Abstract
B-cell maturation antigen (BCMA) is a prominent tumor-associated target for chimeric antigen receptor (CAR)-T cell therapy in multiple myeloma (MM). We describe the case of a MM patient, enrolled in the CARTITUDE-1 trial (NCT03548207), who developed a progressive movement disorder with features of parkinsonism approximately three months after BCMA-targeted ciltacabtagene autoleucel CAR-T cell infusion, associated with CAR-T cell persistence in the blood and cerebrospinal fluid, and basal ganglia lymphocytic infiltration. We demonstrate BCMA expression on neurons and astrocytes in the basal ganglia of the patient. Public transcriptomic datasets further confirm BCMA RNA expression in the caudate of normal human brains, suggesting this may be an on-target effect of anti-BCMA therapy. Given reports of three patients with grade ≥3 parkinsonism on the phase 2 cilta-cel trial and of grade 3 parkinsonism in the idecabtagene vicleucel (ide-cel) package insert, our findings support close neurological monitoring of patients on BCMA-targeted T cell therapies.
Multiple myeloma (MM) is a plasma cell disorder that accounts for ~10% of hematologic malignancies1. MM is considered incurable and characterized by multiple relapses of increasingly refractory disease. Immune therapies (including bispecific antibodies and CAR-T cells) have emerged in clinical trials with promising efficacy in MM2-4. B-cell maturation antigen (BCMA, also referred to as TNFRSF17 or CD269) has attracted interest as a tumor-associated target in MM. BCMA is expressed on mature B lymphocytes (e.g. plasma cells)5-7 and its overexpression and activation are associated with MM progression8. The ligands for BCMA, APRIL and BAFF, are present in the bone marrow and induce differentiation, growth and long-term survival of plasma cells9,10. Two BCMA-targeted CAR-T cells have demonstrated efficacy in phase 2 clinical trials in MM with overall response rates ranging from 73% (idecabtagene vicleucel, ide-cel) to 97% (ciltacabtagene autoleucel, cilta-cel)3,11-13.
Cytokine release syndrome (CRS), a form of systemic inflammatory response syndrome characterized by the elevation of pro-inflammatory cytokines, has been reported in BCMA-targeted CAR-T cell trials. Severity ranges from a mild reaction with flu-like symptoms to a life-threatening cytokine storm and fulminant hemophagocytic lymphohistiocytosis (HLH). The exact timing and duration of CRS in BCMA-targeted CAR-T therapy are variable, but most cases present early (within two weeks of CAR-T infusion) and are of low-grade severity, manageable with supportive care, steroids, tocilizumab, anakinra and other cytokine inhibitors.
Neurotoxicity has been described in BCMA-targeted CAR-T therapy, typically as transient encephalopathy (ICANS, immune effector cell-associated neurotoxicity syndrome)14,15. Reported symptoms include headache, confusion, hallucinations, dysphasia, ataxia, apraxia, tremor and seizures. In a meta-analysis of clinical trials of BCMA-targeted CAR-T, the incidence of neurotoxicity (grade ≥3) was 18%16. ICANS usually presents concurrently or shortly after CRS and management coincides with CRS interventions, including cytokine inhibitors and corticosteroids. Of note, in this case report, the patient displayed delayed neurotoxicity outside the CRS window.
The patient, a 58-year-old male, diagnosed with smoldering MM in 2004 which progressed to active myeloma in 2015, presented with relapsed/refractory disease after six previous lines of therapy. The patient was refractory to multiple drugs, including daratumumab, lenalidomide, pomalidomide and carfilzomib and was a candidate for BCMA-targeted CAR-T therapy. The patient underwent a bridging chemotherapy regimen (melphalan 56.25 mg once and 2 doses of bortezomib 1.3 mg/m2). Prior to CAR-T infusion, he received 300 mg/m2 fludarabine and 30 mg/m2 cyclophosphamide daily for 3 days to induce lymphodepletion. The baseline tumor burden was low (3% plasma cells on bone marrow biopsy), but he had multiple extramedullary plasmacytomas.
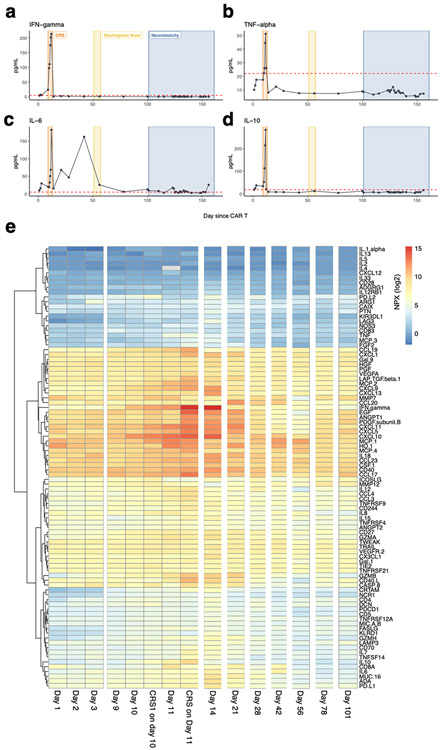
Following cilta-cel infusion (day 1), the patient developed fever on day 9 and hypotension on day 11 (CRS up to maximum grade 3) with the peak of C-reactive protein, ferritin and inflammatory cytokines (TNF-α, IFN-γ, IL-6, IL-18) two weeks after CAR-T infusion. CRS symptoms resolved by day 14 after treatment with tocilizumab and anakinra17. Relevant clinical and biochemical parameters, timing of selected therapeutic agents and additional cytokine profiling are shown in Extended Data Fig. 1-2. The patient remained afebrile until hospitalization on day 51 with neutropenic fever and pneumonia, treated with empiric antibiotics. Cultures remained negative, but PCR detected rhinovirus in respiratory secretions. The patient was discharged at day 57. Disease evaluation (day 79) showed a very good partial response (VGPR) according to IMWG (International Myeloma Working Group) criteria.
At day 101 after CAR-T cell infusion, the patient was evaluated with complaints of increasing fatigue interfering with daily activities. Initially, we observed slow gait and psychomotor retardation. Subsequent evaluation by two independent neurologists confirmed a clinical syndrome with features of parkinsonism including bradykinesia, postural instability, hypophonia, hypomimia, micrographia and a mild right-sided (action and resting) tremor, as well as saccadic intrusions on smooth pursuit and impaired short-term memory. There was no cogwheeling, no focal paresis or atrophy, no pathological reflexes or alterations of deep tendon reflexes, no ataxia and the Romberg was negative. Sensation was intact. The features were progressive over time with development of increasing hypomimia, rigidity and difficulty initiation movements. There were no recent drug changes or toxin exposure to account for the observed clinical phenotype. The patient was on a benzodiazepine for anxiety disorder, which was discontinued without improvement of the clinical features. Labs showed fluctuating neutrophil counts due to regular G-CSF (granulocyte colony stimulating factor) injections (Extended Data Fig. 1c). MRI imaging of the brain with/without contrast only showed small pre-existing foci of T2/FLAIR signal hyperintensity scattered throughout the periventricular and subcortical white matter (Extended Data Fig. 3a) and lumbar puncture findings were non-explanatory (Supplementary Table S1, additional clinical background is provided in the Methods section). A treatment attempt with levodopa because of progressive movement disorder and functional decline was unsuccessful. FDG-PET of the brain for response evaluation indicated a decreased uptake in the caudate nucleus bilaterally in comparison with imaging of two months prior, without any structural abnormalities (Extended Data Fig. 4). Ioflupane (123-I) scan was negative suggesting a disease mechanism different than Parkinson’s disease (Extended Data Fig. 3b).
