Abstract
Automated methods for detecting prostate cancer and distinguishing indolent from aggressive disease on Magnetic Resonance Imaging (MRI) could assist in early diagnosis and treatment planning. Existing automated methods of prostate cancer detection mostly rely on ground truth labels with limited accuracy, ignore disease pathology characteristics observed on resected tissue, and cannot selectively identify aggressive (Gleason Pattern≥4) and indolent (Gleason Pattern=3) cancers when they co-exist in mixed lesions. In this paper, we present a radiology-pathology fusion approach, CorrSigNIA, for the selective identification and localization of indolent and aggressive prostate cancer on MRI. CorrSigNIA uses registered MRI and whole-mount histopathology images from radical prostatectomy patients to derive accurate ground truth labels and learn correlated features between radiology and pathology images. These correlated features are then used in a convolutional neural network architecture to detect and localize normal tissue, indolent cancer, and aggressive cancer on prostate MRI. CorrSigNIA was trained and validated on a dataset of 98 men, including 74 men that underwent radical prostatectomy and 24 men with normal prostate MRI. CorrSigNIA was tested on three independent test sets including 55 men that underwent radical prostatectomy, 275 men that underwent targeted biopsies, and 15 men with normal prostate MRI. CorrSigNIA achieved an accuracy of 80% in distinguishing between men with and without cancer, a lesion-level ROC-AUC of 0.81±0.31 in detecting cancers in both radical prostatectomy and biopsy cohort patients, and lesion-levels ROC-AUCs of 0.82±0.31 and 0.86±0.26 in detecting clinically significant cancers in radical prostatectomy and biopsy cohort patients respectively. CorrSigNIA consistently outperformed other methods across different evaluation metrics and cohorts. In clinical settings, CorrSigNIA may be used in prostate cancer detection as well as in selective identification of indolent and aggressive components of prostate cancer, thereby improving prostate cancer care by helping guide targeted biopsies, reducing unnecessary biopsies, and selecting and planning treatment.
Keywords: Prostate cancer, computer-aided diagnosis, correlated feature learning, radiology-pathology fusion
Graphical Abstract
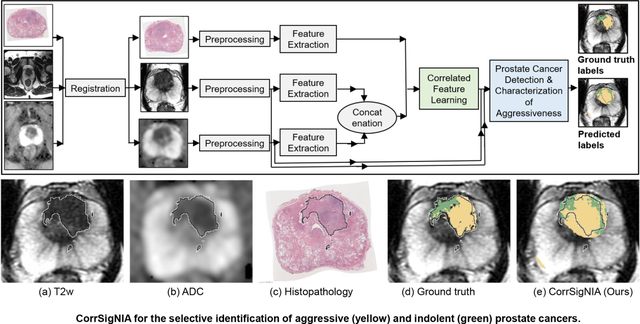
1. Introduction
Prostate cancer causes the second highest number of cancer deaths among American men. The American Cancer Society estimated 191,930 new cases of prostate cancer, and 33,330 deaths from prostate cancer in 2020 (Siegel et al., 2020). Magnetic Resonance Imaging (MRI) is increasingly used in early detection of prostate cancer, deciding who to biopsy, and guiding biopsies and local treatment (Liu et al., 2018). However, interpretation of MR images has many challenges, particularly because of the very subtle visual differences between benign tissue, indolent cancer and aggressive cancer. Radiologist interpretations suffer from false negatives (12% of aggressive cancers missed during screening (Ahmed et al., 2017), 34% of aggressive and 81% of indolent cancers missed in men undergoing prostatectomy (Johnson et al., 2019)), false positives (>35% false-positive rate (Ahmed et al., 2017)), and high inter-reader variability (inter-reader agreement κ = 0.46 − 0.78 (Barentsz et al., 2016; Sonn et al., 2019)). Moreover, it is challenging for radiologists to distinguish and localize the indolent and aggressive components of prostate cancer when they co-exist within the same lesion which occurs commonly (48% of all cancers and 76% of index lesions Johnson et al. (2019)).
Machine learning methods could help radiologists interpret prostate MRI by accurately and robustly detecting, localizing and stratifying the aggressiveness of prostate cancer on MRI. Some automated methods focus on detecting and localizing cancer on prostate MRI (Sumathipala et al., 2018; Cao et al., 2019; Sanyal et al., 2020), while others focus on classifying the aggressiveness of already demarcated lesions on MRI (Viswanath et al., 2019; Hectors et al., 2019). These methods use a variety of techniques including traditional classifiers with hand-crafted and radiomic features (Viswanath et al., 2012; Litjens et al., 2014; Viswanath et al., 2019), and deep learning based models (Sumathipala et al., 2018; Cao et al., 2019; Sanyal et al., 2020; Schelb et al., 2019; De Vente et al., 2020; Seetharaman et al., 2021; Bhattacharya et al., 2020). However, most of these methods suffer from one or more of the following shortcomings:
Inaccurate ground truth labels: Currently, there are three ways of deriving pathology-confirmed ground truth cancer labels for training machine learning models: (1) radiologist demarcated lesions confirmed by biopsy (Wang et al., 2014; Kwak et al., 2015; Armato et al., 2018; Litjens et al., 2014), (2) cognitive manual alignment of pre-operative MRI and post-operative histopathology images (Sumathipala et al., 2018; Cao et al., 2019), and (3) manual or semi-automatic registration between MRI and histopathology images using landmarks, ex-vivo MRI or external fiducials (Penzias et al., 2018; Hurrell et al., 2017; Wu et al., 2019; Ward et al., 2012; Li et al., 2017). All these ground truth labels suffer from various inaccuracies. First, radiologist demarcated lesions substantially underestimate tumor size, miss MRI invisible cancers (Priester et al., 2017), and cannot provide pixel-level cancer grading on the entire prostate as biopsy tracts sample the prostate sparsely. Secondly, cognitive registration provides pixel-level annotations of cancer and Gleason grade for the entire prostate, but the extent of cancer on MRI is often underestimated (Priester et al., 2017), and it is still challenging to outline the ~20% of tumors that are not clearly seen on MRI (Barentsz et al., 2016). Finally, manual and semiautomatic registration approaches are labor-intensive, requiring highly skilled experts in both radiology and pathology (Kalavagunta et al., 2015; Hurrell et al., 2017; Losnegård et al., 2018). Thus, many automated methods focus only on the index lesion (largest lesion with highest Gleason grade) (Sanyal et al., 2020) or only MRI-visible lesions (Sumathipala et al., 2018; Cao et al., 2019), failing to capture the complete extent of the disease.
Agnostic to disease pathology characteristics: Most existing machine learning methods learn from MRI alone (Sumathipala et al., 2018; Cao et al., 2019; Seetharaman et al., 2021; Schelb et al., 2019; De Vente et al., 2020), ignoring disease pathology characteristics observed on resected tissue. Our recent work (Bhattacharya et al., 2020) is the only prior study that seeks to learn MRI features that are correlated to pathology characteristics of prostate cancer. We are seeking to extend our prior work to not only detect prostate cancer on MRI but also to differentiate between aggressive and indolent cancer.
Inability to distinguish between indolent and aggressive cancer in mixed lesions: Identifying and localizing the indolent and aggressive components of mixed lesions on a pixel-level can help guide targeted biopsies and treatment planning. Our prior work SPCNet (Seetharaman et al., 2021) is the only study that seeks to localize indolent and aggressive cancers using pixel-level histologic grade labels, while all other prior studies focus on lesion-level grade labels. Unlike our prior SPCNet work (Seetharaman et al., 2021) that only uses MRI features, in this study we focus on showing the benefits of using the pathology image information in addition to mapping the indolent and aggressive labels from histopathology onto MRI.
Generalizability: Most prior studies focus on patients that either underwent radical prostatectomy or biopsy, but not on both groups together. It is, however, important to test such generalizability, because models trained on prostatectomy patients with advanced disease would be clinically relevant only if they generalize to the biopsy population that shows a different disease distribution, ranging from benign conditions to aggressive cancer.
To address these challenges, we build on our prior studies (Bhattacharya et al., 2020; Seetharaman et al., 2021) and present CorrSigNIA, the Correlated Signature Network for Indolent (Gleason pattern=3) and Aggressive (Gleason pattern ≥4) prostate cancer detection and localization on MRI. CorrSigNIA was trained on men with cancer that underwent radical prostatectomy and others who had no cancer. Including radical prostatectomy patients with confirmed prostate cancer in the training set ensured CorrSigNIA could learn to reliably identify and localize aggressive and indolent cancers on prostate MRI. Including men without prostate cancer in the training set ensured CorrSigNIA could also learn the appearance of normal prostates which is necessary when MRI is used as a screening tool.
While training on men with radical prostatectomy, CorrSigNIA used ground truth cancer labels from an expert pathologist, and pixel-level histologic grading assigned by a pathology deep learning model (Ryu et al., 2019) on whole-mount histopathology images. These post-operative histopathology images were accurately registered with pre-operative MRI using the RAP-SODI registration platform (Rusu et al., 2020), enabling the capture of both the extent and histologic grades of all cancers, including MRI invisible cancers. CorrSigNIA included two major steps. First, it learned correlated features using a common representation learning framework by identifying MRI features that are correlated with corresponding spatially aligned histopathology features. Second, it used the learned correlated features and original MRI intensities in a convolutional neural network model to detect and localize normal tissue, indolent cancer, and aggressive cancer at a pixel-level. The histopathology images were used only in the first step to learn correlations between radiology and pathology features. Once learned, the correlated features can be extracted from MRI alone, without the need for pathology, thereby enabling feature extraction and prediction in new patients. In this study, we sought to evaluate the performance of CorrSigNIA in comparison to existing methods for detecting aggressive and indolent prostate cancer. We also sought to test the generalizability of CorrSigNIA on men who underwent radical prostatectomy, men who underwent biopsies, and men with normal prostate MRI without cancer.
2. Materials and Methods
2.1. Dataset
This retrospective chart review study was approved by the Institutional Review Board (IRB) of Stanford University. As a chart review of previously collected data, patient consent was waived. The study included three cohorts C1, C2, and C3 as detailed in Table 1. Cohort C1 included 129 men with confirmed prostate cancer who underwent MRI followed by radical prostatectomy at Stanford University, and whole-mount histologic processing of the excised prostate tissue. Cohort C2 included 275 men who had an MRI which was examined by an expert radiologist followed by MRI-Ultrasound fusion targeted biopsy at Stanford University to confirm presence or absence of prostate cancer. Cohort C3 included 39 men without cancer, as confirmed by a negative prostate MRI and negative biopsy.
Table 1:
Summary of the three datasets.
| Cohort C1: Radical Prostatectomy | Cohort C2: Targeted Biopsy | Cohort C3: Normal | |||||
| Variable | MRI | Pathology | MRI | MRI | |||
| Patient Number | 129 | 129 | 275 | 39 | |||
| Sequence/Data Type | T2w | ADC | Whole-mount | T2w | ADC | T2w | ADC |
| Acquisition Characteristics | TR: [3.9, 6.3] TE: [122, 130] |
b-vals: 0, 50, 800, 1000, 1200 | H&E staining | TR: [2.0, 7.4] TE: [92, 150] |
b-vals: 0, 25, 50, 800, 1200, 1400 | TR: [1.7, 7.6] TE: [81, 149] |
b-vals: 0, 25, 50, 800, 1200, 1400 |
| Matrix Size | [256, 512] | [50, 256] | [1665, 7556] | [256, 512] | [50, 256] | [512] | [256] |
| Slice Number | [20, 44] | [15, 40] | [3,11] | [20, 43] | [14, 40] | [24, 48] | [24, 48] |
| Pixel Spacing (mm) | [0.27, 0.94] | [0.78, 1.50] | [0.008, 0.016] | [0.39, 0.47] | [0.78, 1.01] | [0.35, 0.43] | [0.78, 1.25] |
| Distance between slices (mm) | [3, 5.2] | [3, 4.50] | [3, 5.2] | [3, 4.2] | [3, 4.6] | [3, 4.2] | [3, 6] |
| Annotations (number of patients) | |||||||
| Prostate | Yes (129) | Yes (129) | Yes (129) | Yes (275) | No | Yes (39) | No |
| Pathologist outlined cancer | Yes (123) | Yes (123) | Yes (123) | No | No | No | No |
| Per-pixel Gleason pattern (Ryu et al., 2019) | Yes (121) | Yes (121) | Yes (121) | No | No | No | No |
| Train-test splits (number of patients) | |||||||
| Train | Yes (74) | Yes (74) | Yes (74) | No | No | Yes (24) | Yes (24) |
| Test | Yes (55) | Yes (55) | No | Yes (275) | Yes (275) | Yes (15) | Yes (15) |
Abbreviations- T2w: T2-weighted MRI, H& E: Hematoxylin & Eosin, TR: Relaxation Time (s), TE: Echo Time (ms), b-vals: b-values (s/mm2). [a, b] indicates range between a and b.
2.1.1. MRI
For patients in all three cohorts, the MR images were acquired using 3 Tesla GE scanners (GE Healthcare, Waukesha, WI, USA) with external 32-channel body array coils without endorectal coils. The imaging protocol included T2-weighted MRI (T2w), diffusion weighted imaging, derived Apparent Diffusion Coefficient (ADC) maps, and dynamic contrast-enhanced imaging sequences. Axial T2w MRI (acquired using a 2D Spin Echo protocol) and ADC maps were used in this study (Table 1).
2.1.2. Histopathology
Histopathology images were only available for cohort C1. For patients in cohort C1, the whole-mount excised prostate tissue was sectioned in the same plane as the T2w MRI using patient-specific 3D printed molds. The whole-mount prostate slices were then stained using Hematoxylin & Eosin (H&E) and digitized at 20x magnification. For patients in cohort C2 and C3, the biopsy samples were stained in H&E for evaluation and the final pathology result was used to label the radiologist demarcated lesions.
