Summary
Resistance can occur in patients receiving adoptive cell therapy (ACT) due to antigen-loss-variant cancer cell (ALV) outgrowth. Here we demonstrate that murine and human Th9 cells, but not Th1/Tc1 or Th17 cells, expressing tumor-specific T-cell-receptors (TCR) or chimeric antigen receptors (CAR), eradicate advanced tumors that contain ALVs. This unprecedented antitumor capacity of Th9 cells is attributed to both enhanced direct tumor cell killing and bystander antitumor effects promoted by intratumor release of IFNα/β. Mechanistically, tumor-specific Th9 cells increase the intratumor accumulation of extracellular ATP (eATP, released from dying tumor cells), because of a unique feature of Th9 cells that lack the expression of ATP degrading ectoenzyme CD39. Intratumor enrichment of eATP promotes the monocyte infiltration and stimulates their production of IFNα/β by inducing eATP-endogenous retrovirus-TLR3/MAVS pathway activation. These results identify tumor-specific Th9 cells as a unique T cell subset endowed with the unprecedented capacity to eliminate ALVs for curative responses.
Graphical Abstract
eTOC
Tumor heterogeneity and antigen loss cause tumor relapse or progression after cancer adoptive cell immunotherapy. Xue et al. uncover that tumor-specific Th9 cells not only kill antigen-positive cancer cells but kill antigen-negative cancer cells in tumors, which hold promise to advance the utility and efficacy of adoptive cell immunotherapy.
Introduction
Drug resistance may cause cancer treatment failure and death in over 90% of patients with advanced tumors. Patients may initially respond to treatment, but recurrence often occurs because of the heterogeneous nature or genetic diversity of cancer cells (Bedard et al., 2013). Few cells carrying specific somatic mutation are iteratively selected and escape during chemotherapy or targeted therapies causing tumor relapse or progression (Bedard et al., 2013).
Similarly, resistance can emerge after cancer adoptive cell immunotherapy. As a result, despite the high initial response rates in patients treated with TCR (T-cell-receptors) or CAR(chimeric antigen receptors) engineered ACT (adoptive cell therapy), durable complete responses are observed in less than 30% of patients in hematological malignancies and ACT has not mediated sustained responses in solid tumors, which display much higher heterogeneity and contain a significant amount of ALVs (antigen-loss-variant cancer cell) (Jacoby et al., 2016; June et al., 2018; Gerbitz et al., 2012). Relapses due to ALVs upon ACT have been frequently reported in patients with melanoma (Mehta et al., 2018), leukemia (Jacoby et al., 2016), multiple myeloma (Adam et al., 2019), and glioblastoma (Brown et al., 2016; O’Rourke et al., 2017). For example, in melanoma patients treated with ACT targeting the melanoma-melanocytic antigen MART1, tumor relapse due to the selection of MART1-loss or multiple melanocytic antigen-loss ALVs was observed (Landsberg et al., 2012; Mehta et al., 2018). Similarly, CD19− ALVs occur in up to 50% of leukemia/lymphoma patients after CD19 CAR-T cell treatment (Park et al., 2016). When the CD19 antigen is lost due to mutation or lineage switch (Velasquez and Gottschalk, 2017), malignant cells become invisible to CD19-specific CAR-T cells (Park et al., 2016). However, such strategy may significantly raise cost and toxicity of ACT, and may only delay the onset of the relapse, but eventually select the outgrowth of multi-antigen “negative” tumor cells (Hegde et al., 2016; Fry et al., 2017; Wang et al., 2017; Zhang et al., 2015). Thus, we currently lack therapeutic approaches that efficiently prevent the emergence of ALVs.
CD4+ T cells generated by TGF-β1 and IL-4 that produce IL-9 are defined as a separate Th subset termed Th9 to distinguish them from classical Th2 cells (Dardalhon et al., 2008; Veldhoen et al., 2008). We (Lu et al., 2012; Lu et al., 2014a; Zhao et al., 2016) and others (Purwar et al., 2012) have characterized tumor-specific Th9 cells as an antitumor T cell subset. Furthermore, we (Lu et al., 2018) have recently reported that tumor-specific Th9 cells are less susceptible to exhaustion, fully cytolytic to tumor cells, and possess long-term persistence capacity because of their unique hyperproliferative T cell feature. However, these observations only reveal how tumor-specific Th9 cells eradicate antigen-positive tumor cells (Lu et al., 2018; Lu et al., 2012; Lu et al., 2014a; Zhao et al., 2016). Given the unmet need to develop effective ACT strategies for solid tumors with the heterogeneity in antigen expression, the role of ACT with tumor-specific Th9 cells in ALV-containing tumors is still elusive. Therefore, we carried out this study to uncover the T cell features of Th9 cells for ALV clearance.
Results
Th9 cells eliminate “natural-occurring” ALVs
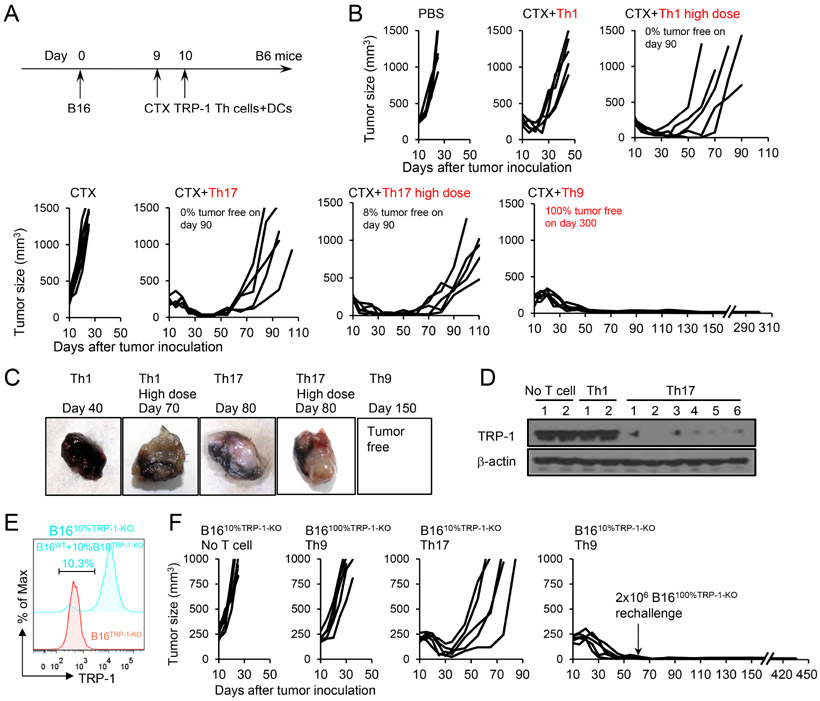
Tyrosinase-related protein-1 (TRP-1) is a native antigen expressed by melanoma. We compared the antitumor effects of Th1 and Th9 cells in an advanced murine B16 melanoma model (Figure 1A). We also included Th17 cells as “an enhanced version of Th1 cells” (Muranski et al., 2011). The characterization of these Th cells was consistent with our prior publication (Lu et al., 2018) and Th9 cells displayed a less-exhausted, fully cytolytic, and hyperproliferative phenotype (Figure S1A-S1F). In advanced B16 melanoma-bearing mice, adoptive transfer of TRP-1-specific Th9 cells eradicated the large established tumor (8×7 mm, established s.c.), and led to long-term tumor-free survival (~300 days) (Figure 1B). In sharp contrast, TRP-1-specific Th1 cells failed in controlling tumor growth, while TRP-1-specific Th17 cells promoted tumor regression, but failed in eradicating the tumor since the majority of mice succumbed after 2 months due to tumor recurrence (Figure 1B).
Figure 1. Transfer of tumor-specific Th9 cells prevents the relapse of variant tumors.
(A) TRP-1-specific Th1, Th17, Th9 cells (CD45.2+, regular dose, 2.5×106), high dose Th1 (2×107), or high dose Th17 cells (7.5×106) were transferred i.v. into CD45.1+ B6 mice bearing 10-day large established B16 tumors (1×106 B16 cells challenged s.c. 10 days before T cell transfer). Adjuvant cyclophosphamide (CTX, i.p.) and DC vaccination (2.5×105, i.v.) were administered to some mice as indicated. (B) Tumor responses to TRP-1 cell transfer are shown (n=5/group). The description of tumor-free survival is summarized from several independent studies. (C-D) Tumors were harvested from No-T-cell-treated mice, Th1 or Th17-cell-treated mice. Images of representative tumors are shown in (C). Tumor tissue lysates were analyzed for TRP-1 expression by western-blot (D). (E) Intracellular staining of TRP-1 in B1610%TRP-1-KO and B16TRP-1-KO tumor cells. (F) B1610%TRP-1-KO tumor (contain 10% B16TRP-1-KO ALVs)-bearing mice were treated similarly to Fig. 1A. Surviving mice in the Th9-cell-treated group were rechallenged with 2×106 ALVs (B16TRP-1-KO) on day 60. Tumor responses are shown (n=5/group). Representative results from one of two repeated experiments are shown (total # of mice/group ≥ 20 in B; ≥ 10 in F).
We performed mechanistic studies to identify the more pronounced antitumor effects of Th9 cells as compared to Th17 cells. Firstly, we investigated T cell persistence and in vivo killing activity. Tumor-bearing mice treated with Th9 and Th17 cells were euthanized ~60 days after treatment when Th17 cell-treated mice started showing tumor regrowth. Persistence of the transferred TRP-1-T cells in the peripheral blood was very similar between Th9 and Th17 cell-treated mice (Figure S1G-S1H). Similarly, no significant differences were observed in the in vivo killing activity against TRP-1 peptide-loaded targets, suggesting that both Th9 and Th17 cells efficiently recognize their cognate targets (Figure S1I-S1J). We also thought to determine if a higher dose of Th1 or Th17 cells may be required to prevent tumor recurrence. Interestingly, high dose Th1 (2×107) or Th17 (7.5×106) cell ACT enhanced antitumor efficacy compared to the regular dose (2.5×106), but still failed to prevent the recurrence in majority of the treated mice (Figure 1B). Of note, these high doses of Th1 and Th17 cells were the maximum dose a mouse can receive, because a further increase of T cell dose was too toxic (>60% of mice died of side effects in the 1st week after ACT).
However, we noted that macroscopically most of the tumors recurrent in mice treated with Th17 cells were white to gray, while tumors growing in mice receiving Th1 cells appeared to be dark black as typically observed in the B16 tumor model (Figure 1C). We thus hypothesized tumors relapsing in Th17 cell treated mice may be B16 ALV tumor cells that lost TRP-1 expression, which is required for the melanin production, and escape TRP-1-mediated recognition (Kobayashi et al., 1994). We confirmed that in the white recurrent tumors, the expression of TRP-1 protein was low or undetectable by Western blot analysis and qPCR (Figure 1D and S1K), whereas the gene expression of Mitf (encodes Melanocyte Inducing Transcription Factor) and Dct (encodes TRP-2) was only slightly decreased (Figure S1L). Thus the adoptive transfer of TRP-1-Th17 cells in B16 tumor-bearing mice faithfully recapitulates the clinical scenario of tumor relapse due to the emergence of ALVs observed after adoptive T cell therapy.
Tumor-specific TCR and CAR Th9 cells eradicate ALV-containing “chimeric tumors”
Although Th9 cell transfer appears to possess the capacity to prevent ALV-mediated relapse, an important question is whether Th9 cells can eliminate a more heterogeneous tumor cell population that already contains a significant fraction of ALVs, as often observed in patient tumors (Klebanoff et al., 2016). We used CRISPR/Cas9 to knock out the expression of TRP-1 in B16 cells (referred as B16TRP-1-KO cells, Figure S1M) and composed chimeric tumor B1610%TRP-1-KO containing 90% of WT-B16 cells and 10% of B16TRP-1-KO cells (Figure 1E). We then used mice bearing large B1610%TRP-1-KO chimeric tumors to test the antitumor effects of TRP-1 T cell-transfer. In this model, we observed rapid tumor relapse upon TRP-1-Th17 T cell transfer, while TRP-1-Th9 cells continued to cause tumor eradication and mice remained tumor-free at 440 days (Figure 1F). Mice transferred with Th9 cells did not display significant systemic toxicity including limited weight loss, no significant pathological change, relatively low proinflammatory cytokines in serum, and moderate vitiligo development (Figure S1N-S1Q). Interestingly, the relapsed tumors in Th17 cell ACT group preferentially lost Tyrp1 but not Mitf and Dct gene expression (Figure S1R). Remarkably, TRP-1-Th9 cell transfer also extends to the protection of mice upon tumor-specific rechallenge in the contralateral flank at 60 days after initial transfer (Figure 1F and Figure S1S), although isolated Th9 cells from treated mice did not display apparent cytotoxicity against B16TRP-1-KO cells (Figure S1T), suggesting that Th9 cells may not directly eradicate ALVs in vivo. Furthermore, B16 tumors containing up to 30% ALVs can be eradicated by TRP-1-Th9 cells (Figure S1U).
