Abstract
Background and Aims:
Fecal microbiota transplantation (FMT) is an emerging treatment modality for ulcerative colitis (UC). Several randomized controlled trials have shown efficacy for FMT in the treatment of UC, but a better understanding of the transferable microbiota and their immune impact is needed to develop more efficient microbiome-based therapies for UC.
Methods:
Metagenomic analysis and strain tracking was performed on 60 donor and recipient samples receiving FMT for active UC. Sorting and sequencing of IgA-coated microbiota (called IgA-seq) was used to define immune-reactive microbiota. Colonization of germ-free or genetically engineered mice with patient-derived strains was performed to determine the mechanism of microbial impact on intestinal immunity.
Results:
Metagenomic analysis defined a core set of donor-derived transferable bacterial strains in UC subjects achieving clinical response, which predicted response in an independent trial of FMT for UC. IgA-seq of FMT recipient samples and gnotobiotic mice colonized with donor microbiota identified Odoribacter splanchnicus as a transferable strain shaping mucosal immunity, which correlated with clinical response and the induction of mucosal regulatory T cells (Tregs). Colonization of mice with O. splanchnicus led to an increase in Foxp3+/RORγt+ Tregs, induction of IL-10, and the production of short chain fatty acids, all of which were required for O. splanchnicus to limit colitis in mouse models.
Conclusions:
This work provides the first evidence of transferable, donor-derived strains that correlate with clinical response to FMT in UC and reveals O. splanchnicus as a key component promoting both metabolic and immune cell protection from colitis. These mechanistic features will help enable strategies to enhance the therapeutic efficacy of microbial therapy for UC.
Keywords: Fecal Microbiota Transplant, Ulcerative colitis, Odoribacter, IgA-seq
Lay Summary
Fecal microbiota transplantation is an emerging therapy to treat ulcerative colitis. These findings define strain transferability associated with clinical response and highlight the mechanisms of their immune impact.
Graphical Abstract

Introduction
Over two million people worldwide suffer from ulcerative colitis (UC)1. While biologic therapies have significantly improved treatment efficacy of UC, nearly two-thirds of patients attenuate response to medical therapy. As a result, new therapeutic modalities are urgently needed that target the underlying pathophysiology of UC. Given the strong correlation of alterations in the gut microbiome with disease activity in inflammatory bowel disease (IBD), microbial-based therapies have emerged with the potential to treat UC. In particular, several recent trials have demonstrated the efficacy for fecal microbiota transplantation (FMT) in the treatment of UC2–4; however, a clear limitation of FMT is the current use of crude donor fecal material. This practice increases potential infectious risk as well as donor-to-donor variations that could alter therapeutic efficacy5. Thus, rational selection and production of specific microbial strains or communities could improve efficacy, minimize the risk of adverse reactions as well as increase the acceptance of microbiome-based therapies.
To accomplish this task, a mechanistic understanding of microbial transferability, engraftment and immune cell impact underlying the efficacy of FMT in UC needs to be defined. In the treatment of recurrent Clostridium difficile infection with FMT, the change in microbial diversity following FMT is a key metric that associates with clinical resolution of C. difficile6, 7. While some studies highlight the potential role for increasing overall diversity in improving the clinical outcome for UC4, microbial diversity did not correlate with clinical response in all studies8, 9. In contrast, several studies indicate that efficacy may be donor-dependent3, 8, 10. However, the composition and function of a core set of transferable strains associated with clinical response in UC have not been defined.
Based on the hypothesis that key strains would be (i) transferable and (ii) immune-reactive, we performed metagenomic sequencing and ‘IgA-seq’ to define a core of transferable and IgA-coated microbiota using samples from a cohort of 20 recipient-participants pairs with active UC treated with FMT. Clinical response based on endoscopic subscore and total Mayo score was assessed at week 4 post-FMT (NCT02516384). Using a strain inference algorithm11, our data reveal a core group of transferable bacterial species that correlate with clinical response and highlight the metabolic and cellular impact of a patient-derived strain of Odoribacter splanchnicus in shaping mucosal immunity in responders. These findings define strain transferability in the context of clinical response of UC to FMT and highlight the potential mechanisms of their immune impact, which will be critical in enhancing the efficacy of microbial-based therapies for UC.
Materials and Methods
Study population
20 participants with active UC received FMT using two-healthy donor fecal microbiota preparation (FMP) (NCT02516384). Details of this cohort have been previously published by our group8. Clinical responders were defined as Δ Mayo score ≥ 3 and a bleeding subscore < 1 (responders, n = 7; non-responders, n = 13). For Mayo endoscopic subscore (MES) analysis, responders were defined as Δ MES ≥ 1 (responders, n=10; non-responders, n = 10).
Metagenomic analysis and strain tracking
The details of DNA extraction and sequencing are described in Supplementary Methods. After quality control, samples averaged 135 million reads (mean = 135,854,707; max = 203,079,049, min = 76,938,232). One out of 60 fecal samples evaluated in the present study failed to pass the metagenomic sequencing quality control. Taxonomic profiling was then determined by MetaPhlAn2 pipeline12 (version 2.7.5). To analyze microbial strains, metagenomic data was separately processed and run through StrainFinder to determine bacterial species single nucleotide polymorphisms (SNPs) and their frequencies based on maximum likelihood estimates, as previously described11.
IgA seq analysis
To define the IgA-coated microbiota, fecal homogenates were processed, labeled and sorted as previously described13. Samples were stained with anti-human IgA (IS11–8E10; Miltenyi Biotec) or anti-mouse IgA (MA-6E1; eBioscience) and sorted on FACS Melody (BD Biosciences). Samples were costained for nucleic acids with SYTO BC (Invitrogen). Sorted samples were processed using PowerMag Soil DNA Isolation Kit (MO BIO), following manufacture instructions. For human samples the 16S rRNA gene V4 region was amplified and for mouse samples the V4 and V5 regions were amplified as previously described (https://earthmicrobiome.org). Amplicons were then sequenced in an Illumina MiSeq platform using the 2 × 250 bp paired-end protocol. Read pairs were processed using DADA214, VSEARCH15, and SILVA Database16 to generate a rarefied amplicon sequence variant (ASV) table (human sequence depth of 7,684 reads, mouse sequence depth of 25,179 reads).
Bacterial isolation and sequencing
Patient-derived bacterial strains were isolated under anaerobic conditions, as previously reported13. Briefly, single cells were sorted to a 96 well plate and recovered anaerobically. O. splanchnicus, Alistipes finegoldii and Blautia producta strains were characterized by 16S Sanger sequencing. Whole genome sequencing of O. splanchnicus was performed using PacBio Sequel v3 SMRTcell. Sequence data was assembled using Canu default settings17 into 6 contigs using minimap2 (v.2.17-r941). Gene prediction and annotation were carried out using RAST and visualized with GCView server18.
