Abstract
Aberrant cell fate decisions due to transcriptional misregulation are central to malignant transformation. Histones are the major constituents of chromatin, and mutations in histone-encoding genes are increasingly recognized as drivers of oncogenic transformation. Mutations in linker histone H1 genes were recently identified as drivers of peripheral lymphoid malignancy. Loss of H1 in germinal center B-cells results in widespread chromatin decompaction, redistribution of core histone modifications, and reactivation of stem cell-specific transcriptional programs. This review explores how linker histones and mutations therein regulate chromatin structure, highlighting reciprocal relationships between epigenetic circuits, and discusses the emerging role of aberrant three-dimensional chromatin architecture in malignancy.
Chromatin, a complex of DNA polymer and associated proteins, is the physiological substrate of all genomic processes, including gene expression, damage repair, replication, and chromosome organization and segregation. Major constituents of chromatin are histone proteins, including four core subtypes, H2A, H2B, H3, and H4 that together with the associated DNA make up the nucleosome, and H1 linker histones - which associate with the nucleosome at or adjacent to the dyad axis, forming the “chromatosome” particle [reviewed in (1,2)]. H1 family members are larger than core histones, are extremely basic (lysine-rich), and contribute to higher-order chromatin organization, in part by electrostatic interaction with the negatively-charged DNA backbone (expanded upon below). Both core and linker histones are heavily modified by so-called “writer” and “eraser” enzymes, with the post-translational modification (PTM) landscape interpreted by the cognate “readers” [reviewed in (3,4)], or PTMs directly affecting charge-based interactions between proteins and DNA (5). Factors operating on chromatin have long counted among the most prevalent oncogenic drivers across many tumor types (6,7) thus representing a promising class of therapeutic targets (8). More recently, a number of mutations in core histone genes have been identified, initially in few specific tumors (9–12), and later broadly across multiple diverse malignancies (13–17). Mechanistically these mutants, aptly termed “oncohistones”, are thought to disrupt chromatin functions, either as direct and potent inhibitors of SET-domain methyltransferases in case of Lys-to-Met and related mutations (17–21) or as destabilizing agents when incorporated into the nucleosomes in context of the chromatin fiber polymer (13,14,22,23). The discovery of driver loss-of-function mutations in linker histone genes, highly prevalent in peripheral B-cell tumors (24–28), adds another oncogenic mechanism to the “oncohistone” repertoire.
Linker histones: distinct isoforms, common functions, diverse mutations.
In the human genome, linker histones are encoded by ten paralogous genes. Five of these (HIST1H1A, HIST1H1B, HIST1H1C, HIST1H1D and HIST1H1E, also annotated as H1–1, H1–5, H1–2, H1–3 and H1–4, respectively) are transcribed from HIST1 locus in a replication-dependent manner and constitute predominant isoforms in the cycling cells, two (H1FX and H1F0, also annotated as H1–10 and H1–0) are replication-independent and are characteristic of terminally differentiated cells, and remaining three (HIST1H1T, H1FNT, and H1FOO, also known as H1–6, H1–7 and H1–8) are gamete-specific. Two additional linker histone family pseudogenes (HILS1 and HIST1H1PS1, also known as H1–9P and H1–12P, respectively) do not encode functional proteins in humans (29), although in mouse, HILS1 protein is accumulated in elongating spermatids and may regulate chromatin condensation in absence of core histones (30). Despite their shared name, linker histones are not structurally related to core histones; they are larger (200–220 amino acids) proteins with characteristic tripartite structure: a short N-terminal domain, a central winged-helix globular domain (GD), and a long, lysine-rich C-terminal domain. The GD anchors H1 molecules at or near the nucleosome dyad axis, and is remarkably conserved across species. Several DNA-binding amino acids are distinct across isoforms and determine the positioning of the GD relative to the nucleosome core particle with implications for higher order chromatin compaction (31). Both N- and C-terminal tails are intrinsically disordered (32), although several conserved motifs within these regions may adopt specific conformations dependent on PTM status, protein partnerships, or chromatin association (33–35). Significant structural similarity between GD and winged-helix DNA binding domains of pioneer transcription factors was hypothesized to facilitate H1 loading into condensed chromatin, yet unlike pioneer factors, linker histones bind DNA indiscriminately (36). Linker histone association with DNA, in turn, facilitates chromatin condensation and has been convincingly linked to transcriptional repression both in vitro and in vivo (37,38). Several regions in the H1 GD and C-terminal tails appear to function cooperatively; a model has been proposed wherein the C-terminal tail facilitates initial association, and positions the GD for stable binding at the nucleosome dyad axis (39,40). Importantly, unlike core histones, which are stably incorporated into chromatin in context of the nucleosome and require significant energy expenditure by specialized factors for remodeling, eviction and exchange (41–43), association of linker histones with the chromatin fiber is dynamic, with in vivo residence time an order of magnitude shorter than that of core histones (27,44,45). Mobility of H1 proteins is thus uniquely suited to facilitate transient and developmentally regulated condensation of facultative heterochromatin, and emerging evidence support this function of linker histones.
Distinct H1 isoforms demonstrate high cross-species conservation, suggesting selective pressure to retain poorly appreciated isoform-specific functions (46). This complexity is supported by biochemical and structural studies suggesting that specific isoforms have distinct capacity for chromatin condensation (31,47), observations of unique interactors for mouse isoforms H1d and H1e (48), and genetic studies, wherein heterozygous H1c/e+/− mouse germinal center (GC) B-cells demonstrated a proliferative advantage over homozygous H1c/e−/−, therefore suggesting that specific functions of H1c or H1e may be important in GC B-cells (27). Additionally, several isoform-specific post-translational modifications have been reported in transcriptional regulation, including activation (49,50) and repression (51,52). On the other hand, extensive rescue experiments demonstrated that functions of replication-dependent linker histones in mouse are largely interchangeable (53,54), and localization studies found overlapping binding profiles of distinct H1 isoforms genome-wide (55,56). Together, these results suggest that while key functions of chromatin condensation and transcriptional repression are shared across isoforms, subtle differences between H1 isoforms exist and may be highlighted in specific developmental or pathological contexts (expanded upon below).
Missense mutations in both GD and C-terminal domain can drastically reduce the H1 capacity for chromatin association or compaction. While initially implemented as theoretical tools to understand the mechanistic basis of H1-nucleosome association (40,57), many of these and similar mutations have been reported recently across multiple cancer types. Prevalence is particularly high among mature B-cell neoplasms such as follicular lymphoma, Hodgkin lymphoma, and diffuse large B-cell lymphoma, all of which derive from GC B-cells (24–27,58). Interestingly, the mutation landscape of linker histones does not neatly conform to the emerging core “oncohistone” dichotomy, wherein stereotypical gain-of-function substitutions at specific residues - such as classical K27M and K36M - occur at high rates in specific tumor types, while more broadly distributed loss-of-function mutations, often thought to destabilize core nucleosome structure, are found, at lower rates, across many cancers (15,22,59–62) (Table 1). Linker histone mutations are nevertheless broadly distributed and affect all replication-dependent H1 isoforms, with the exception of H1A, which is expressed at low levels in mature B-cells (27,63). Therefore, as genetic, biochemical, and in vivo data agree that H1 mutations are loss-of-function, the cell-type specificity strongly implicates that GC B-cells are uniquely dependent on chromatin condensation by H1, adding to the diverse repertoire of epigenetic drivers of mature B-cell neoplasms.