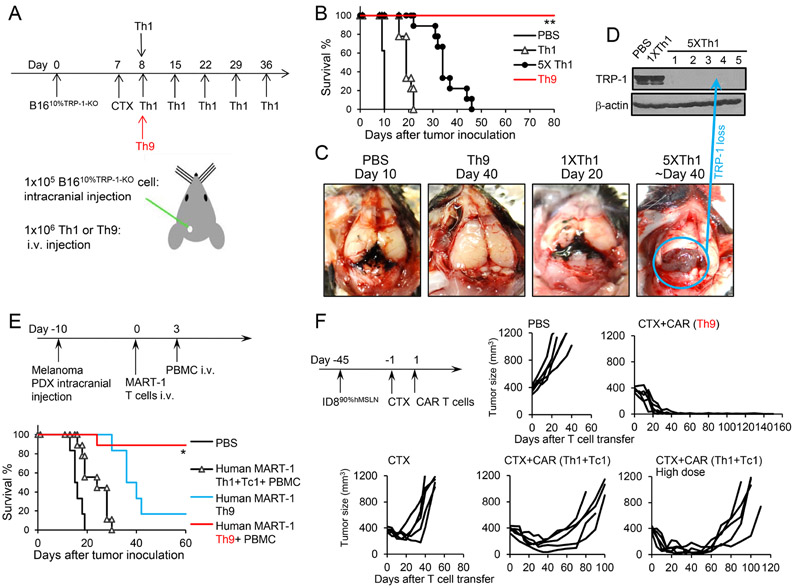
To determine if Th9 cells may also eradicate the ALV-containing metastatic tumors, we took advantage of an extremely aggressive B16 brain metastatic melanoma model (intracranial injection of 1×105 B1610%TRP-1-KO cells, Figure 2A), causing all mice to die on day 9 or 10 after the tumor challenge if untreated. Mice received T cells i.v. on day 8 (just one day before the untreated mice started to die), with the transfer of 1 dose of TRP-1 Th9 cells or Th1 cells, or multiple doses of Th1 cells every week. Again, mice treated with one dose of Th9 cells all survived for at least 80 days, whereas all the mice received one dose of Th1 cells died ~ 20 days. As expected, multiple doses of Th1 cells indeed significantly extended the survival compared to mice receive only one dose; but unfortunately, all mice still died of the disease ~40 days (Figure 2B). Biopsy of the brain B16 tumors revealed some striking features associated with depigmented ALV tumor growth in mice received 5 doses of Th1 cells, whereas brain tumors in PBS or one dose Th1 cell-treated mice are dark black (Figure 2C and 2D). Therefore, providing more tumor-specific Th1 cells is not sufficient to drive a curative response, but eventually, also promotes the escape of ALV tumor outgrowth. To strengthen our findings, we generated human Th9 cells and T1 cells (Th1+Tc1) and found that human Th9 cells were also less-exhausted and hyperproliferative effector T cells by in vitro assays (Figure S2A-S2E). Furthermore, in a human PBMC-engrafted NSG mouse model, human autologous MART-1-Th9 cells display superior antitumor capacity against melanoma patient-derived xenograft (PDX) compared to Th1+Tc1 cells in vivo (Figure 2E). Interestingly, the curative response of human MART-1-Th9 cells requires the engraftment of autologous PBMC, whereas MART-1-loss human melanoma ALV outgrowth was inevitable in non-humanized NSG mice receiving human Th9 cells (Figure S2F).
Figure 2. Effect of TRP-1-specific-Th9 cells and hMesothelin-CAR-Th9 cells.
(A) TRP-1-specific 1.0×106 Th1 (single or multiple doses), or 1.0×106 Th9 cells (CD45.2+) were transferred i.v. into CD45.1+ B6 mice bearing 8-day established intracranially (i.c.) injected B16 tumors (1×105 B1610%TRP-1-KO tumor cells challenged i.c. 8 days before T cell transfer). (B) Survival analysis in response to adoptive cell transfer (n=9-10/group). Data are summarized from two independent studies. **P<0.01, Th9 compared with PBS, Th1, and 5×Th1, survival analysis was conducted by log-rank test. (C-D) Brain tumors were harvested from mice received indicated treatments. Images of representative brain tumors are shown (C). Tumor tissue lysates were analyzed for TRP-1 expression by western blot (D). (E) Human autologous MART-1-specific 1.0×106 Th9, or 1.0×106 Th1+Tc1 cells were transferred i.v. into NSG mice bearing 10-day established intracranially (i.c.) injected melanoma PDX (1×105 cells challenged i.c. 10 days before T cell transfer). Autologous PBMC was administered i.v. to mice on day 3 as indicated. Survival analysis in response to ACT (n=6-9/group). Data are summarized from three independent studies. *P<0.05, Human MART-1 Th9+PBMC compared with any other groups, survival analysis was conducted by log-rank test. (F) hMesothelin-CAR-T cells (5.0×106 GFP+) were transferred i.v. into CD45.1+ B6 mice bearing 45-day-established, s.c. injected ID890%hMesothelin tumors. CAR Th1+Tc1 cells were used in a regular dose of total cell # at 5×106 or in a high dose of 2.5×107 cells. Tumor responses to hMesothelin-CAR-T cell transfer are shown (n=5/group). Data are summarized from two independent studies. Representative results from one of two repeated experiments are shown (total # of mice/group ≥ 10).
To strengthen our findings, we developed another model of hMesothelin-mCAR Th9 cells to target the hMesothelin-expressing murine ID8 ovarian cancer (OvCa). We composed ID890%hMesothelin “chimeric tumor” cells containing 10% of hMesothelin− ID8 ALVs, and inoculated them subcutaneously into female B6 mice. Remarkably, mCAR Th9 cells caused long-term protection with relapse-free responses up to 150 days, whereas a high dose of mCAR Th1+Tc1 cell ACT selected the outgrowth of hMesothelin-loss ID8 ALVs (Figure 2F and S2G). Similarly, human autologous hMesothelin-hCAR Th9 cells but not regular or high doses of Th1+Tc1 CAR T cells eradicate human OvCa PDX in humanized NSG mice (Figure S2H-S2J). Interestingly, human CAR Th9 cell ACT alone is sufficient for the antitumor response, while cotransfer of CAR Tc9 cells may not bring additional benefits (Figure S2I). Therefore, our results highlight a unique feature of tumor-specific Th9 cells with the potential for broad therapeutic exploitation to prevent the relapse of ALVs.
Th9 cells ACT creates a unique tumor microenvironment to overcome ALV outgrowth
Tumor-specific Th9 cells eradicated not only antigen-expressing tumor cells but also chimeric tumors containing ALVs. To investigate the underlying mechanism, we first determined if the unique hyperproliferative Th9 cell feature (Lu et al., 2018), which is indispensable for Th9 elimination of antigen-positive tumor cells and driven by TRAF6-NF-κB signaling, may also dictate the Th9’s eradication of ALVs. Interestingly, although TRAF6 signaling is required for the antitumor response in Th9 ACT (Figure S3A), it seems that TRAF6 signaling does not contribute to the anti-ALV response because tumors at the endpoint show no reduction of Tyrp1 gene expression (Figure S3B). In addition, it appeared that IL-21 did not contribute to Th9-mediated eradication of ALVs (Figure S3C). These results suggest that the role and mechanisms underlying Th9-mediated clearance of antigen-negative ALVs, especially when synergized with lymphodepleting pre-conditioning regimen, may be different from that of elimination of antigen-positive tumor cells by tumor-specific Th9 cells.
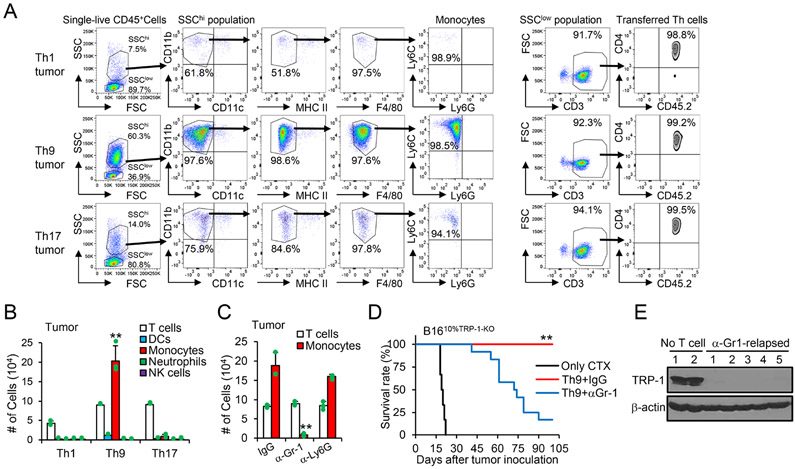
To gain further insight into how Th9 cells eliminate ALVs, we analyzed the tumor-infiltrating immune cells. In mice treated with Th9 cells, we observed a significant increase in the infiltration of SSChiCD11b+CD11c−MHC-IIloF4/80−Ly6G−Ly6Chi inflammatory monocytes (Figure 3A and 3B). We hypothesized that Th9 cells recruit these inflammatory monocytes from the spleen within the tumor because these cells were abundant in the spleens of all CTX-treated mice irrespective of the T cells they have received (Figure S3D). Based on previous studies, these cells seem to be induced as a result of temporary lymphopenia (Becker and Schrama, 2013; Kodumudi et al., 2012).
Figure 3. Tumor inflammatory monocytes recruited by Th9 cells during lymphopenia are crucial to anti-ALV responses.
(A-B) B1610%TRP-1-KO tumor-bearing mice (s.c.) were treated as shown in Fig. 1F. Tumor tissues (200 mg/mice, 10 days after ACT) were harvested and used for FACS analysis. Representative (A) and summarized results (B, n=3-4/group) are shown. Data are mean ± SD. **P<0.01, Th9 compared with Th1 and Th17 groups, one-way ANOVA with Tukey test. (C-E) B1610%TRP-1-KO tumor-bearing mice were treated with Th9 cells as shown in Fig. 1F, together with indicated antibodies every 3 days after ACT. Tumor-infiltrating indicated immune cells are calculated from FACS analysis (C, n=3-4/group). Data are mean ± SD. **P<0.01, α-Gr-1 compared with IgG and α-Ly6G groups, one-way ANOVA with Tukey test. (D) Mice survival curves are shown (n=9-12/group, combined from 2 independent experiments), **P<0.01, compared with any other groups, survival analysis was conducted by log-rank test. (E) Tumor (~day 60) tissue lysates were analyzed for TRP-1 production by western-blot. Representative results from one of two repeated experiments are shown.
The recruitment of inflammatory monocytes within the tumor in mice treated with Th9 cells prompted us to analyze the potential role of these cells. We found that an anti-Gr-1 mAb, but not an anti-Ly6G mAb substantially and preferentially reduced the recruitment of inflammatory monocytes within the tumor in mice treated with Th9 cells (Figure 3C and S3E). Unexpectedly, depletion of these inflammatory monocytes largely precluded Th9 cells from eliminating ALVs because mice relapsed due to the growth of tumor cells lacking TRP-1 expression (Figure 3D and 3E). Collectively, our results pinpoint a pivotal role of a unique Th9 transfer-dependent recruitment of inflammatory monocytes into the tumor that allows the clearance of ALVs.
Th9 cell ACT promotes an eATP-enriched milieu causing the recruitment of monocytes
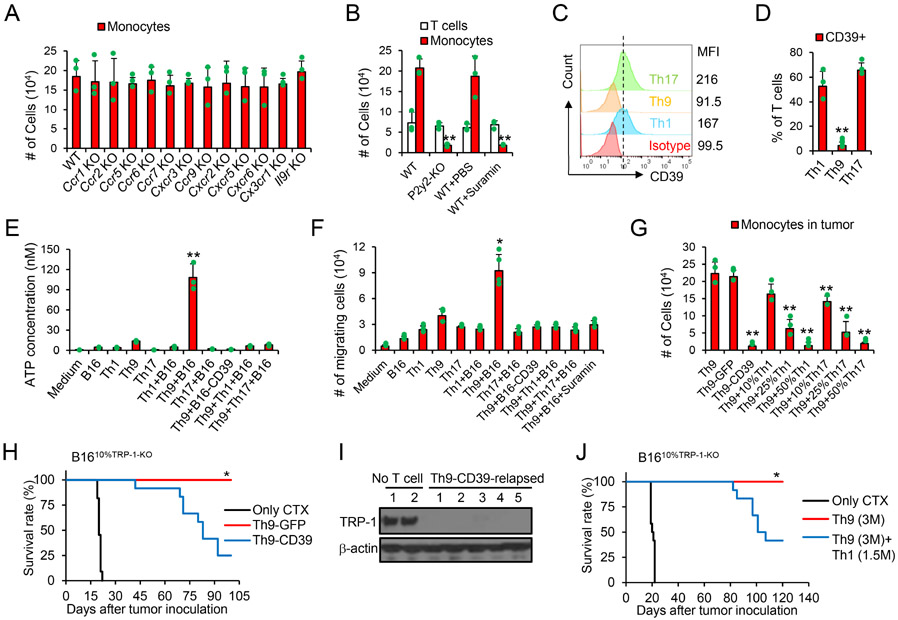
Since monocyte recruitment is crucial to Th9 cell-mediated anti-ALV response, we sought to determine which factors cause the recruitment of monocytes. We found that tumor monocyte infiltration is not mediated by IL-9 or chemokines because host deficiency of Il9r, Ccr1, Ccr2, Ccr5, Ccr6, Ccr7, Cxcr3, Ccr9, Cxcr2, Cxcr5, Cxcr6 and Cx3cr1 did not abrogate the tumor infiltration with monocytes (Figure 4A). We next extend our study to non-classic extracellular nucleotides that promote monocyte chemotaxis towards apoptotic tumor cells by sensing of eATP but not ADP (Elliott et al., 2009). We found that suramin (a pan-purinergic signaling inhibitor) administration remarkably reduced tumor monocyte infiltration, suggesting that extracellular nucleotides may promote monocyte chemotaxis (Figure 4B). The purinergic receptor P2Y2 (encoded by P2ry2) was previously suggested to mediate monocyte chemotaxis towards apoptotic cells by sensing eATP (Elliott et al., 2009). In agreement with this observation, we found that host P2ry2-deficiency prevented monocyte tumor infiltration and abrogated the anti-ALV capacity of Th9 cells, because the relapsed tumors also display largely reduced TRP-1 expression (Figure 4B and S4A-S4C). Reciprocally, reconstituting P2ry2−/− mice with i.v. injection of inflammatory monocytes isolated from WT but not P2ry2−/− mice restored monocyte recruitment to tumors and anti-ALV capacity of Th9 cells (Figure S4D and S4E).