2.1.3. Labels
For 123 of 129 men in cohort C1, our expert pathologist (C.A.K. with 10 years of experience) outlined cancer on all high-resolution whole-mount histopathology slices, generating a per-pixel cancer labeling. For 121 of 129 men in cohort C1, per-pixel Gleason pattern labels on the high resolution histopathology images were derived from a previously validated automated Gleason scoring system trained on histopathology images (Ryu et al., 2019). The automated per-pixel combined Gleason pattern labels achieved an average Dice overlap of 0.80±0.09 with the pathologist annotations of cancer. It may be noted that the moderate Dice overlap metric captured the difference in resolution between the two annotations, i.e., the automated approach outlined glands in great detail, whereas it was impractical for a human annotator to outline each gland at high resolution for a large number of cases. In our study, Gleason pattern 4 and above was considered as aggressive cancer, whereas Gleason pattern 3 was considered indolent cancer. Regions with overlapping Gleason pattern 3 and 4 labels were considered aggressive cancer.
Cancer labels for cohort C2 were derived from radiologist outlined lesions and the highest Gleason grade group of the corresponding targeted biopsies. Cohort C2 included 275 men with radiologist annotated lesions and PI-RADS (Prostate Imaging-Reporting and Data System) (Turkbey et al., 2019) scores of 3 or above. Out of the 275 men, 147 men were confirmed as having cancer. There were a total of 189 cancerous lesions (Gleason grade group ≥ 1), out of which 110 lesions were clinically significant (Gleason grade group ≥ 2).
In addition to cancer labels, prostate segmentations were available on all T2w MRI and histopathology slices for all patients in all cohorts. Prostate segmentations on all T2w slices were initially performed by W.S., J.B.W., S.J.C.S., and N.C.T. (with 6+ months experience in this task) and were carefully reviewed by our experts (C.A.K, G.S. – a urologic oncologist with 13 years of experience, P.G. – a body MR imaging radiologist with 14 years of experience, M.R. – an image analytics expert with 10 years of experience working on prostate cancer).
Train-test splits:
The models were trained with men from cohorts C1 and C3, and evaluated on men from cohorts C1, C2 and C3. The training set included 74 men from cohort C1 and 24 men from cohort C3, used in 5-fold cross validation setting, as detailed in Table 1. In cohort C1-train, 71 men had both pathologist annotations and pixel-level histologic grade labels assigned by (Ryu et al., 2019), while all 74 men in C1-train had pathologist annotated cancer labels. In C1-test, 45 of the 55 men had both pathologist and histologic grade labels, while the remaining 10 men had either one of the two kinds of labels.
2.2. Data Pre-processing
2.2.1. Registration
T2w MRI and digitized histopathology images in cohort C1 were registered using the RAPSODI registration platform (Rusu et al., 2020), thereby enabling mapping of per-pixel cancer and grade labels from histopathology images onto MRI. Fig.1 displays a sample case of registered MRI and histopathology slices.
Fig. 1:
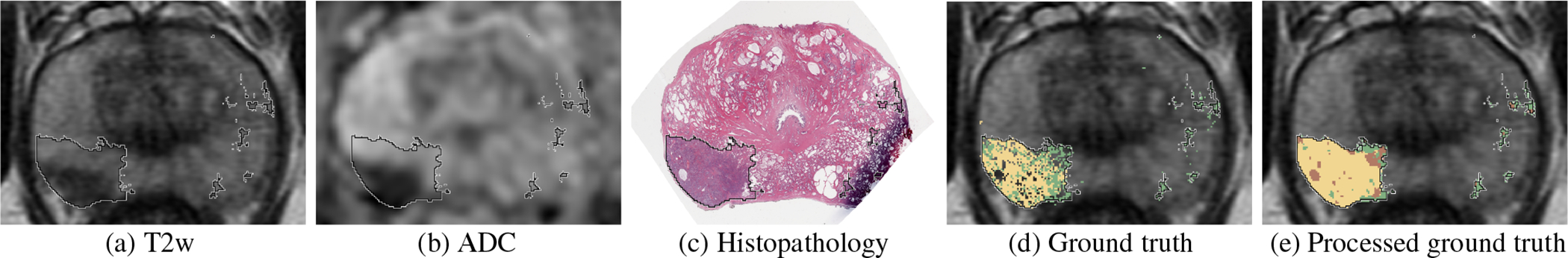
Registered MRI and histopathology slices of a patient in cohort C1, with cancer labels mapped from histopathology to MRI. (a) T2w image, (b) ADC image, (c) Histopathology image, (d) T2w image overlaid with cancer labels from expert pathologist (black outline) and per-pixel histologic grade labels from automated Gleason grading (Ryu et al., 2019): aggressive labels (yellow), indolent labels (green), (e) Processed ground truth labels generated from (d) that were used for training and evaluation of the models (pre-processing of ground truth labels described in Section 2.3.2): aggressive (yellow), indolent (green), aggressive or indolent with equal likelihood (brown).
2.2.2. Histopathology Preprocessing
Smoothing: The histopathology images were smoothed with a Gaussian filter with σ = 0.25 mm to avoid downsampling artifacts. The value of σ was chosen based on visual inspection of downsampled histopathology images after smoothing using different values of σ, namely 0, 0.25, 0.5, 0.75, and 1 mm.
Resampling: The Gaussian-smoothed histopathology images were padded and then downsampled to an X-Y size of 224×224, resulting in an in-plane pixel size of 0.29×0.29 mm2. An image size of 224 × 224 was selected because several deep-learning architectures like the VGG-16 and HED can easily process inputs of this X-Y size.
Intensity normalization: Each RGB channel of the resulting histopathology images were then Z-score normalized.
2.2.3. MRI Preprocessing
Affine Registration: The ADC maps from cohort C1 were manually registered to the T2w images using an affine registration.
Resampling: For patients in cohort C1, the T2w and ADC images, prostate masks, and cancer labels were projected and resampled on the corresponding histopathology images, resulting in images of 224 × 224 pixels with pixel size of 0.29 × 0.29 mm2. For patients in cohort C2 and C3, the T2w and ADC images were cropped around the prostate and resampled to have the same dimensions and pixel size as images in cohort C1.
Intensity standardization: To ensure similar MRI intensity distribution for all patients irrespective of scanners and scanning protocols, a histogram alignment-based intensity standardization method (Nyúl et al., 2000; Reinhold et al., 2019) was applied to the prostate region of T2w and ADC images.
Intensity normalization: Z-score normalization was then applied to the prostate regions of T2w and ADC images.
2.3. The CorrSigNIA Model
CorrSigNIA is comprised of two main modules: (1) correlated feature learning, and (2) prostate cancer detection and characterization of aggressiveness (Fig. 2).
Fig. 2:
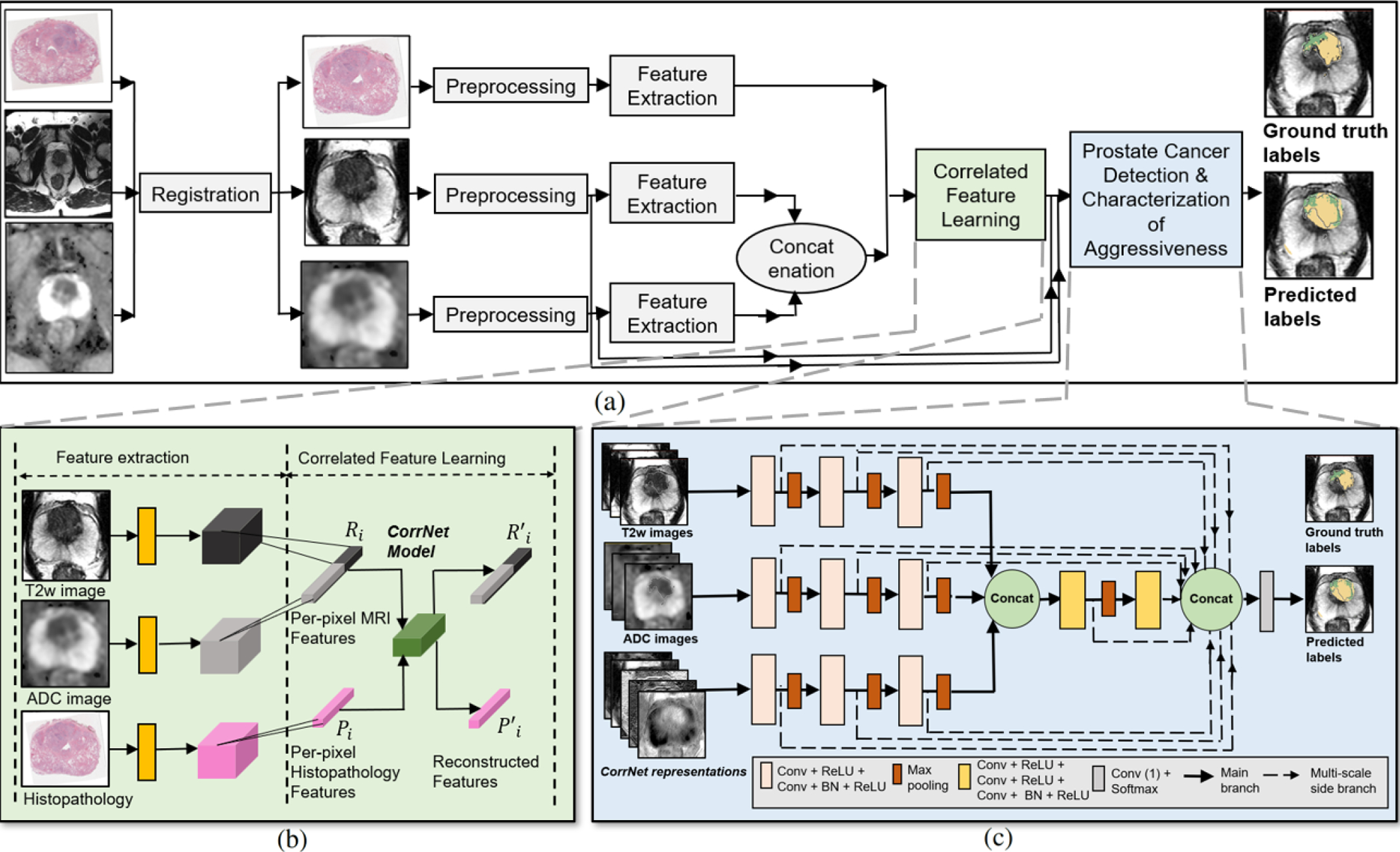
Schematic representation of our approach. (a) Complete flowchart for CorrSigNIA, (b) Correlated feature learning module which uses the CorrNet model to learn correlated feature representations from registered MRI and histopathology features, (c) Prostate cancer detection and characterization of aggressiveness module, which uses a modified HED architecture to selectively identify and localize normal tissue, indolent cancer, and aggressive cancer.
2.3.1. Correlated feature learning
Features from the pre-processed T2w, ADC, and histopathology images of cohort C1-train were extracted by passing them through the first two convolutional and ReLU blocks of a pre-trained VGG-16 architecture (Simonyan and Zisserman, 2014), generating feature maps of size 224 × 224 × 64. Inspired by prior studies that use VGG-16 as a feature extractor for prostate cancer detection on MRI (Abraham and Nair, 2019; Salama and Aly, 2021; Alkadi et al., 2019; Seah et al., 2017), and histopathology images (García et al., 2019), we also used the VGG-16 network to extract similar features on both MRI and histopathology images. This approach allowed the study of correlations between the two modalities in the feature space as the features extracted via the learned kernels capture low-level textural characteristics (Figs. 4 (a–c)).