Table 1.
Major classes of histone mutations in cancer
| Histone | Aberration | Proximal mechanism* | Cancer type | References |
|---|---|---|---|---|
|
| ||||
| H3 | K27M | Strong inhibition of PRC2 methyltransferase activity in trans | Pediatric midline gliomas, AML (infrequent) | (9–11,16,18) |
| K36M | Strong inhibition of H3 K36-specific methyltransferases in trans | Chondroblastoma, undifferentiated sarcoma, chondrosarcoma, head and neck squamous cell carcinoma (subset) | (12,17,19) | |
| H3.3 | G34R/V/W/L | Inhibition of H3 K36-specific methyltransferases in cis | Pediatric glioma (predominantly R/V), osteosarcoma, giant cell tumor of the bone (predominantly W/L) | (12,20,21) |
| H3 | Diverse missense | Unknown, possibly nucleosome destabilization | Many cancers | (14,15,22) |
| H4 | Diverse missense | Unknown, possibly nucleosome destabilization | Many cancers | (15,22) |
| H4G | Overexpression | Unknown | Breast cancer | (23) |
| H2A | Diverse missense | Unknown, possibly nucleosome destabilization | Many cancers, including diffuse large B-cell lymphoma (C4) | (15,22,58) |
| H2A.B | Overexpression | Incorporation may result in nucleosome destabilization | Many cancers, high rate in diffuse large B-cell lymphoma | (78) |
| H2B | E76K and other globular domain missense | Likely nucleosome destabilization | Many cancers, including diffuse large B-cell lymphoma (C4) | (13–15,58) |
| H1B, H1C, H1D, H1E | Diverse missense | Loss of nucleosome association and/or reduced capacity for chromatin compaction | Diffuse large B-cell lymphoma, Hodgkin lymphoma, follicular lymphoma | (24–27,58) |
| H1B | Increased expression | Unclear, may be a marker of proliferative activity | Neuroendocrine tumors and prostate cancer | (59,60) |
| H1.X | Increased expression | Unclear, may be a marker of proliferative activity | Neuroendocrine tumors | (61) |
| H1.0 | loss/reduced expression | Chromatin decompaction, increased self-renewal and tumor heterogeneity | Many cancers, including glioblastoma and breast cancer | (62) |
| Increased expression | Unknown | Lung squamous cell carcinoma (subset) | (120) | |
Simplified, refer to cited works for detailed discussion
Epigenetic misregulation in B-cell malignancy: an extra dimension of complexity.
The extent and impact of epigenetic misregulation in GC B-cell tumors has been extensively reviewed previously (64–66). B-cells undergoing the GC reaction experience a period of critical vulnerability characterized by rapid clonal proliferation, suppression of checkpoint surveillance (67–69), somatic hypermutation by activation-induced cytidine deaminase (AICDA) (70,71), metabolic reprogramming (72,73), and downregulation of terminal differentiation programs (74). Importantly, these canonical “cancer hallmarks”, resulting in rapid diversification and proliferation of B-cells in response to antigen stimulation, are reversibly regulated by epigenetic mechanisms (64,65). It is therefore not surprising that mutations implicated in oncogenic transformation in GC B-cells often arise in epigenetic regulators, such as chromatin modifiers, transcription factors, and histone proteins themselves.
The cell of origin classification of DLBCLs distinguishes two major classes, the GC B-cell-like (GCB) derived from GC B-cells, and the more aggressive activated B-cell-like (ABC) subtype, originating from B-cells that completed the GC transit. Two recent studies developed more detailed classifications based on genetic signatures, yielding roughly similar DLBCL subtypes. One of these two approaches classified DLBCLs into (i) MCD: characterized by MYD88 L265P and CD79B mutations, (ii) BN2: carrying BCL6 structural variants, NOTCH2 pathway mutations, and aberrant NF-κB signaling, (iii) N1: mutations in NOTCH1, (iv) A53: TP53 mutations, (v) ST2: mutations in SGK1, TET2 and others, and (vi) EZB: mutations in epigenetic regulators including EZH2, and BCL2 structural variants (with a subset of these also containing MYC translocations and associated with poor clinical outcomes) (75,76). Although H1 mutations are common across most of these subsets, the highest prevalence was observed among the MCD-DLBCLs, which are mostly overlapping with the ABC-DLBCLs (27). Strikingly, even among ABC-DLBCLs, those with H1 mutations experienced inferior clinical outcomes (26). The second study defined five distinct coordinate genetic signatures, including C1 (BCL6 fusions, NOTCH2 pathway alterations, generally corresponding to BN2), C2 (characterized by loss of checkpoints like TP53, CDKN2A, and RB1, similar to A53), C3 (BCL2-IgH translocations, and mutations in chromatin modifiers including EZH2, KMT2D, and CREBBP, similar to EZB), C4 (SGK1 mutations, and frequent linker and core histone gene mutations), and C5 (BCL2 gain, MYD88 L265P and CD79B mutations - closely resembling MCD class) (58). In this study, cluster C4 tumors were predominantly classified as GCB-type. This apparent discrepancy likely stems from several factors, including potentially non-uniform effects of distinct HIST1H1 mutations, absence of matched non-tumor controls in many samples across both studies, and non-exclusive occurrence of histone mutations across all tumor classes, an observation consistent with previous reports that H1 mutations occur early in B-cell neoplastic transformation (77). Interestingly, cluster C4 tumors were further characterized by increased frequency of mutations in genes encoding core histones H2A and H2B - implicated in nucleosome stability (13,14,22). Together with a recent report that short H2A isoforms, normally restricted to gametogenesis and recapitulating many aspects of H2A “oncohistones”, are upregulated across several cancers, with highest enrichment in DLBCLs (78), these observations suggest that these tumors are particularly sensitive to compromised organization of chromatin fiber (Table 1).
Emerging evidence support a critical role for genome topology in the rapid phenotypic transitions experienced by GC B-cells. Genome-wide chromatin conformation capture studies using Hi-C showed that B-cells undergo extensive reorganization of 3D chromatin architecture as they transition from quiescent naïve B-cells into GC B-cells. For example, GC B-cells undergo significant transitioning of compacted compartment B, into the more open and transcriptionally active compartment A (79). Consistent with this decompaction, GC B-cells also feature merging of boundary-delimited gene domains into larger conglomerates of interacting genes, broad gains in enhancer interactivity, and the formation of intergenic architectural interaction hotspots, at least one of which was essential for formation of GC B-cells and survival of DLBCL cells (79,80). Likewise, exit from the GC to form plasma cells involves extensive architectural remodeling involving massive cohesin-dependent changes in chromatin boundaries and enhancer interactivity (81). Accordingly, haploinsufficiency of cohesin subunit SMC3 caused GC B-cell hyperplasia and acceleration of BCL6-driven lymphomagenesis due to blockade of plasma cell formation, through failure of timely establishment of enhancer-promoter contacts at key differentiation genes (81). Another recent report defined an intermediate chromosomal compartment that featured Polycomb-dependent H3K27 trimethylation, that underwent broad and reversible expansion in GC B-cells (82). While this temporally poised chromatin generally reverts to B compartment upon further differentiation in wild type cells (82), GC B-specific linker histone loss resulted in stable expansion of compartment A at the expense of shrinking H3K27me3-demarcated domains (27,83) Together, these data suggest that unique topological reorganization of metastable GCB chromatin represents significant vulnerability in B-cell differentiation trajectory.