Figure 4. Tumor-specific Th9 cells promote eATP-P2Y2 dependent chemotaxis of tumor monocytes.
(A-B) B1610%TRP-1-KO tumor-bearing WT or transgenic mice were treated as shown in Fig. 1F. Some mice were treated with i.p. injection of suramin. Tumor tissues (200 mg/mice, 10 days after ACT) were harvested and leukocytes were counted and used for FACS analysis. Tumor-infiltrating indicated immune cells are calculated from FACS analysis (B, n=3-4/group). Data are mean ± SD. **P<0.01, compared with WT and WT+PBS groups, one-way ANOVA with Tukey test. (C-D) Surface expression of CD39 on TRP-1 T cells was analyzed by FACS. Data are mean ± SD. **P<0.01, Th9 compared with Th1 and Th17 groups, one-way ANOVA with Tukey test. (E) B16 tumor cells and TRP-1 T cells were cultured alone or cocultured as indicated for 24 hours (n=3). The ATP concentrations in the culture supernatant were determined by ATP Determination Kit. Data are mean ± SD. **P<0.01, Th9+B16 compared with any other groups, one-way ANOVA with Tukey test. (F) Monocytes migration assay. B16-bearing mice were treated with CTX. SpMono were sorted 10 days later and seeded in Transwells with the bottom media from the supernatant of indicated cell cultures. Cells migrated to the bottom after 18 hrs are counted by cell counter. Data are mean ± SD. *P<0.05, Th9 compared with any other groups, one-way ANOVA with Tukey test. (G) B1610%TRP-1-KO tumor-bearing WT mice were treated as shown in Fig. 1F. Tumor tissues (200 mg/mice, 10 days after ACT) were harvested and leukocytes were counted and used for FACS analysis (G, n=3-4/group). Data are mean ± SD. **P<0.01, compared with Th9 and Th9-GFP groups, one-way ANOVA with Tukey test. (H-I) B1610%TRP-1-KO tumor-bearing WT mice were treated as shown in Fig. 1F. (H) Mice survival curves are shown (n=10-12/group, combined from 3 independent experiments), *P<0.05, compared with any other groups, survival analysis was conducted by log-rank test. (I) Tumor (~day 60) tissue lysates were analyzed for TRP-1 production by western-blot. (J) B1610%TRP-1-KO tumor-bearing WT mice were treated as shown in Fig. 1F. Mice survival curves are shown (n=9-12/group, combined from 3 independent experiments), *P<0.05, compared with any other groups, survival analysis was conducted by log-rank test. Representative results from one of two or three repeated experiments are shown.
The above observation suggests that Th9 cells may promote an eATP-enriched milieu allowing monocyte recruitment. However, tumor-specific cytolytic effector T cells and memory T cells express CD39 (Gupta et al., 2015; Moncrieffe et al., 2010; Takenaka et al., 2016), a surface-expressing ectoenzyme that converts pro-inflammatory eATP released from stressed or damaged cells into AMP (Vijayan et al., 2017). Since immune cells other than transferred T cells may express CD39, our results suggested that cyclophosphamide (CTX) should have depleted the major immunosuppressive cell subsets in tumors that may express CD39 (see Figure 3B; ~90% of tumor-infiltrating immune cells are transferred Th1 or Th17 cells on day 10). We thus hypothesized that Th9 cells, but not Th1 and Th17 cells, do not produce CD39 so that eATP remains available within the tumor environment and recruits the monocytes. In line with previous reports (Chalmin et al., 2012; Li et al., 2012), we also found that human and murine Th1, Tc1, and Th17 cells express high levels of CD39, whereas such expression is very low on human and murine Th9 cells (Figure 4C-4D and S4F-S4G), which may be a result of high GFI-1 expression in Th9 cells that suppress CD39 expression (Chalmin et al., 2012). We also tested the presence of eATP in the supernatant of the B16 tumor cells cocultured with TRP-1 T cells. Significantly higher eATP was detected in the coculture of Th9 cells and tumor cells as compared to Th1 and Th17 cells (Figure 4E), and supernatants collected from Th9 cell cocultures caused the migration of monocytes in transwell assays (Figure 4F). The addition of suramin, Th1 cells, Th17 cells, or overexpression of CD39 in tumor cells reduced the Th9 cell-induced eATP enrichment and prevented monocyte recruitment in the cocultures (Figure 4E and 4F). Furthermore, transfer of Th9 cells, but not the CD39-overexpressed Th9 cells, or cotransfer of Th9 with Th1 or Th17 cells effectively promoted tumor monocyte infiltration in tumor-bearing mice (Figure 4G). Meanwhile, knockdown of Entpd1 in Th1 or Th17 cells promoted tumor monocyte infiltration in tumor, and knockdown of Entpd1 in Th1 cell, but not Th17 cells, induced an anti-ALV immunity (Figure S4H-S4L). In addition, overexpression of CD39 in Th9 cells largely abrogated the capacity of Th9 cells to eliminate ALVs and led to the growth of TRP-1 negative tumor cells (Figure 4H and 4I). Similar results were observed by co-transferring Th9 cells and TRP-1 Th1 cells (Figure 4J). Finally, other potential CD39+ immune cells may infiltrate into tumors after lymphodepletion, which may affect the anti-ALV capacity of Th9 cells and contribute to the reduced antitumor response. Our results demonstrated that i.p. administration of CD39 inhibitor POM-1 significantly improved Th9 ACT efficacy when targeting tumors contained ~40% of ALVs (Figure S4M), although POM-1 alone had no antitumor effects (Li et al., 2019). Therefore, our results highlight that Th9 cells promote non-classic eATP-promoted monocyte chemotaxis that allows potent antitumor effects.
Induction of a Type I IFN responsive signature in monocytes by Th9 cells causes the elimination of ALVs
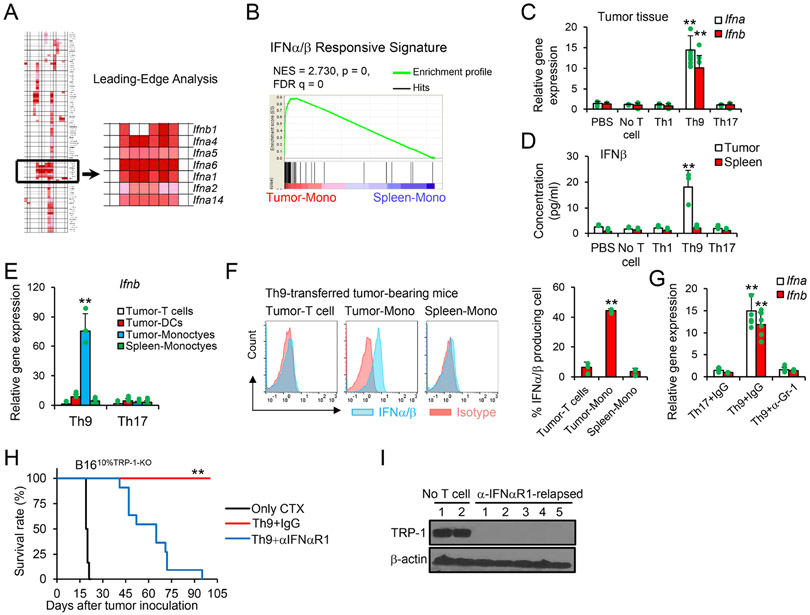
To further elucidate the role of Th9 cells in recruiting inflammatory monocytes, we sorted CD11b+CD11c−Ly6Chi inflammatory monocytes infiltrating the tumor and their counterparts in the spleens of mice treated with Th9 cells. We performed gene enrichment analysis of tumor-infiltrating monocytes (Tumor-Mono) versus splenic monocytes (Spleen-Mono) by Gene Set Enrichment Analysis (GSEA)(Subramanian et al., 2005) for 186 KEGG gene sets. Among the top 10 most altered gene sets, we further conducted a leading-edge analysis to determine which subsets of genes contributed the most to the enrichment signal of a given gene set's leading edge or core enrichment (Subramanian et al., 2005). A cluster of 6 type I IFN genes represented the leading edge among the most altered gene sets (Figure 5A and S5A). Similarly, the IFNα/β Responsive Signature was also significantly enriched in tumor monocyte compared to Spleen-Mono (Figure 5B).
Figure 5. Th9 cell ACT induces type I IFN signatures in tumor for ALV-clearance.
(A-B) Mice were treated as in Fig. 1F, and mice transferred with Th9 cells were euthanized 10 days after transfer. Tumor monocytes and SpMono were sorted for gene array (n=3). Shown is a gene cluster that contributes the most to the enrichment signal of a given gene set's leading edge or core enrichment (A) and IFNα/β responsive signature (B). (C-G) Mice were treated as in Fig. 1F. (C) Tumor tissues were extracted for total RNA and analyzed by qPCR (n=3/group). Data are mean ± SD. **P<0.01, compared with any other groups, one-way ANOVA with Tukey test. (D) Tumor lysate supernatants were analyzed by ELISA for IFNβ (n=3/group). Data are mean ± SD. **P<0.01, compared with any other groups, one-way ANOVA with Tukey test. (E) The indicated cells were sorted for qPCR (n=3/group). Data are mean ± SD. **P<0.01, Th9 compared with Th17 group, one-way ANOVA with Tukey test. (F) Intracellular staining of IFNα/β was performed on the indicated cells obtained from Th9-treated mice (n=3/group). Data are mean ± SD. **P<0.01, tumor-Mono compared with Tumor-T cells or Spleen-Mono groups, one-way ANOVA with Tukey test. (G) Tumor tissues from mice with the indicated treatments were extracted for total RNA and analyzed by qPCR (n=3/group). Data are mean ± SD. **P<0.01, Th9+IgG compared with any other groups, one-way ANOVA with Tukey test. (H-I) B1610%TRP-1-KO tumor-bearing WT mice were treated as shown in Fig. 1F, with antibody injection i.p. every 3 days after ACT. (H) Mice survival curves are shown (n=9-11/group, combined from 2 independent experiments), **P<0.01, compared with any other groups, survival analysis was conducted by log-rank test. (I) Tumor (~day 60) tissue lysates were analyzed for TRP-1 production by western-blot. Representative results from one of two repeated experiments are shown.
The type I IFN signature of tumor monocytes observed after Th9 adoptive transfer was corroborated by the upregulation of mRNA levels of Ifna/Ifnb genes within the tumor and IFNβ protein in tumor tissue lysate supernatants (Figure 5C and 5D). Furthermore, tumor monocytes are the dominant source of type I IFN production in Th9 cell-treated tumors (Figure 5E-5F), and the depletion of tumor monocytes obtained by the administration of the anti-Gr-1 antibody substantially reduced type I IFNs (Figure 5G). Similarly, significantly upregulated mRNA levels of human IFNA/B genes can be detected in human OvCa PDX tissues from NSG mice treated with human Mesothelin-CAR Th9 cells and co-transferred with PBMC but not human monocyte-depleted PBMC (Figure S5B). Finally, to ascertain the contributions of type I IFN in controlling the emergence of ALVs, B1610%TRP-1-KO tumor-bearing mice were treated with TRP-1 Th9 cells together with blocking antibodies targeting the mIFNαR1. We observed tumor relapse in mice receiving mIFNαR1 blocking antibodies, and the relapsed tumor appears to have very limited TRP-1 expression (Figure 5H and 5I). Interestingly, our results also demonstrated that Th9 ACT induced an IFNaR1 signaling-dependent host CD4+ T cell, CD8+ T cell, and NK cell activation which contributes to ALV tumor clearance (Figure S5C and S5D). We next analyzed tumor-infiltrating host T cells activated by Th9 ACT, and found that host T cells expressed moderate levels of PD-1/Lag3 and displayed central memory and effector memory phenotypes (Figure S5E and S5F). In particular, there was an increase in the number of gp100- and TRP-2-tetramer+ CD8+ T cells after Th9 ACT (Figure S5G). In addition, increased IFNγ production in Th9 group was detected by ELISA after restimulation of these CD8+ or CD4+ T cells with gp100, TRP-2, or B16 neo-antigen peptides (Kreiter et al., 2015) compared to control peptides, while Th1 or Th17-ACT had moderate or no effects on the generation of these antigen-specific T cells, respectively (Figure S5H). Furthermore, we also found that the number of activated T cell and DC populations were significantly increased in the tumor-draining lymph nodes (TDLNs) after Th9 ACT (Figure S5I-S5K). Consistent with these results, the deficiency/depletion of all types of host CD4+ T cell, CD8+ T cell, and NK cell subsets almost completely abrogated the anti-ALV effect of Th9 cells (Figure S5L and S5M). Taken together, our results highlight that tumor monocyte-derived type I IFN is required to control the clonal expansion of ALVs, while the elimination of TRP-1+ tumor cells remains exquisitely Th9 cell-dependent.