Metabolomics analysis
Cecal content was collected on day 10 or 21 post-colonization with UC patient-derived O. splanchnicus, A. finegoldi or B. producta. Short chain fatty acids (SCFAs) were detected and measured in cecal contents by liquid chromatography–quadruple–time of flight mass spectrometry (LC–Q–TOF) as previously described19.
Gnotobiotic and SPF Mice
For gnotobiotic experiments, 6- to 8-week-old germ-free C57BL/6 wild type (WT) or TCRβδ−/− mice were gavaged with 200μl of a two-healthy donor FMP. Seven weeks post-colonization feces were collected for IgA-seq analysis. Germ-free C57BL/6 WT, IL-10−/−, Rag1−/−, TCRβδ−/− and heterozygous (het) mice were bred and maintained at WCM20. For mono-colonization experiments, 6- to 8-week-old mice were gavaged with ~1 × 109 CFU of patient-derived A. finegoldii, B. producta and O. splanchnicus, respectively, at log-phase grown under anaerobic conditions in Yeast Casitone Fatty Acids Broth with Carbohydrate media (Anaerobe Systems). Colonization was confirmed by 16S rRNA qPCR from DNA extracted using feces or cecal content.
For colonization of SPF mice, 6- to 8-week-old mice were subjected to one week of antibiotic (ampicillin, vancomycin, metronidazole, neomycin; Sigma-Aldrich) treatment in the drinking water ad libitum up to 1 day prior to gavage. Mice were then gavaged with ~5 × 109 CFU log-phase bacteria. Colonization was confirmed by O. splanchnicus 16S rRNA PCR (F primer: 5’- ATGTAATGATGAGCACTCTAACGG-3’; R primer: 5’- GGCTTTTGAGATTGGCATCC-3’)21. C57BL/6 GPR109a−/− and GPR43−/− mice were obtained from Marcel van den Brink lab.
Colitis Mouse Models
Experimental models were previously described20. For induction of chemical colitis, 2% DSS (w/v) (M.W. 40,000–50,000; Affymetrix) was added to drinking water and administered ad libitum for 6 days. Mice were then monitored daily for weight loss and survival. Mouse Lipocalin-2 was measured in the supernatant by sandwich ELISA (R&D Systems).
Cellular isolation and intracellular cytokine staining
Lamina propria mononuclear cells (LPMC) were isolated from colonic tissue as previously described20. LIVE/DEAD fixable aqua dead cell stain kit (Molecular Probes) was used to select live cells. For analysis related to transcription factors, large intestine LPMC cells were stained with anti-CD3- E780 (eBiosciences 17A2) and anti-CD4-AF700 (eBiosciences GK1.5) before fixation and permeabilization with Cytoperm/Cytofix flowed by intracellular staining against anti-Foxp3-E450 (eBiosciences FJK-16s) and anti-RORγt-PE (eBiosciences B2D). For analysis related to cytokine, LPMC cells were stimulated with phorbol myristate acetate (PMA) and ionomycin with BD GolgiPlug for 3.5 hours. Following surface-marker staining with anti-CD3- E780 (eBiosciences 17A2) and anti-CD4-AF700 (eBiosciences GK1.5), LPMC cells were prepared as per manufacturer’s instruction with Cytoperm/Cytofix (BD Biosciences) for intracellular cytokine evaluation of IL-17A (eBiosciences 17B7) and IFNγ (eBiosciences XMG1.2).
Statistics
Microbiome analysis was performed in R studio (Boston, MA). Alpha and beta diversity was calculated using R package ‘phyloseq’22 while plots were constructed in ‘ggplot2’ or GraphPad Prism (San Diego, CA). Principal coordinate plots used the Monte Carlo permutation test to estimate the P-values. For taxonomic analysis, significance of microbial relative abundance was calculated using the nonparametric Mann- Whitney test and P-values were adjusted for multiple hypothesis with the false discovery rate algorithm. If paired analyses were applicable, Wilcoxon-paired rank test was used followed by Bonferroni multiple comparison correction. Hypothesis testing was done using two- sided test as appropriate at a 95% significance level.
Bacterial strains were quantified based on presence/absence of strain haplotypes (as defined by SNPs) by StrainFinder11. Strains present in WK4 samples were classified into four groups: derived from DON (present in DON but absent in PRE samples), derived from PRE (present in PRE but absent in DON samples), derived from WK4 (absent in DON and PRE samples), and derived from both DON and PRE (present in DON and PRE samples). T- test followed by Tukey multiple comparison correction was applied to strain analysis.
To test the efficiency of the core transferrable microbiota in determining clinical response to FMT, area under the curve (AUC) of receiver operating characteristic (ROC) curves, sensitivity, specificity, and P-values were computed using R (see Supplemental Methods). P-values were calculated from Mann-Whitney test to address the null hypothesis that the area under the ROC curves was 0.5.
Results
Core transferable microbiota (CTM) define FMT clinical response
FMT has emerged as a promising therapy for UC, however defining the characteristics of a core group of transferable bacteria remain a critical need in identifying and developing more efficient microbial-based therapy. To determine the CTM in a cohort of patients with active UC, we performed deep shotgun metagenomic sequencing of 60 samples (20 donor (DON); 20 recipients pre-FMT (PRE); and 20 recipients week 4 post-FMT, (WK4)) from a previously reported open-label trial of FMT for the treatment of UC8. Principal coordinate analysis (PCoA) based on beta diversity at the species-level (Bray-Curtis) showed that treatment groups (DON, PRE and WK4) had significantly different microbial composition (Supplementary Figure 1A). Consistent with our previously reported 16S rRNA analysis8, recipients’ microbial composition markedly shifted towards that observed in donor (Supplementary Figure 1A and B), irrespective of clinical response (Supplementary Figure 1C). To determine microbial transferability and identify the core bacterial species acquired from donors, a Venn diagram approach was used (Figure 1A). Based on a patient prevalence cut off of ≥ 50% and microbial relative abundance cut off of ≥ 0.1%, 17 bacterial species were found to be part of the CTM (Figure 1A). To evaluate the magnitude of the CTM contribution to the recipients’ microbiota, the relative abundance of these 17 species were compared between DON, PRE and WK4. The CTM median relative abundance of WK4 did not differ from that observed in DON, but was significantly higher than that found in recipients’ pre-transplant (Figure 1B). Since species-level analysis cannot determine if donors and recipients post-FMT share bacterial strains, we performed strain haplotype deconvolution using StrainFinder11 to evaluate if this CTM reflected the transfer of donor-derived strains. On average, a total of 22 (standard error of the mean; SEM ± 2.37) distinct bacterial strains belonging to the CTM were detected in WK4 post-FMT samples of which 52% (SEM ± 4.46) were found to be derived from donors and only 12.2% (SEM ± 3.14) from PRE (Figure 1C). Collectively, these results provide evidence of a core set of transferable bacterial strains in patients with active UC following FMT.