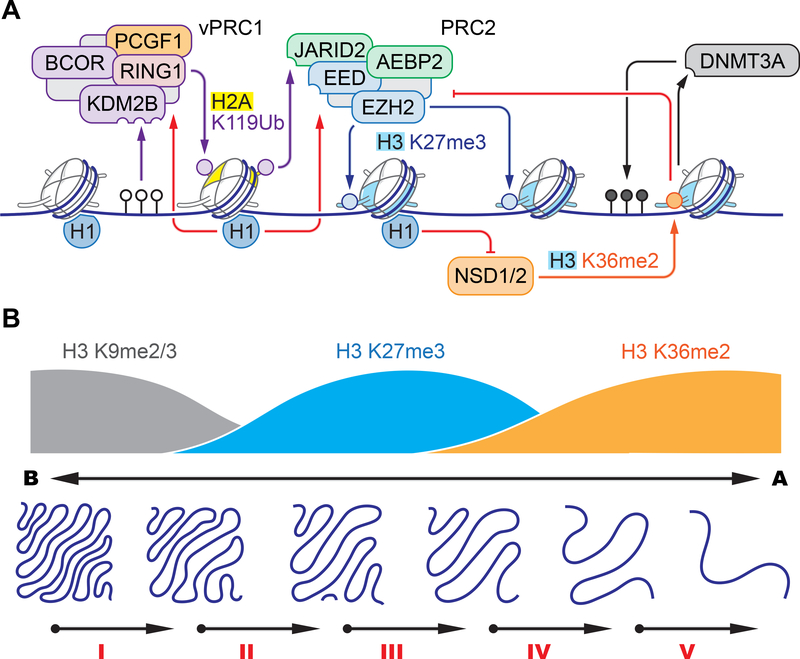
Several features may make GC B-cells particularly sensitive to mutations in linker histone genes. First, only three of seven somatic H1 isoforms account for over 90 per cent of total H1 in lymphoid cells, potentially reducing the overall fault tolerance of the system (63). Second, B-cells transiting the GC reaction employ non-canonical Polycomb repressive complex 1 (PRC1), downregulating BMI1 (PCGF4) and PHC1–3, and upregulating BCOR, PCGF1, and KDM2B subunits associated with vPRC1 (PRC1.1 variant) (83,84). A stepwise model of mutual reinforcement between Polycomb group proteins postulates a central role of H2A K119 ubiquitylation established by canonical PRC1 for subsequent recruitment of PRC2 and H3 K27 methylation (85,86) (Figure 1A). As PRC2 function is additionally stimulated by H1 incorporation in vitro (63,87), it is intriguing to hypothesize that GC B-cells may be uniquely sensitized to H1 mutations in a background lacking sufficient canonical PRC1. Interestingly, a recent study demonstrated that H1-dependent chromatin compaction stimulates propagation of H2A K119Ub by variant PRC1 complexes (88). Further, the potential importance of PRC1 subunit stoichiometry in GC B-cell biology is highlighted by related observations that overexpression of BMI1 or CBX7, occurring frequently in B-cell malignancies, result in ectopic repression of key tumor suppressor genes and is a poor prognostic marker in lymphomas (89,90). Of note, intact and dynamically regulated function of PRC2 complex is necessary for GC formation, and has been linked to repression of cell cycle checkpoint and differentiation genes, including cyclin-dependent kinase inhibitor CDKN1A, in proliferating centroblasts (91). EZH2, the enzymatic subunit of PRC2, is upregulated in centroblasts, and downregulated as B-cells exit the GC reaction (92), highlighting the key reliance of GC B-cell development on epigenetic regulation. Finally, GC reaction is accompanied by reduction in Lamin B1, a nuclear lamina component implicated in organization of repressive chromatin at the nuclear periphery (93). Together we propose that relative downregulation of these repressive and architectural mechanisms is responsible for the unique dependency of GC B-cells on normal function of linker histones.
Figure 1. Mechanisms of linker histone H1 function in chromatin and effects of H1 loss.
A, H1 histone fine-tunes Polycomb and NSD1/2 activities in chromatin. Variant PRC1 (vPRC1) complex, recruited to unmethylated CpG islands by KDM2B subunit, establishes core histone H2A K119Ub, in turn recruiting PRC2 complex, responsible for H3 K27 methylation. H1 incorporation stimulates both vPRC1 and PRC2, and opposes the function of NSD1/2 enzymes, which di-methylate H3 K36 - a modification implicated in recruitment of DNMT3A DNA methyltransferase to broad intergenic regions, and direct inhibition of PRC2 function. Subunit composition of protein complexes is simplified for accessibility, open and filled lollipops indicate unmethylated and methylated CpG sequences, respectively. B, Linker histones integrate chromatin compaction and core histone modifications. Top, schematic distribution of three H3 tail modifications (K9me2/3, K27me3 and K36me2) relative to genome A/B compartment score, with K9me occupying most compact regions, K27me3 found in intermediate regions, and K36me2 demarcating open and highly interactive compartment A. Bottom, while loss of linker histone function universally leads to B-to-A shift, both the degree of decompaction and the trajectories of core histone modifications are distinct and fall into five specific clusters. See text for details.
Chromatin architecture and histone H3 methylations: it takes (more than) two to tango.
The emergence of “intermediate” I-compartment as key dynamic feature of GC B-cell chromatin (82), the observations that A- and I-compartments are reciprocally demarcated by H3K36me2 and H3K27me3 (27,63,82), and the reduction of compartment B chromatin coincident with reorganization of core histone methylations upon linker histone loss in lymphoma (27) together point to mutually interdependent relationship between core histone modifications, genome architecture, and developmental trajectory. The issue of directionality of dependencies is key in chromatin biology - whether epigenetic modifications are instructive to, or merely correlate with transcriptional programs (94,95). While the prevailing body of evidence points to a causative relationship between chromatin modifications and transcriptional state in eukaryotes (96–98), the role of chromatin architecture remains controversial (99,100). To this end, GC B-cells, lacking several compensatory redundancies, provide a mechanistically promising and clinically relevant model to decipher these processes in vivo.