Fig. 4:
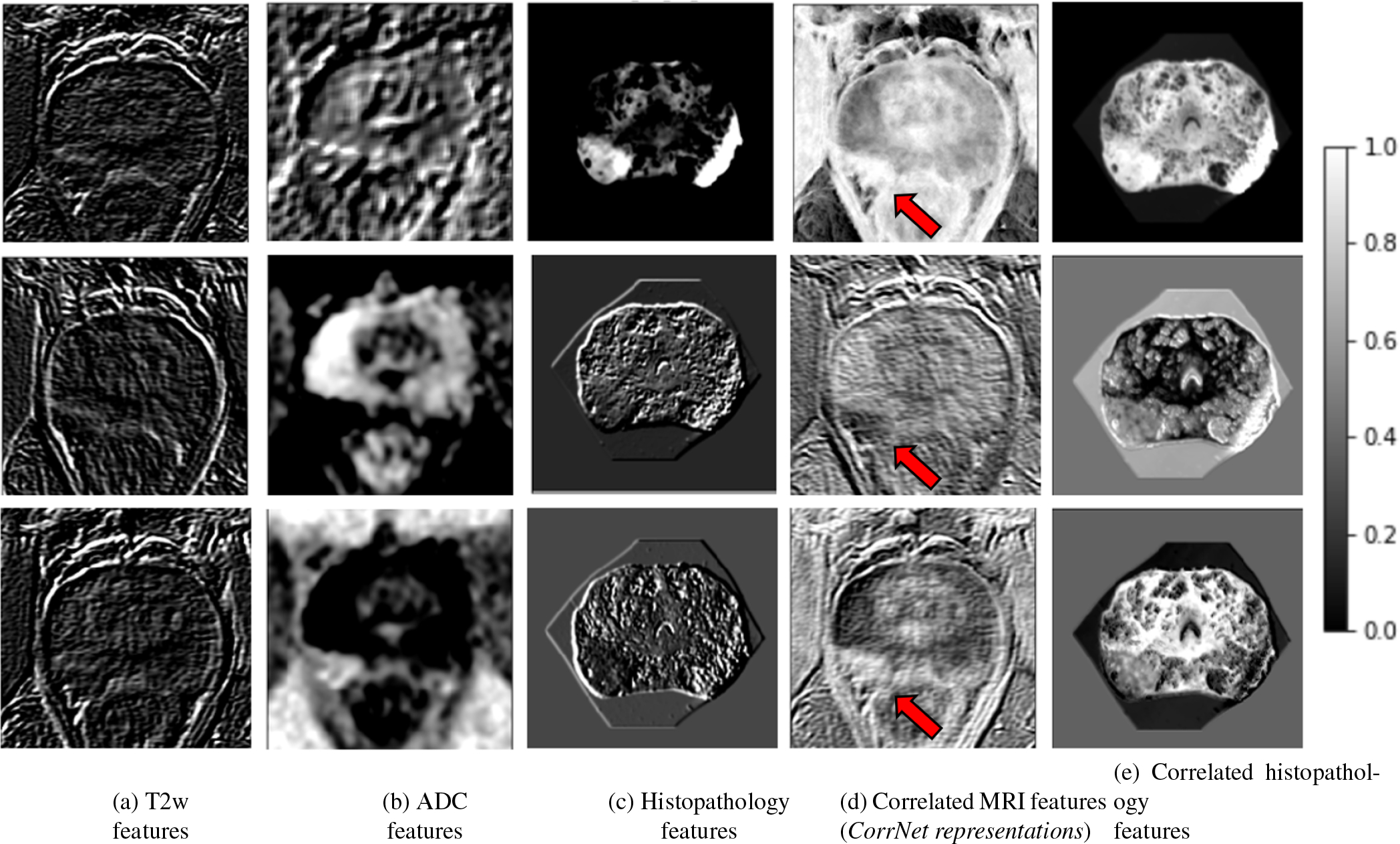
Learning correlated feature representations for prostate cancer for the slice shown in Figure 1. Pre-trained features extracted from (a) T2w image, (b) ADC image, (c) histopathology image. Correlated latent representations formed by projecting (d) from MRI features, and (e) from histopathology features. Column (d) shows the learned correlated MRI feature representations (or CorrNet representations) that are used in the subsequent cancer detection task. Red arrow in column (d) points to cancerous lesion.
The T2w and ADC features for each pixel were concatenated to form an MRI feature representation , and the histopathology features formed a histopathology representation . The pixel-level representations were used to train a Correlational Neural Network (CorrNet) architecture (Chandar et al., 2016) to learn common (or shared latent) representations from MRI and histopathology features. While training CorrNet, the k-dimensional shared latent representation H(Zi) was computed from the input Zi = [Ri, Pi] as:
| (1) |
where , and , and f is the activation function in the hidden layer. The reconstructed output was computed as:
| (2) |
where , and , and g is the activation function at the outer layer. The CorrNet model learned common representations between MRI and histopathology features by learning the parameters of the CorrNet model θ = {W,W′, V, V′, b, b′} through the objective function (Equation 3) that (1) maximized the correlation between the latent representations of both modalities (correlation error), and (2) minimized the reconstruction error of features from both modalities.
| (3) |
where, λ = 2 weighed the correlation error term twice the reconstruction error term.
The correlation error was defined as:
| (4) |
where, H(Ri) = f(WRi + b) represents the latent representation computed from MRI features alone, and H(Pi) = f(VPi + b) represents the latent representations computed from histopathology features alone. and indicate the mean of the MRI and histopathology latent feature representations. The reconstruction error was defined as:
| (5) |
where, Zi = [Ri, Pi] represents the input MRI and histopathology features, indicates the reconstructed features, and L is squared error between original and reconstructed features. Identity was used as both activation functions f and g. This optimum objective function for the CorrNet model was decided based on experiments described in Section 2.4.
For correlated feature learning, an equal number of cancer and non-cancer pixels were sampled from within the prostate. Training the CorrNet on a per-pixel basis enabled learning common representations using a large training sample of ≈ 1.2M pixels. After the CorrNet model was trained, the learned weights W and b were used to compute the latent representations of the input MRI features Ri as H(Ri) = f(WRi + b). These k-dimensional MRI latent representations H(Ri) formed our CorrNet representations of the input MRI. The CorrNet representations (correlated MRI features) were thus a linear weighted combination of T2w and ADC low-level texture features that were maximally correlated with a weighted combination of spatially aligned histopathology low-level texture features (H(Pi)). Once trained, these CorrNet representations (correlated feature maps) can be constructed even in the absence of histopathology images.
The CorrNet architecture was chosen for correlated feature learning because (1) it enables learning correlations between similar kinds of MRI and histopathology features in the latent representation, and (2) ensures the correlated latent representations do not distort the original features.
2.3.2. Prostate cancer detection and characterization of aggressiveness
The SPCNet architecture (Seetharaman et al., 2021) which is a modified version of the Holistically Nested Edge Detection (HED) architecture (Xie and Tu, 2015) (Fig. 2(c)) was considered as the baseline architecture for the detection and localization of normal tissue, indolent cancer and aggressive cancer. Our modifications to the original HED and SPCNet architectures to be used for the CorrSigNIA model involved (1) including three input sequences (normalized T2w and ADC images, and CorrNet representations) processed in three independent branches which allows the learning of individual features for each input sequence, and (2) including three consecutive slices for each input sequence to predict a cancer probability map for the central slice while learning the volumetric continuity of MRI scans. The modified HED architecture leads to 11 side outputs, which are fused using a Conv-1D layer to form a weighted fused output. To reduce internal covariate shift, accelerate training, and reduce over-fitting, batch normalization was added in each block, before ReLU activation. To address the class imbalance problem caused by the fact that aggressive and indolent cancer pixels accounted for ≈ 4% each of the prostate pixels, class-balanced categorical cross-entropy loss functions (Equation 6) were optimized for each side output and final output.
| (6) |
and
| (7) |
where represents the class-balanced cross entropy loss computed for a batch and side output/final output m, m represents one of the 11 side outputs or the final input, N is the number of pixels in the batch, represents the ground truth label for pixel n, represents the predicted label for pixel n, i represents one of the three classes (normal, indolent or aggressive), ϵ = 10−5, and βi represents the class-specific weight assigned to the loss function of class i and is equivalent to the inverse proportion of pixels belonging to class i in each batch. The per-batch losses were averaged across all the batches to compute the loss for each epoch.
The SPCNet architecture was chosen for the prostate cancer detection framework because it enables learning and combining multi-scale and multi-level features, and have been used in prior studies on prostate cancer localization (Bhattacharya et al., 2020; Seetharaman et al., 2021). Moreover, in our prior study, we found that SPCNet outperformed vanilla U-Net (Ronneberger et al., 2015) and DeepLabv3+ (Chen et al., 2018) architectures in aggressive and indolent prostate cancer detection.
While training CorrSigNIA, histologic grading from the automatic grading platform (Ryu et al., 2019) were considered only when they overlapped with cancer annotations from the expert pathologist (C.A.K.), thereby ensuring agreement between the two kinds of labels while emphasizing annotations from the expert pathologist. The difference in resolution between the original histopathology images and the MRI scans resulted in pixelated histologic grading on the MRI scans Fig. 1(d). To account for this, the pixel-level histologic grade labels were smoothed using a median filter of size 3 × 3 pixels. Pixels without cancer annotations by the expert pathologist were considered to be normal tissue. Pixels within the expert pathologist cancer annotations that did not have histologic grade labels were considered to be indolent or aggressive with equal likelihood. Fig. 1 (e) shows the processed ground truth labels used in training and evaluating the models.
Softmax activation function in the last layer of CorrSigNIA generated three probability maps for the prostate, one for each of the normal, indolent, and aggressive classes. These probability maps were used to generate multi-class predicted labels, assigning the label to the class with the highest predicted probability. This is in tune with standard multi-class classification problems. No post processing was performed on the predicted probabilities or the predicted labels.
2.4. Experimental Design
CorrSigNIA used normalized T2w, ADC images and the top five CorrNet representations, where five was chosen based on our prior study (Bhattacharya et al., 2020). The correlated feature learning framework was trained using MRI and histopathology images from patients in cohort C1-train. For training the correlated feature learning framework, a learning rate η = 10−5, and 300 training epochs were used, based on our prior study (Bhattacharya et al., 2020).
The prostate cancer detection framework was trained using men from cohorts C1-train and C3-train. For training the prostate cancer detection framework, an Adam optimizer with an initial learning rate η = 10−3, weight decay α = 0.1, epochs = 100 with early stopping, and a batch size of 8 were used. A patience of 10 epochs was used to reduce learning rate on plateau and a patience of 20 epochs was used for early stopping on no improvement in validation loss. The last model from early stopping was used for evaluation on the test sets. In addition, the prostate cancer detection framework augmented the training data using one random rotation between angle −15 and 15 degrees and left-right flipping.
Several ablation studies and experiments were performed to (a) determine the best strategy to learn and integrate correlated features in CorrSigNIA, and (b) to compare CorrSigNIA performance with existing deep-learning models with MRI-only inputs. All experiments used the same training data with identical pre-processing steps, five-fold cross validation splits, batch size, and data augmentation. Moreover, the prostate cancer detection models were trained using the same class-balanced cross entropy loss function (Equation 6) to have multi-class predictions of normal, indolent and aggressive cancer. The training strategy using learning rate decay and early stopping was identical for all models, with the only exception being in the initial learning rate η. The following experiments were designed to ensure a fair comparison of different approaches with CorrSigNIA:
1. Models with MRI inputs only:
The following models with MRI-only inputs (T2w and ADC images) were trained: (a) SPCNet (Seetharaman et al., 2021), (b) U-Net (Ronneberger et al., 2015), (c) BrU-Net (branched U-Net). All MRI-only models were given three adjacent slices of T2w and ADC images as input, similar to the prostate cancer detection framework of CorrSigNIA. The BrU-Net architecture is a modification of the vanilla U-Net with two independent branches for T2w and ADC images, with each branch taking in three adjacent MRI slices. The BrU-Net incorporates the changes that SPCNet incorporates to the baseline HED architecture (independent branches for T2w and ADC images, and three adjacent slice inputs for each branch). The initial learning rate η was 10−3 for SPCNet, and 10−6 for U-Net and BrU-Net, decided based on experiments in our prior study (Seetharaman et al., 2021).
2. Models with MRI and correlated features as inputs:
(a). Correlated feature learning:
To determine the optimum objective function for the CorrNet model, several individual reconstruction error terms were defined:
is the reconstruction error when pathology features are used alone as input, enabling prediction of both radiology and pathology features from the shared representation of pathology features alone. Please note that H(Pi) = f(VPi + b) indicates that the hidden/shared representation is computed from pathology features alone, in absence of radiology features. The term G(.) is used as an abbreviation for Equation 2.
is the reconstruction error when radiology features are used alone as input, enabling prediction of both radiology and pathology features from the shared representation of radiology features alone.
is the reconstruction error when both radiology and pathology features are used as inputs, enabling prediction of both features from their shared representation.
is the cross reconstruction error, enabling reconstructing one modality features from the other modality features.
Each reconstruction term used a squared error loss function, similar to Equation 5. Then, the following CorrNet objective functions were defined by a combination of the individual reconstruction terms Lrecon1 through Lrecon4 with the correlation error term Lcorr defined in Equation 4:
J123(θ) = Lrecon1 + Lrecon2 + Lrecon3 − λLcorr.
J3(θ) = Lrecon3 − λLcorr.
J4(θ) = Lrecon4 − λLcorr.
The subscripts in each of the CorrNet objective functions indicate the individual reconstruction loss terms (Lrecon1 through Lrecon4) incorporated. We set λ = 2 for all objective functions, based on our prior study (Bhattacharya et al., 2020). For each objective function, the activation functions f and g in Equations 1 and 2 were simultaneously chosen as either identity or sigmoid, resulting in six different CorrNet models. The CorrNet representations learned from these six different CorrNet models were named as below:
CR-123-I trained with objective function J123(θ) and identity activation functions.
CR-123-S trained with objective function J123(θ) and sigmoid activation functions.
CR-3-I trained with objective function J3(θ) with identity activation functions.
CR-3-S trained with objective function J3(θ) with sigmoid activation functions.
CR-4-I trained with objective function J4(θ) with identity activation functions.
CR-4-S trained with objective function J4(θ) with sigmoid activation functions.
(b). Prostate cancer detection and characterization of aggressiveness:
Ablation studies with SPCNet as baseline: CorrNet representations (correlated features) corresponding to each of the six CorrNet models trained in Section 2.4 (2(a)) above were extracted for all patients in all cohorts. Histopathology images were not needed while extracting the CorrNet representations from the already trained CorrNet models. Six models with MRI and correlated features as inputs were trained using the SPCNet model as the baseline, as detailed in rows 1–6 of Table 2. These six MRI+correlated features-based models were evaluated on a lesion-level in detecting cancer and clinically significant cancer on cohort C1 (Tables 3, 4), and the two best performing CorrNet representations were identified.
Table 2:
Models with MRI and correlated features as inputs.
| Model name | Baseline model | CorrNet objective fn. | CorrNet activation fn. | CorrNet representation |
|---|---|---|---|---|
| CorrSigNIA | SPCNet | J3(θ) | Identity | CR-3-I |
| SPCNet + CR-3-S | SPCNet | J3(θ) | Sigmoid | CR-3-S |
| SPCNet + CR-123-I | SPCNet | J123(θ) | Identity | CR-123-I |
| SPCNet + CR-123-S | SPCNet | J123(θ) | Sigmoid | CR-123-S |
| SPCNet + CR-4-I | SPCNet | J4(θ) | Identity | CR-4-I |
| SPCNet + CR-4-S | SPCNet | J4(θ) | Sigmoid | CR-4-S |
| U-Net + CR-3-I | U-Net | J3(θ) | Identity | CR-3-I |
| U-Net + CR-123-I | U-Net | J123(θ) | Identity | CR-123-I |
| BrU-Net + CR-3-I | BrU-Net | J3(θ) | Identity | CR-3-I |
| BrU-Net + CR-123-I | BrU-Net | J123(θ) | Identity | CR-123-I |
Note: CorrNet activation fn. refers to functions f and g in Equations 1 and 2. Abbreviation used: fn. = function.