Figure 1. Clinical response to FMT is associated with a core transferable microbiota.
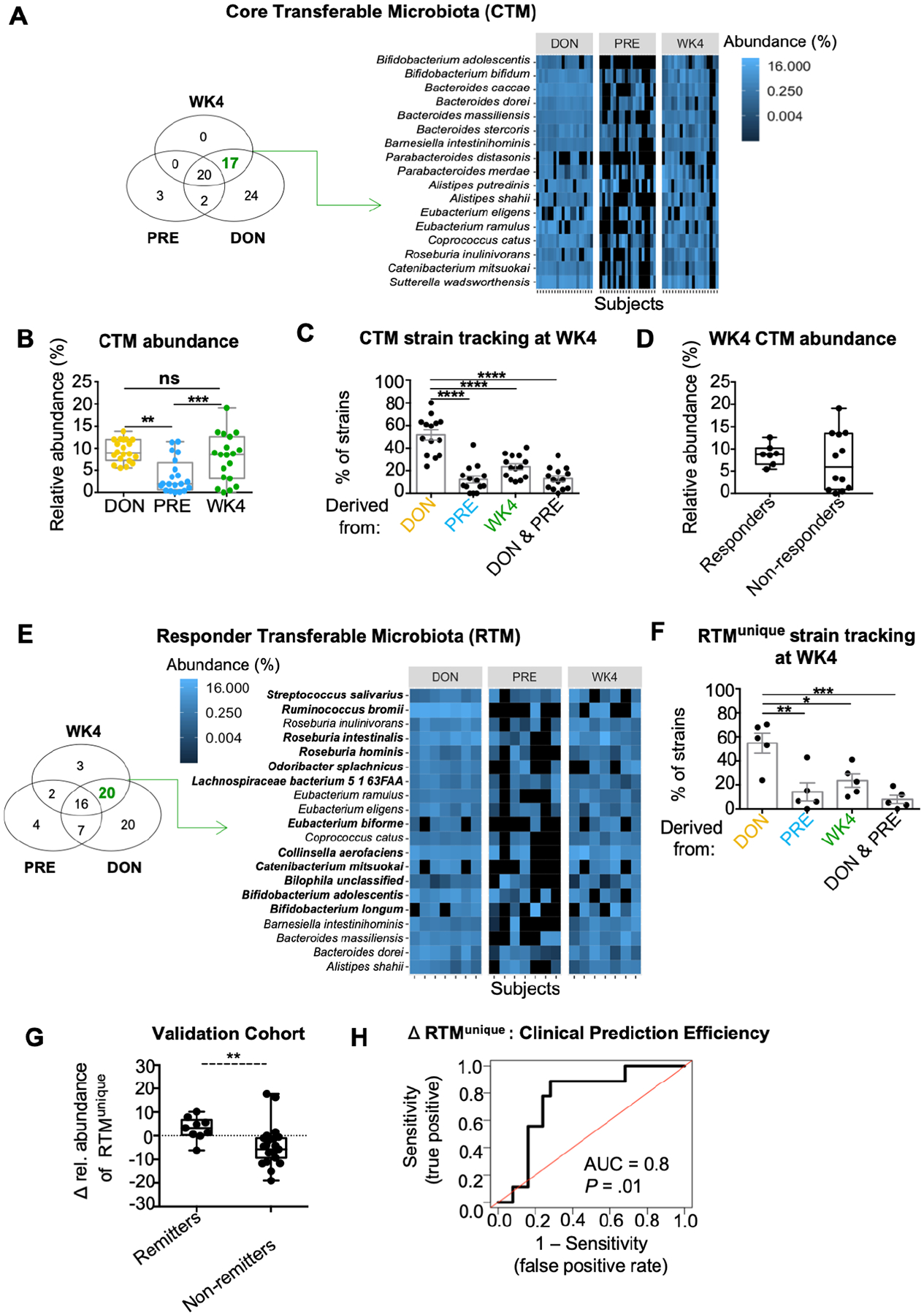
(A) Venn diagram depicting the number of bacterial species unique to a single variable (DON, PRE or WK4) or shared by two, or all three variables. Microbial cut off for prevalence was defined as ≥ 50% and relative abundance as ≥ 0.1%. Heatmap depicting the relative abundance of the 17 species defined by the Venn diagram as being part of the CTM. (B) Boxplot comparing the median relative abundance of the CTM in DON, PRE and WK4 post FMT. **p < 0.01, ***p < 0.001, Wilcoxon-paired test (Bonferroni adjusted). (C) Proportion of bacterial strains detected on WK4 belonging to the CTM (17 bacterial species) shown in figure 1B. Strains detected on WK4 were categorized as derived from DON, PRE, DON and PRE, and WK4. Each dot represents a subject and error bars represent SEM. ****p < 0.0001, t test (Tukey adjusted). (D) Boxplot comparing median relative abundance of the CTM on WK4 between responders and non-responders. Boxplots present the median, 25th and 75th percentiles. (E) Responders Venn diagram depicting the number of bacterial species unique to a single variable (Responders: DON, PRE or WK4) or shared by two, or all three variables. Heatmap depicting the relative abundance of the 20 species defined as being part of the RTM. Species marked in bold belongs to the RTMunique (n = 12) and therefore absent in the NRTM. (F) Proportion of bacterial strains detected on WK4 belonging to the RTMunique (n = 12) shown in figure 1F. Strains detected on WK4 were categorized as derived from DON, PRE, DON and PRE, and WK4. Each dot represents a subject (strain analysis for two patients is missing due to insufficient RTMunique genome coverage) and error bars represent SEM. *p < 0.05, **p < 0.01, ***p < 0.001, t test (Tukey adjusted). (G) Paramsothy et al, 2017 cohort is shown (FMT patients n = 54; Δ based on final follow up time point). Boxplots present the median, 25th and 75th percentiles; **p < 0.01, Mann-Whitney test. (H) Receiver operating characteristics (ROC) curve of the Δ relative abundance shown in G. Prediction on Paramsothy cohort yielded an AUC of 0.8 (80% accuracy, sensitivity = 60%, specificity = 80%). Mann-Whitney test P-value is shown.