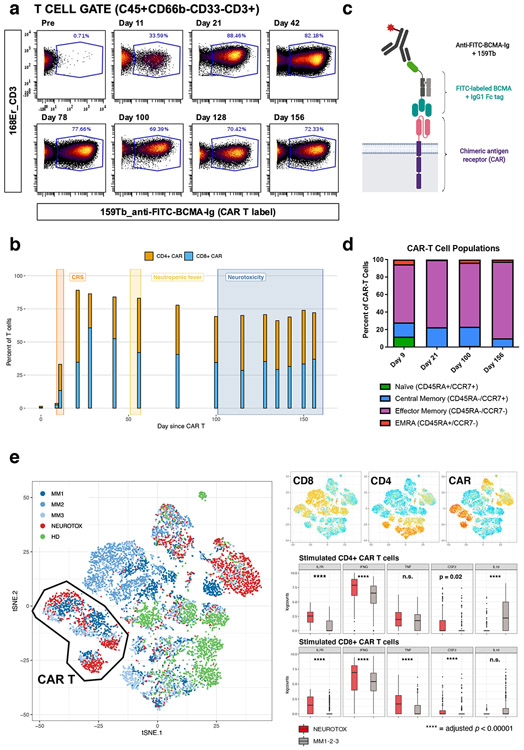
As shown in Fig. 1a, CAR-T cells were detectable in the blood in large numbers, starting at day 11 after infusion, up to day 156. Notably, 70-90% of all T cells in the peripheral blood were CAR-T cells (Fig. 1b). This observation suggested a role for persistent CAR-T cells in the development of the patient’s neurologic complaints. Extensive phenotyping of T cells using a mass cytometry (CyTOF) approach (Fig. 1c), suggested most CAR-T cells had an effector-memory phenotype (i.e. CD45RA-CCR7-) (Fig. 1d and Extended Data Fig. 5). Functional assays of peripheral blood CAR-T cells, isolated 128 days after treatment, confirmed their ability to produce inflammatory cytokines (IFN-γ, TNF-α and GM-CSF) upon PMA/ionomycin stimulation in vitro (Extended Data Fig. 6), highlighting their cytotoxic/pro-inflammatory potential. The full list of cytokines tested and comparison with a healthy donor are shown in Extended Data Fig. 6b. The patient’s CAR-T cells did not exhibit a Th17 phenotype, a T cell subtype previously associated with immunologic neurodegenerative disorders. Comparative single-cell analysis of CAR-T cells of this patient with 3 other patients from the same trial (without parkinsonism) by CITE-seq (Fig. 1e, Extended Data Fig. 7) showed qualitative transcriptomic differences with significantly higher expression of genes associated with long-term survival (e.g. IL7R) and genes encoding inflammatory cytokines (IFNG, TNF, CSF2) and lower expression of anti-inflammatory cytokine genes (e.g. IL10).
Figure 1: Persistence of chimeric antigen receptor (CAR) T cells with an activated effector-memory phenotype in the peripheral blood.
(a) Mass cytometry (CyTOF) plots gated on CD3+ T cells, showing fraction of FITC-BCMA-labeled (i.e. CAR) T cells at different time points after CAR-T infusion. (b) Quantitative representation of data in (a), showing the relative contribution of CD4+ and CD8+ CAR-T cells at different timepoints. The time periods associated with cytokine release syndrome (CRS), neutropenic fever and neurotoxicity are annotated. Each bar corresponds to N=1 sample collected from the patient. (c) Schematic illustration of CyTOF strategy used to detect the CAR on the T cell surface, see Methods for details (figure panel created with BioRender.com). (d) CAR-T cell phenotype, as determined by expression of CCR7 and CD45RA, illustrating a high fraction of effector-memory T cells. Each bar corresponds to N=1 sample collected from the patient. (e) t-SNE plot representation of CITE-seq analysis of peripheral blood mononuclear cells before and after PMA/ionomycin stimulation. Clustering was determined by similarity network fusion (SNF) and Louvain clustering algorithm. Individual cells are colored by subject (healthy donor (HD), neurotoxicity patient (NEUROTOX) and 3 other patients on the same clinical trial without neurotoxicity (MM1, MM2, MM3). Highlighted on side plots is expression level of CD4, CD8 and CAR ADT (antibody-derived tag, representation of protein level, high = red, low = blue) and boxplots (median, Q1 and Q3 quartiles, whiskers up to 1.5 x IQR) showing expression of a subset of differentially expressed genes (*: p < 0.01, n.s.: p ≥ 0.05, two-sided Mann-Whitney U test) in patient with neurotoxicity (NEUROTOX, N = 1, data on 145 stimulated CD4+ CAR-T cells (top, red) and 119 stimulated CD8+ CAR-T cells (bottom, red) total) and the other MM patients (MM1-2-3, N = 3, data on 152 stimulated CD4+ CAR-T cells (top, gray) and 406 stimulated CD8+ CAR-T cells (bottom, gray) total).
Microarray data of healthy human brains of the Allen Brain Atlas project18 confirmed localized RNA expression of BCMA in the basal ganglia and more specifically in the caudate nucleus in 5 out of 6 available specimens (Fig. 2b, Extended Data Fig. 8). We hypothesized that the symptoms could result from CAR-T cell infiltration in the brain targeting BCMA-expressing cells, thereby causing a movement disorder with features of parkinsonism. Analysis of the cerebrospinal fluid (CSF) by fluorescence activated cell sorting (FACS) confirmed the presence of CAR-T cells in the CSF (0.477 CAR-T cells/μL, Extended Data Fig. 9b). Cytokine profiling of CSF and blood plasma of the patient and a healthy control demonstrated overexpression of multiple cytokines in the patient’s CSF associated with T cell chemotaxis (e.g. CXCL5, CXCL10, CXCL11), T cell activation (e.g. granzymes, IFN-γ, CD40-L) and blood-brain barrier dysfunction (e.g. PDGFb, angiopoetin-1) (Extended Data Fig. 9).
Figure 2: BCMA is expressed in the caudate nucleus of healthy donors and post-mortem in the patient following CAR-T cell therapy.
