type AC 3′ splice site was modified to create different spacings. We also generated additional mutations to determine the optimum spacing for this intron. The mutant constructs for these experiments are shown in Fig. 2.
Figure 4 shows the RT-PCR results of in vivo splicing of P120 introns with a variety of branch site-to-3′ splice site distances. The wild-type P120 intron has a distance of 10 nucleotides, the smallest distance seen in natural introns (see Fig. 1). As expected, all splicing was to the AC at the wild-type position (Fig. 4, lane 1). When the AC dinucleotide was moved one nucleotide upstream to a distance of 9 nucleotides (P120 +9 AC, lane 2), a small amount of splicing occurred at the +9 site, but most splicing occurred at the AU dinucleotide at the +12 position. This is similar to the activation of the same +12 AU in the A98 mutations described above. A further translocation of the AC to the +8 position led to even more splicing at the +12 position (lane 3; the band at the position of +10 was not reproducible). An additional effect seen in this mutant was the activation of the cryptic U2-dependent splice sites (11). Activation of these sites was previously observed in mutant introns with debilitated 5′ splice sites (19). The effects of these mutations show that moving the 3′ splice site even a single nucleotide closer to the branch site from the observed natural limit of 10 nucleotides significantly weakens the 3′ splice site.
FIG. 4.
Pattern of in vivo splicing of 3′ splice site spacing mutants. The analysis and presentation are as in Fig. 3.
To test the functional upper limit of this spacing, we constructed modified introns with AC dinucleotides at progressively larger distances. When an AC was positioned 18 nucleotides from the branch site, most splicing was to this position (Fig. 4, lane 5). A small amount of splicing also occurred at the +12 position, where there was a UU dinucleotide (location confirmed by sequencing). An AG dinucleotide positioned at +18 gave similar results (lane 4). To inactivate the +18 splice site, the A at +17 was mutated to C. In this mutant, P120 +27 AC, the closest potential 3′ splice sites become the AG at +25 and the AC at +27 (Fig. 2). The +25 AG is preceded by a consensus C residue, while the AC at +27 is preceded by a less common G residue. As shown in Fig. 4, lane 6, no splicing was observed to either potential 3′ splice site; only splicing to the UU site at +12 was observed. In addition, levels of both the U2-dependent cryptic spliced product and unspliced RNA were increased in this mutant.
We next wanted to determine the preferred distance between the branch site and the 3′ splice site. For this, we inserted a sequence containing five repetitions of AC into the 3′ splice site region so that potential 3′ splice sites with the sequence CAC were available at distances of 10, 12, 14, 16, and 18 residues from the branch site adenosine (P120 oligo AC, Fig. 2). When this construct was assayed in vivo, splicing occurred almost exclusively at the +12 position (Fig. 4, lane 7). Note that in this construct, a completely wild-type CAC 3′ splice site positioned 10 nucleotides from the branch site is ignored in favor of the site located 12 nucleotides from the branch site. A similar result was seen in the A99U mutant above, in which AU sites were present at +10 and +12.
Finally, to test the relative strengths of 3′ splice sites located near the limits of the natural distance window, we mutated G107 to C to generate a new CAC 3′ splice site located 18 nucleotides from the branch site in the presence of the normal 3′ splice site at a distance of 10 nucleotides. Lane 8 in Fig. 4 shows that only a trace amount of splicing to the +18 site was seen, with the vast majority of splicing events using the site at +10. Thus, branch site-proximal AC dinucleotides appear to be dominant over distal ones. However, this is not a strict requirement, since a site at +12 is used preferentially over a site at +10 in both the oligo AC and C99U constructs.
Spacing mutants do not activate cryptic branch sites.
The RT-PCR assay used above to analyze the splicing products of the various mutants detects only spliced RNAs which contain most of exons 6 and 7. Spliced products resulting from splicing to more distal cryptic splice sites would be missed. Our previous experiments with mutations in the branch site region of intron F demonstrated that splicing to the normal 3′ splice site was abolished (17). Subsequent work has shown that the effect of branch site mutations is to shift 3′ splice site usage to a cryptic U12-dependent 3′ splice site 124 nucleotides downstream of the wild-type 3′ splice site (R. A. Dietrich, A. S. Seyboldt, and R. A. Padgett, submitted for publication).