We next sought to determine a potential relationship between the CTM and clinical response to FMT. In our cohort and similar to data from recent randomized clinical trial (RCTs), the primary endpoint of clinical response was achieved in 35% of participants (defined as a Δ Mayo score ≥ 3 with a rectal bleeding score ≤1) by week 4 post-FMT. No significant differences were detected in the median relative abundance of the CTM when WK4 samples were compared between responders and non-responders (Figure 1D). Similarly, no significant correlation was observed between the increase in CTM relative abundance (Δ CTM) and decrease in Mayo score post-FMT (Δ Mayo, Supplementary Figure 1D).
To specifically identify the CTM associated with clinical response, Venn diagram analysis was performed by clinical response (responders, Figures 1E and non-responders, Supplementary Figure 1E). Twenty bacterial species were found to be part of the responders core transferable microbiota (RTM; Figure 1E). Of these 20 species, 12 were unique to responders (RTMunique, species marked in bold; Figure 1E), and therefore absent in the non-responders core transferable microbiota (NRTM, Supplementary Figure 1E). Strain-level analysis detected on average 25 RTMunique strains (SEM ± 4.1) in WK4 samples of which 51 % (SEM ± 4.38) were derived from donors and only 14.04% (SEM ± 7.59) from PRE (Figure 1F). This analysis highlights the transferability of a core set of bacterial strains that correlate with clinical efficacy of FMT for UC.
We next expanded our analysis to the entire cohort to evaluate whether the relative abundance of the RTMunique is associated with UC disease activity. Changes in RTMunique relative abundance (Δ RTMunique) were found to correlate linearly with Δ Mayo (Supplementary Figure 1F) and a tendency for Δ MES (P = 0.055, Supplementary Figure 1G). For internal validation and to test the ability of the Δ RTMunique in determining FMT clinical response, we generated ROC curves for AUC analysis using Δ RTMunique (Supplementary Figure 2A and 2B) as input to our FMT-response models. LOO-CV yielded an AUC of 0.87 (87% accuracy, sensitivity = 60%, specificity = 83%) and 100-by-Split-CV yielded an AUC of 0.82 (82% accuracy, sensitivity = 60%, specificity = 85%). To evaluate the reproducibility of these findings, delta relative abundance of the RTMunique taxa was determined in an independent study with 54 UC subjects receiving FMT therapy4 (Figure 1G and Supplementary Figure 2C). The Δ relative abundance of the RTMunique in remitters was significantly higher than non-remitters at all time-points evaluated (WK4, WK8 and > 8 weeks: final follow up). To test the generalizability of our predictive model, our original cohort was used in a model training for classifying clinical response of the validation cohort (Figure 1H). Δ RTMunique analysis robustly predicted clinical response in this independent cohort, which yielded an AUC of 0.8 (accuracy = 80%, sensitivity = 60%, specificity = 80%).
IgA-coating of RTM-associated Odoribacter correlates with clinical response
Given the transferability of strains associated with clinical response observed above, we next sought to determine their potential contribution to mucosal immunity by sorting and sequencing IgA-coated bacteria (called IgA-seq)13 (Supplementary Figure 3A). The proportion of fecal bacteria coated with IgA did not change post-FMT (Figure 2A) and did not differ between responders and non-responders (Supplementary Figure 3B); however, the IgA-coated community of WK4 post-FMT was more diverse than PRE (Shannon index, Supplementary Figure 3C) and higher in responders than non-responders (Shannon index, Supplementary Figure 3D). Beta diversity analysis, based on unweighted UniFrac, revealed that IgA-coated microbial community of FMT-recipients (WK4) shifted toward that observed in DON (Supplementary Figure 3E). To define the set of IgA-reactive bacteria acquired from donors, the Venn diagram approach was applied to the IgA-coated bacterial community using a patient prevalence cut off of ≥ 50% and taxa relative abundance cut off of ≥ 0.1%. Twenty-nine IgA-coated ASVs were found to be shared by DON and WK4 (Figure 2B). The median relative abundance post-FMT of these ASVs was found to be significantly higher in responders when compared to non-responders (Supplementary Figure 3F). Of these 29 ASVs, Bacteroides (5 ASVs), Coprococcus (2 ASVs), Eubacterium (1 ASV), Lachnospiraceae (4 ASVs), Odoribacter (1 ASV) and Ruminococcus (3 ASVs) were the only taxa to overlap with the RTM (Figure 1E and 2B).
Figure 2. IgA-seq identifies donor-derived Odoribacter as an immune reactive bacteria associated with clinical response to FMT.

(A) Boxplot comparing the median percentage of IgA-coated bacteria in fecal homogenate of DON, PRE and WK4 groups. Boxplots present the median, 25th and 75th percentiles. (B) Venn diagram depicting the number of IgA-coated bacterial taxa unique to a single variable (DON, PRE or WK4) or shared by two, or all three variables. Heatmap depicting the relative abundance of the 29 taxa defined by the Venn diagram as being part of the IgA-coated transferable microbiota. (C) Gnotobiotic mouse model experimental design and IgA-sequencing strategy are shown. Germ-free mice, 6–8 weeks old, received fecal transplant from a 2-donor FMP. Fecal homogenate was filtered, blocked, and stained with cell-permeable DNA dye Syto BC and anti-IgA antibody. Samples were gated on the basis of forward scatter (FSC) and side scatter (SSC). (D) IgA coating index (ICI) was calculated for genera found to be differentially abundant between IgA+ and IgA− microbial communities of the humanized mice. (E) Correlation between IgA-coated Odoribacter and Δ Mayo. Linear regression analysis revealed a significant correlation for Δ relative abundance of IgA-coated Odoribacter genus and Δ Mayo.
To confirm the transferability of these IgA-coated taxa in a model system of FMT, we transferred a 2-donor FMP into germ-free C57BL/6 mice, sorted and sequenced IgA-coated microbiota at week 7 post-transplantation (Figure 2C). Eleven donor-derived genera were differentially abundant in IgA-coated compared to non-coated bacteria (Supplementary Figure 4A), including species from the RTM belonging to Alistipes, Odoribacter and Ruminococcus genera (Figure 1E). Enrichment in the IgA-coated fraction was then assessed by IgA-coating index (ICI) analysis23 (Figure 2D). Of the donor-derived genera, only the relative abundance of Odoribacter at WK4 post-FMT (Supplementary Figure 4B) and its increase post-FMT (Δ Odoribacter, Figure 2E) was found to significantly correlate with decrease in Mayo score. Increase in Odoribacter post-FMT was also found in MES-responders but not in non-responders (Supplementary Figure 4C). Together these data identify a core transferable microbiota that shapes mucosal immunity and highlight the potential role for IgA-coated Odoribacter as a member of the RTM that associates with FMT clinical response.