(a) FDG-PET/CT illustrates decreased uptake in the caudate nucleus after development of neurotoxicity (POST, right, day 134 after CAR-T infusion), compared with previous imaging before development of neurotoxicity symptoms (PRE, left, day 77 after CAR-T infusion). Prior FDG-PET/CT imaging (before CAR-T infusion) was comparable to the pre-neurotoxicity scan. The scatter plot on the right illustrates the normalized Z-score of different regions of the brain before and after CAR-T infusion. The caudate is highlighted. The normalized score is calculated using MIMneuro, comparing the image with a library of 43 FDG neurologic controls (41-80 years old). (b) Visual representation of the expression of DRD1 and TNFRSF17 (BCMA) in a single patient from the Allen Brain Atlas. Expression of both genes (left, red = high) overlaps with the caudate nucleus region shown in 3D (right, purple). Image credit: Allen Institute: © 2010 Allen Institute for Brain Science. Allen Human Brain Atlas; available from: human.brain-map.org. (c) H&E staining of the caudate nucleus subependymal region (10x magnification, scale bar 200 μm). (d) GFAP (glial fibrillary acidic protein) immunohistochemistry of the caudate nucleus subependymal region (10x magnification, scale bar 200 μm) (e) CD3 immunohistochemistry of the caudate nucleus subependymal region (10x magnification, scale bar 200 μm) (f) BCMA immunohistochemistry of the caudate nucleus subependymal region (10x magnification, scale bar 200 μm, inset 40x magnification showing a neuron staining positively). (c-f) Images shown are representative slides from the caudate nucleus from the patient described in this case report (N=1). For each stain, at least 3 slides were available showing similar results.
Due to the sustained proliferation of CAR-T cells with spread beyond the blood-brain barrier and progressive decline in the patient’s general condition, IV cyclophosphamide (300 mg/m2), IT cytarabine (100 mg) and hydrocortisone (50 mg) were given on day 149, after careful consideration, aiming to rapidly reduce circulating CAR-T cells. We observed a decline of the absolute T cell count in the CSF with a stable fraction of CAR-T cells (0.128 CAR-T cells/μL, Extended Data Fig. 9c). A second dose of IV cyclophosphamide (300 mg/m2) and IT cytarabine/hydrocortisone were administered on day 156. The patient subsequently developed neutropenic fever with acute respiratory distress syndrome and multi-organ failure and passed away on day 162.
Post-mortem analysis of the caudate nucleus revealed the presence of focal gliosis as shown on H&E (hematoxylin and eosin) staining and immunohistochemistry (IHC) of glial fibrillary acidic protein (Fig. 2c-d). IHC further showed a T cell infiltrate (CD3+, predominantly CD8+) in the periventricular region of the basal ganglia (Fig. 2e). BCMA staining was performed (see Methods) and we found BCMA expression on a subset of neurons and astrocytes in the caudate nucleus as well as on a layer of neurons in the adjacent frontal cortex (Fig. 2f, Extended Data Fig. 10).
The value of BCMA as a tumor-associated target in MM depends upon the selective expression on (malignant) plasma cells. Even though BCMA expression has been extensively characterized on hematopoietic lineages, studies on other tissues are limited6,7,19. We found that microarray data from the Allen Brain Atlas shows BCMA mRNA expression in the basal ganglia and confirmed this in the patient and in a healthy brain (Extended Data Fig. 10) by IHC. Assessment of BCMA protein expression on a human tissue array was positive on lymph node, spleen, lung and stomach, due to plasma cells present in the bronchus- and mucosa-associated lymphoid tissue respectively5. This tissue array, however, demonstrated some positivity on climbing fibers in the cerebellum and explicitly does not rule out low-density expression in the central nervous system. We acknowledge that this data is to some extent conflicting due to lack of standardized protocols for staining tissues, other than bone marrow. A comprehensive evaluation of brain tissue for BCMA protein expression might be warranted to characterize the prevalence and extent of BCMA expression in the central nervous system and confirm the findings of this case report.
BCMA expression on neurologic tissues in a subset of patients could impact the applicability of BCMA-targeted adoptive cell transfer in MM. Implications for other BCMA-targeted immunotherapies, e.g. antibody-drug conjugates and bispecific antibodies are unknown. Even though therapeutic antibodies are thought not to cross the blood-brain barrier, their permeability into the CSF should be carefully evaluated. Other tumor-associated targets are being currently studied in MM, including bispecific antibodies and CAR constructs targeting GPRC5D, FcRH5, CD19, CD38, CD56, CD138 and SLAMF7, some of which have a broader expression outside of plasma cells and warrant careful monitoring20.
Using chemotherapy to destroy CAR-T cells after infusion is itself associated with toxicity, as this case illustrates, since the patient died of infectious complications. Other strategies include a modification of CAR structure with engineered suicide genes, the incorporation of inhibitory CAR constructs or usage of a small molecule system, as a safety switch to selectively deactivate the CAR-T cells. Recently, CAR natural killer (NK) cells have been proposed as an alternative with off-the-shelf use as a potential advantage20.
This case illustrates the potential of BCMA-targeted CAR-T cells to cross the blood-brain barrier in a subset of patients and cause a progressive neurocognitive and movement disorder possibly through targeting of BCMA-expressing cells of the basal ganglia. Neurotoxicity in general has been observed in 23/128 patients on ide-cel11 and 20/97 patients on cilta-cel13. Non-ICANS neurotoxicity was not addressed specifically in the ide-cel study, but was reported in 12/97 patients from the phase 2 study of cilta-cel, of which 5 patients with a cluster of movement and neurocognitive adverse events (3 with ≥ grade 3 parkinsonism)13. The development of this toxicity in the cilta-cel trial was associated with presence of two or more risk factors (including high tumor burden, previous grade ≥2 CRS, previous ICANS, and high CAR-T cell expansion and persistence). The ide-cel package insert also mentions that grade 3 parkinsonism has occurred after treatment, suggesting that this complication is not necessarily specific to one BCMA-targeted CAR-T cell product. We acknowledge that important questions remain unanswered. Our patient developed neutropenic fever at day 51; it is not well studied whether infections post-CAR-T infusion might activate CAR-T cells in vulnerable patients and whether more stringent prophylaxis of infection is warranted. Additional studies to confirm the proposed mechanism of neurotoxicity could help delineate the fraction of patients at risk. In conclusion, our findings suggest anti-BCMA CAR-T cell therapies although effective in MM, warrant close monitoring for neurotoxicity especially as such treatments acquire more widespread implementation in MM patients.
METHODS
Trial design.