Th9 cells promote viral mimicry in tumor monocytes
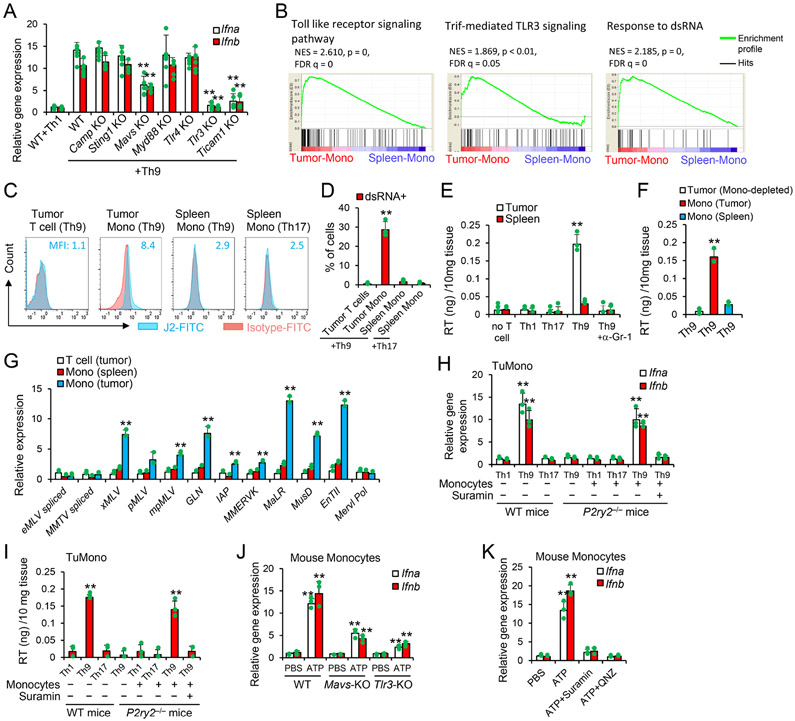
We performed a screening on the pattern recognition receptors that may control the induction of type I IFN pathways in tumor monocytes. Our analysis reveals that the expression of Ifna/b in monocytes was predominantly due to the TLR3-TRIF-dependent pathway, and to a lesser extent, to the mitochondrial antiviral signaling protein (MAVS)-dependent pathway (Figure 6A). Tumor monocytes are highly enriched in Toll-like receptor signaling and TRIF-mediated TLR3 signaling signatures (Figure 6B), and it is thus plausible that monocytes may sense the presence of double-strand RNA (dsRNA) via TLR3 and MAVS (Chattopadhyay and Sen, 2014). Indeed, gene expression profile of tumor monocytes isolated from Th9 cell-treated mice was significantly enriched in the “Response to dsRNA” gene signature as compared to SpMono (Figure 6B). Furthermore, dsRNA was detected in tumor monocytes by the intracellular staining with the J2 antibody (Weber et al., 2006) (Figure 6C and 6D).
Figure 6. Viral mimicry in tumor monocytes is required for Th9 anti-ALV responses.
(A) B1610%TRP-1-KO tumor-bearing WT or transgenic mice were treated as shown in Fig. 1F. Tumor tissues (10 days after ACT) were extracted for total RNA and analyzed by qPCR (n=3/group), Data are mean ± SD. **P<0.01, compared with WT+Th9 group, one-way ANOVA with Tukey test. (B) GSEA of the gene profiles as in Fig. 5A was performed for the indicated gene signatures. (C-D) Intracellular staining of dsRNA (n=3/group) was performed in the indicated cells 10 days after treatments similar as in Fig. 1F. Data are mean ± SD. **P<0.01, compared with any other groups, one-way ANOVA with Tukey test. (E-F) B1610%TRP-1-KO tumor-bearing WT mice were treated as shown in Fig. 1F. Tumor tissues (E) or sorted cells (F) 10 days after ACT were lysed by 1% Triton-X-100. Data are mean ± SD. **P<0.01, compared with any other groups, one-way ANOVA with Tukey test. (G) Tumor-infiltrating cells sorted from Th9-treated mice 10 days after transfer (n=3/group) were tested for transcript levels of the indicated murine ERVs measured by qPCR. Data are mean ± SD. **P<0.01, compared with T cell (tumor) and Mono (spleen) groups, one-way ANOVA with Tukey test. (H-I) WT SpMono sorted from CTX-treated mice were injected into tumor of P2ry2−/− mice (10 days after tumor-bearing P2ry2−/− mice treated with CTX+T cells w/o suramin). The tumor tissues were analyzed by qPCR (H) and RT activity (I) 3 days later (n=3-4/group). Data are mean ± SD. **P<0.01, Th9 cells compared with Th1, Th17 cells, and spleen-mono injected into tumor receiving Th9 cells and suramin, one-way ANOVA with Tukey test. (J) SpMono sorted from CTX-treated WT, Tlr3 KO, or Mavs KO mice were treated as indicated and performed for qPCR analysis (n=3/group). Data are mean ± SD. **P<0.01, compared with PBS or WT/ATP groups, one-way ANOVA with Tukey test. (K) WT SpMono sorted from CTX-treated mice were treated as indicated and performed for qPCR (n=3/group). Data are mean ± SD. **P<0.01, compared with any other groups, one-way ANOVA with Tukey test. Representative results from one of two repeated experiments are shown.
Both TLR3 and MAVS are involved in the recognition of dsRNAs associated with dsRNA-virus infections (Chattopadhyay and Sen, 2014). As endogenous retrovirus (ERVs) represent up to 10% of mouse and human genomes and monocytes are the reservoir for multiple endogenous retroviruses (Johnston et al., 2001), we thus hypothesized that the intracellular dsRNA is a result of viral mimicry by endogenous transcripts from murine endogenous retroviral (mERV) elements. We found that tumor monocytes isolated from Th9 cell-treated mice displayed upregulated reverse transcriptase (RT) activity, and that depletion of tumor monocytes completely reduced the RT activity within the tumor (Figure 6E and 6F). We further confirmed the expression of multiple murine ERVs by qPCR that include xMLV, pMLV, mpMLV, GLN, IAP, MMERVK, MaLR, MusD and EnTII (Figure 6G).
To assess the presence of viral mimicry effects in tumor monocytes upon Th9 cell treatment we performed studies using P2ry2−/− mice that lack monocytes (Figure 4B) to determine if the enforced infiltration of monocytes into tumor engrafted in P2ry2−/− mice (intratumor injection of monocytes) may induce type I IFNs. Injection of sorted Spleen-Mono into tumor in P2ry2−/− mice receiving Th9 cells, but not Th1 or Th17 cells, caused production of type I IFNs and RT activity, which is abrogated by the administration of suramin (Figure 6H and 6I). Notably, when murine Spleen-Mono were exposed to 100 nM ATP in vitro, we detected > 10-fold increase of Ifna and Ifnb mRNA expression, whereas the deficiency in Mavs or Tlr3 partially abrogated ATP-mediated Ifna and Ifnb induction (Figure 6J). Similarly, when human monocytic THP-1 cells were stimulated with 100 nM ATP in culture, we detected significantly increased expression of human type I IFNs and multiple human ERVs, compared to PMA that promotes hERVs expression in human monocytes (Johnston et al., 2001) (Figure S6A-S6C).
ATP activates NF-κB signaling (Franke et al., 2012) that is the master transcriptional factor for ERV transcription (Zeng et al., 2014). Specific inhibition of NF-κB signaling abrogated the ERV production in both murine and human monocytes treated with ATP (Figure 6K and S6B-S6C). Further, we found that silencing p65 and, to a lesser extend, p50, significantly reduced ERV induction in monocytes injected into tumors of Th9 cell-treated P2ry2−/− mice (Figure S6D). Importantly, reconstitution of P2ry2−/− mice by intratumoral injection with WT inflammatory monocytes transduced with control-siRNA but not RelA-siRNA restored host T cell response against tumor-specific antigens (Figure S6E). Finally, we tested whether the host expression of TLR3 and/or MAVS is required to control the emergence of ALVs. Remarkably, the deficiency in Mavs or Tlr3 reduced the capacity of Th9 cells in controlling ALV-containing tumors (Figure S6F). Collectively, our data indicate that tumor-specific Th9 cells activate the molecular program of dsRNA-TLR3/MAVS in tumor monocytes and that this molecular program is essential in controlling the growth of ALVs (Figure S6G).
Discussion
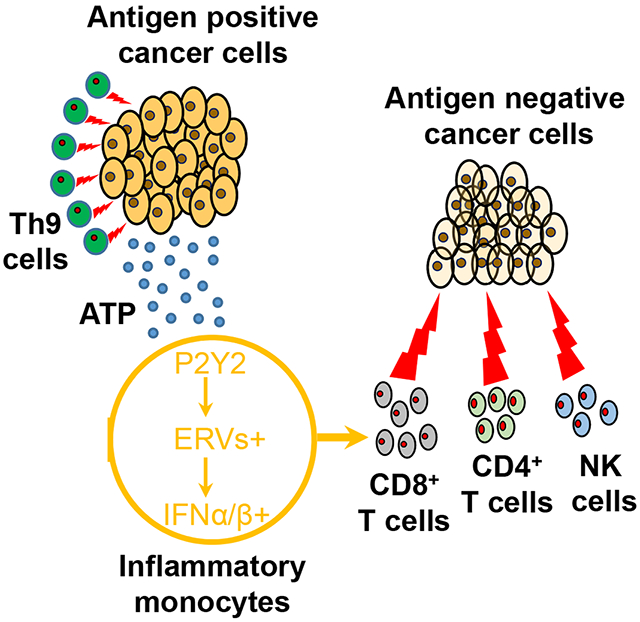
Acquired resistance is an important mechanism by which tumors escape immune attacks including those caused by ACT (Hegde et al., 2016; Fry et al., 2017; Wang et al., 2017). Recent studies targeted acquired resistance by using CAR-T cells to overexpress CD40L (Kuhn et al., 2019), IL-12 (Chmielewski et al., 2011), Flt3L (plus poly I:C and anti-4-1BB coinjection) (Lai et al., 2020), or CCL19 and IL-7 (Adachi et al., 2018). Although these approaches may also reduce/delay ALV outgrowth, curative response rates are relatively low and whether they can successfully eliminate poorly immunogenic ALVs (e.g. B16 ALVs) in late-stage tumors are still elusive. In the current study, we observed that ACT based on tumor-specific Th9 cells prevents tumor escape due to antigen loss in tumor models in which Th9 cells are either expressing a tumor-specific TCR or a CAR. Tumor eradication caused by tumor-specific Th9 cells is achieved via direct killing of Th9 cells of tumor cells expressing the targeted antigen and an indirect effect of Th9 cells that causes the elimination of ALVs via recruitment of inflammatory monocytes and production of type I IFNs. Our study thus demonstrated that Th9 cells possess another critical antitumor property that further differentiates these cells from classical Th1, Tc1, and Th17 cells.
We discovered that ACT based on tumor-specific Th9 brings together the adaptive and innate immunity, which combination allows tumor eradication. Th9 cells retain the strong antigen-specific antitumor effects that are characteristic of the Th1 cells of the adaptive immune responses, but also create a unique modification of the immune landscape of the tumor microenvironment that is reminiscent of an active innate immune response. Th9 cells recruit inflammatory monocytes within the tumor during the immune cell recovery occurring upon myelosuppressive chemotherapy that is normally used in the clinical protocols of ACT. Our results also indicate that the specific recruitment of inflammatory monocytes as well as the type I IFN milieu they provide play a vital role in clearing tumor clones that lack the targeted antigen and thus escape classical TCR or CAR-mediated cytolytic effect.
We mechanistically dissected how Th9 cells cause the monocyte recruitment within the tumor and how they imprint a type I IFN signature. Monocyte recruitment within the tumor mediated by Th9 is not due to IL-9, which is the all marker of Th9 cells, nor to classical chemokine-mediated pathways. In contrast, we discovered that the purinergic receptor P2ry2 deficiency reduces the tumor monocyte infiltration, indicating that this effect may rely on the eATP-promoted chemotaxis. Under physiological conditions, ATP is detected intracellularly at high concentrations ranging from 1 to 10 mM (Takenaka et al., 2016), while in the extracellular milieu ATP concentrations are in the nanomolar range (Takenaka et al., 2016). However, ATP can be released upon cell necrosis and apoptosis and serve as a “danger signal” or “find me” signal that promotes the monocyte chemotaxis (Elliott et al., 2009). Due to the incapacity of Th9 cells to metabolize ATP, it accumulates within the tumor microenvironment and specifically recruits monocytes within the tumor.
Another critical finding is that monocytes recruited within the tumor microenvironment by Th9 cells become proinflammatory since they display a type I IFN signature. Monocyte polarization to proinflammatory function was predominantly TLR3-TRIF-dependent, and to a lesser extent, MAVS pathway-dependent. Both pathways sense dsRNA, and we discovered that endogenous retrovirus is the main source of dsRNA in tumor monocytes, recapitulating previous finding that monocytes are reservoir for multiple endogenous retroviruses (Johnston et al., 2001). In line with reported by others (Chiappinelli et al., 2015), ERV-derived dsRNA can activate both TLR3 and MAVS signalings. The detection of dsRNA by TLR3 is likely because TLR3 recognizes extracellular dsRNA released from activated or damaged cells (Dela Justina et al., 2020; Vercammen et al., 2008; Chattopadhyay and Sen, 2014), and we indeed detected a significant amount of damaged/dying monocytes in Th9-treated tumors. Interestingly, monocyte infiltration of tumor was insufficient for dsRNA-type I IFN production, whereas eATP enrichment was indispensable for this response.
In a recent clinical trial with 74 metastatic melanoma patients following TIL ACT, baseline serum levels of IL-9 was able to predict response to TIL ACT, while TIL persistence and mutation burden did not correlate with outcome (Forget et al., 2018). Similarly, an increase in physiological Th9 cell counts but not Th1 or Th17 counts during the treatment with Nivolumab (anti-PD-1 Abs) has been reported to be associated with all 18 clinical responders with metastatic melanoma (46 patients in total) (Nonomura et al., 2016). In this study, human Th9 cells also seem to be endowed with robust anti-ALV activities. These results will provide the basis for our future clinical trial of ACT with human tumor-specific Th9 cells.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yong Lu (yolu@waekhealth.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The data reported in this paper is deposited in the Gene Expression Omnibus (GEO) database under accession number GSE151712.