Regulation of histone H3 lysine 27 methylation by PRC2
Loss of linker histone in GC B-cells predominantly resulted in transcriptional de-repression of genes silenced by PRC2 during hematopoietic cell differentiation. H3K27 trimethylation was likewise reduced, while H3K36 mono- and dimethylation were upregulated (27,63). H3K27me is generally associated with transcriptional repression in developmentally regulated “facultative heterochromatin”, and is established by a PRC2 complex, containing core Enhancer of zeste homolog 1/2 (EZH1/2) methyltransferase subunit, Embryonic ectoderm development (EED) subunit carrying a K27 methylation-sensitive pocket important for allosteric activation and propagation of PRC2 on chromatin fiber substrate (101), and Suppressor of zeste 12 (SUZ12) and Retinoblastoma-binding protein p46/p48 (RBBP4/7) implicated in structural integrity and substrate specificity of the complex (102). Activation of PRC2 involves two additional subunits, including Jmj/ARID domain containing protein 2 (JARID2) (102,103), and Adipocyte enhancer binding protein 2 (AEBP2) (104). JARID2 carries a conserved Arg-Lys (RK) motif in its N-terminal part; when methylated at K116 by EZH2 it binds EED, mimicking H3 K27 methylated substrate for allosteric activation of PRC2 (103). AEBP2, in turn, appears to facilitate PRC2 activity by enhanced nucleosome binding, independent of allosteric activation (105). Recent structural studies further suggest that both JARID2 and AEBP2 interact with H2A K119Ub, a modification installed by PRC1 complex, to stimulate PRC2 function (106) (Figure 1A). Together, these studies highlight the complexity of PRC2 regulation and implicate allosteric effects, nucleosome spacing, and nucleosome affinity as key factors affecting H3 K27me levels. While the specific mechanisms of PRC2 regulation affected by H1 loss in vivo are not known, allosteric regulation of PRC2 is an emerging field in cancer therapy and may be of particular interest in lymphoma (107). Along these lines it is notable that H1 deficiency did not affect GC -specific facultative repression by EZH2 through bivalent chromatin at gene promoters, and instead mostly affected sets of PRC2 targets that are stably silenced through H3K27me3 during earlier stages of development (27).
Histone H3 K36 dimethylation is reciprocal to PRC2-dependent H3 K27me
A reciprocal modification, H3 lysine 36 dimethylation (K36me2), shows significant increase upon H1 loss (27,63). The landscape of H3K36me2 writers is complex and includes a family of related Nuclear receptor SET Domain (NSD) enzymes NSD1, NSD2 and NSD3, and Absent, small, or homeotic discs 1-like (ASH1L) enzyme [reviewed in (108)]. Importantly, H3 K36me3, a distinct lysine 36 modification produced by a separate enzymatic system (SETD2) within gene bodies (109) is not affected by H1 loss (27). Biochemical and genome-wide studies have demonstrated that H3K36me inhibits PRC2 function locally on cognate nucleosomes in vitro (110). Several lines of evidence support extensive biological connection between K27 and K36 methylation systems. First, broad increase of H3K36me2 is associated with shrinking of H3K27me-demarkated domains in multiple myeloma (111). Second, reduction of H3K36me2, either due to H3K36M oncohistone incorporation (19,20), or NSD enzyme loss (112), coincides with reciprocal expansion of H3K27me (Figure 1A). Third, H3 K36me2 expansion in PRC2-deficient diffuse intrinsic pontine glioma was recently described as a potential tumor vulnerability, demonstrating a therapeutically relevant relationship between H3K27 and K36 methylation (113). Interestingly, NSD2 is both the highest expressed H3K36 di-methyltransferase in GC B-cells (27) and the only di-methyltransferase that carries distinct High Mobility Group (HMG) domain (114) - bearing similarity to HMG family proteins, which compete with H1 for association with chromatin substrate (115). It is intriguing to speculate that in vivo, NSD2 may rely on HMG domain to compete with H1 for the nucleosome substrate, with reduced H1 dose resulting in expansion of H3 K36me2. Additionally, chromatin decompaction upon H1 loss may result in increased accessibility and occupancy of transcription factor binding sites - including GATA6, which was recently shown to directly recruit NSD2 to chromatin (116), and OCT2, which mediates key GC-specific architectural functions, and the motifs for which are enriched at key regulatory elements that acquire H3K36me2 in H1-deficient GC B-cells (63,80). NSD2 overexpression in a multiple myeloma model, driving excess K36me2, results in broad chromatin decompaction (117), and activating mutations in NSD2 methyltransferase domain have been reported in several hematopoietic malignancies, including B-cell acute lymphoblastic leukemia (B-ALL), chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (118). Interestingly, gene sets upregulated by NSD2 mutation in B-ALL overlap significantly with genes derepressed in mouse H1c/e−/− GC B-cells, suggesting a common mechanism may underlie these distinct malignancies (27). Further, a recent study implicated the related methyltransferase NSD3 as an oncogene in a non-lymphoid malignancy, lung squamous cell carcinoma (LUSC), with increased levels of H3K36me2 driving cancer progression (119). Of note, a non-overlapping subset of LUSC samples is characterized by an amplification encompassing H1F0 gene, resulting in overexpression of a replication-independent H1 variant (120), a relatively poor compactor with distinct chromatosome association (31,47). Whether such overexpression can effectively displace replication-dependent H1 variants, effectively employing a dominant negative mechanism, remains to be tested. We predict that, in the future, causative mutations in genes encoding linker histones will be identified across other tumors, specifically ones with known dependencies on the H3 K27/K36 methylation circuit.
The genomic function of H3K36 dimethylation is unclear. In vertebrates, de novo methyltransferase DNMT3A PWWP domain acts as a “reader” of H3K36me2, directing intergenic DNA methylation (121). Interestingly, deficiency of either the TET2 dioxygenase, involved in DNA hydroxymethylation and cytosine demethylation, or the DNMT3A methyltransferase may lead to GC hyperplasia, a pre-malignant phenotype also observed in H1 deficient B-cells (26,122,123). In contrast, NSD2 deficiency results in impaired GC formation (124). It is hence intriguing to speculate that H3K36me2 expansion in HIST1H1 mutant lymphomas has a broad effect on intergenic DNA methylation, with consequences ranging from potential alterations in spatial organization of chromosomal territories (125) to local variations in nucleosome positioning and sequence-specific transcription factor association (126,127). Together, these observations point to an emerging role of broad intergenic domains and distal elements in transcriptional regulation and cancer biology.