The CARTITUDE-1 trial (https://clinicaltrials.gov/ct2/show/NCT03548207) is an open-label, single-arm phase 1b/2 trial that evaluates safety and efficacy of JNJ-68284528 (ciltacabtagene autoleucel, cilta-cel), a CAR-T cell therapy directed against BCMA in participants with relapsed or refractory MM. Here, we provide the case report of a patient with neurotoxicity enrolled on the CARTITUDE-1 trial. Analysis and reporting follow the CARE guidelines. The CITE-seq experiment includes data on three additional MM patients enrolled on the CARTITUDE-1 trial (61-year-old female, 67-year-old male, 67-year-old female). Furthermore, all MM patients included in this work consented to participation in the Multiple Myeloma Biorepository (HSM:18-00456). All subjects provided written informed consent for the evaluations. All study protocols were approved by the Program for the Protection of Human Subjects (PPHS) and the Institutional Review Board (IRB) at the Icahn School of Medicine at Mount Sinai and adhere to the 2008 Declaration of Helsinki.
Sample collection and tissue processing.
Peripheral blood was collected in heparin anticoagulated green tops (10 mL) via venipuncture throughout the course of his treatment in concordance with standard of care lab draws. Plasma was isolated from peripheral blood. Peripheral blood mononuclear cells (PBMC) were Ficoll density separated and cryopreserved. Cryopreserved PBMC samples were used for flow cytometry, mass cytometry and other assays as detailed below. Cerebrospinal fluid (CSF) was collected by lumbar puncture in concordance with standard of care. Each sample of 8 mL was centrifuged 300g at 4 °C, for 10 min. 0.5 mL of supernatant was divided into aliquots and frozen at −80 °C. About 200 μL of CSF and plasma from peripheral blood were used for O-link and Ella proteomics analysis as detailed below. Cells from CSF were used immediately for flow cytometry.
Statistics and reproducibility.
Experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. No statistical method was used to predetermine sample size for the analyses. For clinical and cytokine assays no data points were removed from the analysis. Cytometry data were gated to relevant population as shown in Extended Data Figure 5a. CITE-seq data were filtered to remove multiplets based on the crossSampleDoublets() and withinSampleDoublets() functions of the Citefuse package (v1.2.0) in R (v3.6.1). No other cells were excluded from the analysis. The non-parametric Mann-Whitney U test was used to compare gene expression values were appropriate. The Pearson correlation coefficient was used to characterize correlation of cytokine expression between blood and CSF. For all analyses, a two-sided p value < 0.05 was considered significant.
Additional clinical information on the patient.
There was no documented family history of movement disorders for the patient of the case report. In terms of neuro-psychiatric history, the patient had a remote history of migraines, documented in 2009, for which he received sumatriptan 100 mg as needed. In 2014, the patient was diagnosed with a mood disorder and started on sertraline 150 mg. He had been taking lorazepam and alprazolam; these were discontinued at that time and clonazepam 1 mg daily was started instead. No anti-dopaminergic medications were taken by the patient around this episode or later. Due to recurring anxiety with panic attacks the sertraline dose was increased to 200 mg and clonazepam was gradually increased to a maximum daily dose of 4 mg as needed. Sertraline was discontinued in 2016. The clonazepam dose was maintained for recurring anxiety with panic attacks. In addition, the patient was seen at an outside hospital in 2018 after a traffic accident. All documented neurological exams at that time were normal. He received an MRI of the brain which noted nonspecific punctate foci T2/FLAIR hyperintensity in the periventricular and subcortical white matter, likely secondary to chronic microvascular ischemic disease but no other intracranial abnormalities. Six months prior to the CAR-T trial, the patient was seen by a neurologist for the evaluation of weakness in the right hand. The neurological exam and tests of motor function in the limbs were normal with exception of portions of the right arm. Symptoms were thought to be suggestive of a radial nerve irritation at the spiral groove related to a work-related overuse problem. Electromyography (EMG) confirmed mild acute denervation demonstrating a radial nerve injury with mild acute axonal involvement and the patient’s symptoms resolved with rest. During the screening visit for the CAR-T trial, the patient reported grade 1 fatigue, a remote syncope (around 2011), as well as grade 1 peripheral sensory neuropathy (which affects soles of the feet, toes, calves and fingers), described as cramping without numbness. The neuropathy complaints did not interfere with walking, balance, or fine motor movements and developed after bortezomib treatment. During the hospitalization at the time of CAR-T cell infusion, which includes the CRS period, the patient received a neurological evaluation every day. The patient was specifically monitored for neurotoxicity according to the specifications of the clinical trial protocol. Neurological exam was documented at every subsequent study visit (every 28 days) after CAR-T infusion. Additionally, handwriting logs for dexterity were performed as specified. In conclusion, the patient was evaluated by a neurologist six months prior to CAR-T therapy (by the same physician who evaluated the patient when he presented with the described neurotoxicity after CAR-T). The received other neurological exams immediately before and after CAR-T cell infusion according to the clinical trial protocol. There were no pre-existing signs of parkinsonism present during evaluation prior to CAR-T infusion.
Flow cytometry.
Cryopreserved Ficoll density separated peripheral blood mononuclear cells were thawed by standard technique. Cells in the CSF were used within 3 hours of collection after isolation. CD3+/CD4+/CD8+ T cell, CD19+ B cell, and anti-BCMA directed T cells were measured by multiple-color flow cytometry with human monoclonal Acrobiosystems anti-BCMA (FITC) (cat# BCA-HF254-25ug) and BioLegend human monoclonal anti-CD3 (cat# 300472 and cat# 344842), human monoclonal anti-CD4 (cat# 317434), human monoclonal anti-CD8 (cat# 344742), human monoclonal anti-CD19 (cat# 561121). All cell surface antibodies were used at a 1:20 dilution following the manufacturer’s recommendation. The FITC-labeled human BCMA as used at a 1:100 dilution. The samples were acquired on a BD FACS LSR Fortessa flow cytometry system (BD Biosciences). Data was visualized and analyzed using Cytobank21.
Olink multiplex proteomics assay.
Relative protein expression was measured in the CSF and peripheral blood plasma using Olink proximity extension technology, a high throughput multiplex proteomic immunoassay following the manufacturer’s protocols. The commercially available Immuno_Oncology (Article number 95310) includes 92 immune and oncology related proteins was used. A table with all cytokines measured is included below as Supplementary Table S2. Olink uses marker-specific binding and hybridization of a set of paired oligonucleotide antibody probes that is subsequently amplified using a quantitative PCR. Protein expression values are reported as normalized protein expression (NPX) values on a log2 scale. Analysis was conducted in R (v3.6.1) and figures were produced using the package pheatmap22.
Ella cytokine detection.
The protein simple Ella cytokine detection system uses microfluidics ELISA assays in a multianalyte chip that were run within cartridges in triplicate following the manufacturer’s instructions. Human analytes of IL-6, IL-8, TNF-α, IL18, IFNg, and IL-10 were performed by the Mount Sinai Human Immune Monitoring Center (HIMC) using 25-30uL of plasma or CSF from subject. Analysis was conducted in R and figures were generated using the package ggplot223.
Mass cytometry.