To determine if such alternative splicing events were being activated in the mutant constructs discussed above, an RT-PCR analysis using a downstream distal primer was performed. Figure 5 shows that while the branch site element mutant P120 TC84/85AG (lane 2) activated splicing to the +124 cryptic 3′ splice site, none of the spacing mutants used in this study significantly activated this cryptic 3′ splice site. In particular, the +27 AC construct, which is unable to splice using the normal branch site, was also unable to use the +124 cryptic branch site.
In vitro effects of alterations of the 3′ splice site.
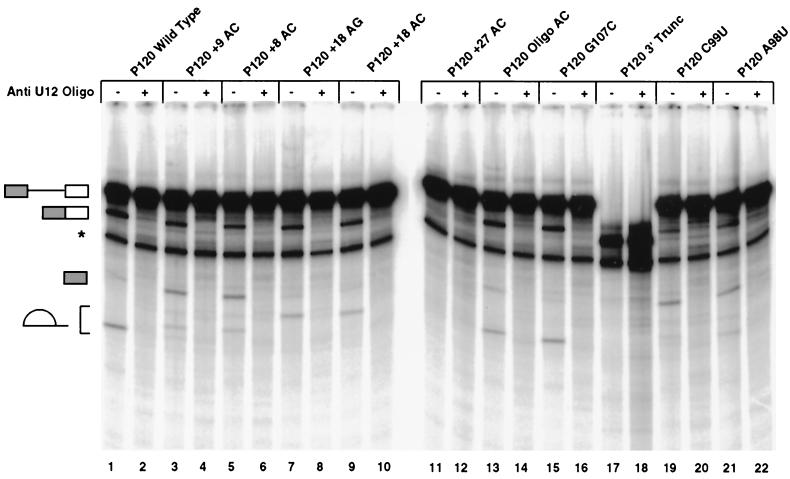
The in vivo splice site selection experiments discussed above defined the range of distances between the branch site and the 3′ splice site that are compatible with U12-dependent splicing. Since such experiments only assay the final sites of splicing, however, they cannot determine the effects of splice site alterations on spliceosome formation or the individual steps of splicing.
To investigate the effects of 3′ splice site alterations on individual splicing and spliceosome assembly steps, we prepared in vitro RNA transcripts of the P120 intron F and portions of the adjacent exons from the wild-type and mutant constructs used above. These RNAs were spliced in an in vitro HeLa cell nuclear extract system in the presence of an antisense 2′-O-methyl oligonucleotide against U2 snRNA. This reagent blocks the activity of the U2-dependent splicing system and activates the U12-dependent splicing system on this precursor RNA (11, 41).
Figure 6 shows the pattern of spliced RNA after 3 h of reaction. Three RNA products of U12-dependent splicing can be seen: the spliced exons and the excised lariat intron, which are the products of the second step of splicing, and the 5′ exon intermediate RNA produced by the first step of splicing. The appearance of all these RNAs is sensitive to the addition of an antisense oligonucleotide against U12 snRNA (even-numbered lanes). The other product of the first step of splicing, the lariat intron-exon 2 RNA, is obscured due to comigration with another band.
FIG. 6.
In vitro splicing patterns of 3′ splice site constructs. Templates for in vitro transcription of the indicated 3′ splice site constructs were produced by PCR amplification from the minigene constructs tested in vivo. Equal amounts of transcribed precursor RNAs were spliced in vitro. All reactions contained an anti-U2 snRNA 2′-O-methyl oligonucleotide which inhibits U2-dependent splicing. An anti-U12 snRNA 2′-O-methyl oligonucleotide was also added to even-numbered lanes to inhibit U12-dependent splicing. The structures of the various RNA products are shown on the left and correspond, from top to bottom, to the unspliced precursor, spliced exon product, the exon 1 intermediate generated by the first step of splicing, and the excised lariat intron generated by the second step of splicing. The lariat introns from different constructs migrate differently due to the varying length of RNA 3′ of the branch. A degradation product which migrates below the position of spliced exons is labeled with an asterisk. The splicing reactions shown in lanes 17 and 18 used a truncated precursor RNA that terminated 7 nucleotides 3′ of the branch site and thus was missing the 3′ splice site and the downstream exon.