While high affinity IgA coating of certain key pathobionts are T cell dependent (TD)24, IgA coating of gut commensals can be T cell independent (TI)24, 25. To evaluate the role for T cells in IgA-coating of the transferable bacteria, donor microbiota was transferred to germ-free TCR βδ−/− and het littermate control mice. Microbial engraftment of donor material was similarly achieved in both TCR βδ−/− and het mice (Supplementary Figure 5). RTM taxa enriched in IgA-coated fraction in het controls were similarly enriched in IgA-coated fraction of TCR βδ−/− mice (Figure 1E and 2D as well as Supplementary Figure 4A). These results suggest that the RTM induce recipient IgA responses in a TI manner.
O. splanchnicus induces colonic iTregs and limits DSS colitis
To evaluate the potential mechanistic role for O. splanchnicus in the clinical efficacy of FMT for UC, O. splanchnicus was isolated by cloning single cell, IgA-coated isolates from recipient samples. The complete genome of a patient-derived O. splanchnicus was assembled de novo into seven contigs (4,712,004 base pairs) and most closely aligned to the reference strain O. splanchnicus DSM (Supplementary Figure 6).
To test the impact of O. splanchnicus in colitis, wild-type (WT) germ-free mice were mono-colonized with patient-derived O. splanchnicus followed by 2% dextran sodium sulfate ad lib (Figure 3A). To evaluate the specificity of O. splanchnicus effects in colitis, germ-free mice were also colonized with recipient-derived isolates of either B. producta (non-IgA inducer) or A. finegoldii species (Alistipes sp. were found to be members of the CTM detected in both responders and non-responders; Figure 1E, 2C and 2E). Colonization was assessed on day 7 post-colonization (Supplementary Figure 7A). While WT germ-free and mice mono-colonized with either A. finegoldii or B. producta succumbed to DSS-induced colitis, O. splanchnicus colonization was sufficient to prevent significant weight loss and reduced fecal lipocalin (Figure 3A). These results show the specificity of O. splanchnicus, but not A. finegoldii nor B. producta, in protecting mono-colonized mice from the severity of DSS induced colitis.
Figure 3. O. splanchnicus induces iTreg and limits DSS colitis.

(A) Germ-free C57BL/6 WT mice were colonized with patient-derived A.finegoldii (Alistipes), B. producta (Blautia) and O. splanchnicus (Odori) for seven days and exposed to 2% DSS ad libitum for 6 days. Weight loss and levels of lipocalin in cecal contents are shown. Graphs show data from two independent experiments. Error bars represent SEM; ****p < 0.0001, ANOVA test. Boxplots present the median, 25th and 75th percentiles; *p < 0.05, **p < 0.01, Mann–Whitney test (Tukey adjusted). (B) Flow cytometry of live CD4+ T cells was used to evaluate RORγt and Foxp3 expression in colonic lamina propria 21 days post-colonization. Mean percentages of RORγt, RORγt+Foxp3+, or RORγt+Foxp3+/ RORγt+ CD4+ T cells per colon is shown in germ-free or mice mono-colonized with patient-derived bacteria. Data are from one representative out of four independent experiments. Error bars represent SEM; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, t test (Tukey adjusted). (C) Correlation between IgA-coated Odoribacter and Δ Foxp3. Linear regression analysis revealed a significant correlation for Δ IgA-coated Odoribacter genus and Δ Foxp3. Lamina propria mononuclear cells (LPMCs) were isolated from rectal endoscopic biopsies taken pre and 4 weeks after FMT. Flow cytometry of live, CD4+ T cells was used to evaluate Foxp3. (D) Germ-free C57BL/6 WT or RAG1−/− mice were colonized with patient-derived O. splanchnicus for seven days and exposed to 2% DSS ad libitum for 6 days. Weight loss and levels of lipocalin in cecal contents are shown. Graphs show data from two out of three independent experiments. Error bars represent SEM; ****p < 0.0001, anova test. Boxplots present the median, 25th and 75th percentiles; *p < 0.05, **p < 0.01, Mann–Whitney test (Tukey adjusted).
Next, we assessed the impact and specificity of O. splanchnicus isolate on intestinal immunity by mono-colonizing 6- to 8-week old WT germ-free mice with patient-derived O. splanchnicus, A. finegoldii or B. producta for 21 days (Figure 3B and Supplementary Figure 7B). Analysis of colonic CD4+ T cells revealed a significant induction of RORγt+ Foxp3+ by O. splanchnicus as well as an increased ratio of Foxp3+ RORγt+ to RORγt+ CD4+ T cells following 21 days of colonization. While B. producta induces RORγt+, but not Foxp3+ RORγt+, A. finegoldii induces Foxp3+ RORγt+ CD4+ T cells to equivalent levels of that observed in O. splanchnicus (Figure 3B).
We next evaluated if mucosal T cell responses from patients’ rectal biopsies correlated with the relative abundance of Odoribacter-IgA+. Foxp3+ CD4+ T cells positively correlated with both Odoribacter-IgA+ relative abundance on WK4 post-FMT (Supplementary Figure 8A), and also with increase in Odoribacter-IgA+ post-FMT (Δ Odoribacter, Figure 3C). No significant correlation was found for Δ relative abundance of Alistipes- or Blautia ASVs identified in IgA analysis (Supplementary Figure 8B). Additional analyses were done for Odoribacter, but no significant correlation was found for RORγt or IL-17 CD4+ T cells (Supplementary Figure 8C).
To test if lymphocytes played a critical role in O. splanchnicus mediated protection from colitis, WT and RAG1−/− germ-free mice were mono-colonized with O. splanchnicus for 7 days followed by 6 days exposure to 2% DSS. Despite equal colonization between WT and RAG1−/− mice (Supplementary Figure 9A), O. splanchnicus failed to prevent weight loss and gut inflammation as measured by fecal lipocalin in the absence of lymphocytes (Figure 3D).