Table 3:
Detecting cancer: Lesion-level evaluation on cohorts C1-test and cohort C2.
| Evaluation on cohort C1-test | ||||||||||
| Patients with cancer lesion volumes ≥ 250 mm3 N = 37, number of cancerous lesions = 44 | ||||||||||
|
| ||||||||||
| Model | ROC AUC | PR AUC | Sensitivity | Specificity | Precision | NPV | F1 Score | Dice | Accuracy | Final Rank |
|
| ||||||||||
| SPCNet | 0.82±0.30 | 0.76±0.36 | 0.62±0.47 | 0.75±0.30 | 0.44±0.41 | 0.84±0.22 | 0.48±0.40 | 0.27±0.26 | 0.72±0.24 | 5 |
| U-Net | 0.82±0.34 | 0.81±0.34 | 0.84±0.35 | 0.30±0.31 | 0.35±0.27 | 0.57±0.47 | 0.45±0.25 | 0.30±0.20 | 0.43±0.26 | 8 |
| BrU-Net | 0.80± 0.32 | 0.78±0.34 | 0.82±0.35 | 0.39±0.38 | 0.37±0.29 | 0.57±0.46 | 0.47±0.28 | 0.27±0.17 | 0.52±0.27 | 9 |
|
| ||||||||||
| CorrSigNIA | 0.81±0.31 | 0.73±0.38 | 0.73±0.43 | 0.72±0.33 | 0.50±0.41 | 0.86±0.25 | 0.55±0.39 | 0.30±0.24 | 0.72±0.26 | 1 |
| SPCNet + CR-3-S | 0.79±0.31 | 0.72±0.38 | 0.62±0.46 | 0.80±0.21 | 0.45±0.39 | 0.88±0.15 | 0.49±0.38 | 0.27±0.23 | 0.75±0.17 | 4 |
| SPCNet + CR-123-I | 0.79±0.33 | 0.72±0.37 | 0.66±0.47 | 0.77±0.29 | 0.48±0.42 | 0.87±0.21 | 0.52±0.41 | 0.29±0.24 | 0.75±0.23 | 2 |
| SPCNet + CR-123-S | 0.73±0.35 | 0.70±0.38 | 0.55±0.48 | 0.82±0.24 | 0.41±0.41 | 0.83±0.21 | 0.44±0.40 | 0.27±0.24 | 0.75±0.20 | 11 |
| SPCNet + CR-4-I | 0.80±0.34 | 0.77±0.36 | 0.58±0.47 | 0.80±0.30 | 0.44±0.42 | 0.82±0.25 | 0.47±0.42 | 0.28±0.23 | 0.73±0.24 | 7 |
| SPCNet + CR-4-S | 0.79±0.34 | 0.75±0.37 | 0.69±0.44 | 0.72±0.32 | 0.50±0.41 | 0.82±0.28 | 0.54±0.39 | 0.30±0.23 | 0.71±0.27 | 3 |
| U-Net + CR-3-I | 0.80±0.34 | 0.77±0.34 | 0.89±0.29 | 0.27±0.33 | 0.34±0.23 | 0.46±0.48 | 0.47±0.24 | 0.29±0.17 | 0.44±0.26 | 10 |
| U-Net + CR-123-I | 0.80±0.31 | 0.72±0.38 | 0.88±0.32 | 0.41±0.37 | 0.40±0.29 | 0.65±0.46 | 0.51±0.28 | 0.31±0.21 | 0.55±0.28 | 6 |
| BrU-Net + CR-3-I | 0.77±0.38 | 0.75±0.38 | 0.88±0.32 | 0.10±0.25 | 0.30±0.22 | 0.20±0..9 | 0.42±0.23 | 0.27±0.17 | 0.32±0.23 | 13 |
| BrU-Net + CR-123-I | 0.72±0.39 | 0.66±0.40 | 0.69±0.44 | 0.58±0.38 | 0.43±0.38 | 0.70±0.38 | 0.49±0.37 | 0.27±0.23 | 0.62±0.30 | 12 |
|
| ||||||||||
| Evaluation on cohort C2 | ||||||||||
| Patients with cancer N = 147, number of cancerous lesions = 189 | ||||||||||
|
| ||||||||||
| Model | ROC AUC | PR AUC | Sensitivity | Specificity | Precision | NPV | F1 Score | Dice | Accuracy | Final Rank |
|
| ||||||||||
| SPCNet | 0.75±0.36 | 0.76±0.35 | 0.50±0.47 | 0.77±0.36 | 0.42±0.43 | 0.69±0.33 | 0.43±0.42 | 0.25±0.25 | 0.69±0.24 | 5 |
| U-Net | 0.78±0.33 | 0.77±0.34 | 0.76±0.39 | 0.57±0.39 | 0.50±0.35 | 0.66±0.41 | 0.56±0.33 | 0.33±0.23 | 0.63±0.26 | 3 |
| BrU-Net | 0.79± 0.35 | 0.79±0.34 | 0.89±0.29 | 0.27±0.36 | 0.41±0.26 | 0.41±0.47 | 0.53±0.25 | 0.33±0.20 | 0.48±0.26 | 4 |
|
| ||||||||||
| CorrSigNIA | 0.81±0.31 | 0.79±0.33 | 0.58±0.46 | 0.76±0.36 | 0.49±0.43 | 0.71±0.33 | 0.49±0.41 | 0.30±0.26 | 0.71±0.25 | 1 |
| SPCNet + CR-123-I | 0.77±0.33 | 0.76±0.34 | 0.55±0.47 | 0.81±0.33 | 0.47±0.44 | 0.75±0.29 | 0.48±0.42 | 0.30±0.27 | 0.74±0.22 | 2 |
| U-Net + CR-3-I | 0.49±0.43 | 0.54±0.39 | 0.40±0.46 | 0.59±0.40 | 0.30±0.38 | 0.56±0.36 | 0.31±0.37 | 0.19±0.22 | 0.53±0.29 | 9 |
| U-Net + CR-123-I | 0.67±0.38 | 0.66±0.38 | 0.52±0.47 | 0.67±0.39 | 0.41±0.41 | 0.65±0.35 | 0.42±0.39 | 0.24±0.24 | 0.62±0.28 | 7 |
| BrU-Net + CR-3-I | 0.81±0.33 | 0.81±0.33 | 0.90±0.27 | 0.12±0.26 | 0.36±0.20 | 0.22±0.40 | 0.49±0.20 | 0.32±0.19 | 0.39±0.21 | 6 |
| BrU-Net + CR-123-I | 0.61±0.40 | 0.62±0.39 | 0.41±0.46 | 0.78±0.34 | 0.37±0.44 | 0.68±0.30 | 0.36±0.41 | 0.21±0.25 | 0.66±0.25 | 7 |
Mean and standard deviation values for each metric in each cohort is reported. Column “Final rank” represents rank of model based on sum of individual metric ranks (detailed table with individual metric ranks in Supplementary table S1). The first three rows above the dotted lines in both cohorts represent the MRI-only models, whereas the rows below the dotted lines represent the MRI + correlated features based models. CorrSigNIA outperforms all models in cancer detection.
Table 4:
Detecting clinically significant cancer: Lesion-level evaluation on cohorts C1-test and cohort C2.
| Evaluation on cohort C1-test | |||||||||||
| Patients with clinically significant cancer with lesion volume ≥ 250 mm3 N = 34, number of clinically significant cancerous lesions = 37 | |||||||||||
|
| |||||||||||
| Model | ROC AUC | PR AUC | Sensitivity | Specificity | Precision | NPV | F1 Score | Dice | Accuracy | Agg cancer overlap | Final Rank |
|
| |||||||||||
| SPCNet | 0.83±0.30 | 0.77±0.37 | 0.68±0.47 | 0.76±0.29 | 0.47±0.41 | 0.88±0.21 | 0.52±0.41 | 0.29±0.25 | 0.75±0.24 | 0.18±0.28 | 4 |
| U-Net | 0.84±0.32 | 0.81±0.34 | 0.88±0.34 | 0.33±0.31 | 0.35±0.28 | 0.65±0.46 | 0.46±0.27 | 0.30±0.20 | 0.46±0.28 | 0.30±0.33 | 7 |
| Branched U-Net | 0.81 ± 0.32 | 0.75±0.38 | 0.84±0.36 | 0.39±0.37 | 0.34±0.28 | 0.62±0.46 | 0.45±0.28 | 0.27±0.18 | 0.51±0.27 | 0.20±0.26 | 12 |
|
| |||||||||||
| CorrSigNIA | 0.82±0.31 | 0.74±0.38 | 0.79±0.40 | 0.72±0.33 | 0.53±0.40 | 0.88±0.25 | 0.59±0.39 | 0.32±0.24 | 0.73±0.27 | 0.20±0.29 | 2 |
| SPCNet + CR-3-S | 0.79±0.31 | 0.70±0.39 | 0.66±0.47 | 0.79±0.22 | 0.45±0.39 | 0.91±0.13 | 0.51±0.40 | 0.28±0.23 | 0.77±0.18 | 0.14±0.26 | 8 |
| SPCNet + CR-123-I | 0.83±0.32 | 0.76±0.37 | 0.71±0.46 | 0.76±0.29 | 0.50±0.41 | 0.89±0.21 | 0.55±0.41 | 0.31±0.23 | 0.76±0.23 | 0.18±0.28 | 1 |
| SPCNet + CR-123-S | 0.77±0.34 | 0.73±0.39 | 0.60±0.48 | 0.81±0.25 | 0.44±0.41 | 0.86±0.20 | 0.48±0.42 | 0.29±0.24 | 0.77±0.21 | 0.17±0.27 | 9 |
| SPCNet + CR-4-I | 0.82±0.33 | 0.78±0.36 | 0.63±0.47 | 0.80±0.30 | 0.47±0.43 | 0.85±0.25 | 0.51±0.43 | 0.29±0.23 | 0.76±0.25 | 0.16±0.26 | 6 |
| SPCNet + CR-4-S | 0.81±0.32 | 0.76±0.37 | 0.72±0.44 | 0.72±0.32 | 0.50±0.41 | 0.85±0.29 | 0.55±0.39 | 0.31±0.23 | 0.72±0.27 | 0.21±0.29 | 3 |
| U-Net + CR-3-I | 0.82±0.32 | 0.75±0.36 | 0.93±0.25 | 0.24±0.28 | 0.31±0.19 | 0.48±0.49 | 0.44±0.20 | 0.28±0.18 | 0.41±0.23 | 0.36±0.29 | 10 |
| U-Net + CR-123-I | 0.83±0.29 | 0.75±0.37 | 0.90±0.29 | 0.41±0.37 | 0.39±0.29 | 0.66±0.46 | 0.50±0.27 | 0.31±0.21 | 0.53±0.29 | 0.46±0.34 | 5 |
| BrU-Net + CR-3-I | 0.80±0.36 | 0.77±0.37 | 0.91±0.28 | 0.09±0.20 | 0.26±0.17 | 0.22±0.41 | 0.39±0.19 | 0.26±0.16 | 0.29±0.18 | 0.53±0.30 | 13 |
| BrU-Net + CR-123-I | 0.73±0.41 | 0.70±0.40 | 0.72±0.44 | 0.56±0.38 | 0.41±0.37 | 0.70±0.39 | 0.49±0.37 | 0.27±0.23 | 0.60±0.31 | 0.42±0.36 | 11 |
|
| |||||||||||
| Evaluation on cohort C2 | |||||||||||
| Patients with clinically significant cancer N = 90, number of clinically significant cancerous lesions = 110 | |||||||||||
|
| |||||||||||
| Model | ROC AUC | PR AUC | Sensitivity | Specificity | Precision | NPV | F1 Score | Dice | Accuracy | Agg cancer overlap | Final Rank |
|
| |||||||||||
| SPCNet | 0.79±0.34 | 0.79±0.34 | 0.58±0.47 | 0.74±0.37 | 0.48±0.42 | 0.71±0.34 | 0.49±0.41 | 0.29±0.25 | 0.70±0.25 | 0.14±0.24 | 7 |
| U-Net | 0.83±0.30 | 0.81±0.32 | 0.81±0.37 | 0.58±0.39 | 0.53±0.34 | 0.68±0.41 | 0.60±0.32 | 0.38±0.23 | 0.67±0.26 | 0.26±0.28 | 3 |
| Branched U-Net | 0.84± 0.31 | 0.82±0.33 | 0.94±0.22 | 0.26±0.35 | 0.42±0.25 | 0.41±0.48 | 0.55±0.23 | 0.35±0.19 | 0.48±0.25 | 0.24±0.24 | 4 |
|
| |||||||||||
| CorrSigNIA | 0.86±0.26 | 0.83±0.31 | 0.67±0.43 | 0.74±0.38 | 0.55±0.41 | 0.72±0.36 | 0.57±0.39 | 0.35±0.25 | 0.74±0.25 | 0.24±0.31 | 1 |
| SPCNet + CR-123-I | 0.81±0.30 | 0.79±0.33 | 0.65±0.45 | 0.80±0.34 | 0.54±0.42 | 0.77±0.31 | 0.56±0.41 | 0.36±0.21 | 0.76±0.21 | 0.21±0.29 | 2 |
| U-Net + CR-3-I | 0.55±0.44 | 0.59±0.40 | 0.46±0.47 | 0.62±0.40 | 0.34±0.39 | 0.61±0.36 | 0.36±0.39 | 0.22±0.24 | 0.58±0.29 | 0.15±0.24 | 9 |
| U-Net + CR-123-I | 0.72±0.37 | 0.70±0.37 | 0.61±0.46 | 0.70±0.37 | 0.48±0.42 | 0.70±0.35 | 0.50±0.41 | 0.28±0.25 | 0.67±0.28 | 0.26±0.31 | 6 |
| BrU-Net + CR-3-I | 0.86±0.31 | 0.85±0.30 | 0.93±0.24 | 0.12±0.23 | 0.35±0.17 | 0.25±0.42 | 0.49±0.18 | 0.35±0.19 | 0.39±0.19 | 0.62±0.30 | 5 |
| BrU-Net + CR-123-I | 0.67±0.39 | 0.67±0.38 | 0.49±0.47 | 0.80±0.33 | 0.44±0.45 | 0.73±0.30 | 0.44±0.43 | 0.25±0.26 | 0.71±0.25 | 0.23±0.30 | 8 |
Mean and standard deviation values for each metric in each cohort is reported. Column “Final rank” represents rank of model based on sum of individual ranks (details in Supplementary material table S2). CorrSigNIA ranks second in cohort C1-test, closely following another CorrSigNIA variant, whereas it ranks first in cohort C2.