Cells were stained with either CyTOF antibody Panel 1 or Panel 2 listed in Supplementary Tables S3 and S4. Antibodies used were either purchased pre-conjugated with metals from Fluidigm or purchased unconjugated and metal conjugated in-house at the at the Human Immune Monitoring Center, Icahn School of Medicine at Mount Sinai, New York. All in-house conjugated antibodies were tittered and validated on healthy donor PBMC. All antibodies for CyTOF listed in Supplementary Tables S3 and S4 were used at a dilution of 1:100. For longitudinal monitoring of phenotypic changes, cells from selected timepoints were thawed and labeled with Rh103 intercalator (Fluidigm) as a viability dye and cell proliferation marker IdU (Cell-ID™ 127 5-Iodo-2′-deoxyuridine, Fluidigm). Cells were initially stained with a cocktail of surface antibodies that included BCMA-FITC (AcroBiosystems) (Panel 1). Surface-stained cells were further stained with polyclonal Anti-FITC −159Tb (source) and fixed with 1.6% formaldehyde. Each timepoint was then barcoded with CyTOF Cell-ID 20-Plex Palladium Barcoding Kit (Fluidigm). Barcoded cells were fixed and permeabilized with Fix-Perm buffer (BD Biosciences) and stained with the remaining intracellular antibodies from CyTOF Panel 1. Intracellular cytokine expression was monitored using CyTOF Panel 2. Cells from selected timepoints were activated with PMA and Ionomycin (Biolegend) in the presence of Brefeldin-A (Biolegend) for 6 hours. Post activation cells were stained with Rh103 intercalator and stained with BCMA-FITC and fixed with 1.6% formaldehyde. Fixed cells were Palladium barcoded with CyTOF Cell-ID 20-Plex Palladium Barcoding Kit, pooled and stained with surface markers from CyTOF Panel 2 including polyclonal Anti-FITC-169Tm. Cells stained with surface antibodies were fixed and permeabilized with Fix-Perm buffer and stained with cytokine antibodies. Samples stained with either CyTOF antibody Panel 1 or Panel 2 were finally fixed in freshly diluted 2.4% formaldehyde containing 125nM intercalator-Ir (Fluidgm) and 300nM OsO4 (ACROS Organics) and stored at 4°C in cell staining buffer containing (Fluidigm) 125nM intercalator-Ir until acquisition. Samples for CyTOF acquisition were washed with CAS buffer (Fluidigm) and re-suspended in CAS buffer containing EQ normalization beads (Fluidigm) and acquired on CyTOF2 (Fluidigm). Post-acquisition the data was normalized using bead-based normalization algorithm in the CyTOF software (Fluidigm). Normalized data was debarcoded using methods and software developed in Gary Nolan’s group at the Stanford University School of Medicine24. Normalized and debarcoded data was uploaded to Cytobank21 for final analysis as detailed below.
Mass cytometry data analysis.
Data in .fcs file format was downloaded from Cytobank21. For analysis of mass cytometry data, we used a workflow based on the example by Nowicka et al.25 using the diffcyt26 and CATALYST27 packages in R (v3.6.1). Briefly, data was imported and transformed for analysis using the read.flowSet() function from the flowCore package28 and the prepData(…, cofactor = 5) function from the CATALYST package respectively. Clustering was based on the FlowSOM algorithm29 using all protein markers from the panel on a 10x10 grid size with maximum K = 20 clusters. These clusters were visualized using UMAP dimension reduction and subsequently annotated based on canonical protein markers and the FITC-BCMA tag to identify chimeric antigen receptor (CAR) T cells.
CITE-seq.
For each sample, cell suspensions were split and barcoded using “hashing antibodies” staining beta-2-microglobulin and CD298 and conjugated to “hash-tag” oligonucleotides (HTOs). Prior to hashing, each of the 5 samples were split into two aliquots and were either "stimulated" or "unstimulated". Stimulated aliquots were incubated for 3hrs at 37°C with PMA/Ionomycin. Unstimulated aliquots were incubated for 3hrs at 37C with cRPMI. After these incubations the 10 aliquots were hashed and pooled. Hashed samples were pooled and stained with CITE-seq antibodies purchased from the BioLegend TotalSeq catalog; the FITC antibody was a custom conjugate from BioLegend. All commercial antibodies were diluted at 1:100 according to the manufacturer’s instructions. The custom conjugate is tittered to find the optimal volume to stain PBMC. The CITE-seq panel is detailed in Supplementary Table S5. Stained cells were then encapsulated for single-cell reverse transcription using the 10X Chromium platform (5’ v1.0) and libraries were prepared according to the manufacturer’s instructions with minor modifications summarized hereafter. Briefly, cDNA amplification was performed in the presence of 2pM of an antibody oligo specific primer to increase yield of antibody derived tags (ADTs) and 3pM of specific primer to increase yield of HTOs. The amplified cDNA was then separated by SPRI size selection into cDNA fractions containing mRNA derived cDNA (>300bp) and ADT-derived cDNAs (<180bp), which were further purified by additional rounds of SPRI selection. Independent sequencing libraries were generated from the mRNA and ADT cDNA fractions, which were quantified, pooled and sequenced together on an Illumina NextSeq/NovaSeq to a targeted depth of at 25-750 million reads per gene expression library and 1,000-30,000 targeted reads/cell.
CITE-seq data analysis.
Illumina sequencer base call files were demultiplexed using into FASTQ files using the cellranger (v3.0.1) mkfastq and count pipeline. CITE-seq data was analyzed using R (v3.6.1) and the CiteFuse package30, using the proposed analysis pipeline with minor modifications. Briefly, matrices with counts representing RNA, ADT and HTO data respectively were read into R separately and combined into a SingleCellExperiment object31 using the preprocessing() function. Metadata (incl. patient ID and experimental condition (stimulated vs. unstimulated)) were added based on known experimental design and corresponding hashtag oligonucleotides (HTOs). HTO expression was normalized using the log transform method and the normaliseExprs() function. Cross-sample doublets and within-sample doublets were identified and removed using the crossSampleDoublets() and withinSampleDoublets(…, minPts = 10) function respectively. Similarity network fusion (SNF) was used to integrate RNA and ADT matrices after calculating log-transformed normalized expression values with the Citefuse() function. Both spectral clustering with K = 25 and Louvain clustering were attempted and t-SNE embedding was used to visualize dimension reduction. Manual inspection and canonical gene/protein expression were used to identify clusters corresponding to CD4+ and CD8+ chimeric antigen receptor (CAR) T cells. These cells were isolated into distinct SingleCellExperiment objects for downstream analysis. SNF, clustering and dimension reduction of CAR T cells was done in a similar fashion as detailed above. The DEgenesCross() function with standard parameters was used to determine differentially expressed genes between the patient with neurotoxicity and all other patients. Differential expression is determined with a two-sided Mann-Whitney U test, p values are corrected using the Benjamini-Hochberg method.
Immunohistochemistry.