Experimental models and subject details
Mice
C57BL/6, CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ), Ccr1−/− (B6.129S-Ccr1tm1Gao/AdlJ ), Ccr2−/– (B6.129S4-Ccr2tm1Ifc/J ), Ccr5−/− (B6;129P2-Ccr5tm1Kuz/J ), Ccr6−/− (B6.129P2-Ccr6tm1Dgen/J ), Ccr7−/− (B6.129P2(C)-Ccr7tm1Rfor/J ), Ccr9−/− (B6N.129-Ccr9tm1Lov/JmfJ), Cxcr2−/− (B6.129S2(C)-Cxcr2tm1Mwm/J), Cxcr3−/− (B6.129P2-Cxcr3tm1Dgen/J), Cxcr5−/− (B6.129S2(Cg)-Cxcr5tm1Lipp/J), Cxcr6−/− (B6.129P2-Cxcr6tm1Litt/J), Cx3cr1−/− (B6.129P2(Cg)-Cx3cr1tm1Litt/J), P2ry2−/− (B6.129P2-P2ry2tm1Bhk/J), Ifnar1−/− (B6(Cg)-Ifnar1tm1.2Ees/J), Camp−/− (B6.129X1-Camptm1Rlg/J), Sting−/− (B6(Cg)-Sting1tm1.2Camb/J), Mavs−/− (B6;129-Mavstm1Zjc/J), Myd88−/− (B6.129P2(SJL)-Myd88tm1.1Defr/J), Tlr4−/− (B6(Cg)-Tlr4tm1.2Karp/J), Tlr3−/− (B6;129S1-Tlr3tm1Flv/J), Trif−/− (C57BL/6J-Ticam1Lps2/J), TRP-1 (B6.Cg-Rag1tm1Mom Tyrp1B-wTg(Tcra,Tcrb)9Rest/J), MHCII−/− NSG (NOD.Cg-PrkdcscidH2-Ab1tm1DoiIl2rgtm1Wjl/SzJ), Cd4−/− (B6.129S2-Cd4tm1Mak/J) and MHCI−/− NSG (NOD.Cg-B2mtm1UncPrkdcscidIl2rgtm1Wjl/SzJ) mice were purchased from Jackson Laboratory. Il9r−/− mice were a gift from Dr. Jean-Christophe Renauld at Ludwig Institute for Cancer Research. Male and female 6- to 8-week-old mice were used for each animal experiment. The studies were approved by the Institutional Animal Care and Use Committee and Institutional Review Board of the Wake Forest School of Medicine.
Cell lines
Murine melanoma cell line B16 and human ovarian cancer cell line SK-OV-3 were purchased from ATCC. Murine ovarian cancer cell line ID8 was a gift from Dr. Neveen Said at Wake Forest School of Medicine. B16TRP-1-KO cell line was generated using CRISPR/Cas9 for TRP-1 deletion. ID8 cancer cells were KO of p53 gene by CRISPR/Cas9 to recapitulate the human high-grade serous OvCa(Walton et al., 2016). hMesothelin-expressing ID8 cancer cells were generated by transduction with lentivirus vectors encoding human hMesothelin to ID8p53KO cells. CD39-expressing B16 cancer cells were generated by transduction with lentivirus vectors encoding murine CD39. Cells (including T cells) were cultured in RPMI 1640 Medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Thermo Scientific), and 100 U/ml penicillin-streptomycin and 2 mM L-glutamine (Invitrogen).
Reagents
αGr-1 (clone RB-8C5), αLg6G (clone 1A8), αIFNAR-1 (clone MAR1-5A3), αIL-4 (clone 11B11), αCD8 (clone 2.43), αNK1.1 (clone PK136), αIL-21R (clone 4A9), and αIFNγ (clone XMG1.2) were purchased from BioXCell. Mouse cytokines IL-4, IL-6, IL-12, and human IL-4, TGFβ, IL-2 and IL-6 were purchased from R&D Systems. eATP signaling inhibitor Suramin (Catalog# sc-200833), CD39 inhibitor POM-1 (Catalog# sc-203205) and NF-κB inhibitor QNZ (Catalog# sc-200675) were purchased from Santa Cruz Biotechnology.
Method details
Viral production and T cell transduction
The cDNA sequence of human MSLN-(SS1)(Ho et al., 2011)-mBBZ or hMSLN-(SS1) (Ho et al., 2011)-hBBZ was synthesized by GenScript and then cloned into MigR1 retroviral vector for murine T cell transduction or pRRLSIN.EF-1a-EGFP-2A.WPRE lentiviral vector for human T cell transduction, respectively. Human HLA-A*02-restricted MART-127-35 specific TCR lentivector (DMF5)(Hughes et al., 2005) is a gift from Dr. Steven Rosenberg. Control, Entpd1, or Traf6 targeting shRNA were constructed on PMKO.1 vector. Viruses were packaged in 293T cells transfected with Lipofectamine 2000 (Life Science). Viral supernatant was harvested from day 1 to day 3, filtered with a 0.45-μm filter, concentrated with PEG-itVirus Precipitation Solution, and stored at −80°C until use.
Murine TRP-1 T cells preparation
Naive CD4+CD62L+ T cells were purified from spleens of TRP-1 mice using (EasySep™ Mouse Naïve CD4+ T Cell Isolation Kit, Catalog#19765) and differentiated into Th1, Th9, or Th17 cells according to established methods(Downs-Canner et al., 2017; Ghoreschi et al., 2010; Lu et al., 2012; Muranski et al., 2011). Briefly, TRP-1-specific naïve CD4+ T cells were cultured for 3 days with irradiated splenic APCs from C57BL/6 mice (2 ml culture medium per well in 24-well plate) in the presence of TRP-1106-133 peptide (5 μg/ml) with:
Th1-polarizing medium supplemented with mIL-2 (30 ng/ml), mIL-12 (4 ng/ml), and anti-IL-4 mAb (10 μg/ml);
Th9-polarizing medium supplemented with mIL-4 (10 ng/ml), hTGF-β1 (1 ng/ml), and anti-IFN-γ monoclonal antibodies (mAbs; 10 μg/ml);
Th17-polarizing medium supplemented with mIL-6 (30 ng/ml), mIL-1β (20 ng/ml), hTGF-β1 (2.5 ng/ml), mIL-21 (100 ng/ml), anti-IL-4 mAbs (10 μg/ml), anti-IL-2 mAbs (10 μg/ml) and anti-IFN-γ mAbs (10 μg/ml)(Muranski et al., 2011);
After an initial 3-day culture, cells were provided with mIL-2 (5 ng/ml), except Th17 cells which received IL-2 (5 ng/ml) plus IL-23 (50 ng/ml)(Muranski et al., 2011). After culturing for a total of 5 days, differentiated Th cells were depleted of dead cells using Dead Cell Removal kit (Miltenyi, Catalog# 130-090-101) and then used in animal studies.
Murine CAR T cell preparation
Murine T cells were activated by priming CD4+ or CD3+ T cells isolated from the spleens of C57BL/6 (B6) mice with mouse αCD3/CD28 Dynabeads (ThermoFisfer, Catalog# 11453D) in the presence of mIL-4 (10 ng/ml), hTGF-β1 (1 ng/ml), and anti-mIFN-γ monoclonal antibodies (mAbs; 10 pg/ml) for Th9 cells; or 10 ng/ml mIL-7 and 10 ng/ml mIL-15 for Th1+Tc1 cells (Bead-to-cell ratio was 1:3, 2 ml culture medium per well in 24-well plate). During activation, T cells were transduced with hMSLN-(SS1) (Ho et al., 2011)-mBBZ-CAR encoding γ-retrovirus to make T cells targeting MSLN in the presence of 10 μg/ml protamine sulfate (Sigma) by centrifugation for 2 hrs at 1,800 rpm at room temperature. The general transduction rate of CAR vector was 30-40% based on GFP expression. CAR+ (GFP+) T cells were sorted on day 3 and then expanded in the presence of 5 ng/ml mIL-2 (Th9) or 10 ng/ml mIL-7 and 10 ng/ml mIL-15 (T1: Th1+Tc1) cells for an additional 4 days.
Human T cell preparation
Human autologous T cells were activated by priming human CD4+ or CD3+ T cells from PBMC of melanoma patient (HLA-A2+) with human αCD3/CD28 Dynabeads (ThermoFisfer, Catalog# 11132D) in the presence of hIL-4 (10 ng/ml), TGF-β1 (1 ng/ml), and anti-IFN-γ monoclonal antibodies (mAbs; 10 μg/ml) for Th9 cells or 10 ng/ml hIL-7 and 10 ng/ml hIL-15 for Th1+Tc1 cells (Bead-to-cell ratio was 1:3, 2 ml culture medium per well in 24-well plate). After 24 hrs of stimulation, T cells were transduced with human HLA-A*02-restricted MART-127-35 specific TCR lentivector (DMF5, MOI=3:1) to make T cells targeting melanoma-associated antigen MART-1 (Hughes et al., 2005) in the presence of 10 μg/ml protamine sulfate (Sigma) by centrifugation for 2 hrs at 1,800 rpm at room temperature. T cells were washed 48 hrs after transfection and then expanded in the presence of 100 U/ml hIL-2 (Th9) or 10 ng/ml hIL-7 and 10 ng/ml hIL-15 (T1: Th1+Tc1) cells for an additional 4 days before use. The general transduction rate of MART-1-TCR vector was >80% based on tetramer staining.
In some studies, human autologous CAR T cells were generated by priming human CD4+ or CD3+ T cells from PBMC of OvCa patient with human αCD3/CD28 Dynabeads (ThermoFisfer, Catalog# 11132D) in the presence with hIL-4 (10 ng/ml), hTGF-β1 (1 ng/ml), and anti-hIFN-γ monoclonal antibodies (mAbs; 10 μg/ml) for Th9 cells; or 10 ng/ml hIL-7 and 10 ng/ml hIL-15 for Th1+Tc1 cells (Bead-to-cell ratio was 1:3, 2 ml culture medium per well in 24-well plate). After 24 hrs of stimulation, T cells were transduced with hMSLN-(SS1) (Ho et al., 2011)-hBBZ-CAR-GFP encoding lentivector (MOI=3:1) to make T cells targeting human MSLN in the presence of 10 μg/ml protamine sulfate (Sigma) by centrifugation for 2 hrs at 1,800 rpm at room temperature. T cells were washed 48 hrs after transfection and then expanded in the presence of 100 U/ml hIL-2 (Th9) or 10 ng/ml hIL-7 and 10 ng/ml hIL-15 (T1: Th1+Tc1) cells for an additional 4 days before use. The general transduction rate of CAR vector was >85% based on GFP expression.
Real-time PCR
Total RNA was extracted from cells using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Genes were expressed with specific primers and analyzed by using SYBR green real-time PCR (Applied Biosystems). Expression was normalized to the expression of the housekeeping gene GAPDH. Primers: mIfna 5′-TCAGTCTTCCCAGCACATTG-3′, mIfna R: 5′-GAGAAGAAACACAGCCCCTG-3′; mIfnb F: 5 ′-CAGCTCCAAGAAAGGACGAAC-3′, mIfnb R: 5′-GGCAGTGTAACTCTTCTGCAT-3′; eMLV spliced F: 5′-CCAGGGACCACCGACCCACCG-3′, eMLV spliced R: 5′-TAGTCGGTCCCGGTAGGCCTCG-3–; MMTV spliced F: 5′-AGAGCGGAACGGACTCACCA-3′, MMTV spliced R: 5′-TCAGTGAAAGGTCGGATGAA-3′; xMLV F: 5′-TCTATGGTACCTGGGGCTC-3′, xMLV R: 5′-GGCAGAGGTATGGTTGGAGTAG-3′; pMLV/mpMLV common F: 5′-CCGCCAGGTCCTCAATATAG-3′, pMLV R: 5′-AGAAGGTGGGGCAGTCT-3′, mpMLV R: 5′-CGTCCCAGGTTGATAGAGG-3′; GLN F: 5′-TGTGTAAGTCCAGACGCAG-3′, GLN R: 5′-CCAACCTACTCCAAAAACAG-3′; IAP F: 5′-AAGCAGCAATCACCCACTTTGG-3′, IAP R: 5′-CAATCATTAGATGCGGCTGCCAAG-3′; MMERVK F: 5′-CAAATAGCCCTACCATATGTCAG-3′, MMERVK R: 5′-AGCCCCAGCTAACCAGAAC-3′; MusD/EnTII common F: 5′-GTGCTAACCCAACGCTGGTTC-3′, MusD R: 5′-CTCTGGCCTGAAACAACTCCTG-3′, ETnII R: 5′-ACTGGGGCAATCCGCCTATTC-3′; Mervl Pol F: 5′-ATCTCCTGGCACCTGGTATG-3′, Mervl Pol R: 5′-AGAAGAAGGCATTTGCCAGA-3′; mGAPDH F: 5′-TTGATGGCAACAATCTCCAC-3’, mGAPDH R: 5’-CGTCCCGTAGACAAAATGGT-3’. hIFNa F: 5′-GCCTCGCCCTTTGCTTTACT-3′, hIFNa R: 5′-CTGTGGGTCTCAGGGAGATCA-3′; hIFNb F: 5′-ATGACCAACAAGTGTCTCCTCC-3′, hIFNb R: 5′-GGAATCCAAGCAAGTTGTAGCTC-3′, hMSLN F: 5′-CCAACCCACCTAACATTTCCAG-3′, hMSLN R: 5′-CAGCAGGTCCAATGGGAGG-3′, HERV-F F: 5′-CCTCCAGTCACAACAACTC-3′, HERV-F R: 5′-TATTGAAGAAGGCGGCTGG-3′; HERV-E F: 5′-GGTGTCACTACTCAATACAC-3′, HERV-E R: 5′-GCAGCCTAGGTCTCTGG-3′; HERV-W F: 5′-TGAGTCAATTCTCATACCTG-3′, HERV-W R: 5′-AGTTAAGAGTTCTTGGGTGG-3′; ERV-F2B F: 5′-AAAAAGGAAGAAGTTAACAGC-3′, ERV-F2B R: 5′-ATATAAAGACTTAGGTCCTGC-3′; HERV-K(HML-2) F: 5′-AAAGAACCAGCCACCAGG-3′, HERV-K(HML-2) R: 5′-CAGTCTGAAAACTTTTCTCTC-3′; HERV-K(HML-5) F: 5′-TGAAAGGCCAGCTTGCTG-3′, HERV-K(HML-5) R: 5′-CAATTAGGAAATTCTTTTCTAC-3′; hLINE ORF1 F: 5′-TTGGAAAACACTCTGCAGGATATTAT-3′, hLINE ORF1 R: 5′-TTGGCCTGCCTTGCTAGATT-3′; hGAPDH F: 5′-AGTCAACGGATTTGGTCGTATTGGG-3′, hGAPDH R: 5′-ACGTACTCAGCGCCAGCATCG-3′.