Ablation studies with U-Net models as baseline: The two best performing CorrNet representations for the SPCNet baseline model were then added as inputs (along with T2w and ADC images) to the vanilla U-Net and BrU-Net architectures, resulting in four U-Net model variants with CorrNet representations, as detailed in rows 7–10 of Table 2.
All the models were evaluated on cohorts C1, C2, and C3 using a lesion-level and patient-level evaluation approach as detailed in Section 2.5.
2.5. Evaluation Methods
2.5.1. Lesion-level evaluation
The models were evaluated for their ability to detect and localize (1) cancerous (aggressive and indolent) lesions and (2) clinically significant cancerous lesions. For cohort C1, clinically significant lesions were defined as cancerous lesions with at least 1% aggressive ground truth pixels labeled by the histologic grading platform (Ryu et al., 2019). For cohort C2, clinically significant lesions were defined as lesions with targeted biopsy Gleason grade group ≥ 2.
To evaluate the models on a lesion-level, the per-pixel ground truth labels in cohort C1 were converted to connected lesion labels, continuous in the MRI volume, using a morphological closing operation with a 3D structuring element. The 3D structuring element was formed by stacking three disks of sizes 0.5 mm, 1.5 mm and 0.5 mm respectively. This structuring element was chosen based on visual inspection of generated lesions from several randomly selected patients, after experimenting with various structuring elements of different sizes. Visual inspection was done to ensure that the generated lesions were continuous in 3D and faithfully represented the original data.
Lesion labels less than 250 mm3 in volume were discarded in our lesion-level evaluation because: (1) the PI-RADS v2 (Turkbey et al., 2019) guidelines state a lesion volume threshold of 500 mm3 for a lesion to be clinically significant cancer, and (2) previous studies (Matoso and Epstein, 2019; Seetharaman et al., 2021) have considered such small volume lesions (≈ 6 mm×6 mm ×6 mm) as clinically insignificant as they are often highly likely to be MRI invisible. Discarding these small lesions from our lesion-level analysis also helps in dealing with the radiology-pathology resgistration errors (~2 mm on prostate border, ~3 mm inside the prostate) from our RAPSODI registration platform (Rusu et al., 2020). The models were then evaluated on a lesion-level as follows:
True positives and false negatives: If the 90th percentile of the predicted labels within the ground truth lesion was cancer, the lesion was considered to be detected (true positive), otherwise it was considered not detected (false negative).
True negatives and false positives: The prostate was split into sextants, by first dividing it into left and right halves, and then dividing each half into 3 roughly equal regions, base, mid and apex along the Z-axis (Fig. 3). A sextant was labeled as ground truth negative if it had less than 5% ground truth cancer pixels. If the 90th percentile of the predicted labels for this sextant was normal, the sextant was considered to be correctly classified as a negative (true negative), otherwise it was considered to be wrongly classified as a positive (false positive). A sextant with more than 5% ground truth cancer pixels was evaluated as per step (1) above. This sextant-based approach for evaluating lesions was based on how biopsies are performed clinically (the most common biopsy protocol includes 12-core needle samples with 2 cores from each sextant) and has been used in prior studies (Seetharaman et al., 2021).
Fig. 3:
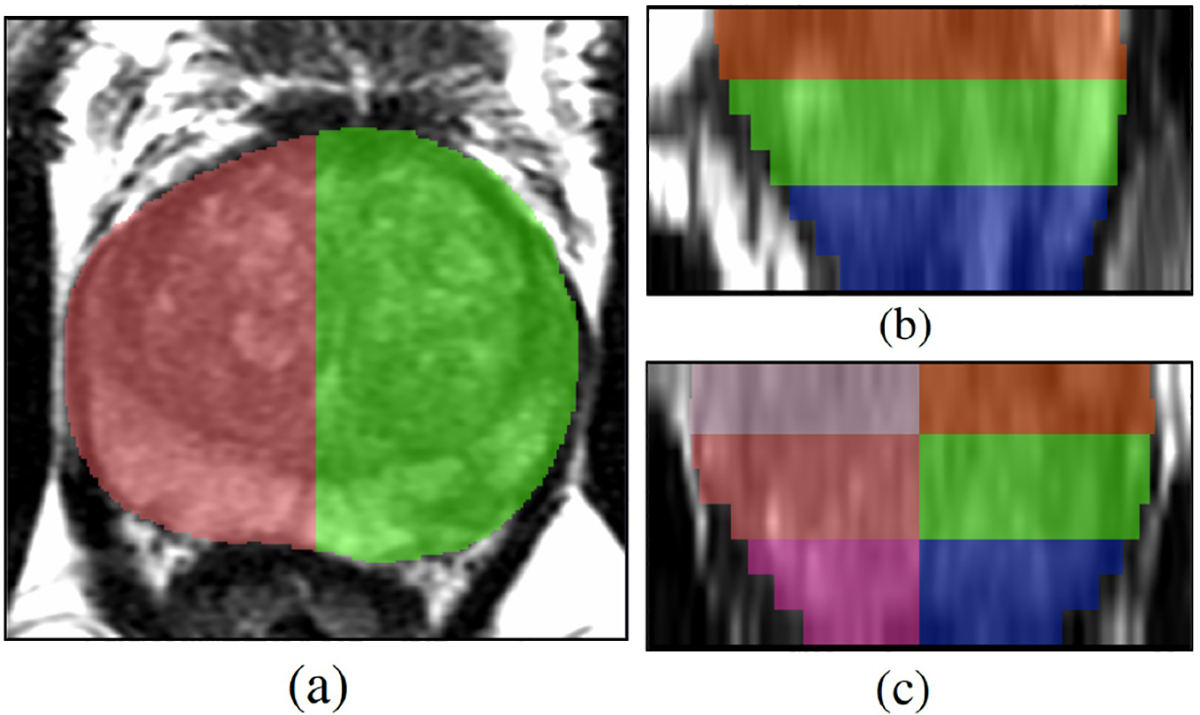
Sextants for lesion-level evaluation. (a) Axial, (b) Sagittal, and (c) Coronal views. The sextant-based approach for evaluating lesions was based on how biopsies are performed clinically. It is very common to do 12-core needle sampling with 2 cores from each sextant.
Metrics:
The predicted labels and the ground truth labels were used to compute true and false positives, and true and false negatives, which were then used to compute Sensitivity (Se), Specificity (Sp), Precision (Pr), Negative Predictive Value (NPV), F1-score, Dice, and Accuracy (Acc) on a perpatient basis. The 90th percentile of the predicted cancer probability maps derived by summing the indolent and aggressive cancer probabilities were used to compute the Area under the Receiver Operating Characteristic curve (ROC-AUC) and the Precision-Recall curve (PR-AUC). For clinically significant lesions, an additional metric “aggressive cancer overlap” was defined, which was computed as the percentage of overlap between predicted and ground truth aggressive labels. This aggressive cancer overlap metric measures how well the models selectively identified and localized aggressive cancer in mixed lesions.
For lesion-level evaluation on cohort C1-test, only patients having both pathologist labels and histologic grade labels (Ryu et al., 2019) and lesion volumes ≥ 250 mm3 were considered. For cohort C2, all biopsy confirmed radiologist lesions were considered, irrespective of size.
2.5.2. Patient-level Evaluation
All patients in cohort C1 were confirmed to have prostate cancer, while all patients in cohort C3 were confirmed to be normal. The models were evaluated on a patient-level in their ability to distinguish between normal and cancerous patients. For a patient with cancer, the lesion-level approach described in Section 2.5.1 was used. If the models correctly detected even one lesion, it was considered a true positive patient, otherwise a false negative patient. For normal patients without cancer, the model predicted labels were morphologically processed to produce predicted lesions, in a way similar to the ground truth lesion labels in Section 2.5.1. If the predicted lesions were less than 250 mm3 in volume, the patient was considered to be a true negative, otherwise the patient was a false positive.
Metrics:
All patients from C1-test (N=55) and C3-test (N=15) were combined to compute Sensitivity (Se), Specificity (Sp), Precision (Pr), Negative Predictive Value (NPV), F1-score, and Accuracy (Acc) in patient-level evaluation. Patients from cohort C2 were not used for patient-level evaluation because even if an MRI scan did not have radiologist outlined lesions and positive targeted biopsies, it was possible that systematic biopsies were positive. Thus, it was not possible to confirm whether a targeted biopsy negative man in cohort C2 was actually normal without cancer.
2.5.3. Ranking of models
The models were evaluated on a per-patient basis (mean and standard deviation values were computed for the test set patients for each evaluation metric). All models were then assigned a rank based on their performance in individual metrics, with the highest rank assigned to the model with the highest mean metric performance. If two models had the same mean metric performance, the model with the smaller standard deviation was assigned the higher rank. If both mean and standard deviation values were identical for more than one model, they were all assigned the same rank. Then, all individual metric ranks were summed up to derive an aggregate model rank. The model with the highest aggregate rank was considered to be the best model. This ranking-based evaluation scheme was motivated by the need to find clinically relevant models that perform consistently across all evaluation metrics. This ranking-based evaluation scheme has been used in prior studies to rank classification models (Brazdil and Soares, 2000; Nai et al., 2021), including ranking prostate segmentation models (Nai et al., 2021).
3. Results
3.1. Correlated feature learning
3.1.1. Qualitative evaluation
For the sample patient slice in Fig. 1, Fig. 4 shows VGG-16 low-level features extracted from the T2w slice (column (a)), ADC slice (column (b)), histopathology slice (column (c)), correlated latent feature representations when projected from MRI (H(Ri) (column (d)), also known as the CorrNet representations), and when projected from histopathology (H(Pi)) (column (e)). Column (d) shows a linear combination of the 64 textural feature maps from the T2w image and the 64 textural feature maps from the ADC image, while column (e) shows a linear combination of the 64 textural feature maps from the histopathology image. The linear combinations were learned by the CorrNet model in a way that columns (d) and (e) are highly correlated in the latent space. Rows in Fig. 4 show the top feature maps from each modality (columns (a)-(c)) that contributes to the corresponding correlated representations (columns (d)-(e)). It may be noted that in the CorrNet representations (Column (d)), the cancerous lesion on the left side of the image (pointed by the red arrow, and corresponding to the black outline in Fig.1) shows less textural variations than the rest of the image, thus indicating how correlated representations help to emphasize cancerous regions over normal tissue.
It may also be noted that although pixel-level correlations were learned, the correlated feature maps still capture spatial relationships and maintain spatial integrity. For example, the correlated MRI features (Fig. 4 (d)) have different signatures for the peripheral zone and central gland of the prostate, which are visually distinguishable in the correlated MRI features. Another visual indication that spatial relationships are not lost is that the signatures of the cancerous lesion (shown by the red arrow) are very different from the rest of the prostate, and the entire lesion stands out visually from the normal prostate tissue.
3.1.2. Ablation studies
Ablation studies to determine the best way to learn and integrate correlated features to the baseline SPCNet model showed that the CorrNet representations CR-3-I, obtained by minimizing CorrNet objective function J3(θ) with identity activation functions f and g performed the best in cancer detection on a lesion-level (Table 3) and in distinguishing between men with and without prostate cancer (Table 5). The second-best correlated feature in lesion-level evaluation was CR-123-I, while the correlated feature CR-4-S also showed improvements over MRI-only SPCNet model in lesion and patient level evaluations (Tables 3, 4, and 5). The two best-performing correlated features for lesion-level analysis (CR-3-I and CR-123-I) were selected for building U-Net + correlated feature models described in Section 2.4, which were used for comparative evaluation.