The missing link(er): effects of H1 loss on chromatin structure and function
H1 incorporation has distinct effects on specific chromatin regulatory systems. On one hand, in vitro studies in single chromatosome particle demonstrated that H1 association repositions the histone H3 tail in a spatially inaccessible conformation and restricts the activity of “writer” complexes (128). This is consistent with genome-wide localization of H1 to gene-poor, largely inactive domains (55,129) and observations that H1-compacted chromatin does not preclude coactivator binding but remains resistant to histone acetyltransferase activity until H1 is evicted (130). However, it does not address how repressive PTMs are successfully propagated across H1-compacted fibers. To this end, biochemical studies in di- and oligonucleosomal templates demonstrated that H1 incorporation both promotes PRC2-dependent H3 lysine 27 methylation, and inhibits NSD2-dependent lysine 36 dimethylation (63,87). Recent advances in cryo-EM provide mechanistic basis for these findings, as PRC2 appears to be sensitive to linker DNA length as it engages two neighboring nucleosomes (101). Likewise, as NSD association results in unwrapping of nucleosomal DNA from entry-exit site (131), and H1 incorporation reduces linker DNA flexibility adjacent to the nucleosome dyad axis (31), linker histones may act as direct competitive inhibitors of NSD enzymes in chromatin. Additionally, PRC2 may reach out to non-adjacent nucleosomes in context of a chromatin fiber (132); as H1 incorporation facilitates nucleosome contacts both in cis and in trans (133), it is likely that H1-condensed chromatin would be more amenable to processive H3K27 methylation. Further, H1 may directly facilitate positioning of H2A tail for accessibility by PRC1, as the C-terminal tail of H2A interacts with H1 (31,134) and variant PRC1 complexes exhibit preference for H1-compacted chromatin substrate (88). Observations of frequent ectopic expression of potentially oncogenic short H2A isoforms lacking the C-terminal tail in lymphomas (78) further suggest that the H1-PRC1 axis may represent an additional oncogenic vulnerability on GC B-cells. Finally, chromatin effects of H1 loss may be indirectly mediated by additional factors, as SMARCA2 was recently identified as interactor of NSD2 (135). Whether K36me2 expansion drives increased chromatin accessibility by direct recruitment of chromatin remodeler remains to be tested, yet interestingly, several subunits of the BAF complex are mutated at a high rate across GC B-cell neoplasms (136). While the mechanistic effects of these mutations remain to be elucidated, these results further implicate alterations of chromatin fiber fidelity in B-cell malignancy.
Integrative analyses of chromatin compartment trajectories and core histone modifications in H1 knock-out GC B-cells demonstrate distinct effects of H1 loss on differentially compacted genomic regions (27). While H1 loss resulted in overall chromatin decompaction manifested as global B-to-A compartment shift, hierarchical clustering genomic regions based on mapped core histone modifications revealed five distinct classes, strongly associated with transcriptional effects of decompaction upon H1 loss (Figure 1B). (I) compartment B regions that undergo decompaction but remain in B upon H1 loss, characterized by very low levels of either H3 K27me3 or H3 K36me2; (II) regions that gain modest levels of H3 K27me3 or H3 K36me2, which start in compartment B and transition into intermediate compartment upon H1 loss; (III) regions that start in intermediate compartment and gain moderate levels of H3 K36me2 as they transition into compartment A; (IV) regions that start in compartment A and decompact further, gaining significant H3 K36me2 levels; and (V) compartment A regions that demonstrate most significant decompaction, coincident with dramatic loss of H3 K27me3 (27). Together, these observations support the notion that activity of distinct core histone modifiers is optimal at specific chromatin compaction states. Further, as genes upregulated upon H1 loss were located almost exclusively within regions III-V - associated with significant gain of H3 K36me2 or loss of H3 K27me3 - it is likely that changes in core histone modifications rather than decompaction per se mediate transcriptional effect of H1 loss. Meanwhile, several non-exclusive mechanisms may explain how most compacted compartment B regions remain largely resistant to H1 loss. First, in addition to linker histone, heterochromatin is compacted by H3 K9me “reader” HP1, providing a redundant mechanism possibly compensating for the local loss of H1. Second, linker histone levels are decreased but not abolished in either mouse knock-out model or lymphoma patients, and as heterochromatin is generally compartmentalized within the nucleus, it is conceivable that a residual population of H1 is preferentially retained in these dense nuclear territories. These possibilities are supported by distinct dynamics of H1 in heterochromatin and euchromatin compartments (44) and observations that H1 is critical for spatial organization of nucleosome nanodomains in euchromatin but is dispensable for silencing of heterochromatin-associated transposable elements in Arabidopsis (137). Together, these observations underscore the complex relationship between chromatin structure, core histone modification landscape, and gene activity, highlighting the need for cross-disciplinary approaches in future studies.
Concluding remarks
The dynamic nature of H1 association with chromatin fibers is a defining property of linker histone biology (44). Where core histones are incorporated in context of nucleosome particles, allowing for post-translational modifications and variants to stably demarcate specific genomic regions (138), linker histones are highly mobile, with residence time in chromatin on the order of minutes (27,44), and thus are well-suited for rapid and reversible chromatin condensation and gene repression. While significant redundancy compensates for partial linker histone loss under normal developmental conditions (53), the extreme environment comprised of rapid cell cycles coupled with reduced activity of canonical repressors during the GC reaction highlight the dependency of peripheral B-cells on a full repertoire of linker histones.
Emerging evidence that many nuclear processes employ intrinsically disordered proteins for compartmentalization of biological activities by liquid-liquid phase separation (LLPS) [reviewed in (139,140)] place linker histones center stage. While the role of LLPS in active expression remains a subject of debate (141), condensation of chromatin fibers is a defining feature of transcriptional repression (142). Recent studies implicate HP1 proteins in sequestering heterochromatin via LLPS (143–145), and provide structural insights into how HP1α binding reshapes core nucleosome particles to facilitate phase transition (146). Of note, developmental phenotypes of HP1α loss in mouse appear limited to lymphoid lineage (147). Importantly, while heterochromatin provides robust and visually striking example of repression by condensation, subtle transitions at the intermediate compartment, corresponding to PRC2-repressed genes, are understood in much less detail. We envision that linker histone association provides both a robust and dynamic means of chromatin condensation. This is supported by both recent and long-standing observations that linker histones may self-associate, a hallmark of LLPS processes (148,149). As recent studies implicate C0T-1 RNAs in euchromatic chromosome organization (150), and linker histones readily associate with single-stranded nucleic acids (151), complete understanding of chromatin structure will require a synthesis of many such lines of evidence. While the consequences of specific tumor-associated missense mutations in linker histones are yet to be investigated, we expect that mechanistic understanding of these processes at mesoscale will emerge as the next great frontier in both fundamental cell biology and cancer research.
Acknowledgements
We thank members of the Allis, Cesarman, and Melnick laboratories for discussions during the preparation of the manuscript and apologize to colleagues whose work could not be cited due to space constraints. Collaborative work in the Allis, Cesarman and Melnick laboratories is funded through NIH/NCI R01 CA234561, LLS SCOR 17403-19 and STARR I9-A9-062. Allis laboratory is also supported by the NCI P01 CA196539, Leukemia and Lymphoma Society (LLS-SCOR 7006-13), The Rockefeller University and St Jude Children’s Research Hospital Collaborative on Chromatin Regulation in Pediatric Cancer, and Robertson Therapeutic Development Fund. A.M.M. is also funded by NIH/NCI P01 CA229086, NIH/NCI R35 CA220499, LLS TRP 6572, LLS SCOR 7012, the Follicular Lymphoma Consortium, the Samuel Waxman Cancer Research Foundation, and the Chemotherapy Foundation. A.A.S. was funded by the Damon Runyon Cancer Research Foundation (DRG-2185-14).
Authors’ Disclosures
A.M.M. has research funding from Janssen Pharmaceuticals, Epizyme, Sanofi and Daiichi Sankyo, is on the advisory board for KDAC Pharma, has consulted for Astra Zeneca, Daiichi Sankyo, Epizyme and Constellation, and has been on advisory boards for BMS and ExoTherapeutics. C.D.A is a co-founder of Chroma Therapeutics and Constellation Pharmaceuticals, and a Scientific Advisory Board member of EpiCypher.