Enzyme-linked immunosorbent assay
Cell culture supernatants were tested by ELISA using Mouse IFN Beta ELISA Kit (High Sensitivity, pbl Assay Science) according to the manufacturer’s protocol.
ATP quantification assay
ATP concentration in the cell culture supernatants were tested by ATP Determination Kit (ThermoFisher) according to the manufacturer’s protocol.
Tumor models and adoptive transfer
Mice received subcutaneous (s.c.) injection with 1×106 B16 tumor cells or 1×106 B1610%TRP-1-KO (contains 90% ofWT-B16 cells and 10% of B16TRP-1-KO cells) tumor cells. At 10 days after tumor injection, mice were treated with adoptive transfer of 2.5×106 Th1, Th9, or Th17 cells, high dose Th1 (2×107), or high dose Th17 cells (7.5×106) followed by intravenous (i.v.) injection of 2.5×105 TRP-1 peptide-pulsed bone marrow-derived dendritic cells generated as previously described (Hong et al., 2012). Cyclophosphamide (CTX, Sigma) was administrated intraperitoneally (i.p.) as a single dose at 120 mg/kg 1 day before T-cell transfer. Mice were euthanized at indicated days. In some experiments, surviving mice were s.c. rechallenged with 2×106 B16TRP-1-KO tumor cells on the contralateral flank. In some experiments, mice have been injected i.p. with 150 μg mAbs (e.g. αGr-1, αIL-21R, αNK1.1, αCD8, or αIFNaR1 mAbs) every 3 days starting from one day before T cell transfer for a total of 8 times (αGr-1 and αIFNaR1), a total 15 times (αIL-21R), or until the endpoint (αNK1.1 and αCD8). In some experiments, mice have been injected i.p. with 3 mg suramin every 3 days one day before ACT until mice were used for indicated tests. In some experiments, mice were injected i.p. with 125 μg POM-1 every 3 days starting from day 5 after ACT for a total of 15 doses.
Mice received intracranial injection of 1×105 B1610%TRP-1KO tumor cells. At 8 days after tumor injection, mice were treated with adoptive transfer of 1×106 Th1 or Th9, or multiple injections of 1×106 Th1 as long as mice still alive. CTX was administrated intraperitoneally (i.p.) as a single dose at 120 mg/kg 1 day before T-cell transfer. For human Th9 cell study, MART-1+/HLA-A2+ melanoma patient-derived xenografts (PDX) were injected into MHC I−/−/II−/− double KO-NSG mice (DKO-NSG, crossed from class T−/− and class II−/− NSG). Mice were treated with MART-specific T cells and/or autologous PBMC (1×107) on day 10. Human T cells were transduced with human HLA-A*02-restricted MART-127-35 specific TCR lentivector (DMF5) (Johnson et al., 2006) to make T cells targeting MART-1 (upregulated in most melanomas)(Borbulevych et al., 2011). These human MART-1-specific CD4+ T cells have been reported to exhibit multifunctional effector and helper responses, and can mediate HLA-A2-restricted MART-1-specific cytolytic function of comparable efficacy to that of CD8+ CTL (Ray et al., 2010). Mice were euthanized at endpoints.
Female B6 mice received subcutaneous (s.c.) injection with 1×107 ID890%hMesothelin (containing 90% of hMesothelin-expressing ID8 cells and 10% of hMesothelin− ID8 ALVs) tumor cells. At 45 days after tumor injection, mice were treated with adoptive transfer of 5×106 CAR Th1+Tc1, 5×106 CAR Th9 or high dose of 2.5×107 Th1+Tc1cells. CTX was administrated intraperitoneally (i.p.) as a single dose at 120 mg/kg 1 day before T-cell transfer. For human CAR Th9 cell study, OvCa patient-derived xenografts (PDX) were injected into MHC I−/−/II−/− double KO-NSG mice (DKO-NSG, crossed from class I−/− and class II−/− NSG). Mice were treated with Mesothelin-specific CAR T cells (5×106 CAR Th1+Tc1, 5×106 CAR Th9, 5×106 CAR Th9+Tc9 or high dose of 2.5×107 Th1+Tc1 cells) and autologous PBMC (1×107) on day 10. Human T cells were transduced with human Mesothelin CAR vector to make T cells targeting human Mesothelin (highly expressed on >65% of OvCa patients (Morello et al., 2016)). Mice were euthanized at endpoints.
In some experiments, monocytes were sorted from spleens of WT or P2ry2−/− tumor-free mice 10 days after CTX treatment. P2ry2−/− mice bearing 10-day established B1610%TRP-1-KO tumors (1×106 B1610%TRP-1-KO cells injected s.c.) were treated with CTX on day 9, followed by Th9 cell and DC transfer as described above. Monocytes (1×107) were transferred i.v. into mice on day 12 and day 22. In some experiments, monocytes (1×106) were transduced with siRNA and intratumorally injected into tumors 7 and/or 14 days after Th9 ACT.
The enzyme-linked immunosorbent spot (ELISpot) assay
Mice bearing 10-day established B1610%TRP1KO tumors were treated by Th1, Th9, or Th17 cell ACT as described above. Mice were sacrificed on day 20 after ACT and tumor tissues were minced and digested by tumor dissociation kit (Miltenyi Biotec). Each host immune cell subset in about 200 mg tumor tissues was isolated by a bead positive selection kit (CD8, NK1.1, or CD4). Isolated cells per 200 mg tumor tissues were cocultured with irradiated B16TRP-1KO tumor cells on IFNγ ELISpot Kit plates (Mouse IFN-gamma ELISpot Kit, R&D Systems) for 48 hours following the manufacturer's instructions. The plates were imaged and evaluated by Cellular Technology Limited ELISPOT Analyzer.
Host T cell response assay
TRP-1-specific Th9 cells (2.5×106) were transferred i.v. into WT mice bearing 10-day established B1610%TRP-1-KO tumors (1×106 B1610%TRP-1-KO cells challenged s.c. 10 days before T cell transfer). Adjuvant cyclophosphamide (CTX, i.p.) and DC vaccination (2.5×105, i.v.) were administered to mice. B1610%TRP-1-KO tumors 20 days after Th9 cell ACT were harvested, minced, and digested by tumor dissociation kit (Miltenyi Biotec). CD8+ or CD4+ T cells in about 200 mg tumor tissues were isolated by a bead positive selection kit (CD8, or CD4). T cells were then restimulated with peptide (10 μg/ml)-loaded DCs for 72 hrs in the presence of 5 ng/ml mIL-2. Peptide sequence: MHC-I peptide for TRP-2180-188 (SVYDFFVWL) (Overwijk and Restifo, 2001), MHC-I peptide for gp10025-33 (EGSRNQDWL) (Overwijk and Restifo, 2001), and neoantigens (Kreiter et al., 2015) B16-M27 (MHC-I peptide: REGVELCPGNKYEMRRHGTTHSLVIHD), B16-M30 (MHC-II peptide: PSKPSFQEFVDWENVSPELNSTDQPFL), and B16-M48 (MHC-II peptide: SHCHWNDLAVIPAGVVHNWDFEPRKVS). OVA peptides were used as controls: OVA MHC-II peptide (ISQAVHAAHAEINEAGR) and OVA MHC-I peptide (SIINFEKL).
Flow cytometry and western blot analysis
FITC-, PE-, APC- or Brilliant Violet-conjugated mAbs (1:100 dilution) were used for staining after Fc blocking, and analyzed using a FACS Fortessa flow cytometer.
For Western blot, TRP-1 mAbs (#sc-166857) from Santa Cruz Biotechnology were used at a 1:500 dilution. β-Actin mAbs (#3700T) from Cell Signaling and used at a 1:1000 dilution. Full unedited scans of blots shown in Figures are shown in Supplementary Figure 12.
CFSE dilution assay and in vitro cytotoxicity assay
In some experiments, Th cells were incubated for 5 minutes at 37°C with 1 mM CFSE in PBS, and then washed extensively. The proliferation of T cells by the relative CFSE dilution after stimulation was determined by FACS. In the cytotoxicity assay, B16 target cells or MC38 non-target cells for TRP-1 T cells were labeled with 5 mM CFSE. B16 target cells or MC38 non-target cells were incubated alone in triplicate with TRP-1 T cells at a 1:10 tumor-to-T cell ratio. After 48 hour culture, CFSE+ tumor cells from each target and control wells were stained using FVD and analyzed by FACS. FVD+ tumor cells were considered dead cells. The percent specific lysis was calculated as (FVD+ target - FVD+ control)×100%.
In vivo T Lymphocyte cytotoxicity assay
In vivo CTL assay was performed as described previously with some modifications (Xue et al., 2019; Keller et al., 2008; Lu et al., 2014b). Spleen cells from C57BL/6 mice were pulsed with 5 μg/ml TRP-1 peptide or OT-II peptide. Pulsed splenocytes were labeled with CFSE at a final concentration of 1 μM (CFSEhi) or at a final concentration of 0.1 μM (CFSElo). Two cell populations were mixed at 1:1 ratio, and 2 × 107 total cells were injected intravenously into treated mice. The percentage of antigen-specific lysis was determined 36 h later in T cell treated mice (TRP1 peptide-pulsed CFSEhi target + OT-II peptide-pulsed CFSElo non-target), by analyzing the relative proportion of CFSEhi target cells and CFSElo non-target cells in splenocytes based on CFSE-labeling intensity. The percentage of specific lysis was calculated as follows: (1–R × M2/M1) × 100, where M1 = non-target events, M2 = target events, R = M1 in untreated mice/M2 in untreated mice.
Microarray analysis
Mice were treated with Th9 cells, and 10 days after Th9 transfer, tumor tissue and spleens were harvested and dissociated for cell sorting. CD11b+CD11c−Ly6Chi inflammatory monocytes were flow-sorted and total RNA was extracted with the RNeasy Mini kit (Qiagen). RNA samples were sent to the Genomic Core of Case Western Reserve University for quality evaluation using an Agilent Bioanalyzer. Samples with intact 18S and 28S ribosomal RNA bands with RIN >8.5 were processed for microarray analysis, which was performed with a Mouse Gene 2.0 ST Kit at the Case Western Reserve University Genomic Core. Gene set enrichment analysis (GSEA) of the gene expression profiles of the 5 cell types was implemented using GSEA software (gsea-v3.0, http://software.broadinstitute.org/gsea/downloads.jsp). P-values were calculated with the Kolmogorov-Smirnov test (threshold = 0.01). The false discovery rate (FDR), q value, is the estimated probability that a gene set with a given NES represents a false-positive finding. The threshold for q value in GSEA is 0.25. Gene sets for the mature effector were derived from a publicly available study of the genes differentially expressed by >2 fold in quaternary versus primary cells(Gu et al., 2016).