As shown in Fig. 6, the in vitro splicing activity of the spacing mutants is similar to that observed in vivo. The Oligo AC and G107C mutants spliced with similar efficiency to the wild type, while the +18 AG and +18 AC mutants were slightly impaired. In contrast, the +27 AC mutant was inactive for both steps of splicing.
An interesting result was observed with the +9 AC and +8 AC mutant RNAs. These could proceed through the first step of splicing to produce the 5′ exon intermediate RNA but were partially blocked prior to the second step of splicing, as indicated by the decreased abundance of the lariat intron and spliced exon product RNAs. This second-step defect could be due either to the AC's being too close to the branch site or to the use of the AU 3′ splice site. To differentiate between these possibilities, the splicing abilities of the C99U and A98U mutants were also tested. These mutants showed an even more pronounced defect in the second step of splicing than the +9 AC and +8 AC mutants, suggesting that an AU 3′ splice site is a poor substrate for attack by the upstream exon in U12-dependent splicing.
To ensure that these mutant RNAs were using the same 3′ splice sites in vitro as we observed in vivo, splicing reactions similar to those in Fig. 6 were analyzed by RT-PCR (Fig. 7). In all cases, the major spliced products were the same as seen in the in vivo analysis shown in Fig. 4.
FIG. 7.
RT-PCR analysis of the products of in vitro splicing. RNA was isolated from the position of spliced exons in a preparative version of the experiment shown in Fig. 6, reverse transcribed using a primer near the 3′ end of the precursor, and PCR amplified using a nested 3′ primer and a 5′ primer in the first exon, followed by denaturing gel electrophoresis to determine the sites of splicing. Due to differences in RNA recovery and amplification, differences in band intensities between lanes are not quantitatively meaningful.
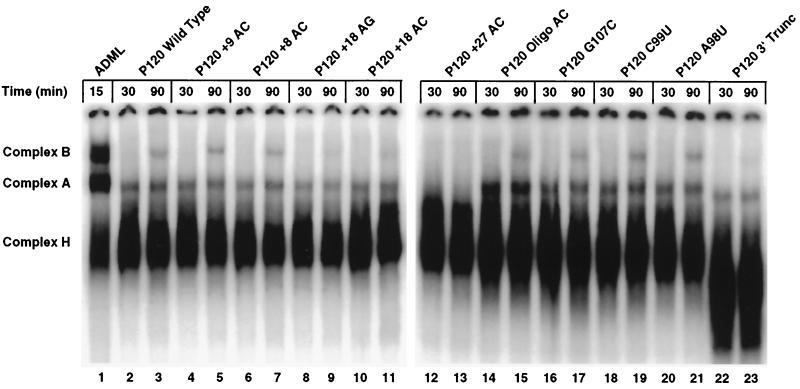
These precursor RNAs were also assayed for their ability to form spliceosomal complexes using non denaturing gel electrophoresis. Two splicing-specific complexes are resolved in this system: complex A represents an early-forming prespliceosome containing U11 and U12 snRNPs, while complex B is the product of the further addition of U4atac/U6atac and U5 snRNPs and the concomitant loss of the U11 snRNP (40, 41). As shown in Fig. 8, both splicing complexes formed on all the RNAs except the +27 AC mutant. The +18 AG and +18 AC mutant RNAs formed a smaller amount of both complexes than the wild type, consistent with their somewhat diminished splicing activity in vitro. The Oligo AC construct appeared to form complex A more efficiently than the wild type, but neither complex B formation nor splicing was stimulated. The +27 AC mutant RNA did not form either complex to a detectable degree.
FIG. 8.
Spliceosome formation activity of 3′ splice site constructs. Equal amounts of the in vitro-transcribed RNA used in Fig. 6 were incubated under U12-dependent splicing conditions for the times indicated, treated with heparin, and separated on nondenaturing gels. The splicing-specific complexes A and B are well resolved from the nonspecific complex H. Lane 1 shows complexes formed on a U2-dependent adenovirus major late intron precursor incubated in the absence of the anti-U2 snRNA oligonucleotide.