References
- 1.McGinty RK, Tan S. Nucleosome structure and function. Chem Rev 2015;115:2255–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol 2018;19:192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soshnev AA, Josefowicz SZ, Allis CD. Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome. Mol Cell 2016;62:681–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan JC, Maze I. Nothing Is Yet Set in (Hi)stone: Novel Post-Translational Modifications Regulating Chromatin Function. Trends Biochem Sci 2020;45:829–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, et al. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019;179:470–84 e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;173:371–85 e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett RL, Licht JD. Targeting Epigenetics in Cancer. Annu Rev Pharmacol Toxicol 2018;58:187–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012;44:251–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 2012;124:439–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31 [DOI] [PubMed] [Google Scholar]
- 12.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 2013;45:1479–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett RL, Bele A, Small EC, Will CM, Nabet B, Oyer JA, et al. A Mutation in Histone H2B Represents a New Class of Oncogenic Driver. Cancer Discov 2019;9:1438–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arimura Y, Ikura M, Fujita R, Noda M, Kobayashi W, Horikoshi N, et al. Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome. Nucleic Acids Res 2018;46:10007–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nacev BA, Feng L, Bagert JD, Lemiesz AE, Gao J, Soshnev AA, et al. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 2019;567:473–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehnertz B, Zhang YW, Boivin I, Mayotte N, Tomellini E, Chagraoui J, et al. H3(K27M/I) mutations promote context-dependent transformation in acute myeloid leukemia with RUNX1 alterations. Blood 2017;130:2204–14 [DOI] [PubMed] [Google Scholar]
- 17.Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet 2017;49:180–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013;340:857–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016;352:844–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain SU, Khazaei S, Marchione DM, Lundgren SM, Wang X, Weinberg DN, et al. Histone H3.3 G34 mutations promote aberrant PRC2 activity and drive tumor progression. Proc Natl Acad Sci U S A 2020;117:27354–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koelsche C, Schrimpf D, Tharun L, Roth E, Sturm D, Jones DTW, et al. Histone 3.3 hotspot mutations in conventional osteosarcomas: a comprehensive clinical and molecular characterization of six H3F3A mutated cases. Clin Sarcoma Res 2017;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagert JD, Mitchener MM, Patriotis AL, Dul BE, Wojcik F, Nacev BA, et al. Oncohistone mutations enhance chromatin remodeling and alter cell fates. Nat Chem Biol 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long M, Sun X, Shi W, Yanru A, Leung STC, Ding D, et al. A novel histone H4 variant H4G regulates rDNA transcription in breast cancer. Nucleic Acids Res 2019;47:8399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011;476:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 2014;46:176–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017;171:481–94 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yusufova N, Kloetgen A, Teater M, Osunsade A, Camarillo JM, Chin CR, et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 2021;589:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichel J, Chadburn A, Rubinstein PG, Giulino-Roth L, Tam W, Liu Y, et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015;125:1061–72 [DOI] [PubMed] [Google Scholar]
- 29.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016;44:D733–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan W, Ma L, Burns KH, Matzuk MM. HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc Natl Acad Sci U S A 2003;100:10546–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou BR, Feng H, Kale S, Fox T, Khant H, de Val N, et al. Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms. Mol Cell 2021;81:166–82 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Dalal Y, Papoian GA. Binding Dynamics of Disordered Linker Histone H1 with a Nucleosomal Particle. J Mol Biol 2021:166881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Churchill ME, Suzuki M. ‘SPKK’ motifs prefer to bind to DNA at A/T-rich sites. EMBO J 1989;8:4189–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy KJ, Cutter AR, Fang H, Postnikov YV, Bustin M, Hayes JJ. HMGN1 and 2 remodel core and linker histone tail domains within chromatin. Nucleic Acids Res 2017;45:9917–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roque A, Iloro I, Ponte I, Arrondo JL, Suau P. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J Biol Chem 2005;280:32141–7 [DOI] [PubMed] [Google Scholar]
- 36.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 2011;25:2227–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballare C, et al. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev 2011;25:845–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Structure Widom J., dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct 1998;27:285–327 [DOI] [PubMed] [Google Scholar]
- 39.Stasevich TJ, Mueller F, Brown DT, McNally JG. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J 2010;29:1225–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol 2006;13:250–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nodelman IM, Bowman GD. Biophysics of Chromatin Remodeling. Annu Rev Biophys 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pardal AJ, Fernandes-Duarte F, Bowman AJ. The histone chaperoning pathway: from ribosome to nucleosome. Essays Biochem 2019;63:29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou K, Gaullier G, Luger K. Nucleosome structure and dynamics are coming of age. Nat Struct Mol Biol 2019;26:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature 2000;408:877–81 [DOI] [PubMed] [Google Scholar]
- 45.Bernas T, Brutkowski W, Zarebski M, Dobrucki J. Spatial heterogeneity of dynamics of H1 linker histone. Eur Biophys J 2014;43:287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prendergast L, Reinberg D. The missing linker: emerging trends for H1 variant-specific functions. Genes Dev 2021;35:40–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osunsade A, Prescott NA, Hebert JM, Ray DM, Jmeian Y, Lorenz IC, et al. A Robust Method for the Purification and Characterization of Recombinant Human Histone H1 Variants. Biochemistry 2019;58:171–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang SM, Kim BJ, Norwood Toro L, Skoultchi AI. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc Natl Acad Sci U S A 2013;110:1708–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao R, Mizzen CA. Site-specific regulation of histone H1 phosphorylation in pluripotent cell differentiation. Epigenetics Chromatin 2017;10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saha A, Seward CH, Stubbs L, Mizzen CA. Site-Specific Phosphorylation of Histone H1.4 Is Associated with Transcription Activation. Int J Mol Sci 2020;21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 2004;14:183–93 [DOI] [PubMed] [Google Scholar]
- 52.Weiss T, Hergeth S, Zeissler U, Izzo A, Tropberger P, Zee BM, et al. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin 2010;3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 2005;123:1199–212 [DOI] [PubMed] [Google Scholar]
- 54.Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol 2001;21:7933–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao K, Lailler N, Zhang Y, Kumar A, Uppal K, Liu Z, et al. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet 2013;9:e1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Izzo A, Kamieniarz-Gdula K, Ramirez F, Noureen N, Kind J, Manke T, et al. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep 2013;3:2142–54 [DOI] [PubMed] [Google Scholar]