Slides with 5um sections from paraffin embedded tissues from autopsies were stained with CD3 (LN10) and GFAP (GA5) pre-diluted Bond reagents from Leica Microsystems, HEIR (Heat Induced Epitope Retrieval) for 20 minutes with ER2 (Bond Epitope Retrieval Solution 2), MSMC DAB detection, counterstained per established staining protocol on the automated Leica Bond Biosystems III platform (USA, Buffalo Grove, IL). Immunohistochemistry for BCMA was performed using VENTANA DISCOVERY ULTRA from Roche. This system allows for automated baking, deparaffinization and cell conditioning. Semiautomatic staining was performed using BCMA antibody (cat#B0807 fromUSBiological) at 1:10 dilution during 60 min. As secondary antibody Discovery OMNIMap anti-rabbit-HRP from Roche (760-4310) was used and the signal was obtained using Discovery ChromoMap DAB RUO from Roche (760-2513) (brown signal) Tissues were counterstained with Hematoxylin (in blue).
Analysis of public datasets.
The mRNA expression data from the Allen Brain Atlas was last accessed on April 25, 2021. The heatmap can be found at http://human.brain-map.org/ (Human Brain data) when doing a Gene Search for TNFRSF17 and DRD1 and selecting “View Selection Thumbnails”. Raw expression data of the heatmap used as part of the figures have been downloaded from the Allen Brain Atlas data portal by the authors and are included as Source Data of Extended Data Figure 8.
Extended Data
Extended Data Fig. 1. Clinical course and biochemical parameters after CAR-T cell treatment.
The time periods associated with cytokine release syndrome (CRS), neutropenic fever and neurotoxicity are annotated in the individual subplots. All cytokine levels were determined in the peripheral blood. (a) Temperature curve. (b) Administration of relevant pharmacologic treatments during the period after CAR-T treatment. (c) Total leukocyte, lymphocyte, and neutrophil counts. (d) Time course of CRP level (mg/L). (e) Time course of ferritin level (ng/mL). (f) Time course of IL-18 level (pg/mL). (g) Time course of IL-2Ra (CD25) level (pg/mL).
Extended Data Fig. 2. Cytokine levels and time course after CAR-T cell treatment.
The time periods associated with cytokine release syndrome (CRS), neutropenic fever and neurotoxicity are annotated in the individual subplots. (a) Time course of IFN-gamma level (pg/mL). (b) Time course of TNF-alpha level (pg/mL). (c) Time course of IL-6 level (pg/mL). (d) Time course of IL-10 level (pg/mL). All measurements are from peripheral blood plasma. (e) Olink cytokine profiling of peripheral blood plasma at different time points after chimeric antigen receptor (CAR) T cell therapy. Values shown are normalized protein expression (NPX) values according the Olink protocol in log2 scale (high protein levels in red, low protein levels in blue).
Extended Data Fig. 3. MRI and Ioflupane single photon emission computed tomography imaging after the onset of neurotoxicity.
(a) MRI axial FLAIR (left) and T2 (right) images at the level of the deep brain nuclei (top) and the cerebral cortex (top), conducted at day 101 after CAR-T infusion. Images demonstrate small punctuate hyperintensities present on imaging prior to CAR-T therapy and putatively due to pre-existing microvascular damage. (b) Ioflupane (123-I) scan images, conducted at day 155 after CAR-T infusion, show normal uptake at the level of the basal ganglia.
Extended Data Fig. 4. Quantitative analysis of FDG-PET/CT images confirms decreased metabolism in caudate nucleus after CAR-T cell therapy.
(a) FDG-PET axial splash images pre (top) and post (bottom) CAR-T infusion. Shown is a spectral scale with high metabolism/perfusion in red, to low metabolism/perfusion in dark blue. (b) Quantitative analysis showing normalized Z-score for all available regions of the brain before (blue) and after (red) CAR-T infusion. The caudate is highlighted. The normalized score was calculated using MIMneuro, comparing the image with a library of 43 FDG neurologic controls (41-80 years old).
Extended Data Fig. 5. Mass cytometry characterizes the effector-memory phenotype of CAR-T cells over time.
(a) Representative mass cytometry (CyTOF) plots illustrating the gating strategy for identifying CAR-T cells and T cell subsets as shown in Figure 1a, 1d and Extended Data Figure 5e. (b) UMAP representation of peripheral blood mononuclear cells (PBMC) collected at time points shown in (a) shows the clustering of major immune cell types. (c) Relative contribution of major immune cell types in samples at different time points. (d) Expression of canonical markers, showing accurate classification of major immune celI types. (e) CAR-T cell phenotype, as determined by expression of CCR7 and CD45RA, illustrating a high fraction of effector-memory T cells at all time points. Each bar corresponds to N=1 sample collected from the patient. The UMAP plots visually illustrate the clustering of T cells and confirm low CCR7 and CD45RA expression on CAR-T cells.
Extended Data Fig. 6. Cytokine expression of peripheral blood CAR-T cells isolated at day 128 after treatment vs. healthy donor T cells.
(a) CAR-T cells isolated at day 128 after CAR-T infusion were stimulated with PMA/ionomycin and cytokine production was assessed with mass cytometry. Shown here is high expression of TNF-alpha, interferon-gamma and GM-CSF and lack of expression of IL-17 in CD4+ (left) and CD8+ (right) CAR-T cells. (b) Percentage of CAR-T cells (orange) and healthy donor (HD) T cells (blue) expressing the full set of cytokines tested before (UNSTIM) or after (STIM) stimulation with PMA/ionomycin. Each bar represents N=1 sample analyzed from the patient or healthy donor.
Extended Data Fig. 7. Expression of canonical markers on CITE-seq data identifies and clusters major immune cell types.
(a) t-SNE plot representation of CITE-seq analysis of peripheral blood mononuclear cells before and after PMA/ionomycin stimulation. Clustering was determined by similarity network fusion (SNF) and Louvain clustering algorithm. Individual cells are colored by subject (healthy donor (HD), neurotoxicity patient (NEUROTOX) and 3 other patients on the same clinical trial without neurotoxicity (MM1, MM2, MM3). Highlighted are the major immune cell types (B cells, NK cells, CD8+ T cells, CD4+ T cells, CAR-T cells and monocytes). There is a small cluster of events that corresponds to multiplets or debris (centrally, not highlighted). (b) Expression level of canonical genes: CD8A, CD4, CD14, FCGR3A (CD16), CD19 and NCAM1 (CD56). In each case showing both mRNA (top) and ADT (antibody-derived tag, representation of protein level) (high = red, low = blue). Expression levels are normalized as described in the Methods.
Extended Data Fig. 8. Expression of BCMA in healthy donors of the Allen Brain Atlas and presence of CAR-T cells in CSF of patient.