Table 5:
Patient-level evaluation on cohorts C1-test and C3-test (Total number of subjects N = 70, including 55 men with cancer from C1-test and 15 men with normal MRI from C3-test. Column “Final rank” represents rank of model based on sum of individual ranks (detailed in Supplementary material table S3). CorrSigNIA outperforms all other models in patient-level evaluation.
| Model | Sensitivity | Specificity | Precision | NPV | F1 score | Accuracy | Final Rank |
|---|---|---|---|---|---|---|---|
|
| |||||||
| SPCNet | 0.71 | 0.80 | 0.93 | 0.43 | 0.80 | 0.73 | 3 |
| U-Net | 0.87 | 0.00 | 0.76 | 0.00 | 0.81 | 0.69 | 13 |
| Branched U-Net | 0.87 | 0.13 | 0.79 | 0.22 | 0.83 | 0.71 | 9 |
|
| |||||||
| CorrSigNIA | 0.78 | 0.87 | 0.96 | 0.52 | 0.86 | 0.80 | 1 |
| SPCNet + CR-3-S | 0.73 | 0.53 | 0.85 | 0.35 | 0.78 | 0.69 | 12 |
| SPCNet + CR-123-I | 0.71 | 0.80 | 0.93 | 0.43 | 0.80 | 0.73 | 3 |
| SPCNet + CR-123-S | 0.67 | 0.87 | 0.95 | 0.42 | 0.79 | 0.71 | 7 |
| SPCNet + CR-4-I | 0.67 | 0.80 | 0.93 | 0.40 | 0.78 | 0.70 | 10 |
| SPCNet + CR-4-S | 0.78 | 0.73 | 0.91 | 0.48 | 0.84 | 0.77 | 2 |
| U-Net + CR-3-I | 0.91 | 0.07 | 0.78 | 0.17 | 0.84 | 0.73 | 8 |
| U-Net + CR-123-I | 0.89 | 0.07 | 0.78 | 0.14 | 0.83 | 0.71 | 10 |
| BrU-Net + CR-3-I | 0.91 | 0.00 | 0.77 | 0.00 | 0.83 | 0.71 | 5 |
| BrU-Net + CR-123-I | 0.78 | 0.53 | 0.86 | 0.40 | 0.82 | 0.73 | 5 |
3.2. Prostate cancer detection and characterization of aggressiveness
3.2.1. Qualitative evaluation
Fig. 5 shows all slices of a test set patient in cohort C1 from apex to base, with ground truth labels (column (d)) and CorrSigNIA predicted labels (column (e)). CorrSigNIA successfully predicted the majority of cancerous pixels, and identified the lesion as aggressive. CorrSigNIA predictions also had volumetric continuity in three dimensions. Fig. 6 shows all the slices of a test set patient in cohort C3 from apex to base. CorrSigNIA correctly predicted the patient as normal, without any false positive cancer predictions.
Fig. 5:
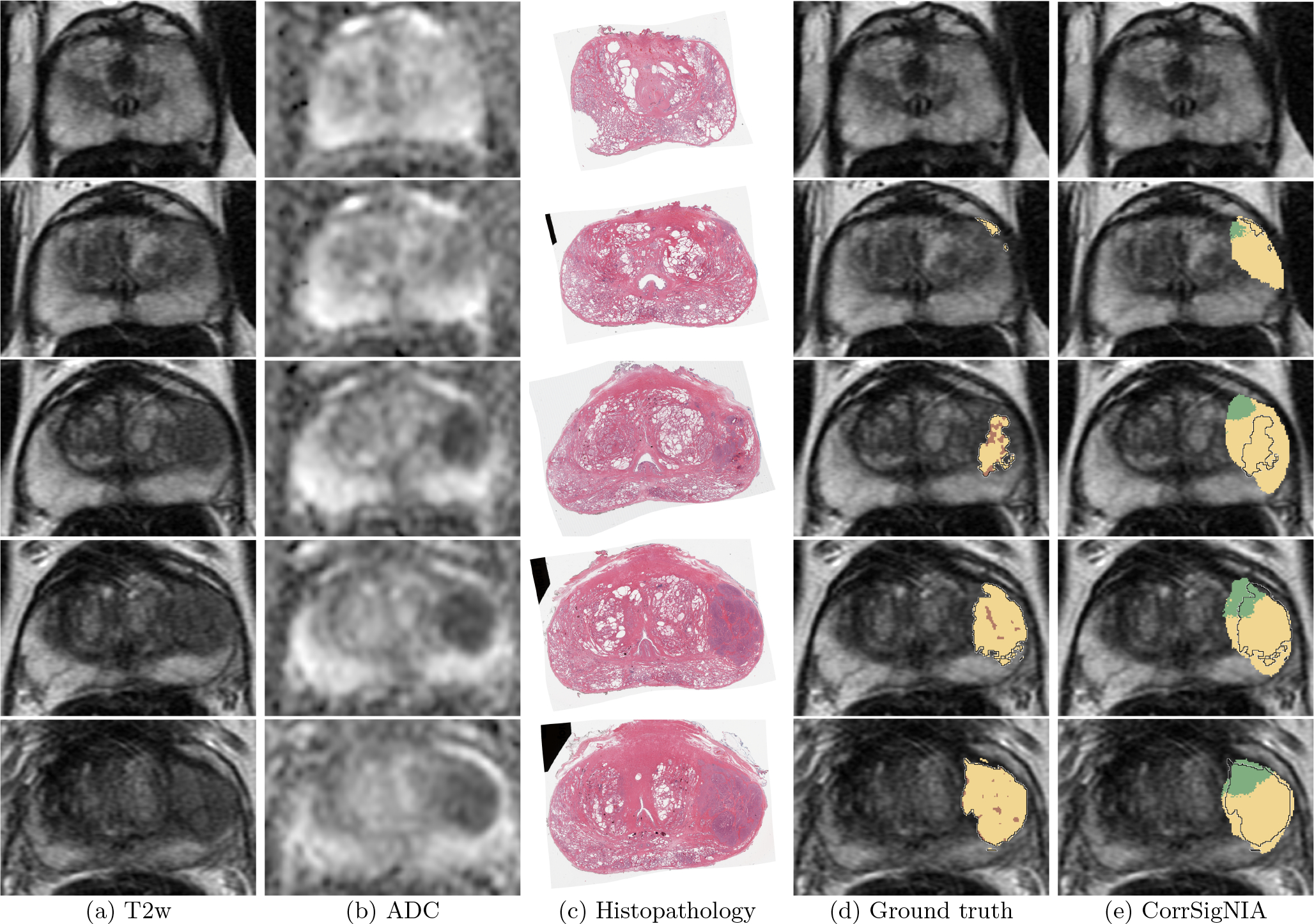
Detection and localization of aggressive cancer in a patient in cohort C1 shown from apex (top row) to base (bottom row). Registered (a) T2w image, (b) ADC image, (c) histopathology image, (d) T2w image overlaid with ground truth labels: cancer from expert pathologist (black outline), aggressive cancer (yellow) and indolent cancer (green) histologic grading from (Ryu et al., 2019), pixels within pathologist outline without automated histologic grade labels shown in brown, (e) T2w image overlaid with predicted labels from CorrSigNIA: predicted aggressive cancer (yellow) and predicted indolent cancer (green), black outline represents ground truth pathologist cancer outline.
Fig. 6:
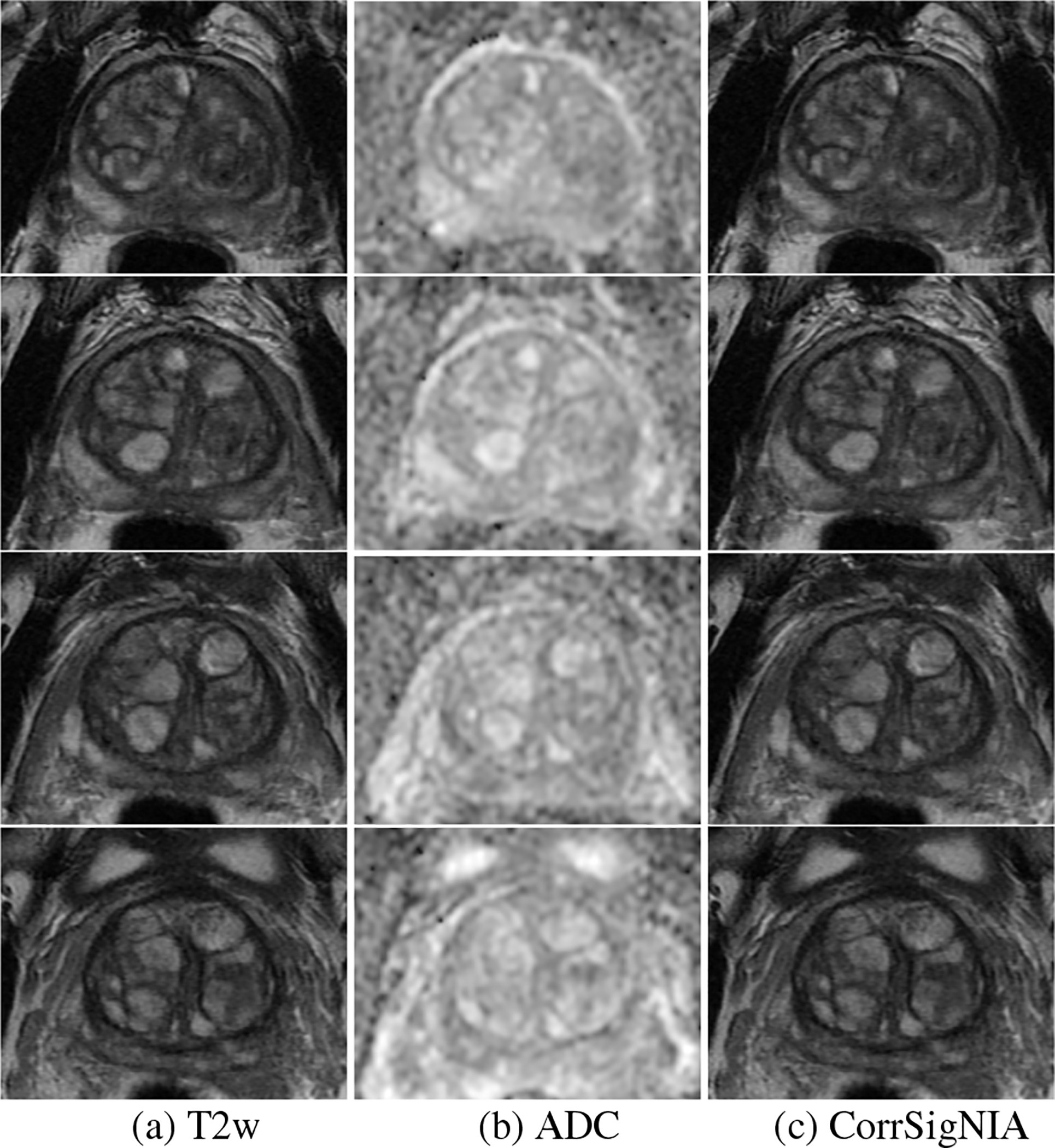
Prediction in a man from cohort C3 (without cancer) shown in 4 consecutive slices in the mid gland. (a) T2w, (b) ADC, and (c) CorrSigNIA predictions.
Fig. 7 shows comparative performance among the MRI-only models SPCNet (column (d)), U-Net (column (e)), BrU-Net (column (f)) and CorrSigNIA (column (g)) in six different patients, two from the test set in each cohort. Rows 1 and 2 are patients from cohort C1. CorrSigNIA detected the lesions in both these patients, with the least number of false positive predictions. CorrSigNIA also correctly identified the general areas of aggressive (yellow) and indolent (green) pixels in both patients. Rows 3 and 4 are patients from cohort C2. While all models detected the aggressive lesion in row 3, CorrSigNIA achieved the best localization, with fewer false positives. The patient on row 3 had an indolent lesion, which was only detected and correctly identified as indolent by CorrSigNIA. Although for this patient, CorrSigNIA had false positive predictions, when compared to the other models, the false positives were the least for CorrSigNIA. Rows 5 and 6 are men from cohort C3, without cancer. Only CorrSigNIA could correctly identify these patients as normal, while all other models had false positive predictions.