The lack of spliceosome formation activity of the +27 mutant was surprising, since Frilander and Steitz (14) have reported that an in vitro transcript which was truncated between the branch site and the 3′ splice site was able to form both A and B complexes. We repeated this spliceosome complex result (P120 3′ Trunc; Fig. 8, lanes 22 and 23) and also confirmed that this RNA was unable to carry out the first step of splicing to a detectable level (Fig. 6, lane 17). Thus, this truncated RNA, which is missing sequences downstream of the branch site, can still form spliceosome complexes, while the +27 AC mutant RNA, which has RNA but no functional splice sites downstream of the branch site, could not.
DISCUSSION
Any nucleotide can function at a U12-dependent 3′ splice site.
U12-type introns that begin with the dinucleotide AU usually end with the dinucleotide AC. In a few cases, AA and AG can also serve as 3′ splice sites. When we experimentally mutated the 3′ nucleotide of an AU-AC intron, any nucleotide could function at the end of the intron in vivo. From the pattern of use of other nearby sites, a rough idea of the strength of the various sites can be deduced. The normal AC site and the AA mutant site were used preferentially over all other sites, including the AU and AG dinucleotides located two and eight nucleotides downstream, respectively. When the normal AC was mutated to AG, the AG at +18 was also used to a detectable level, while the AU was not. Finally, when the normal site was changed to AU, the downstream AU was used significantly more than the AU at the normal site. This preference for a 3′ splice site close to the +12 position was also demonstrated in the Oligo AC construct. Thus, the in vivo data suggest that while any nucleotide can serve as the 3′ end of a U12-type AU intron, there is a preference, with the order AC ≈ AA > AG > AU. This order is supported by the in vitro splicing data showing that splicing to an AU 3′ splice site causes a defect in the second step of splicing.
In contrast to the 3′-terminal nucleotide, the penultimate A residue appears to play a major role in specifying the 3′ splice site. When this residue was mutated to any other nucleotide, an alternative AU 3′ splice site was selected. Mutation of the A residues at the alternative 3′ splice sites likewise inactivated them.
The ability of the U12-dependent splicing system to use any nucleotide at the 3′ splice site contrasts sharply with the very strong requirement for a G residue at the 3′ splice site seen in U2-dependent introns. In mammalian U2-type introns, it appears that this requirement is due to at least two types of interactions. First, the small subunit of U2AF has been shown to bind to the 3′ splice site AG but not to an AC mutant (46). Second, selection of the 3′ splice site in the in vitro trans-splicing reaction, in which the first step of splicing takes place in the absence of a 3′ splice site, also shows a strong bias to AG 3′ splice sites (2). This second type of selection could be due to interactions with the 5′ splice site G residue, as seen in both the yeast (7, 29) and mammalian (36) U2-dependent systems.
In U12-type introns of the AU-AC class used here, it seems unlikely that either subunit of U2AF is involved in 3′ splice site selection or activation. The absence of a polypyrimidine tract between the branch site and the 3′ splice site suggests that the large subunit does not bind to this region, and the demonstrated inability of the small subunit to bind to AC sequences suggests that it is not involved either (46, 48). However, biochemical attempts to address this question directly have so far been unsuccessful (44; unpublished results).
The differences in efficiency seen with different 3′ splice site nucleotides in the U12-dependent system imply that there is some specificity in its selection. Both earlier data (11) and work to be published elsewhere shows that in the U12-type as in the U2-type splicing system, the identity of the first intron nucleotide influences the choice of the last nucleotide. For instance, in both splicing systems, an AU-AC pairing is functional, while a GU-AC pairing is not (7, 11, 29, 36). The data here show that a first intron nucleotide A residue, while most active with a terminal C residue, can functionally splice to any residue in U12-dependent introns. For U2-type introns in both the yeast (7, 29) and mammalian (36) systems, only an AU-AC pairing is active. In agreement with this, all of the recently identified natural U2-type introns which begin with AU also end with AC (11, 43, 45). Thus, while both splicing systems have similar preferences for pairs of first and last nucleotides, the U12-dependent system appears to be more flexible in its choice of a 3′ splice site than the U2-dependent system.