IL-10 induced by O. splanchnicus is required to limit inflammatory Th17 cells and colitis
IL-10 is a key regulatory cytokine for intestinal homeostasis and regulatory T cell function. Mono-colonization of germ-free WT mice for 21 days with patient-derived O. splanchnicus was sufficient to induce il10 expression in colonic lamina propria cells and was significantly reduced in the absence of lymphocytes (Figure 4A). To assess the functional role for O. splanchnicus-induced IL-10 in regulating colitis, WT and IL10−/− mice were mono-colonized with patient-derived O. splanchnicus for 7 days followed by 6 days exposure to 2% DSS. In contrast to WT mice, O. splanchnicus failed to prevent weight loss and reduce lipocalin in IL10−/− mice (Figure 4B and C) despite equal colonization (Supplementary Figure 9B). Analysis of colonic lamina propria from IL10−/− mice also revealed an increase in single positive RORγt, IL-17+, and IFNγ+ CD4+ T cells and decrease in Foxp3+ T cells, as well as the ratio of Foxp3+ RORγt+ over RORγt+ CD4+ T cells compared to that observed in WT mice (Figure 4D and E). Together this data provides evidence that lymphocyte-dependent induction of IL-10 by O. splanchnicus restrains inflammatory T cells and is required for reducing the severity of DSS-induced colitis.
Figure 4. O. splanchnicus induces IL-10 to limit T cell inflammation and colitis.
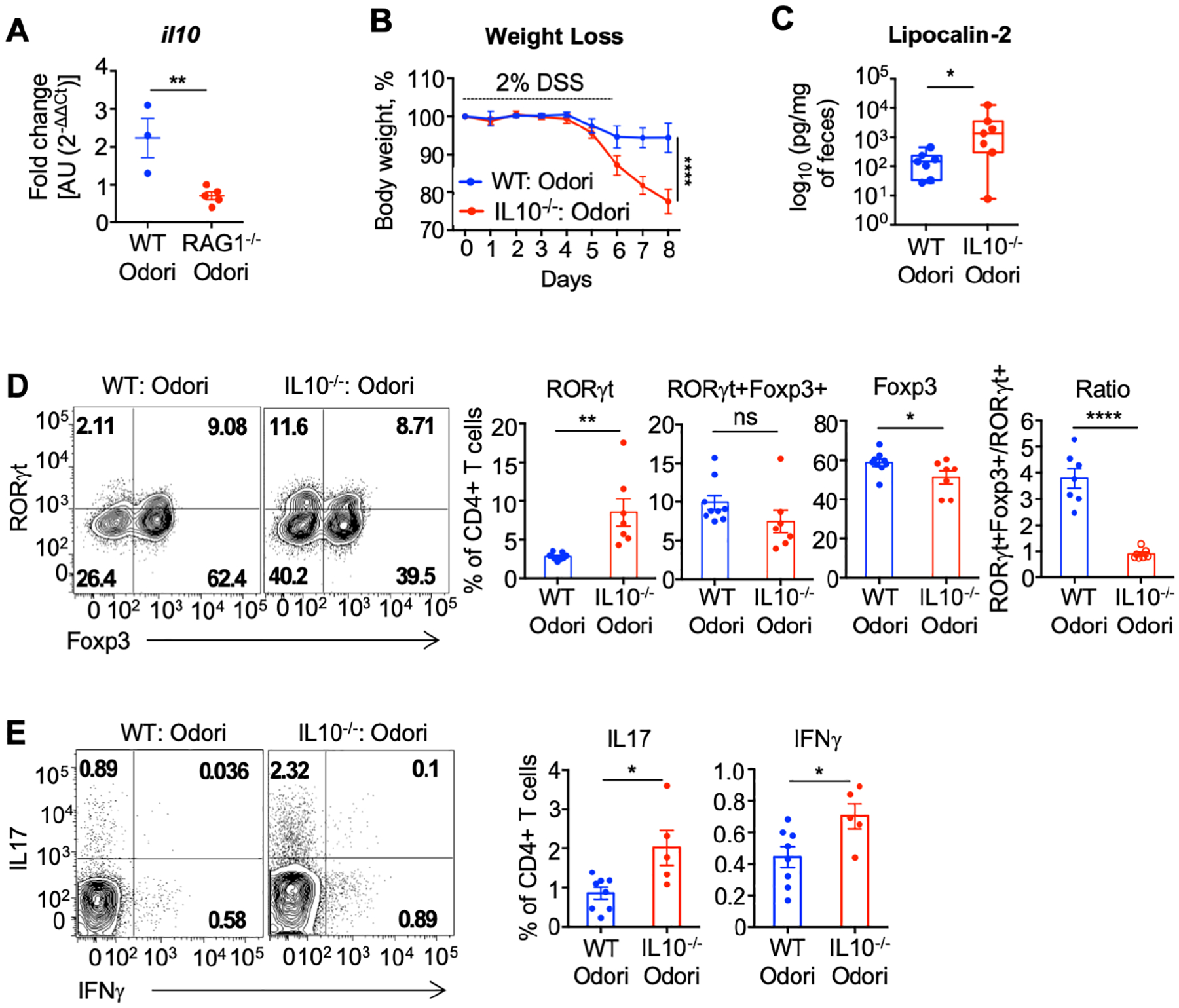
(A) Germ-free C57BL/6 mice were colonized with patient-derived O. splanchnicus and Il10 expression in the colonic lamina propria were measured 21 days after colonization. Graph shows data from one out of three independent experiments. Error bars represent the SEM. **p ≤ 0.01, t test. (B - E) Germ-free C57BL/6 WT or IL10−/− mice were colonized with patient-derived O. splanchnicus. Seven days post-colonization, mice were exposed to 2% DSS ad libitum for 6 days. Weight loss (B), and fecal levels of lipocalin (C) are shown. Flow cytometry of live CD4+ T cells was used to evaluate RORγt, Foxp3, RORγt+Foxp3+ (D) and IL17 as well as IFNγ (E) expression. Graphs show data from two independent experiments (WT, n = 8; IL10−/−, n =7). Error bars represent SEM; ****p < 0.0001, ANOVA test. Boxplots present the median, 25th and 75th percentiles; *p < 0.05, **p < 0.01, Mann–Whitney test. Bar plot error bars represent the SEM. *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001 t test.
SCFA production by O. splanchnicus protects against colitis
SCFAs are bacterial metabolic products known to play a critical role in host-intestinal immunity and function26, 27 and previous reports have highlighted O. splanchnicus as a significant SCFA-producer28, 29. To determine the metabolic function of our clinical isolates, germ-free mice were mono-colonized with patient-derived O. splanchnicus, A. finegoldii or B. producta for 10 days. As expected, a significantly higher abundance of acetate, butyrate and propionate was detected in cecal contents of O. splanchnicus mono-colonized mice compared to germ-free controls (Figure 5A). While B. producta mono-colonized mice had similar SCFA levels to that observed in germ-free mice, mono-colonization with A. finegoldii led to significantly higher production of acetate and lower butyrate and propionate compared to O. splanchnicus (Figure 5A).