(a) Microarray data on top illustrates the expression of TNFRSF17 (BCMA) in the caudate nucleus of 5 healthy brain donors. The bottom shows that regions of TNFRSF17 (BCMA) expression coincides with DRD1 (dopamine receptor D1) expression, a protein know to be highly specific for the caudate nucleus. Image credit: Allen Institute: © 2010 Allen Institute for Brain Science. Allen Human Brain Atlas; available from: human.brain-map.org. (b) Schematic representation showing the log2 intensity of TNFRSF17 (BCMA) RNA expression in a single patient from the Allen Brain Atlas. Image credit: Allen Institute: © 2010 Allen Institute for Brain Science. Allen Human Brain Atlas; available from: human.brain-map.org. (c) Quantitative representation of the Allen Brain Atlas data with boxplots (median, Q1 and Q3 quartiles, whiskers up to 1.5 x IQR) showing normalized expression (z-score) across all six donors for different brain structures (N = 6, total of 3,702 probes across 27 brain regions). The p-values shown correspond to a two-sided Mann-Whitney U test of striatum versus any other region (**: p < 0.001, ***: p < 0.0001, n.s.: p ≥ 0.05).
Extended Data Fig. 9. Presence and persistence of CAR-T cells in CSF of patient and cytokine profiling in peripheral blood plasma versus CSF after development of neurotoxicity.
(a) Representative plots showing the gating strategy on CSF to get to the T cell gate. (b) Flow cytometric data of cerebrospinal fluid from day 148 after CAR-T cell infusion, showing presence of CD4+ and CD8+ CAR-T cells. (c) Flow cytometric data of cerebrospinal fluid from day 155 after CAR-T cell infusion (i.e. after administration of intravenous cyclophosphamide and intrathecal cytarabine), showing persistent presence of CD4+ and CD8+ CAR-T cells. (d) Normalized protein expression (NPX) log2 values of all cytokines in the Olink Immuno-Oncology panel, in serum (top) and CSF (bottom) (high protein levels in red, low protein levels in blue). (e) Scatter plot showing overall correlation of cytokine levels in plasma versus CSF (Pearson correlation coefficient r = 0.70, two-sided p < 0.001). (f) The log2 fold change (FC) of CSF versus blood plasma in a healthy control (along x-axis) and the patient who developed neurotoxicity (along y-axis). Highlighted are a selection of cytokines that are overrepresented in the patient’s CSF compared to the healthy control data. Among the cytokines that are overrepresented, we note a set of cytokines suggesting T cell activation (e.g. GZMB, GZMA, IFN-γ, CD40L, CD8A, CD27, FASLG), cytokines that are induced by IFN-γ (e.g. CXCL5, CXCL10, CXCL11) and that are known to act as chemo-attractants for T cells (among other immune cell types), and cytokines that point to possible involvement of cells in the blood-brain barrier (BBB) (e.g. PDGFb, EGF and ANGPT1).
Extended Data Fig. 10. Immunohistochemistry showing BCMA protein expression in brain tissue of the patient and in a control brain.
(a) BCMA immunohistochemistry of the caudate nucleus subependymal region (10x magnification, left, scale bar 200 μm). Inset (40x magnification, right, scale bar 50 μm) shows high magnification image of astrocytes (top) and a neuron (bottom) that stained positive for BCMA, whereas surrounding cells were negative. Images shown are representative slides from the caudate nucleus from the patient described in this case report (N=1). For each region stained, at least 3 slides were available. (b) BCMA immunohistochemistry of selected brain regions as annotated in the patient of interest (left) versus a control brain (right) from a subject who died due to non-neurologic illness (10x magnification (top), scale bar 200 μm and 20x magnification (middle, bottom), scale bar 100 μm). Images shown are representative slides from the patient described in this case report (N=1), as well as a single control brain (N = 1). For each region stained, at least 3 slides were available. The experiment was repeated in a second control brain with similar results.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Travis Dawson, Hui Xie, Manishkumar Patel and the rest of the staff at the Human Immune Monitoring at the Icahn School of Medicine for sample management and their help conducting omics assays. Furthermore, we would like to thank Monika Garcia-Barros, and Rachel Brody from the Biorepository and Pathology CoRE at the Icahn School of Medicine for immunohistochemistry staining.
Samir Parekh acknowledges support by the National Cancer Institute (NCI) (R01 CA244899, CA252222) and receives research funding from Amgen, Celgene/BMS and Karyopharm. Miriam Merad acknowledges support by the National Institute of Allergy and Infectious Diseases (NIAID) (U24 AI118644-05S1, U19 AI128949, U19 AI118610), by the NCI (R01 CA254104, R01 CA257195, P30 CA196521-05S2), by a Fast Grant (George Mason University), by the Gates Foundation and by the Samuel Waxman Cancer Research Foundation. Joshua Brody acknowledges support by the NCI (R01 CA246239-01).
Footnotes
COMPETING INTERESTS STATEMENT
OVO has no relevant conflicts to disclose. AA has no relevant conflicts to disclose. BU has no relevant conflicts to disclose. SS has no relevant conflicts to disclose. SS is currently employed by Sema4 but was not working for the company at the time of preparation of the manuscript. DM has worked as consultant for Bristol Myers Squibb, Celgene, Foundation Medicine, GSK, Janssen, Kinevant, Sanofi and has received grant/research support from Allogene, Amgen, Bristol Myers Squibb, Celgene, Janssen and Regeneron. DM is currently employed by Johnson & Johnson but was not working for the company at the time of preparation of the manuscript. SG has no relevant conflicts to disclose. JTF has no relevant conflicts to disclose. JFC has no relevant conflicts to disclose. CBS is a member of the ciltacabtagene autoleucel Risk Evaluation and Mitigation Strategy (REMS) advisory board. SKS has no relevant conflicts to disclose. AR is currently employed by Immunai but was not working for that company at the time of preparation of the manuscript. AL has no relevant conflicts to disclose. JDB has received consulting fees from Celldex, Genentech, Gilead, Janssen, Kite, Merck and research funding or reagents provided by Celldex, Genentech, Janssen, Kite and Merck. MM reports no competing interests. SJ is a consultant for Bristol Myers Squibb, Janssen, Karyopharm Therapeutics, Merck, Sanofi, and Takeda Pharmaceuticals. SP receives research funding from Amgen, Celgene/BMS, Karyopharm and consulting fees from Foundation Medicine. All other authors declare no competing interests.