Branch site-to-3′ splice site distance is functionally constrained in U12-type introns.
U12-dependent introns appear to be very dependent on a properly situated 3′ splice site for activity. The branch site-to-3′ splice site distances of natural introns fall in the range of 10 to 20 nucleotides. When we reduced the distance from the 10 nucleotides found in the wild-type human P120 intron F to either 9 or 8 nucleotides, we found that splicing in vivo occurred instead at a cryptic AU 3′ splice site at the +12 position. In vitro, these mutants showed only weak splicing to any 3′ splice sites but were able to form spliceosomes with close to wild-type efficiency and progressed through the first step of splicing. The second step of splicing was significantly inhibited in these mutants, probably due to the use of a 3′ splice site U residue, since the same second-step defect was seen with the C99U and A98U mutants. These results show that 10 nucleotides represents a significant functional minimum for the branch site-to-3′ splice site distance in U12-dependent introns, consistent with the data on natural introns.
When we moved the 3′ splice site to 18 nucleotides from the branch site, we observed that this weakened use of the 3′ splice site. In vivo, a 3′ splice site at +18 activated use of a cryptic UU 3′ splice site located at +12. In vitro, the +18 3′ splice site mutants formed spliceosomes less efficiently than wild-type RNA and were less efficiently spliced. When placed in competition with the wild-type AC at +10, an AC at +18 was inactive in vivo and in vitro. These results suggest that moving the 3′ splice site to near the 20-nucleotide maximum observed in normal introns causes significant defects in splice site function.
Finally, when we moved the first available 3′ splice site beyond the 20-nucleotide limit, no U12-dependent splicing was observed either in vivo or in vitro. Furthermore, this mutant was unable to form spliceosome complexes in vitro. From these results, it appears that there is a maximum functional limit for the branch site-to-3′ splice site of between 18 and 27 nucleotides.
3′ splice sites at a spacing of 12 to 13 nucleotides are highly preferred over proximal and distal sites.
To determine the optimal distance between the branch site and the 3′ splice site, we constructed an intron in which CAC motifs were located at 10, 12, 14, 16, and 18 nucleotides and tested it in vivo and in vitro. The clear result was that over 90% of splicing was to the AC at a spacing of 12 nucleotides. Similarly, in the C99U construct with AU dinucleotides at +10 and +12, most splicing events used the +12 position. These results establish that the optimal spacing is about 12 nucleotides.
The idea of an optimal distance for the 3′ splice site downstream from the branch site led us to examine the 3′ splice site regions of U12-dependent introns for natural examples of such 3′ splice site choice. The mouse cdk5 gene contains an intron with good matches to U12-dependent splice site sequences of the AU-AC class (28). The 3′ end of this intron contains the sequence GACAC/AC, where the slash denotes the 3′ splice site located 13 nucleotides from the branch site. This intron has AC dinucleotides located at spacings of 11, 13, and 15 nucleotides but only uses the AC at +13. This natural example differs slightly from our experimental construct in that the AC at +11 is preceded by a G residue rather than the consensus pyrimidine.
Another interesting example is provided by the human E2F1 intron 4, a putative U12-dependent AU-AC intron which has a 3′ splice site sequence of GCCAAC/CC, with the 3′ splice site located at a spacing of 11 (28). In this case a CAA is located at +10, where it was functional in the P120 C99A mutant but is skipped in favor of the AAC located at +11. A final example is the sixth intron of the human GT335 gene, a putative U12-dependent AU-AC intron which ends with the sequence CACAC (21). The functional 3′ splice site is located at a spacing of 11, while a nonfunctional CAC sequence is at a spacing of 9 nucleotides.
U12-dependent 3′ splice sites are not selected by a simple scanning mechanism.
The results discussed above suggest that 3′ splice site selection in the U12-dependent system is not dictated solely by a scanning mechanism. If a simple scanning mechanism were used, the CAC located at a distance of 10 nucleotides in the Oligo AC construct would have been used exclusively. In fact, the CAC at +12 was used in preference to the +10 position. The combination of the small range of acceptable distances, the pronounced preference for a spacing of about 12 nucleotides, and the selection of a downstream over an upstream 3′ splice site strongly argue against a scanning model for 3′ splice site selection.