Figure 5. SCFAs production by O. splanchnicus limits colitis.

(A) Germ-free C57BL/6 WT mice were colonized with patient-derived A.finegoldii (Alistipes), B. producta (Blautia) and O. splanchnicus (Odori), respectively. Acetate, butyrate and propionate levels in cecal contents were assessed 10 days post-colonization. Error bars represent SEM. *p < 0.05, ***p < 0.001, ****p < 0.0001, t test (Tukey adjusted). (B-E) SPF GPR43−/−, GPR109a−/− and het littermate controls were colonized with patient-derived O. splanchnicus and then exposed to 2% DSS ad libitum for 6 or 3 (GPR43−/−) days. Weight loss (B-D), percent survival (B-D) and lipocalin in the cecal contents (E) are shown. Graph shows data from one representative experiment (Het control, n = 10; Het Odori n = 7; GPR109a−/− control, n = 6; GPR109a−/− Odori, n = 6; GPR43−/− Odori, n = 8; GPR43−/− Odori, n = 7). Error bars in weight loss graphs represent SEM; ****p < 0.0001, repeated measures ANOVA test. Survival analysis; *p < 0.05, **p < 0.01, log-rank (Mantel-Cox) test. Boxplots present the median, 25th and 75th percentiles, Kruskal-Wallis test.
To evaluate if O. splanchnicus production of SCFAs contributes to its protective effect in colitis, we colonized mice deficient in G protein-coupled SCFA receptors 43 (GPR43−/−) and 109a (GPR109a−/−) as well as het littermate controls with O. splanchnicus for 14 days and subjected them to 2% DSS treatment (Figure 5B, 5C, 5D and 5E). O. splanchnicus colonization in the presence of a complex microbiota limited the severity of DSS-induced colitis in het mice compared to littermates not colonized with O. splanchnicus (Figure 5B and 5E). This protection was significantly reduced in GPR109a−/− mice and completely lost in GPR43−/− mice as demonstrated by reduced survival and higher lipocalin (Figure 5C, 5D and 5E). No differences in O. splanchnicus colonization were observed between GPR109a−/−, GPR43−/− and het littermate mice (Supplementary Figure 9C). Despite the GRP43 and GPR109 dependent protection (Figure 5C, 5D and 5E), SPF mice colonized with O. splanchnicus had similar levels of SCFAs in cecal contents compared to that of the littermate controls not colonized with O. splanchnicus in steady state (Supplementary Figure 9D). Collectively, these results support a role for both metabolic and immune cell impact of O. splanchnicus in reducing the severity of colitis.
Discussion
Although FMT is an emerging therapy for the treatment of UC, there remains a critical need to define the composition and mechanism of the transferable microbiota associated with clinical response to optimize safety and efficacy of this therapy. The results presented here provide the first strain level analysis of FMT in UC participants and define a transferable microbiota associated with clinical response. Using deep metagenomic sequencing and strain tracking algorithms, we define a core set of donor-derived strains transferred to the recipient. Although engraftment has been suggested to prevent C. difficile recurrence through a mechanism of colonization resistance7, 30, engraftment alone is not a robust predictor of clinical response in UC8. Consistent with this, the overall transferred microbiota did not distinguish responders from non-responders. However, analysis stratified by clinical response allowed us to identify a unique set of 12 donor-derived transferable bacterial species that correlates with clinical response and robustly predicts response in the largest RCT of FMT for UC to date4. Our work highlights the potential for a core set of transferable bacterial markers to serve as a prognostic biomarker that can be evaluated in future studies.
In addition to transferability, rational design of microbial therapy for UC should focus on microbes that engage the immune system and help to restore intestinal homeostasis. IgA-seq has been developed as a method to identify sentinel microbiota that interact with the mucosal immune system8, 23, 31, 32. Adding this approach to our analysis, we identified the core set of donor-derived taxa coated with IgA in the recipient. Of these microbes, O. splanchnicus is the only microbe within the responders core that correlates with clinical response and highlights the potential impact of this taxa seen in independent cohorts33, 34 as well as mouse models of colitis and colorectal cancer35. In addition to revealing immune recognition of these taxa, IgA coating of transferable microbial strains may promote durable engraftment as well as spatial location in a complex microbiota to enable immune cell activation36, 37.
Our findings reveal a mechanistic role for O. splanchnicus in resolving UC, working through both cellular and metabolic effectors in limiting colitis. IL-10 is a key regulatory pathway in IBD and our findings reveal a role for O. splanchnicus induction of IL-10 in promoting iTreg and restraining inflammatory T cells in the lamina propria. With the efficacy of biologic therapy targeting IL-23 and inflammatory Th17 cells for UC38, 39, these findings may offer a durable non-biologic approach for this pathway of disease. In addition to the IL-10- and lymphocyte-dependent regulation, our results define a key functional contribution of O. splanchnicus production of SCFAs acting through GPR43 and GPR109a receptors. The function of SCFAs can have a pleiotropic impact in promoting healing during colitis by acting directly on epithelial cells and by modulating regulatory T cell responses40, 41. Adding to this complexity, the production and consumption of SCFAs are likely regulated by local factors including the binding of IgA. These characteristics of O. splanchnicus along with other unique aspects including outer membrane vesicles42, bile acid metabolism34, and antigen specific immunity may account for the protective ability of O. splanchnicus in contrast to other commensals that also induce IL10 and SCFAs. Emerging studies have begun to elucidate how IgA binding may regulate the metabolic function of the gut microbiota43, 44. The impact of metabolic effectors of the transferable bacteria raises the possibility that dietary manipulation may potentiate the clinical efficacy of fecal microbiota transplantation for IBD as recently demonstrated in a clinical trial targeting subjects with metabolic dysfunction45.
Collectively, this work provides the first evidence of transferrable, donor-derived strains that correlate with clinical response to FMT in UC and reveals O. splanchnicus as a key component, which mechanistically promotes protection through both cellular and metabolic function. These mechanistic features will help enable desperately needed strategies to enhance therapeutic efficacy of microbial therapy for UC.
Supplementary Material
What you need to know.
BACKGROUND AND CONTEXT
Fecal microbiota transplantation (FMT) is a promising therapy for ulcerative colitis (UC), but little is known about the transferable bacterial strains and their relevance to clinical response.