Quantification and statistical analysis
For statistical analysis, Student’s t-test or ANOVA was used. A P value less than 0.05 was considered statistically significant. Results are presented as mean ± s.d. unless otherwise indicated.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| αGr-1 | BioXcell | Cat#BE0075 |
| αLy-6G | BioXcell | Cat#BE0075-1 |
| αIFNAR-1 | BioXcell | Cat#BE0241 |
| αIL-4 | BioXcell | Cat#BE0045 |
| αCD8 | BioXcell | Cat#BE0061 |
| αNK1.1 | BioXcell | Cat#BE0036 |
| αIL-21R | BioXcell | Cat#BE0258 |
| αIFNγ | BioXcell | Cat#BE0055 |
| αIL-2 | BioXcell | Cat#BE0043 |
| TRP-1 | Santa Crus | Cat#sc-166857 |
| β-actin | Cell Signaling | Cat#3700T |
| BV421 anti-mouse CD11b | BioLegend | Cat#101251 |
| PerCP anti-mouse CD11c | BioLegend | Cat#117326 |
| BV605 anti-mouse F4/80 | BioLegend | Cat#123133 |
| BV570 anti-mouse Ly6G | BioLegend | Cat#127629 |
| PE anti-mouse Ly6C | BioLegend | Cat#128008 |
| FITC anti-mouse CD4 | BioLegend | Cat#100405 |
| APC anti-mouse CD45.2 | BioLegend | Cat#109814 |
| PE anti-mouse CD3 | BioLegend | Cat#100206 |
| PE anti-mouse CD39 | BioLegend | Cat#143804 |
| PE anti-mouse PD-1 | BioLegend | Cat#135206 |
| APC anti-mouse Lag3 | BioLegend | Cat#125210 |
| APC anti-mouse IFNγ | BioLegend | Cat#505810 |
| APC anti-mouse IL-9 | BioLegend | Cat#514106 |
| APC anti-mouse IL-17 | BioLegend | Cat#v506916 |
| PE anti-human IL-9 | BioLegend | Cat#507603 |
| FITC anti-human CD4 | BioLegend | Cat#357406 |
| PE anti-human CD39 | BioLegend | Cat#328208 |
| PE anti-mouse CD44 | BioLegend | Cat#103024 |
| FITC anti-mouse CD62L | BioLegend | Cat#104406 |
| PE anti-mouse CD8 | BioLegend | Cat#100708 |
| APC anti-mouse CD11c | BioLegend | Cat#117310 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Patient-derived xenografts (PDX) | Wake Forest Baptist Comprehensive Cancer Center | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| The NF-κB-specific inhibitor QNZ | Santa Cruz Biotechnology | Cat#sc-200675 |
| eATP signaling inhibitor Suramin | Santa Cruz Biotechnology | Cat#sc-200833 |
| CD39 inhibitor POM-1 | Santa Cruz Biotechnology | Cat#sc-203205 |
| MHC-I peptide for TRP-2180-188 (SVYDFFVWL) | GenScript | N/A |
| MHC-I peptide for gp10025-33 (EGSRNQDWL) | GenScript | N/A |
| B16-M27 (MHC-I peptide: REGVELCPGNKYEMRRHGTTHSLVIHD) | GenScript | N/A |
| B16-M30 (MHC-II peptide: PSKPSFQEFVDWENVSPELNSTDQPFL) | GenScript | N/A |
| B16-M48 (MHC-II peptide: SHCHWNDLAVIPAGVVHNWDFEPRKVS) | GenScript | N/A |
| OVA MHC-II peptide (ISQAVHAAHAEINEAGR) | GenScript | N/A |
| OVA MHC-I peptide (SIINFEKL) | GenScript | N/A |
| MHC class II–restricted TRP1 (SGHNCGTCRPGWRGAACNQKILTVR) | GenScript | N/A |
| Mouse IL-1β | R&D Systems | Cat#NP_032387 |
| Mouse IL-4 | R&D Systems | Cat#P07750 |
| Mouse IL-6 | R&D Systems | Cat#P08505 |
| Mouse IL-2 | R&D Systems | Cat#P04351 |
| Mouse IL-12 | R&D Systems | Cat#P43432 |
| Mouse IL-23 | R&D Systems | Cat#P43432 |
| Mouse IL-21 | R&D Systems | Cat#Q9ES17.1 |
| Human TGF-β1 | R&D Systems | Cat#P01137 |
| Human IL-4 | R&D Systems | Cat#204-IL-050/CF |
| Critical commercial assays | ||
| ATP Determination Kit | ThermoFisher | Cat#A22066 |
| ELISA kits mouse TNF-α | eBioscience | Cat#50-112-8899 |
| ELISA kits mouse IFN-γ | eBioscience | Cat#50-112-9023 |
| ELISA kits mouse IL-6 | eBioscience | Cat#50-112-8808 |
| ELISA kits mouse IL-1β | eBioscience | Cat#50-112-8706 |
| ELISA kits mouse IFN-β | R&D | Cat#424001 |
| Annexin V-FITC Apoptosis Detection Kit | eBioscience | Cat#BMS500FI-100 |
| Deposited data | ||
| CD11b cell gene array | NCBI Gene Expression Omnibus | GSE151712 |
| Experimental models: Cell lines | ||
| B16 | ATCC | Cat#CRL-6323 |
| ID8 | A gift from Dr. Neveen Said at Wake Forest School of Medicine | N/A |
| SK-OV-3 | ATCC | Cat#HTB-77 |
| 293T | ATCC | Cat#CRL-3216 |
| Experimental models: Organisms/strains | ||
| C57BL/6 | The Jackson Laboratory | Cat#000664 |
| CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) | The Jackson Laboratory | Cat#002014 |
| Ccr1−/− (B6.129S-Ccr1tm1Gao/AdlJ ) | The Jackson Laboratory | Cat#032932 |
| Ccr2−/− (B6.129S4-Ccr2tm1Ifc/J ) | The Jackson Laboratory | Cat#004999 |
| Ccr5−/− (B6;129P2-Ccr5tm1Kuz/J ) | The Jackson Laboratory | Cat#005427 |
| Ccr6−/− (B6.129P2-Ccr6tm1Dgen/J ) | The Jackson Laboratory | Cat#005793 |
| Ccr7−/− (B6.129P2(C)-Ccr7tm1Rfor/J ) | The Jackson Laboratory | Cat#006621 |
| Ccr9−/− (B6N.129-Ccr9tm1Lov/JmfJ) | The Jackson Laboratory | Cat#027041 |
| Cxcr2−/− (B6.129S2(C)-Cxcr2tm1Mwm/J) | The Jackson Laboratory | Cat#006848 |
| Cxcr3−/− (B6.129P2-Cxcr3tm1Dgen/J) | The Jackson Laboratory | Cat#005796 |
| Cxcr5−/− (B6.129S2(Cg)-Cxcr5tm1Lipp/J) | The Jackson Laboratory | Cat#006659 |
| Cxcr6−/− (B6.129P2-Cxcr6tm1Litt/J) | The Jackson Laboratory | Cat#005693 |
| Cx3cr1−/− (B6.129P2(Cg)-Cx3cr1tm1Litt/J) | The Jackson Laboratory | Cat#005582 |
| P2ry2−/− (B6.129P2-P2ry2tm1Bhk/J) | The Jackson Laboratory | Cat#009132 |
| Ifnar1−/− (B6(Cg)-Ifnar1tm1.2Ees/J) | The Jackson Laboratory | Cat#028288 |
| Camp−/− (B6.129X1-Camptm1Rlg/J) | The Jackson Laboratory | Cat#017799 |
| Sting−/− (B6(Cg)-Sting1tm1.2Camb/J) | The Jackson Laboratory | Cat#025805 |
| Mavs−/− (B6;129-Mavstm1Zjc/J) | The Jackson Laboratory | Cat#008634 |
| Myd88−/− (B6.129P2(SJL)-Myd88tm1.1Defr/J) | The Jackson Laboratory | Cat#009088 |
| Tlr4−/− (B6(Cg)-Tlr4tm1.2Karp/J) | The Jackson Laboratory | Cat#029015 |
| Tlr3−/− (B6;129S1-Tlr3tm1Flv/J) | The Jackson Laboratory | Cat#005217 |
| Trif−/− (C57BL/6J-Ticam1Lps2/J) | The Jackson Laboratory | Cat#005037 |
| TRP-1 (B6.Cg-Rag1tm1Mom Tyrp1B-wTg(Tcra,Tcrb)9Rest/J) | The Jackson Laboratory | Cat#008684 |
| MHCII−/− NSG (NOD.Cg-PrkdcscidH2-Ab1tm1DoiIl2rgtm1Wjl/SzJ) | The Jackson Laboratory | Cat#021885 |
| Cd4−/− (B6.129S2-Cd4tm1Mak/J) | The Jackson Laboratory | Cat#002663 |
| MHCI−/− NSG (NOD.Cg-B2mtm1UncPrkdcscidIl2rgtm1Wjl/SzJ) | The Jackson Laboratory | Cat#010636 |
| Il9r−/− | A gift from Dr. Jean-Christophe Renauld at Ludwig Institute for Cancer Research | N/A |
| Oligonucleotides | ||
| mIFNa F: 5′-TCAGTCTTCCCAGCACATTG-3′ | Sigma | N/A |
| mIFNa R: 5′- GAGAAGAAACACAGCCCCTG-3′ | Sigma | N/A |
| mIFNb F: 5′-CAGCTCCAAGAAAGGACGAAC-3′ | Sigma | N/A |
| mIFNb R: 5′-GGCAGTGTAACTCTTCTGCAT-3′ | Sigma | N/A |
| eMLV spliced F: 5′-CCAGGGACCACCGACCCACCG-3′ | Sigma | N/A |
| eMLV spliced R: 5′-TAGTCGGTCCCGGTAGGCCTCG-3′ | Sigma | N/A |
| MMTV spliced F: 5′-AGAGCGGAACGGACTCACCA-3′ | Sigma | N/A |
| MMTV spliced R: 5′-TCAGTGAAAGGTCGGATGAA-3′ | Sigma | N/A |
| xMLV F: 5′-TCTATGGTACCTGGGGCTC-3′ | Sigma | N/A |
| xMLV R: 5′-GGCAGAGGTATGGTTGGAGTAG-3′ | Sigma | N/A |
| pMLV/mpMLV common F: 5′-CCGCCAGGTCCTCAATATAG-3′ | Sigma | N/A |
| pMLV R: 5′-AGAAGGTGGGGCAGTCT-3′ | Sigma | N/A |
| mpMLV R: 5′-CGTCCCAGGTTGATAGAGG-3′ | Sigma | N/A |
| GLN F: 5′-TGTGTAAGTCCAGACGCAG-3′ | Sigma | N/A |
| GLN R: 5′-CCAACCTACTCCAAAAACAG-3′ | Sigma | N/A |
| IAP F: 5′-AAGCAGCAATCACCCACTTTGG-3′ | Sigma | N/A |
| IAP R: 5′-CAATCATTAGATGCGGCTGCCAAG-3′ | Sigma | N/A |
| MMERVK F: 5′-CAAATAGCCCTACCATATGTCAG-3′ | Sigma | N/A |
| MMERVK R: 5′-GTATACTTTCTTCTTCAGGTCCAC-3′ | Sigma | N/A |
| MaLR F: 5′-ATGTTTTGGGGAGGACTGTG-3′ | Sigma | N/A |
| MaLR R: 5′-AGCCCCAGCTAACCAGAAC-3′ | Sigma | N/A |
| MusD/EnTII common F: 5′-GTGCTAACCCAACGCTGGTTC-3′ | Sigma | N/A |
| MusD R: 5′-CTCTGGCCTGAAACAACTCCTG-3′ | Sigma | N/A |
| ETnII R: 5′-ACTGGGGCAATCCGCCTATTC-3′ | Sigma | N/A |
| Mervl Pol F: 5′-ATCTCCTGGCACCTGGTATG-3′ | Sigma | N/A |
| Mervl Pol R: 5′-AGAAGAAGGCATTTGCCAGA-3′ | Sigma | N/A |
| mGAPDH F: 5’-TTGATGGCAACAATCTCCAC-3’ | Sigma | N/A |
| mGAPDH R: 5’-CGTCCCGTAGACAAAATGGT-3’ | Sigma | N/A |
| hIFNα F: 5′-GCCTCGCCCTTTGCTTTACT-3′ | Sigma | N/A |
| hIFNα R: 5′-CTGTGGGTCTCAGGGAGATCA-3′ | Sigma | N/A |
| hIFNβ F: 5′-ATGACCAACAAGTGTCTCCTCC-3′ | Sigma | N/A |
| hIFNβ R: 5′-GGAATCCAAGCAAGTTGTAGCTC-3′ | Sigma | N/A |
| hMSLN F: 5′-CCAACCCACCTAACATTTCCAG-3′ | Sigma | N/A |
| hMSLN R: 5′-CAGCAGGTCCAATGGGAGG-3′ | Sigma | N/A |
| HERV-F F: 5′-CCTCCAGTCACAACAACTC-3′ | Sigma | N/A |
| HERV-F R: 5′-TATTGAAGAAGGCGGCTGG-3′ | Sigma | N/A |
| HERV-E F: 5′-GGTGTCACTACTCAATACAC-3′ | Sigma | N/A |
| HERV-E R: 5′-GCAGCCTAGGTCTCTGG-3′ | Sigma | N/A |
| HERV-W F: 5′-TGAGTCAATTCTCATACCTG-3′ | Sigma | N/A |
| HERV-W R: 5′-AGTTAAGAGTTCTTGGGTGG-3′ | Sigma | N/A |
| ERV-F2B F: 5′-AAAAAGGAAGAAGTTAACAGC-3′ | Sigma | N/A |
| ERV-F2B R: 5′-ATATAAAGACTTAGGTCCTGC-3′ | Sigma | N/A |
| HERV-K(HML-2) F: 5′-AAAGAACCAGCCACCAGG-3′ | Sigma | N/A |
| HERV-K(HML-2) R: 5′-CAGTCTGAAAACTTTTCTCTC-3′ | Sigma | N/A |
| HERV-K(HML-5) F: 5′-TGAAAGGCCAGCTTGCTG-3′ | Sigma | N/A |
| HERV-K(HML-5) R: 5′-CAATTAGGAAATTCTTTTCTAC-3′ | Sigma | N/A |
| hLINE ORF1 F: 5′-TTGGAAAACACTCTGCAGGATATTAT-3′ | Sigma | N/A |
| hLINE ORF1 R: 5′-TTGGCCTGCCTTGCTAGATT-3′ | Sigma | N/A |
| hGAPDH F: 5′-AGTCAACGGATTTGGTCGTATTGGG-3′ | Sigma | N/A |
| hGAPDH R: 5′-ACGTACTCAGCGCCAGCATCG-3′ | Sigma | N/A |
| Recombinant DNA | ||
| MART-127-35 specific TCR lentivector (DMF5) | a gift from Dr. Steven Rosenberg | N/A |
| PMKO.1 | Addgene | Cat#10676 |
| MigR1 | Addgene | Cat#27490 |
| pUltra | Addgene | Cat#24129 |
| lentiCRISPR v2 | Addgene | Cat#52961 |
| Software and algorithms | ||
| GSEA v2.2.2 | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| Other | ||
Highlights.