The data appear much more compatible with a model in which the pre-mRNA is held into the spliceosome by interactions at the branch site at a position near the active site for step two of splicing. The RNA extending from the branch site must span this distance and present a functional 3′ splice site to the active site for step two of splicing. If the distance is made too short, downstream cryptic splice sites become active. If the distance is made too long, even highly unfavorable sequences such as UU can be used as 3′ splice sites if they are located at the optimum distance. When the 3′ splice site is moved to 27 nucleotides, splicing to all sites is dramatically reduced. The in vitro results show that this intron cannot form prespliceosomes or spliceosomes, suggesting that proper 3′ splice site spacing plays a role in the early events of spliceosome formation as well as in the final stages of splicing.
Interestingly, an example of a U2-type intron which appears to violate the first AG rule came out of our analysis of U12-type introns. The human cardiac beta myosin heavy chain (GenBank accession no. X52889) intron 9 is a U2-type intron that is in an identical position to the U12-type intron 5 in the smooth muscle homolog (GenBank accession no. AF001548), a common finding in gene families that we have discussed previously (5). In the cardiac U2-type intron, the 3′ end of the intron is TTCAGCAG/ATC, indicating that a fully consensus CAG is skipped in favor of an immediately downstream CAG. While the exact U2-type branch site cannot be determined, a 90% pure pyrimidine region of about 35 nucleotides is located upstream of this sequence. In addition, the 5′ splice site is a U2-type GTGAGT, and no U12-type branch site is present. A similar result was obtained in an experimental intron with closely spaced AGs (38). It appears, then, that a mechanism other than scanning for the first AG 3′ of the branch site may apply in at least some U2-type introns as well.
U12-type splicing complex formation requires an active 3′ splice site.
The results of in vitro spliceosome formation assays on the various 3′ splice site constructs shows that an active 3′ splice site is required for assembly of the A complex. When the 3′ splice site is moved to +18, spliceosome formation efficiency is reduced. When no 3′ splice site is available, as in the case of the +27 AC mutant, no specific complexes form. However, the lack of complexes detectable with the gel shift assay does not mean that the branch site is not being recognized by the splicing machinery. Evidence that the branch site is still active comes from the demonstration that the downstream cryptic branch site-3′ splice site at +124 is not used in any of the constructs, even when no other 3′ splice site is used. From this, it appears that the wild-type branch site is recognized, possibly by U12 snRNP and other factors, in such a way as to limit the ability of the 5′ splice site and its associated factors to interact with another branch site-3′ splice site. Nevertheless, this interaction with the wild-type branch site is unable to proceed to the A complex without the participation of a functional 3′ splice site. Frilander and Steitz (14) showed that A complex formation in U12-dependent introns required the participation of both the 5′ splice site and the branch site. Here we extend this result to show that A complex formation is also dependent on a properly positioned 3′ splice site.
In light of these results, an additional finding reported by Frilander and Steitz (14) that an RNA truncated between the branch site and the 3′ splice site can form spliceosome-like complexes appears to be in contradiction. These complexes, however, appeared to be unable to carry out the first step of splicing. We have repeated these experiments under our conditions and seen similar results. The complexes migrate approximately the same as authentic A and B complexes and form under the same conditions yet do not carry out the first step of splicing to a detectable level. This is in contrast to the +27 mutant, which cannot form either A or B complexes. The apparent difference between these two RNAs is that the +27 construct has nucleotides in the region where a 3′ splice site should be located, while the truncated RNA has nothing.
A possible way to reconcile these results is to suggest that complexes can form transiently at potential U12-dependent branch site sequences but are destabilized by the absence of a properly positioned 3′ splice site. This destabilization could be an active proofreading function which is not triggered in the case of the truncated RNA due to the absence of enough RNA downstream of the branch site. In support of this idea, the lack of activation of the +124 cryptic branch site-3′ splice site in the +27 mutant suggests that the normal branch site is still being recognized in some manner and this is causing the sequestration of the 5′ splice site so that it cannot productively engage the +124 cryptic site.