- 57.Goytisolo FA, Gerchman SE, Yu X, Rees C, Graziano V, Ramakrishnan V, et al. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J 1996;15:3421–9 [PMC free article] [PubMed] [Google Scholar]
- 58.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hechtman JF, Beasley MB, Kinoshita Y, Ko HM, Hao K, Burstein DE. Promyelocytic leukemia zinc finger and histone H1.5 differentially stain low- and high-grade pulmonary neuroendocrine tumors: a pilot immunohistochemical study. Hum Pathol 2013;44:1400–5 [DOI] [PubMed] [Google Scholar]
- 60.Khachaturov V, Xiao GQ, Kinoshita Y, Unger PD, Burstein DE. Histone H1.5, a novel prostatic cancer marker: an immunohistochemical study. Hum Pathol 2014;45:2115–9 [DOI] [PubMed] [Google Scholar]
- 61.Warneboldt J, Haller F, Horstmann O, Danner BC, Fuzesi L, Doenecke D, et al. Histone H1x is highly expressed in human neuroendocrine cells and tumours. BMC Cancer 2008;8:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torres CM, Biran A, Burney MJ, Patel H, Henser-Brownhill T, Cohen AS, et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 2016;353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willcockson MA, Healton SE, Weiss CN, Bartholdy BA, Botbol Y, Mishra LN, et al. H1 histones control the epigenetic landscape by local chromatin compaction. Nature 2021;589:293–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duy C, Beguelin W, Melnick A. Epigenetic Mechanisms in Leukemias and Lymphomas. Cold Spring Harb Perspect Med 2020;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mlynarczyk C, Fontan L, Melnick A. Germinal center-derived lymphomas: The darkest side of humoral immunity. Immunol Rev 2019;288:214–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venturutti L, Melnick AM. The Role of Epigenetic Mechanisms in B Cell Lymphoma Pathogenesis. Annual Review of Cancer Biology 2021;5:311–30 [Google Scholar]
- 67.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol 2007;8:705–14 [DOI] [PubMed] [Google Scholar]
- 68.Ranuncolo SM, Polo JM, Melnick A. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B-cells. Blood Cells Mol Dis 2008;41:95–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 2004;432:635–9 [DOI] [PubMed] [Google Scholar]
- 70.Teater M, Dominguez PM, Redmond D, Chen Z, Ennishi D, Scott DW, et al. AICDA drives epigenetic heterogeneity and accelerates germinal center-derived lymphomagenesis. Nat Commun 2018;9:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet 2008;40:108–12 [DOI] [PubMed] [Google Scholar]
- 72.Mendoza P, Martinez-Martin N, Bovolenta ER, Reyes-Garau D, Hernansanz-Agustin P, Delgado P, et al. R-Ras2 is required for germinal center formation to aid B cells during energetically demanding processes. Sci Signal 2018;11 [DOI] [PubMed] [Google Scholar]
- 73.Finkin S, Hartweger H, Oliveira TY, Kara EE, Nussenzweig MC. Protein Amounts of the MYC Transcription Factor Determine Germinal Center B Cell Division Capacity. Immunity 2019;51:324–36 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 2000;13:199–212 [DOI] [PubMed] [Google Scholar]
- 75.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med 2018;378:1396–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 2020;37:551–68 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell Rep 2014;6:130–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chew GL, Bleakley M, Bradley RK, Malik HS, Henikoff S, Molaro A, et al. Short H2A histone variants are expressed in cancer. Nat Commun 2021;12:490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bunting KL, Soong TD, Singh R, Jiang Y, Beguelin W, Poloway DW, et al. Multi-tiered Reorganization of the Genome during B Cell Affinity Maturation Anchored by a Germinal Center-Specific Locus Control Region. Immunity 2016;45:497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chu CS, Hellmuth JC, Singh R, Ying HY, Skrabanek L, Teater MR, et al. Unique Immune Cell Coactivators Specify Locus Control Region Function and Cell Stage. Mol Cell 2020;80:845–61 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rivas MA, Meydan C, Chin CR, Challman MF, Kim D, Bhinder B, et al. Smc3 dosage regulates B cell transit through germinal centers and restricts their malignant transformation. Nat Immunol 2021;22:240–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vilarrasa-Blasi R, Soler-Vila P, Verdaguer-Dot N, Russinol N, Di Stefano M, Chapaprieta V, et al. Dynamics of genome architecture and chromatin function during human B cell differentiation and neoplastic transformation. Nat Commun 2021;12:651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beguelin W, Teater M, Gearhart MD, Calvo Fernandez MT, Goldstein RL, Cardenas MG, et al. EZH2 and BCL6 Cooperate to Assemble CBX8-BCOR Complex to Repress Bivalent Promoters, Mediate Germinal Center Formation and Lymphomagenesis. Cancer Cell 2016;30:197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piunti A, Shilatifard A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol 2021 [DOI] [PubMed] [Google Scholar]
- 85.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LLP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014;157:1445–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A, Klose RJ. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol Cell 2020;77:857–74 e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martin C, Cao R, Zhang Y. Substrate preferences of the EZH2 histone methyltransferase complex. J Biol Chem 2006;281:8365–70 [DOI] [PubMed] [Google Scholar]
- 88.Zhao J, Wang M, Chang L, Yu J, Song A, Liu C, et al. RYBP/YAF2-PRC1 complexes and histone H1-dependent chromatin compaction mediate propagation of H2AK119ub1 during cell division. Nat Cell Biol 2020;22:439–52 [DOI] [PubMed] [Google Scholar]
- 89.Scott CL, Gil J, Hernando E, Teruya-Feldstein J, Narita M, Martinez D, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci U S A 2007;104:5389–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang GG, Konze KD, Tao J. Polycomb genes, miRNA, and their deregulation in B-cell malignancies. Blood 2015;125:1217–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beguelin W, Rivas MA, Calvo Fernandez MT, Teater M, Purwada A, Redmond D, et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat Commun 2017;8:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 2013;23:677–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klymenko T, Bloehdorn J, Bahlo J, Robrecht S, Akylzhanova G, Cox K, et al. Lamin B1 regulates somatic mutations and progression of B-cell malignancies. Leukemia 2018;32:364–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet 2011;27:389–96 [DOI] [PubMed] [Google Scholar]
- 95.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000;403:41–5 [DOI] [PubMed] [Google Scholar]
- 96.Rossi MJ, Kuntala PK, Lai WKM, Yamada N, Badjatia N, Mittal C, et al. A high-resolution protein architecture of the budding yeast genome. Nature 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 2013;339:698–9 [DOI] [PubMed] [Google Scholar]
- 98.Lau MS, Schwartz MG, Kundu S, Savol AJ, Wang PI, Marr SK, et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science 2017;355:1081–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet 2019;51:1272–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ing-Simmons E, Vaid R, Bing XY, Levine M, Mannervik M, Vaquerizas JM. Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nature Genetics 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Poepsel S, Kasinath V, Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol 2018;25:154–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ge EJ, Jani KS, Diehl KL, Muller MM, Muir TW. Nucleation and Propagation of Heterochromatin by the Histone Methyltransferase PRC2: Geometric Constraints and Impact of the Regulatory Subunit JARID2. J Am Chem Soc 2019;141:15029–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol Cell 2015;57:769–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, Stjepanovic G, et al. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 2018;359:940–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee CH, Holder M, Grau D, Saldana-Meyer R, Yu JR, Ganai RA, et al. Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol Cell 2018;70:435–48 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kasinath V, Beck C, Sauer P, Poepsel S, Kosmatka J, Faini M, et al. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 2021;371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qi W, Zhao K, Gu J, Huang Y, Wang Y, Zhang H, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol 2017;13:381–8 [DOI] [PubMed] [Google Scholar]