Editor summary:
A progressive movement disorder in a patient with multiple myeloma treated with anti-BCMA CAR T cells that may have been related to on-target activity in the brain supports prospective neurologic monitoring following BCMA-targeting therapies
Editor recognition statement:
Saheli Sadanand was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Reviewer recognition statement:
Nature Medicine thanks Leo Rasche, Eric Smith, Daniel Rubin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
DATA AVAILABILITY STATEMENT
All requests for raw and analyzed data and materials will be promptly reviewed by the Icahn School of Medicine at Mount Sinai and the Mount Sinai Hospital to verify if the request is subject to any confidentiality and data protection obligations. Requests for data should be addressed to the corresponding author via e-mail and will receive a reply within 10 business days. Any data and materials that can be shared will be released via a material transfer agreement. Raw and analyzed CITE-seq data are available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (accession no. GSE182527). Mass cytometry and intracellular cytokine data are available through the FlowRepository website (ID FR-FCM-Z4KB). The images derived from the Allen Human Brain Atlas (© 2010 Allen Institute for Brain Science; Allen Human Brain Atlas) can be accessed from: human.brain-map.org. Specific URLs to recreate the following figures are provided: Figure 2b (https://human.brain-map.org/static/brainexplorer), Extended Data Figure 8a (https://human.brain-map.org/microarray/search/show?search_type=user_selections&user_selection_mode=1) and Extended Data Figure 8b (https://human.brain-map.org/microarray/gene/show/605) and source data has been made available. For all clinical measurements and cytokine levels (Extended Data Figure 1-2, Extended Data Figure 6, Extended Data Figure 9d-f), source data has been made available.
REFERENCES
- 1.Kumar SK, et al. Multiple myeloma. Nat Rev Dis Primers 3, 17046 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Madduri D, Dhodapkar MV, Lonial S, Jagannath S & Cho HJ SOHO State of the Art Updates and Next Questions: T-Cell-Directed Immune Therapies for Multiple Myeloma: Chimeric Antigen Receptor-Modified T Cells and Bispecific T-Cell-Engaging Agents. Clin Lymphoma Myeloma Leuk 19, 537–544 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Shah N, Chari A, Scott E, Mezzi K & Usmani SZ B-cell maturation antigen (BCMA) in multiple myeloma: rationale for targeting and current therapeutic approaches. Leukemia (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah UA & Mailankody S Emerging immunotherapies in multiple myeloma. BMJ 370, m3176 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Bu DX, et al. Pre-clinical validation of B cell maturation antigen (BCMA) as a target for T cell immunotherapy of multiple myeloma. Oncotarget 9, 25764–25780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter RO, et al. B-cell Maturation Antigen Is a Promising Target for Adoptive T-cell Therapy of Multiple Myeloma. Clinical Cancer Research 19, 2048–2060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khattar P, et al. B- Cell Maturation Antigen Is Exclusively Expressed in a Wide Range of B-Cell and Plasma Cell Neoplasm and in a Potential Therapeutic Target for Bcma Directed Therapies. Blood 130, 2755–2755 (2017). [Google Scholar]
- 8.Sanchez E, et al. Serum B-cell maturation antigen is elevated in multiple myeloma and correlates with disease status and survival. Br J Haematol 158, 727–738 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Sanchez E, et al. Soluble B-Cell Maturation Antigen Mediates Tumor-Induced Immune Deficiency in Multiple Myeloma. Clinical cancer research : an official journal of the American Association for Cancer Research 22, 3383–3397 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Tai YT, et al. APRIL and BCMA promote human multiple myeloma growth and immunosuppression in the bone marrow microenvironment. Blood 127, 3225–3236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munshi NC, et al. Idecabtagene Vicleucel in Relapsed and Refractory Multiple Myeloma. New England Journal of Medicine 384, 705–716 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Raje N, et al. Anti-BCMA CAR T-Cell Therapy bb2121 in Relapsed or Refractory Multiple Myeloma. New England Journal of Medicine 380, 1726–1737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berdeja JG, et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): a phase 1b/2 open-label study. Lancet 398, 314–324 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Lee DW, et al. ASTCT Consensus Grading for Cytokine Release Syndrome and Neurologic Toxicity Associated with Immune Effector Cells. Biol Blood Marrow Transplant 25, 625–638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neelapu SS, et al. Chimeric antigen receptor T-cell therapy — assessment and management of toxicities. Nature Reviews Clinical Oncology 15, 47–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagelmann N, Ayuk F, Atanackovic D & Kroger N B cell maturation antigen-specific chimeric antigen receptor T cells for relapsed or refractory multiple myeloma: A meta-analysis. Eur J Haematol 104, 318–327 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Jatiani SS, et al. Myeloma CAR-T CRS Management With IL-1R Antagonist Anakinra. Clin Lymphoma Myeloma Leuk 20, 632–636.e631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee L, et al. Evaluation of B cell maturation antigen as a target for antibody drug conjugate mediated cytotoxicity in multiple myeloma. Br J Haematol 174, 911–922 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Shah UA & Mailankody S CAR T and CAR NK cells in multiple myeloma: expanding the targets. Best Practice & Research Clinical Haematology, 101141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 21.Kotecha N, Krutzik PO & Irish JM Web-based analysis and publication of flow cytometry experiments. Current protocols in cytometry Chapter 10, Unit10.17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolde R pheatmap: Pretty Heatmaps. R package version 1.0.12 (2019). [Google Scholar]
- 23.Wickham H ggplot2: Elegant Graphics for Data Analysis, (Springer-Verlag; New York, 2016). [Google Scholar]
- 24.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 10, 316–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowicka M, et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 6, 748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber LM diffcyt: Differential discovery in high-dimensional cytometry via high-resolution clustering. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crowell HL, Zanotelli VRT, Chevrier S & Robinson MD CATALYST: Cytometry dATa anALYSis Tools. (2020). [Google Scholar]
- 28.Ellis B, et al. flowCore: flowCore: Basic structures for flow cytometry data. (2019). [Google Scholar]
- 29.Van Gassen S, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry Part A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Lin Y, Geddes TA, Yang JYH & Yang P CiteFuse enables multi-modal analysis of CITE-seq data. Bioinformatics (2020). [DOI] [PubMed] [Google Scholar]
- 31.Amezquita RA, et al. Orchestrating single-cell analysis with Bioconductor. Nat Methods (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All requests for raw and analyzed data and materials will be promptly reviewed by the Icahn School of Medicine at Mount Sinai and the Mount Sinai Hospital to verify if the request is subject to any confidentiality and data protection obligations. Requests for data should be addressed to the corresponding author via e-mail and will receive a reply within 10 business days. Any data and materials that can be shared will be released via a material transfer agreement. Raw and analyzed CITE-seq data are available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (accession no. GSE182527). Mass cytometry and intracellular cytokine data are available through the FlowRepository website (ID FR-FCM-Z4KB). The images derived from the Allen Human Brain Atlas (© 2010 Allen Institute for Brain Science; Allen Human Brain Atlas) can be accessed from: human.brain-map.org. Specific URLs to recreate the following figures are provided: Figure 2b (https://human.brain-map.org/static/brainexplorer), Extended Data Figure 8a (https://human.brain-map.org/microarray/search/show?search_type=user_selections&user_selection_mode=1) and Extended Data Figure 8b (https://human.brain-map.org/microarray/gene/show/605) and source data has been made available. For all clinical measurements and cytokine levels (Extended Data Figure 1-2, Extended Data Figure 6, Extended Data Figure 9d-f), source data has been made available.