For this mechanism to play a role in 3′ splice site fidelity, there must be evidence that an active branch site-3′ splice site can compete with an inactive one. We have shown elsewhere that the +124 cryptic site can compete with the wild-type site when a single nonconsensus nucleotide in the branch site upstream of +124 is corrected to the consensus residue (Dietrich et al., submitted.). Therefore, a single 5′ splice site can productively interact with more than one potential 3′ splice site, implying that mechanisms must exist to choose the correct site and discriminate against incorrect sites. We would argue that one aspect of this discrimination is to check for the presence of a functional 3′ splice site within 10 to 20 nucleotides of a potential branch site.
U12-type 3′ splice site appears to be identified by a local diffusion process.
An alternative to linear scanning is a process in which the 3′ splice site binds to a site on the forming spliceosome that is located at a fixed distance from the branch site binding site. The pre-mRNA bound to the spliceosome through the U12 snRNP interaction at the branch site would encounter the 3′ splice site binding site by local diffusion. The physical properties of the surrounding structure would then determine both the minimum distance that must be bridged to bind both sites and the amount of extra RNA sequence that could fit. These structural constraints would then determine the minimum and maximum distance between the branch site and the 3′ splice site. The sharp spacing optimum of about 12 residues could be interpreted as supplying a distance measurement between the branch site adenosine and the step-two active site within the spliceosome. This would correspond to a distance of about 80 to 85 Å between these two points on the U12-dependent spliceosome. This could be easily accommodated in the 40- to 60-nm U2-dependent spliceosome (32). Such a distance would correspond to a mean or lowest energy state. Shorter and longer distances could be accommodated by energetically costly deformations of the spliceosome or the pre-mRNA.
A rather similar distance for the mammalian U2-dependent spliceosome can be inferred from studies which showed that an AG at +4 was inactive but AGs at +12 and +19 could splice in vitro (38) or that an AG at +11 was inactive but AGs at +15 to +24 were active (10 and references therein). There is also evidence that the S. cerevisiae U2 spliceosome can splice to 3′ splice sites as few as 11 nucleotides from the branch site (12).
From the data presented here, it appears that the U12-dependent spliceosome cannot scan for a 3′ splice site over many tens of bases as the U2-dependent spliceosome can. This might be reflected in the absence from the U12 spliceosome of specific protein factors that promote scanning in the U2 spliceosome. Candidate factors might include the mammalian homologs of the yeast second-step splicing factors Prp16, Prp17, Prp18, and Slu7 (42). In S. cerevisiae, Slu7 is thought to promote splicing to downstream AGs (4, 12). However, so far there is no evidence that the human homolog of Slu7 is dispensable for a subset of introns (9, 10).
ACKNOWLEDGMENT
This work was supported by grant GM55105 from the National Institutes of Health.