Fig. 7:
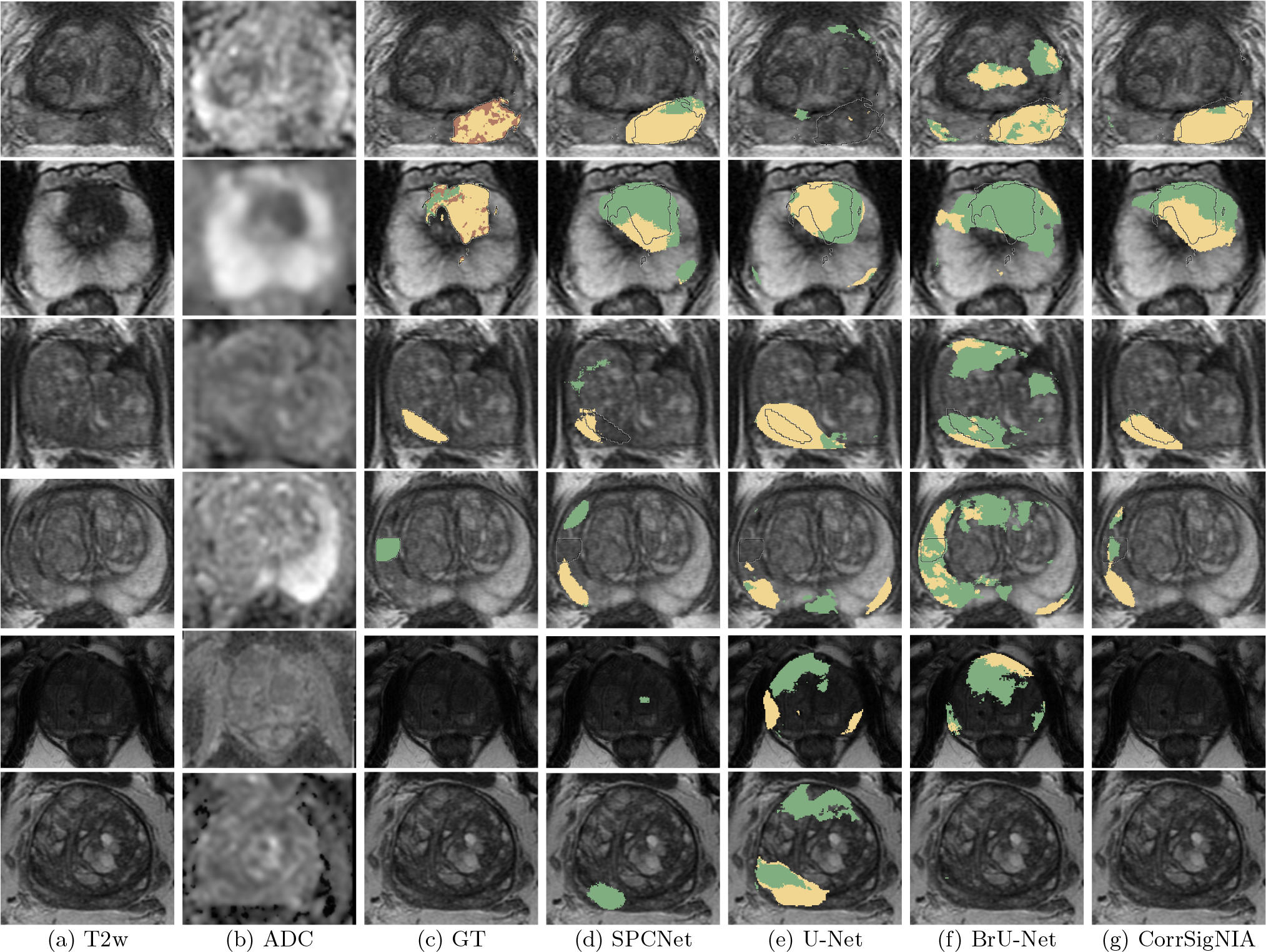
Qualitative performance comparison between SPCNet (Seetharaman et al., 2021), U-Net, BrU-Net and CorrSigNIA in six different patients from the test set of cohorts C1, C2, and C3. (a) T2w image and (b) ADC image. T2w image overlaid with (c) ground truth (GT) labels: cancer from expert pathologist (black outline), aggressive cancer (yellow) and indolent cancer (green) histologic grade labels from (Ryu et al., 2019). Predicted labels, aggressive cancer (yellow) and indolent cancer (green) from (d) SPCNet, (e) U-Net, and (f) branched U-Net and (g) CorrSigNIA (ours).
3.2.2. Quantitative evaluation
Tables 3, 4, and 5 show the lesion-level and patient level evaluations of CorrSigNIA in comparison with other models. Tables S1, S2, and S3 in the Supplementary material show the detailed versions of these tables, with individual metric ranks and sum of ranks, that were used to derive the final rank for each model.
In cancer detection, CorrSigNIA outperformed all other methods in both cohorts C1-test and C2 (Table 3). A paired sample t-test on patients with cancer from cohort C2 (N=147) showed that CorrSigNIA significantly improved Dice coefficient in cancer detection over the baseline MRI-only SPCNet (p = 0.0001). In clinically significant cancer detection, CorrSigNIA ranked second in cohort C1-test, closely following the CorrSigNIA variant, SPCNet + CR-123-I. CorrSigNIA ranked first in clinically significant cancer detection in cohort C2 (Table 4). The average overlap of CorrSigNIA aggressive cancer predictions with aggressive ground truth pixels were 0.20±0.29 for cohort C1-test and 0.24±0.31 for cohort C2. In patient-level evaluation, CorrSigNIA outperformed all other methods in differentiating between patients with and without cancer (Table 5).
Based on quantitative evaluation presented in Tables 3, 4, and 5, CorrSigNIA ranked first in 4 out of 5 evaluations. CorrSigNIA performed consistently better than all models across different metrics and cohorts, proving to be the best performing model and demonstrating the utility of adding correlated features to MRI-only models.
3.2.3. Analyzing false negative and false positive predictions
To assess the clinical applicability of CorrSigNIA in cancer detection and localization, false negative and false positive predictions were analyzed quantitatively and qualitatively. Cohorts C1-test and C3-test were used for this analysis as the ground truth labels in these two cohorts were the most complete (i.e., pixel-level annotations).
In patient-level evaluation, CorrSigNIA had false negative predictions in 12/55 patients in cohort C1-test. In lesion-level evaluation of clinically significant lesions, CorrSigNIA could detect 29/37 lesions, missing 8 lesions in 7 patients from cohort C1-test. The median volume of the missed lesions was 627 mm3, whereas the median volume of all the clinically significant lesions in C1-test was 1262 mm3, suggesting that the missed lesions were relatively small in size. Experienced radiologists had also missed 4 out of these 8 lesions missed by CorrSigNIA, suggesting that some of the missed lesions are hardly visible on MRI. Figure S1 in the supplementary material shows a patient from C1-test with false negative predictions from CorrSigNIA.
In cohort C3, on a patient level, CorrSigNIA had false positive predictions in 2/15 normal men without cancer. Fig. S2 in the supplementary material shows the normal MRI of a man without cancer with false positive predictions from CorrSigNIA. False positive predictions in clinically significant cancer localization were assessed using the sextant-based lesion-level evaluation in cohort C1-test. Of the 124 negative sextants, CorrSigNIA correctly predicted 85 sextants as completely benign, while it generated false positive predictions in 39 sextants. The false positive predictions in the 39 sextants had a median volume of 743.5 mm3, of which false positive aggressive cancer volume was 291.7 mm3, suggesting that the false positive predictions were predominantly indolent. False positive aggressive predictions accounted for a median 9.4% of the sextant volumes, further suggesting that they were relatively small in size, and the majority of the sextant was still correctly predicted benign, although the sextant as a whole was considered false positive.
4. Discussion
In this study, we presented CorrSigNIA, a radiology-pathology fusion model to selectively identify and localize aggressive and indolent prostate cancer on MRI. Our experiments showed that CorrSigNIA successfully distinguished between men with and without prostate cancer, detected and localized cancer and clinically significant cancer. Moreover, unlike prior studies, CorrSigNIA identified aggressive cancer components in mixed cancerous lesions. These encouraging results are due in part to the inclusion of correlated features in our model, using a strategy defined via extensive ablation studies. The ablation studies showed that optimizing an objective function that minimizes the reconstruction errors of both radiology and pathology features, and maximizes the correlation error between their shared representations, with identity activations in the CorrNet model, enabled learning the best correlated features. These correlated features when combined with the baseline SPCNet ar chitecture achieved the best performance in aggressive and indolent cancer detection.
Moreover, we compared CorrSigNIA with prior studies on prostate cancer detection using MRI-only models. While a direct comparison with published results from prior studies is not possible due to unavailability of data, we retrained the prior methods on our cohort, and designed these comparison experiments as fairly as possible, while not repeating experiments already performed in our prior studies (Seetharaman et al., 2021; Bhattacharya et al., 2020). The MRI-only models used in prior studies on prostate cancer detection and aggressiveness characterization are the U-Net (Sanyal et al., 2020; Schelb et al., 2019; De Vente et al., 2020; Ronneberger et al., 2015), the HED (Sumathipala et al., 2018; Xie and Tu, 2015) and the DeepLabv3+(Cao et al., 2019; Chen et al., 2017). In contrast to our study, these prior studies classified entire lesions into Gleason grade groups, without selectively identifying and localizing the aggressive components in mixed lesions. SPCNet (Seetharaman et al., 2021) was the only prior study that sought to selectively identify aggressive and indolent prostate cancer using pixel-level labels. We compared CorrSigNIA performance with three MRI-only models, the baseline SPCNet (Seetharaman et al., 2021), the vanilla U-Net (Ronneberger et al., 2015), and the branched U-Net (BrU-Net). Our experiments showed that CorrSigNIA performs consistently better than other models across different evaluation metrics and cohorts, suggesting robust and generalizable performance. This comparison proves the utility of correlated features over MRI-derived features alone in prostate cancer detection and in selective identification of indolent and aggressive cancer.
Although CorrSigNIA outperforms existing methods, we only found a moderate overlap between the ground truth and predicted aggressive pixels in mixed lesions. This moderate overlap is in part due to the pixelated nature of the histologic grade labels obtained by the automated Gleason scoring platform (Fig. 1(d)) when projecting them onto MRI. Despite these findings, CorrSigNIA enables identifying the aggressive cancer components of mixed lesions with a high confidence (Fig. 7, rows 1–2). Thus, CorrSigNIA may potentially help address the clinical unmet need of selectively identifying aggressive and indolent cancer and guiding targeted biopsies to aggressive cancer components in mixed lesions. Only one prior work, our SPCNet, sought to selectively identify the aggressive cancer components of mixed lesions, and was shown to be inferior to the proposed method, CorrSigNIA. We expect the overlap to improve as more training data is incorporated in our future work.
We chose a two-phase approach, that first identifies correlated features and second trains a detection model, as it allows us to solve the clinically relevant question of detecting aggressive cancer on pre-surgical MRI in the absence of histopathology images (e.g., prior to biopsy or surgery). Although training our two-step approach requires patients that have both MRI and histopathology images, in clinical settings (at test time) our CorrSigNIA network does not require the histopathology images. The ability of CorrSigNIA to operate with only MRI as input allows its usage to detect aggressive and indolent cancers on MRI before any pathology sample is collected. Thus, CorrSigNIA could be used to guide the clinical decision of whether to proceed with biopsy, radical prostatectomy, active surveilance, or any other form of treatment.
In comparison to existing methods of prostate cancer detection and characterization of aggressiveness, CorrSigNIA provides four important contributions. First, CorrSigNIA was trained using radical prostatectomy patients with accurate ground truth labels derived from registration between radiology and pathology images, enabling learning the complete cancer extent on MRI, including cancers that are invisible to most radiologists. Second, CorrSigNIA was trained using pixel-level cancer and histologic grade labels on prostate MRI, enabling selective identification of indolent and aggressive cancers in lesions that contain both histological subtypes (48% of all cancerous lesions, and 76% of index lesions). Third, CorrSigNIA used correlated feature learning to identify MRI features that are correlated with the corresponding histopathology features, enabling the formation of correlated feature maps capturing disease pathology characteristics on MRI. Fourth, CorrSigNIA combined the correlated features with MRI-derived features to selectively identify and localize normal tissue, indolent cancer and aggressive cancer on prostate MRI. To the best of our knowledge, CorrSigNIA is the first approach that leverages radiology-pathology fusion and correlated feature learning for both prostate cancer detection and for distinguishing aggressive from indolent cancer.
Our study has five notable limitations. First, the training cohort was relatively small (N = 98). This is a consequence of the uniqueness of cohort C1, which includes registered MRI and histopathology images of radical prostatectomy patients and pixel-level cancer and histologic grade labels for the entire prostate. The consistent performance of CorrSigNIA on the test set patients from all three cohorts demonstrate its generalizability, despite the limited number of training samples. Second, pixel-level histologic grade labels were derived from an automated method of grading on histopathology images (Ryu et al., 2019), rather than by expert genitourinary pathologists. However, automated methods of Gleason grading on histopathology (a) have excellent performance (Ryu et al., 2019; Bulten et al., 2020b), and (b) have shown to significantly improve Gleason grading by pathologists (Bulten et al., 2020a). The automated labels from (Ryu et al., 2019) have good overlap (Dice: 0.80 ± 0.09) with the pathologist cancer labels on the histopathology images in our dataset. This is sufficient to label MRI scans which are much lower resolution than the histopathology images. Moreover, it is impractical for genitourinary pathologists to manually annotate all prostate pixels with Gleason patterns for a sufficiently large population of patients to train machine learning models. To obviate error introduced by the automated method, we only used automated Gleason pattern labels that overlap with cancer labels from the expert pathologist. The automated method also improves uniformity in grading by reducing inter- and intra-pathologist variation in Gleason Grade group assignment. Third, the MRI-histopathology registration platform has registration errors (~2 mm on the prostate border and ~3 mm inside the prostate) (Rusu et al., 2020). These misalignment errors, which are more likely to affect small lesions, were diminished in our study by discarding these small, clinically insignificant lesions from our sextant-based lesion evaluation. Fourth, the data used for this study is from a single institution (Stanford University) and single manufacturer (GE Healthcare). We intend to include multi-institution and multiscanner data in future studies to test the generalizability of our models on external data. Fifth, our study used retrospective data and has not been validated in clinical settings. In future work, we plan to conduct prospective studies to assess the utility of CorrSigNIA in assisting radiologists in MRI interpretation.
The primary goals of prostate cancer diagnosis and treatment planning are to identify and treat aggressive cancer and to reduce over treatment of indolent cancer. Selective identification and localization of indolent and aggressive cancer on prostate MRI is an unmet clinical need. CorrSigNIA has the potential of improving prostate cancer care by (1) helping detect and target aggressive cancers that are currently missed (Ahmed et al., 2017), (2) eliminating unnecessary biopsies in men without cancer or with indolent cancers (Loeb et al., 2011; van der Leest et al., 2019), and (3) reducing the number of biopsy samples needed to detect aggressive cancers.
5. Conclusion
We have demonstrated the utility and performance of CorrSigNIA, a radiology-pathology fusion based algorithm for prostate cancer detection and for distinguishing indolent and aggressive cancers. CorrSigNIA learns from patients who underwent surgery with registered radiology and pathology images, and performs well in new patients without pathology, thereby making it a useful tool for diagnosis in clinical settings. CorrSigNIA outperformed existing models in prostate cancer detection, localization, and in selective identification of indolent and aggressive components of prostate cancer. Clinically, CorrSigNIA may improve prostate cancer care by helping target biopsies to aggressive cancer, reducing unnecessary biopsies, and deciding treatment plans.
Supplementary Material
Highlights.