Th9 cells eradicate advanced tumors containing antigen-loss-variant cancer cells
Lack of CD39 on Th9 cells increase the intratumor accumulation of eATP
eATP-ERVs-TLR3/Mavs-Type I IFN pathway is activated in the recruited monocytes
Type I IFN induced in the recruited monocytes cells causes the elimination of ALVs
Acknowledgments
This work was supported by National Cancer Institute (NCI, 4R00CA190910-03, 1R37CA251318-01, 1R01CA248111-01A1, R01 CA258477-01, 1R01CA264102-01, 1R01CA200539-01A1, 1R01CA239255-01A1, 3P30CA012197-44S5), Elsa u. Pardee Foundation Award, and CPRIT Scholar (RR180044, RR210067).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Adachi K, Kano Y, Nagai T, Okuyama N, Sakoda Y, and Tamada K (2018). IL-7 and CCL19 expression in CAR-T cells improves immune cell infiltration and CAR-T cell survival in the tumor. Nat. Biotechnol [DOI] [PubMed] [Google Scholar]
- Cohen Adam D., MD, Garfall Alfred L., MD, Stadtmauer Edward A., MD1, Melenhorst JJ, PhD, Lacey Simon F., PhD, Lancaster Eric, MD, Vogl Dan T., MD, Weiss BM, MD1, Dengel Karen, RN, Nelson Annemarie, RN, Plesa Gabriela, MD, PhD, Chen F, PhD, and, Davis Megan M., P. (2019). B cell maturation antigen-specific CAR T cells are clinically active in multiple myeloma. J Clin Invest March 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JC, and Schrama D (2013). The Dark Side of Cyclophosphamide: Cyclophosphamide-Mediated Ablation of Regulatory T Cells. J. Invest. Dermatol 133, 1462–1465. [DOI] [PubMed] [Google Scholar]
- Bedard PL, Hansen AR, Ratain MJ, and Siu LL (2013). Tumour heterogeneity in the clinic. Nature 501, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbulevych OY, Santhanagopolan SM, Hossain M, and Baker BM (2011). TCRs Used in Cancer Gene Therapy Cross-React with MART-1/Melan-A Tumor Antigens via Distinct Mechanisms. J. Immunol 187, 2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, Ostberg JR, Blanchard MS, Kilpatrick J, Simpson J, et al. (2016). Regression of Glioblastoma after Chimeric Antigen Receptor T-Cell Therapy. N. Engl. J. Med 375, 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmin F, Mignot G, Bruchard M, Chevriaux A, Végran F, Hichami A, Ladoire S, Derangère V, Vincent J, Masson D, et al. (2012). Stat3 and Gfi-1 Transcription Factors Control Th17 Cell Immunosuppressive Activity via the Regulation of Ectonucleotidase Expression. Immunity 36, 362–373. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, and Sen GC (2014). DsRNA-activation of TLR3 and RLR signaling: Gene induction-dependent and independent effects. J. Interf. Cytokine Res 34, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. (2015). Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 162, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielewski M, Kopecky C, Hombach AA, and Abken H (2011). IL-12 release by engineered T cells expressing chimeric antigen receptors can effectively muster an antigen-independent macrophage response on tumor cells that have shut down tumor antigen expression. Cancer Res. [DOI] [PubMed] [Google Scholar]
- Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, et al. (2008). IL-4 inhibits TGF-β-induced Foxp3+T cells and, together with TGF-β, generates IL-9+IL-10+Foxp3-effector T cells. Nat. Immunol 9, 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen J-F, Enjyoji K, Linden J, Oukka M, et al. (2007). Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med 204, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs-Canner S, Berkey S, Delgoffe GM, Edwards RP, Curiel T, Odunsi K, Bartlett DL, and Obermajer N (2017). Suppressive IL-17A+ Foxp3+ and ex-Th17 IL-17Aneg Foxp3+ Treg cells are a source of tumour-associated Treg cells. Nat. Commun [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadi A, Walk SF, Park D, Woodson RI, Ostankovich M, Sharma P, et al. (2009). Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461, 282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget MA, Haymaker C, Hess KR, Meng YJ, Creasy C, Karpinets T, Fulbright OJ, Roszik J, Woodman SE, Kim YU, et al. (2018). Prospective analysis of adoptive TIL therapy in patients with metastatic melanoma: Response, impact of anti-CTLA4, and biomarkers to predict clinical outcome. Clin. Cancer Res 24, 4416–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke H, Verkhratsky A, Burnstock G, and Illes P (2012). Pathophysiology of astroglial purinergic signalling. Purinergic Signal. 8, 629–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, Wolters P, Martin S, Delbrook C, Yates B, et al. (2017). CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbitz A, Sukumar M, Helm F, Wilke A, Friese C, Fahrenwaldt C, Lehmann FM, Loddenkemper C, Kammertoens T, Mautner J, et al. (2012). Stromal interferon-γ signaling and cross-presentation are required to eliminate antigen-loss variants of B cell lymphomas in mice. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. (2010). Generation of pathogenic TH17 cells in the absence of TGF-β 2 signalling. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Godec J, Wolski D, Adland E, Yates K, Pauken KE, Cosgrove C, Ledderose C, Junger WG, Robson SC, et al. (2015). CD39 Expression Identifies Terminally Exhausted CD8+ T Cells. PLoS Pathog. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde M, Mukherjee M, Grada Z, Pignata A, Landi D, Navai SA, Wakefield A, Fousek K, Bielamowicz K, Chow KKH, et al. (2016). Tandem CAR T cells targeting HER2 and IL13Rα2 mitigate tumor antigen escape. J. Clin. Invest 126, 3036–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M, Feng M, Fisher RJ, Rader C, and Pastan I (2011). A novel high-affinity human monoclonal antibody to mesothelin. Int. J. Cancer [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Lee SH, Song XT, Jones L, Machida K, Huang XF, and Chen SY (2012). A Super TLR Agonist to Improve Efficacy of Dendritic Cell Vaccine in Induction of Anti-HCV Immunity. PLoS One. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MS, Yu YYL, Dudley ME, Zheng Z, Robbins PF, Li Y, Wunderlich J, Hawley RG, Moayeri M, Rosenberg SA, et al. (2005). Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum. Gene Ther [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby E, Nguyen SM, Fountaine TJ, Welp K, Gryder B, Qin H, Yang Y, Chien CD, Seif AE, Lei H, et al. (2016). CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat. Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Heemskerk B, Powell DJ, Cohen CJ, Morgan RA, Dudley ME, Robbins PF, and Rosenberg SA (2006). Gene Transfer of Tumor-Reactive TCR Confers Both High Avidity and Tumor Reactivity to Nonreactive Peripheral Blood Mononuclear Cells and Tumor-Infiltrating Lymphocytes. J. Immunol 177, 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JB, Silva C, Holden J, Warren KG, Clark AW, and Power C (2001). Monocyte activation and differentiation augment human endogenous retrovirus expression: Implications for inflammatory brain diseases. Ann. Neurol 50, 434–442. [DOI] [PubMed] [Google Scholar]
- June CH, O’Connor RS, Kawalekar OU, Ghassemi S, and Milone MC (2018). CAR T cell immunotherapy for human cancer. Science (80-. ). 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- Dela Justina V, Giachini FR, Priviero F, and Webb RC (2020). Double-stranded RNA and Toll-like receptor activation: A novel mechanism for blood pressure regulation. Clin. Sci 134, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller AM, Schildknecht A, Xiao Y, van den Broek M, and Borst J (2008). Expression of Costimulatory Ligand CD70 on Steady-State Dendritic Cells Breaks CD8+ T Cell Tolerance and Permits Effective Immunity. Immunity. [DOI] [PubMed] [Google Scholar]
- Klebanoff CA, Rosenberg SA, and Restifo NP (2016). Prospects for gene-engineered T cell immunotherapy for solid cancers. Nat. Med 22, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Urabe K, Winder a, Jiménez-Cervantes C, Imokawa G, Brewington T, Solano F, García-Borrón JC, and Hearing VJ (1994). Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO J. 13, 5818–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodumudi KN, Weber A, Sarnaik AA, and Pilon-Thomas S (2012). Blockade of Myeloid-Derived Suppressor Cells after Induction of Lymphopenia Improves Adoptive T Cell Therapy in a Murine Model of Melanoma. J. Immunol 189, 5147–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter S, Vormehr M, Van De Roemer N, Diken M, Löwer M, Diekmann J, Boegel S, Schrörs B, Vascotto F, Castle JC, et al. (2015). Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn NF, Purdon TJ, van Leeuwen DG, Lopez AV, Curran KJ, Daniyan AF, and Brentjens RJ (2019). CD40 Ligand-Modified Chimeric Antigen Receptor T Cells Enhance Antitumor Function by Eliciting an Endogenous Antitumor Response. Cancer Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Mardiana S, House IG, Sek K, Henderson MA, Giuffrida L, Chen AXY, Todd KL, Petley EV, Chan JD, et al. (2020). Adoptive cellular therapy with T cells expressing the dendritic cell growth factor Flt3L drives epitope spreading and antitumor immunity. Nat. Immunol [DOI] [PubMed] [Google Scholar]
- Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, Fatho M, Lennerz V, Wölfel T, Hölzel M, et al. (2012). Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 490, 412–416. [DOI] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, and Leonard WJ (2012). BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Moesta AK, Xiao C, Nakamura K, Casey M, Zhang H, Madore J, Lepletier A, Aguilera AR, Sundarrajan A, et al. (2019). Targeting CD39 in cancer reveals an extracellular ATP- and inflammasome-driven tumor immunity. Cancer Discov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Hong S, Li H, Park J, Hong B, Wang L, Zheng Y, Liu Z, Xu J, He J, et al. (2012). Th9 cells promote antitumor immune responses in vivo. J. Clin. Invest 122, 4160–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Hong B, Li H, Zheng Y, Zhang M, Wang S, Qian J, and Yi Q (2014a). Tumor-specific IL-9–producing CD8 + Tc9 cells are superior effector than type-I cytotoxic Tc1 cells for adoptive immunotherapy of cancers. Proc. Natl. Acad. Sci 111, 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhang M, Wang S, Hong B, Wang Z, Li H, Zheng Y, Yang J, Davis RE, Qian J, et al. (2014b). p38 MAPK-inhibited dendritic cells induce superior antitumour immune responses and overcome regulatory T-cell-mediated immunosuppression. Nat. Commun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang Q, Xue G, Bi E, Ma X, Wang A, Qian J, Dong C, and Yi Q (2018). Th9 Cells Represent a Unique Subset of CD4+T Cells Endowed with the Ability to Eradicate Advanced Tumors. Cancer Cell 33, 1048–1060.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Kim YJ, Robert L, Tsoi J, Comin-Anduix B, Berent-Maoz B, Cochran AJ, Economou JS, Tumeh PC, Puig-Saus C, et al. (2018). Immunotherapy resistance by inflammation-induced dedifferentiation. Cancer Discov. 8, 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrieffe H, Nistala K, Kamhieh Y, Evans J, Eddaoudi A, Eaton S, and Wedderburn LR (2010). High expression of the ectonucleotidase CD39 on T cells from the inflamed site identifies two distinct populations, one regulatory and one memory T cell population. J. Immunol 185, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello A, Sadelain M, and Adusumilli PS (2016). Mesothelin-targeted CARs: Driving T cells to solid Tumors. Cancer Discov. 6, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, et al. (2011). Th17 Cells Are Long Lived and Retain a Stem Cell-like Molecular Signature. Immunity 35, 972–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura Y, Otsuka A, Nakashima C, Seidel JA, Kitoh A, Dainichi T, Nakajima S, Sawada Y, Matsushita S, Aoki M, et al. (2016). Peripheral blood Th9 cells are a possible pharmacodynamic biomarker of nivolumab treatment efficacy in metastatic melanoma patients. Oncoimmunology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJD, Martinez-Lage M, Brem S, Maloney E, Shen A, et al. (2017). A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overwijk WW, and Restifo NP (2001). B16 as a Mouse Model for Human Melanoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Geyer MB, and Brentjens RJ (2016). CD19-targeted CAR T-cell therapeutics for hematologic malignancies: Interpreting clinical outcomes to date. Blood 127, 3312–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, et al. (2012). Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat. Med 18, 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Chhabra A, Chakraborty NG, Hegde U, Dorsky DI, Chodon T, von Euw E, Comin-Anduix B, Koya RC, Ribas A, et al. (2010). MHC-I-restricted melanoma antigen specific TCR-engineered human CD4+ T cells exhibit multifunctional effector and helper responses, in vitro. Clin. Immunol 136, 338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka MC, Robson S, and Quintana FJ (2016). Regulation of the T Cell Response by CD39. Trends Immunol. 37, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez MP, and Gottschalk S (2017). Targeting CD19: The good, the bad, and CD81. Blood 129, 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, and Stockinger B (2008). Transforming growth factor-beta “reprograms” the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol 9, 1341–1346. [DOI] [PubMed] [Google Scholar]
- Vercammen E, Staal J, and Beyaert R (2008). Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin. Microbiol. Rev 21, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan D, Young A, Teng MWL, and Smyth MJ (2017). Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 2017 Nullnull nrc.2017.86. [Google Scholar]
- Walton J, Blagih J, Ennis D, Leung E, Dowson S, Farquharson M, Tookman LA, Orange C, Athineos D, Mason S, et al. (2016). CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res. 76, 6118–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hu Y, and Huang H (2017). Acute lymphoblastic leukemia relapse after CD19-targeted chimeric antigen receptor T cell therapy. J. Leukoc. Biol jlb.5RU0817-315R. [DOI] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, and Paludan SR (2006). Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses. J. Virol 80, 5059–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G, Jin G, Fang J, and Lu Y (2019). IL-4 together with IL-1β induces antitumor Th9 cell differentiation in the absence of TGF-β signaling. Nat. Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Hu Z, Shi X, Li X, Zhan X, Li X-D, Wang J, Choi JH, Wang K., -w., Purrington T, et al. (2014). MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science (80-. ). 346, 1486–1492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang L, Morgan RA, Beane JD, Zheng Z, Dudley ME, Kassim SH, Nahvi AV, Ngo LT, Sherry RM, Phan GQ, et al. (2015). Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chu X, Chen J, Wang Y, Gao S, Jiang Y, Zhu X, Tan G, Zhao W, Yi H, et al. (2016). Dectin-1-activated dendritic cells trigger potent antitumour immunity through the induction of Th9 cells. Nat. Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper is deposited in the Gene Expression Omnibus (GEO) database under accession number GSE151712.