REFERENCES
- 1.Anderson K, Moore M J. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K, Moore M J. Bimolecular exon ligation by the human spliceosome bypasses early 3′ splice site AG recognition and requires NTP hydrolysis. RNA (NY) 2000;6:16–25. doi: 10.1017/s1355838200001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boudvillain M, de Lencastre A, Pyle A M. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- 4.Brys A, Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA (NY) 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 5.Burge C B, Padgett R A, Sharp P A. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 6.Burge C B, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by the spliceosome. In: Gestland R F, Cech T, Atkins J F, editors. The RNA world II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 7.Chanfreau G, Legrain P, Dujon B, Jacquier A. Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3′ splice site selection in S. cerevisiae. Nucleic Acids Res. 1994;22:1981–1987. doi: 10.1093/nar/22.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Anderson K, Moore M J. Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc Natl Acad Sci USA. 2000;97:593–598. doi: 10.1073/pnas.97.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chua K, Reed R. Human step II splicing factor hSlu7 functions in restructuring the spliceosome between the catalytic steps of splicing. Genes Dev. 1999;13:841–850. doi: 10.1101/gad.13.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich R C, Incoravia R, Padgett R A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 12.Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- 13.Frendewey D, Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985;42:355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- 14.Frilander M J, Steitz J A. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gozani O, Patton J G, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall S L, Padgett R A. Conserved sequences in a class of rare eukaryotic introns with non-consensus splice sites. J Mol Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 17.Hall S L, Padgett R A. Requirement of U12 snRNA for the in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 18.Helfman D M, Ricci W M. Branch point selection in alternative splicing of tropomyosin pre-mRNA. Nucleic Acids Res. 1989;17:5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolossova I, Padgett R A. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) introns. RNA (NY) 1997;3:227–233. [PMC free article] [PubMed] [Google Scholar]
- 20.Konarska M M. Analysis of splicing complexes and small nuclear ribonucleoprotein particles by native gel electrophoresis. Methods Enzymol. 1989;180:442–453. doi: 10.1016/0076-6879(89)80116-8. [DOI] [PubMed] [Google Scholar]
- 21.Lafreniere R G, Rochefort D L, Kibar Z, Fon E A, Han F, Cochius J, Kang X, Baird S, Korneluk R G, Andermann E, Rommens J M, Rouleau G A. Isolation and characterization of GT335, a novel human gene conserved in Escherichia coli and mapping to 21q22.3. Genomics. 1996;38:264–272. doi: 10.1006/geno.1996.0627. [DOI] [PubMed] [Google Scholar]
- 22.Luukkonen B G M, Seraphin B. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 24.Moore M J. Intron recognition comes of AGe. Nat Struct Biol. 2000;7:14–16. doi: 10.1038/71207. [DOI] [PubMed] [Google Scholar]
- 25.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson K K, Green M R. Mammalian U2 snRNP has a sequence-specific RNA-binding activity. Genes Dev. 1989;3:1562–1571. doi: 10.1101/gad.3.10.1562. [DOI] [PubMed] [Google Scholar]
- 27.Neuman E, Sellers W R, McNeil J A, Lawrence J B, Kaelin W G., Jr Structure and partial genomic sequence of the human E2F1 gene. Gene. 1996;173:163–169. doi: 10.1016/0378-1119(96)00184-9. [DOI] [PubMed] [Google Scholar]
- 28.Ohshima T, Nagle J W, Pant H C, Joshi J B, Kozak C A, Brady R O, Kulkarni A B. Molecular cloning and chromosomal mapping of the mouse cyclin-dependent kinase 5 gene. Genomics. 1995;28:585–588. doi: 10.1006/geno.1995.1194. [DOI] [PubMed] [Google Scholar]
- 29.Parker R, Siliciano P G. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 30.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 31.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 32.Reed R, Griffith J, Maniatis T. Purification and visualization of native spliceosomes. Cell. 1988;53:949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 33.Ruskin B, Green M R. Role of the 3′ splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985;317:732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- 34.Rymond B C, Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 35.Rymond B C, Rosbash M. Yeast pre-mRNA splicing. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1992. pp. 143–192. [Google Scholar]
- 36.Scadden A D J, Smith C W J. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith C W, Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- 38.Smith C W J, Chu T T, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith C W J, Porro E B, Patton J G, Nadal-Ginard B. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 40.Tarn W-Y, Steitz J A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 41.Tarn W-Y, Steitz J A. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 42.Umen J G, Guthrie C. The second catalytic step of pre-mRNA splicing. RNA (NY) 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Q, Krainer A R. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol Cell Biol. 1999;19:3225–3236. doi: 10.1128/mcb.19.5.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Q, Krainer A R. Purine-rich enhancers function in the AT-AC pre-mRNA splicing pathway and do so independently of intact U1 snRNP. RNA (NY) 1998;4:1664–1674. doi: 10.1017/s1355838298981432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Q, Krainer A R. Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA (NY) 1997;3:586–601. [PMC free article] [PubMed] [Google Scholar]
- 46.Wu S, Romfo C M, Nilsen T W, Green M R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 47.Zhuang Y, Weiner A M. The conserved dinucleotide AG of the 3′ splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene. 1990;90:263–269. doi: 10.1016/0378-1119(90)90189-x. [DOI] [PubMed] [Google Scholar]
- 48.Zorio D A R, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]