Distinguishing indolent from aggressive prostate cancer on MRI is a clinical need
An automated method for distinguishing indolent from aggressive cancer is presented
Correlated feature learning is used to capture pathology characteristics on MRI
Deep learning is used for cancer detection and characterization of aggressiveness
Our method can improve prostate cancer care by guiding treatment planning
6. Acknowledgements
We acknowledge the following funding sources: Departments of Radiology and Urology, Stanford University, Radiology Science Laboratory (Neuro) from the Department of Radiology at Stanford University, GE Blue Sky Award, Mark and Mary Stevens Interdisciplinary Graduate Fellowship, Wu Tsai Neuroscience Institute (to JBW), National Cancer Institute, National Institutes of Health (U01CA196387, to JDB)
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham B, Nair MS, 2019. Automated grading of prostate cancer using convolutional neural network and ordinal class classifier. Informatics in Medicine Unlocked 17, 100256. [Google Scholar]
- Ahmed HU, Bosaily AES, Brown LC, Gabe R, Kaplan R, Parmar MK, Collaco-Moraes Y, Ward K, Hindley RG, Freeman A, et al. , 2017. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. The Lancet 389, 815–822. [DOI] [PubMed] [Google Scholar]
- Alkadi R, Taher F, El-Baz A, Werghi N, 2019. A deep learning-based approach for the detection and localization of prostate cancer in t2 magnetic resonance images. Journal of digital imaging 32, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armato SG, Huisman H, Drukker K, Hadjiiski L, Kirby JS, Petrick N, Redmond G, Giger ML, Cha K, Mamonov A, et al. , 2018. PROSTATEx Challenges for computerized classification of prostate lesions from multiparametric magnetic resonance images. Journal of Medical Imaging 5, 044501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barentsz JO, Weinreb JC, Verma S, Thoeny HC, Tempany CM, Shtern F, Padhani AR, Margolis D, Macura KJ, Haider MA, et al. , 2016. Synopsis of the PI-RADS v2 guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. European Urology 69, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya I, Seetharaman A, Shao W, Sood R, Kunder CA, Fan RE, Soerensen SJC, Wang JB, Ghanouni P, Teslovich NC, et al. , 2020. Corrsignet: Learning correlated prostate cancer signatures from radiology and pathology images for improved computer aided diagnosis, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 315–325. [Google Scholar]
- Brazdil PB, Soares C, 2000. A comparison of ranking methods for classification algorithm selection, in: European conference on machine learning, Springer. pp. 63–75. [Google Scholar]
- Bulten W, Balkenhol M, Belinga JJA, Brilhante A, Çakır A, Farré X, Geronatsiou K, Molinié V, Pereira G, Roy P, et al. , 2020a. Artificial intelligence assistance significantly improves gleason grading of prostate biopsies by pathologists. arXiv preprint arXiv:2002.04500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulten W, Pinckaers H, van Boven H, Vink R, de Bel T, van Ginneken B, van der Laak J, Hulsbergen-van de Kaa C, Litjens G, 2020b. Automated deep-learning system for gleason grading of prostate cancer using biopsies: a diagnostic study. The Lancet Oncology 21, 233–241. [DOI] [PubMed] [Google Scholar]
- Cao R, Bajgiran AM, Mirak SA, Shakeri S, Zhong X, Enzmann D, Raman S, Sung K, 2019. Joint prostate cancer detection and gleason score prediction in mp-MRI via FocalNet. IEEE Transactions on Medical Imaging 38, 2496–2506. [DOI] [PubMed] [Google Scholar]
- Chandar S, Khapra MM, Larochelle H, Ravindran B, 2016. Correlational neural networks. Neural Computation 28, 257–285. [DOI] [PubMed] [Google Scholar]
- Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL, 2017. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence 40, 834–848. [DOI] [PubMed] [Google Scholar]
- Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H, 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation, in: Proceedings of the European conference on computer vision (ECCV), pp. 801–818. [Google Scholar]
- De Vente C, Vos P, Hosseinzadeh M, Pluim J, Veta M, 2020. Deep learning regression for prostate cancer detection and grading in bi-parametric mri. IEEE Transactions on Biomedical Engineering. [DOI] [PubMed] [Google Scholar]
- García G, Colomer A, Naranjo V, 2019. First-stage prostate cancer identification on histopathological images: Hand-driven versus automatic learning. Entropy 21, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hectors SJ, Cherny M, Yadav KK, Beksaç AT, Thulasidass H, Lewis S, Davicioni E, Wang P, Tewari AK, Taouli B, 2019. Radiomics features measured with multiparametric magnetic resonance imaging predict prostate cancer aggressiveness. The Journal of urology 202, 498–505. [DOI] [PubMed] [Google Scholar]
- Hurrell SL, McGarry SD, Kaczmarowski AL, Iczkowski KA, Jacobsohn KM, Hohenwalter MD, Hall WA, See WA, Banerjee A, Charles DK, et al. , 2017. Optimized b-value selection for the discrimination of prostate cancer grades, including the cribriform pattern, using diffusion weighted imaging. Journal of Medical Imaging 5, 011004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Raman SS, Mirak SA, Kwan L, Bajgiran AM, Hsu W, Maehara CK, Ahuja P, Faiena I, Pooli A, et al. , 2019. Detection of individual prostate cancer foci via multiparametric magnetic resonance imaging. European urology 75, 712–720. [DOI] [PubMed] [Google Scholar]
- Kalavagunta C, Zhou X, Schmechel SC, Metzger GJ, 2015. Registration of in vivo prostate mri and pseudo-whole mount histology using local affine transformations guided by internal structures (latis). Journal of Magnetic Resonance Imaging 41, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JT, Xu S, Wood BJ, Turkbey B, Choyke PL, Pinto PA, Wang S, Summers RM, 2015. Automated prostate cancer detection using t2-weighted and high-b-value diffusion-weighted magnetic resonance imaging. Medical physics 42, 2368–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Leest M, Cornel E, Israel B, Hendriks R, Padhani AR, Hoogenboom M, Zamecnik P, Bakker D, Setiasti AY, Veltman J, et al. , 2019. Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naïve men with elevated prostatespecific antigen: a large prospective multicenter clinical study. European urology 75, 570–578. [DOI] [PubMed] [Google Scholar]
- Li L, Pahwa S, Penzias G, Rusu M, Gollamudi J, Viswanath S, Madabhushi A, 2017. Co-registration of ex vivo surgical histopathology and in vivo t2 weighted mri of the prostate via multi-scale spectral embedding representation. Scientific reports 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H, 2014. Computer-aided detection of prostate cancer in MRI. IEEE transactions on medical imaging 33, 1083–1092. [DOI] [PubMed] [Google Scholar]
- Liu W, Patil D, Howard DH, Moore RH, Wang H, Sanda MG, Filson CP, 2018. Adoption of prebiopsy magnetic resonance imaging for men undergoing prostate biopsy in the united states. Urology 117, 57–63. [DOI] [PubMed] [Google Scholar]
- Loeb S, Carter HB, Berndt SI, Ricker W, Schaeffer EM, 2011. Complications after prostate biopsy: data from seer-medicare. The Journal of urology 186, 1830–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losnegård A, Reisæter L, Halvorsen OJ, Beisland C, Castilho A, Muren LP, Rørvik J, Lundervold A, 2018. Intensity-based volumetric registration of magnetic resonance images and whole-mount sections of the prostate. Computerized Medical Imaging and Graphics 63, 24–30. [DOI] [PubMed] [Google Scholar]
- Matoso A, Epstein JI, 2019. Defining clinically significant prostate cancer on the basis of pathological findings. Histopathology 74, 135–145. [DOI] [PubMed] [Google Scholar]
- Nai YH, Teo BW, Tan NL, ODoherty S, Stephenson MC, Thian YL, Chiong E, Reilhac A, 2021. Comparison of metrics for the evaluation of medical segmentations using prostate mri dataset. Computers in Biology and Medicine, 104497. [DOI] [PubMed] [Google Scholar]
- Nyúl LG, Udupa JK, Zhang X, 2000. New variants of a method of MRI scale standardization. IEEE Transactions on Medical Imaging 19, 143–150. [DOI] [PubMed] [Google Scholar]
- Penzias G, Singanamalli A, Elliott R, Gollamudi J, Shih N, Feldman M, Stricker PD, Delprado W, Tiwari S, Böhm M, et al. , 2018. Identifying the morphologic basis for radiomic features in distinguishing different gleason grades of prostate cancer on mri: Preliminary findings. PloS one 13, e0200730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priester A, Natarajan S, Khoshnoodi P, Margolis DJ, Raman SS, Reiter RE, Huang J, Grundfest W, Marks LS, 2017. Magnetic resonance imaging underestimation of prostate cancer geometry: Use of patient specific molds to correlate images with whole mount pathology. The Journal of Urology 197, 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold JC, Dewey BE, Carass A, Prince JL, 2019. Evaluating the impact of intensity normalization on MR image synthesis, in: Medical Imaging 2019: Image Processing, International Society for Optics and Photonics. p. 109493H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical image computing and computer-assisted intervention, Springer. pp. 234–241. [Google Scholar]
- Rusu M, Shao W, Kunder CA, Wang JB, Soerensen SJ, Teslovich NC, Sood RR, Chen LC, Fan RE, Ghanouni P, et al. , 2020. Registration of presurgical mri and histopathology images from radical prostatectomy via rapsodi. Medical Physics 47(9), 4177–4188. doi:0.1002/mp.14337.Epub2020Jul18.PMID:32564359;PMCID:PMC7586964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HS, Jin MS, Park JH, Lee S, Cho J, Oh S, Kwak TY, Woo JI, Mun Y, Kim SW, et al. , 2019. Automated gleason scoring and tumor quantification in prostate core needle biopsy images using deep neural networks and its comparison with pathologist-based assessment. Cancers 11, 1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama WM, Aly MH, 2021. Prostate cancer detection based on deep convolutional neural networks and support vector machines: a novel concern level analysis. Multimedia Tools and Applications, 1–13. [Google Scholar]
- Sanyal J, Banerjee I, Hahn L, Rubin D, 2020. An automated two-step pipeline for aggressive prostate lesion detection from multi-parametric mr sequence. AMIA Summits on Translational Science Proceedings 2020, 552. [PMC free article] [PubMed] [Google Scholar]
- Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S, Kuder TA, Stenzinger A, Hohenfellner M, Schlemmer HP, et al. , 2019. Classification of cancer at prostate mri: deep learning versus clinical pi-rads assessment. Radiology 293, 607–617. [DOI] [PubMed] [Google Scholar]
- Seah JC, Tang JS, Kitchen A, 2017. Detection of prostate cancer on multiparametric mri, in: Medical Imaging 2017: Computer-Aided Diagnosis, International Society for Optics and Photonics. p. 1013429. [Google Scholar]
- Seetharaman A, Bhattacharya I, Chen LC, Kunder CA, Shao W, Soerensen SJ, Wang JB, Teslovich NC, Fan RE, Ghanouni P, et al. , 2021. Automated detection of aggressive and indolent prostate cancer on magnetic resonance imaging. Medical Physics. doi: 10.1002/mp.14855.PMID:33760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2020. Cancer statistics, 2020. CA: a cancer journal for clinicians 70, 7–30. [DOI] [PubMed] [Google Scholar]
- Simonyan K, Zisserman A, 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- Sonn GA, Fan RE, Ghanouni P, Wang NN, Brooks JD, Loening AM, Daniel BL, Too KJ, Thong AE, Leppert JT, 2019. Prostate magnetic resonance imaging interpretation varies substantially across radiologists. European urology focus 5, 592–599. [DOI] [PubMed] [Google Scholar]
- Sumathipala Y, Lay N, Turkbey B, Smith C, Choyke PL, Summers RM, 2018. Prostate cancer detection from multi-institution multiparametric MRIs using deep convolutional neural networks. Journal of Medical Imaging 5, 044507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ, Tempany CM, Choyke PL, Cornud F, Margolis DJ, et al. , 2019. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. European urology 76, 340–351. [DOI] [PubMed] [Google Scholar]
- Viswanath SE, Bloch NB, Chappelow JC, Toth R, Rofsky NM, Genega EM, Lenkinski RE, Madabhushi A, 2012. Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 Tesla endorectal, in vivo T2-weighted MR imagery. Journal of Magnetic Resonance Imaging 36, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanath SE, Chirra PV, Yim MC, Rofsky NM, Purysko AS, Rosen MA, Bloch BN, Madabhushi A, 2019. Comparing radiomic classifiers and classifier ensembles for detection of peripheral zone prostate tumors on T2-weighted MRI: A multi-site study. BMC Medical Imaging 19, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Burtt K, Turkbey B, Choyke P, Summers RM, 2014. Computer aided-diagnosis of prostate cancer on multiparametric mri: a technical review of current research. BioMed research international 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AD, Crukley C, McKenzie CA, Montreuil J, Gibson E, Romagnoli C, Gomez JA, Moussa M, Chin J, Bauman G, et al. , 2012. Prostate: registration of digital histopathologic images to in vivo mr images acquired by using endorectal receive coil. Radiology 263, 856–864. [DOI] [PubMed] [Google Scholar]
- Wu HH, Priester A, Khoshnoodi P, Zhang Z, Shakeri S, Afshari Mirak S, Asvadi NH, Ahuja P, Sung K, Natarajan S, et al. , 2019. A system using patient-specific 3d-printed molds to spatially align in vivo mri with ex vivo mri and whole-mount histopathology for prostate cancer research. Journal of Magnetic Resonance Imaging 49, 270–279. [DOI] [PubMed] [Google Scholar]
- Xie S, Tu Z, 2015. Holistically-nested edge detection, in: Proceedings of the IEEE international conference on computer vision, pp. 1395–1403. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