NEW FINDINGS
Our study reveals a core set of donor-derived transferable bacterial strains associated with clinical response that was predictive of response in an independent cohort of UC patients receiving FMT. Odoribacter splanchnicus, a critical member of the responder transferable bacteria and enriched in the IgA-coated microbiota, promoted both metabolic and immune cell protection from colitis.
LIMITATIONS
The set of transferable bacteria was defined based on clinical response from a pilot cohort, but robustly predicted response in a larger independent cohort. While the results presented her define the role for lymphocytes, IL-10 and short chain fatty acids in mediating O. splanchnicus protection, additional features that confer mechanistic specificity still need to be defined.
IMPACT
These findings reveal new potential strategies for improving microbial therapy for UC.
Grant Support:
Boeringer Ingelheim, NIH R01DK114252-01A1 (R.S.L), Kenneth Rainin Foundation, and the Charina Foundation
Conflict of interest statement:
R.S.L. received grant support from Boeringer Ingelheim for this study. Y.G. is an employee of Finch Therapeutics. S-E.B. and J.H. are employees of Boeringer Ingelheim.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol 2015;12:720–7. [DOI] [PubMed] [Google Scholar]
- 2.Rossen NG, Fuentes S, van der Spek MJ, et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 2015;149:110–118 e4. [DOI] [PubMed] [Google Scholar]
- 3.Moayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled Trial. Gastroenterology 2015;149:102–109 e6. [DOI] [PubMed] [Google Scholar]
- 4.Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 2017. [DOI] [PubMed] [Google Scholar]
- 5.Kelly CR, Laine LA, Wu GD. Monitoring fecal microbiota transplantation practice in a rapidly evolving health and regulatory environment. Gastroenterology 2020;159:2004–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Gao X, Hu H, et al. Clinical Efficacy and microbiome changes following fecal microbiota transplantation in children with recurrent clostridium difficile infection. Front Microbiol 2018;9:2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. The New England journal of medicine 2013;368:407–15. [DOI] [PubMed] [Google Scholar]
- 8.Jacob V, Crawford C, Cohen-Mekelburg S, et al. Single delivery of high-diversity fecal microbiota preparation by colonoscopy is safe and effective in increasing microbial diversity in active ulcerative colitis. Inflamm Bowel Dis 2017;23:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duvallet C, Zellmer C, Panchal P, et al. Framework for rational donor selection in fecal microbiota transplant clinical trials. PLoS One 2019;14:e0222881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olesen SW, Gerardin Y. Re-Evaluating the evidence for faecal microbiota transplantation ‘super-donors’ in inflammatory bowel disease. J Crohns Colitis 2021;15:453–461. [DOI] [PubMed] [Google Scholar]
- 11.Smillie CS, Sauk J, Gevers D, et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 2018;23:229–240 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 2015;12:902–3. [DOI] [PubMed] [Google Scholar]
- 13.Viladomiu M, Kivolowitz C, Abdulhamid A, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 2017;27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grin I, Linke D. GCView: the genomic context viewer for protein homology searches. Nucleic Acids Res 2011;39:W353–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Yao D, Chen C. 2-Hydrazinoquinoline as a Derivatization Agent for LC-MS-Based Metabolomic Investigation of Diabetic Ketoacidosis. Metabolites 2013;3:993–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viladomiu M, Metz ML, Lima SF, et al. Adherent-invasive E. coli metabolism of propanediol in Crohn’s disease regulates phagocytes to drive intestinal inflammation. Cell Host Microbe 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong J, Liu C, Summanen P, et al. Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe 2011;17:64–8. [DOI] [PubMed] [Google Scholar]
- 22.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunker JJ, Flynn TM, Koval JC, et al. Innate and adaptive humoral responses Coat distinct commensal bacteria with immunoglobulin A. Immunity 2015;43:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pabst O New concepts in the generation and functions of IgA. Nat Rev Immunol 2012;12:821–32. [DOI] [PubMed] [Google Scholar]
- 26.Vinolo MA, Rodrigues HG, Nachbar RT, et al. Regulation of inflammation by short chain fatty acids. Nutrients 2011;3:858–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corrêa-Oliveira R, Fachi JL, Vieira A, et al. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology 2016;5:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014;5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Werner H, Rintelen G, Kunstek-Santos H. A new butyric acid-producing bacteroides species: B. splanchnicus n. sp. Zentralbl Bakteriol Orig A 1975;231:133–44. [PubMed] [Google Scholar]
- 30.Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 2014;146:1547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kau AL, Planer JD, Liu J, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 2015;7:276ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunker JJ, Bendelac A. IgA Responses to Microbiota. Immunity 2018;49:211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato Y, Atarashi K, Plichta DR, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 2021. [DOI] [PubMed] [Google Scholar]
- 35.Xing C, Wang M, Ajibade AA, et al. Microbiota regulate innate immune signaling and protective immunity against cancer. Cell Host Microbe 2021;29:959–974.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donaldson GP, Ladinsky MS, Yu KB, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018;360:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu ND, Crothers JW, Nguyen LT, et al. Dynamic colonization of microbes and their functions after fecal microbiota transplantation for inflammatory bowel disease. bioRxiv 2019:649384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almradi A, Hanzel J, Sedano R, et al. Clinical Trials of IL-12/IL-23 Inhibitors in Inflammatory Bowel Disease. BioDrugs 2020;34:713–721. [DOI] [PubMed] [Google Scholar]
- 39.Sands BE, Sandborn WJ, Panaccione R, et al. Ustekinumab as Induction and Maintenance Therapy for Ulcerative Colitis. N Engl J Med 2019;381:1201–1214. [DOI] [PubMed] [Google Scholar]
- 40.Sun M, Wu W, Liu Z, et al. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol 2017;52:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun M, Wu W, Chen L, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun 2018;9:3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiippala K, Barreto G, Burrello C, et al. Novel Odoribacter splanchnicus strain and its outer membrane vesicles exert immunoregulatory effects in vitro. Front Microbiol 2020;11:575455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joglekar P, Ding H, Canales-Herrerias P, et al. Intestinal IgA regulates expression of a fructan polysaccharide utilization locus in colonizing gut commensal Bacteroides thetaiotaomicron. mBio 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakajima A, Vogelzang A, Maruya M, et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med 2018;215:2019–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mocanu V, Zhang Z, Deehan EC, et al. Fecal microbial transplantation and fiber supplementation in patients with severe obesity and metabolic syndrome: a randomized double-blind, placebo-controlled phase 2 trial. Nat Med 2021;27:1272–1279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