- 108.Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol 2019;26:880–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo R, Zheng L, Park JW, Lv R, Chen H, Jiao F, et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell 2014;56:298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jani KS, Jain SU, Ge EJ, Diehl KL, Lundgren SM, Muller MM, et al. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc Natl Acad Sci U S A 2019;116:8295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Zhang Q, Ezponda T, et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet 2014;10:e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rajagopalan KN, Chen X, Weinberg DN, Chen H, Majewski J, Allis CD, et al. Depletion of H3K36me2 recapitulates epigenomic and phenotypic changes induced by the H3.3K36M oncohistone mutation. Proc Natl Acad Sci U S A 2021;118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu JR, LeRoy G, Bready D, Frenster JD, Saldana-Meyer R, Jin Y, et al. The H3K36me2 writer-reader dependency in H3K27M-DIPG. Sci Adv 2021;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kang HB, Choi Y, Lee JM, Choi KC, Kim HC, Yoo JY, et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett 2009;583:1880–6 [DOI] [PubMed] [Google Scholar]
- 115.Gerlitz G, Hock R, Ueda T, Bustin M. The dynamics of HMG protein-chromatin interactions in living cells. Biochem Cell Biol 2009;87:127–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tian TV, Di Stefano B, Stik G, Vila-Casadesus M, Sardina JL, Vidal E, et al. Whsc1 links pluripotency exit with mesendoderm specification. Nat Cell Biol 2019;21:824–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lhoumaud P, Badri S, Rodriguez-Hernaez J, Sakellaropoulos T, Sethia G, Kloetgen A, et al. NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains. Nat Commun 2019;10:4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Swaroop A, Oyer JA, Will CM, Huang X, Yu W, Troche C, et al. An activating mutation of the NSD2 histone methyltransferase drives oncogenic reprogramming in acute lymphocytic leukemia. Oncogene 2019;38:671–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yuan G, Flores NM, Hausmann S, Lofgren SM, Kharchenko V, Angulo-Ibanez M, et al. Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 2019;573:281–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dominguez PM, Ghamlouch H, Rosikiewicz W, Kumar P, Beguelin W, Fontan L, et al. TET2 Deficiency Causes Germinal Center Hyperplasia, Impairs Plasma Cell Differentiation, and Promotes B-cell Lymphomagenesis. Cancer Discov 2018;8:1632–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barwick BG, Scharer CD, Martinez RJ, Price MJ, Wein AN, Haines RR, et al. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat Commun 2018;9:1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen J, Li N, Yin Y, Zheng N, Min M, Lin B, et al. Methyltransferase Nsd2 Ensures Germinal Center Selection by Promoting Adhesive Interactions between B Cells and Follicular Dendritic Cells. Cell Rep 2018;25:3393–404 e6 [DOI] [PubMed] [Google Scholar]
- 125.Zhang L, Xie WJ, Liu S, Meng L, Gu C, Gao YQ. DNA Methylation Landscape Reflects the Spatial Organization of Chromatin in Different Cells. Biophys J 2017;113:1395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010;466:388–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schubeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 2015;528:575–9 [DOI] [PubMed] [Google Scholar]
- 128.Stutzer A, Liokatis S, Kiesel A, Schwarzer D, Sprangers R, Soding J, et al. Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Mol Cell 2016;61:247–59 [DOI] [PubMed] [Google Scholar]
- 129.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 2010;143:212–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shimada M, Chen WY, Nakadai T, Onikubo T, Guermah M, Rhodes D, et al. Gene-Specific H1 Eviction through a Transcriptional Activator-->p300-->NAP1-->H1 Pathway. Mol Cell 2019;74:268–83 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li W, Tian W, Yuan G, Deng P, Sengupta D, Cheng Z, et al. Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leicher R, Ge EJ, Lin X, Reynolds MJ, Xie W, Walz T, et al. Single-molecule and in silico dissection of the interaction between Polycomb repressive complex 2 and chromatin. Proc Natl Acad Sci U S A 2020;117:30465–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Adhireksan Z, Sharma D, Lee PL, Davey CA. Near-atomic resolution structures of interdigitated nucleosome fibres. Nat Commun 2020;11:4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bonner WM, Stedman JD. Histone 1 is proximal to histone 2A and to A24. Proc Natl Acad Sci U S A 1979;76:2190–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chong PSY, Chooi JY, Lim JSL, Toh SHM, Tan TZ, Chng WJ. SMARCA2 is a novel interactor of NSD2 and regulates pro-metastatic PTP4A3 through chromatin remodeling in t(4;14) multiple myeloma. Cancer Res 2021 [DOI] [PubMed] [Google Scholar]
- 136.Ma MCJ, Tadros S, Bouska A, Heavican T, Yang H, Deng Q, et al. Subtype-specific and co-occurring genetic alterations in B-cell non-Hodgkin lymphoma. Haematologica 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rutowicz K, Lirski M, Mermaz B, Teano G, Schubert J, Mestiri I, et al. Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis. Genome Biol 2019;20:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Escobar TM, Oksuz O, Saldana-Meyer R, Descostes N, Bonasio R, Reinberg D. Active and Repressed Chromatin Domains Exhibit Distinct Nucleosome Segregation during DNA Replication. Cell 2019;179:953–63 e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sabari BR. Biomolecular Condensates and Gene Activation in Development and Disease. Dev Cell 2020;55:84–96 [DOI] [PubMed] [Google Scholar]
- 140.Sabari BR, Dall’Agnese A, Young RA. Biomolecular Condensates in the Nucleus. Trends Biochem Sci 2020;45:961–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Trojanowski J, Frank L, Rademacher A, Grigaitis P, Rippe K. Transcription activation is enhanced by multivalent interactions independent of liquid-liquid phase separation. bioRxiv 2021:2021.01.27.428421 [DOI] [PubMed] [Google Scholar]
- 142.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487–500 [DOI] [PubMed] [Google Scholar]
- 143.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature 2017;547:241–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 2017;547:236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Keenen MM, Brown D, Brennan LD, Renger R, Khoo H, Carlson CR, et al. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. Elife 2021;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 2019;575:390–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 2012;487:249–53 [DOI] [PubMed] [Google Scholar]
- 148.Turner AL, Watson M, Wilkins OG, Cato L, Travers A, Thomas JO, et al. Highly disordered histone H1-DNA model complexes and their condensates. Proc Natl Acad Sci U S A 2018;115:11964–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McGhee JD, Felsenfeld G. Nucleosome structure. Annu Rev Biochem 1980;49:1115–56 [DOI] [PubMed] [Google Scholar]
- 150.Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, et al. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell 2014;156:907–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Leicher R, Osunsade A, Latham AP, Chua GNL, Watters JW, Christodoulou-Rubalcava S, et al. Single-stranded nucleic acid sensing and coacervation by linker histone H1. bioRxiv 2021:2021.03.17.435841 [DOI] [PMC free article] [PubMed] [Google Scholar]