Abstract
Infectious disease outbreaks are causing widespread declines of marine invertebrates including corals, sea stars, shrimps, and molluscs. Dermo is a lethal infectious disease of the eastern oyster Crassostrea virginica caused by the protist Perkinsus marinus. The Pacific oyster Crassostrea gigas is resistant to Dermo due to differences in the host-parasite interaction that is not well understood. We compared transcriptomic responses to P. marinus challenge in the two oysters at early and late infection stages. Dynamic and orchestrated regulation of large sets of innate immune response genes were observed in both species with remarkably similar patterns for most orthologs, although responses in C. virginica were stronger, suggesting strong or over-reacting immune response could be a cause of host mortality. Between the two species, several key immune response gene families differed in their expansion, sequence variation and/or transcriptional response to P. marinus, reflecting evolutionary divergence in host-parasite interaction. Of note, significant upregulation of inhibitors of apoptosis (IAPs) was observed in resistant C. gigas but not in susceptible C. virginica, suggesting upregulation of IAPs is an active defense mechanism, not a passive response orchestrated by P. marinus. Compared with C. gigas, C. virginica exhibited greater expansion of toll-like receptors (TLRs) and positive selection in P. marinus responsive TLRs. The C1q domain containing proteins (C1qDCs) with the galactose-binding lectin domain that is involved in P. marinus recognition, were only present and significantly upregulated in C. virginica. These results point to previously undescribed differences in host defense genes between the two oyster species that may account for the difference in susceptibility, providing an expanded portrait of the evolutionary dynamics of host-parasite interaction in lophotrochozoans that lack adaptive immunity. Our findings suggest that C. virginica and P. marinus have a history of coevolution and the recent outbreaks may be due to increased virulence of the parasite.
Keywords: oyster, comparative transcriptomics, dermo disease, innate immune response, host-parasite, interaction, gene expansion, adaptation
Introduction
Marine diseases can be caused by a variety of factors, such as parasite infections, biological toxins and environmental stress (Bossart, 2007; Burge et al., 2014; Wilson and Ho, 2015). Environmental stress caused by climate change and other human activities contributes to the development of marine diseases. Global warming has caused range extension of some parasites (Ford, 1996; Studer et al., 2010), and ocean acidification may weaken the immune defense of calcifying hosts (Bibby et al., 2008). Human activities have facilitated rapid transmission of some marine infectious diseases (Harvell, 1999; Harvell et al., 2004; Guo and Ford, 2016). Outbreaks of marine infectious disease have become more frequent and severe in recent decades (Groner et al., 2016; Sutherland et al., 2016).
Infectious diseases can cause mass mortalities of diverse marine organisms, affecting their abundance, changing community structure, and threatening the health of marine ecosystems (Harvell et al., 2004; Tim, 2007). They may impact major fishery and aquaculture species and cause immeasurable economic losses. Molluscs are a major group of marine animals, many of which are economically important as major fishery and aquaculture species (Guo et al., 1999; Shumway, 2021). Molluscan populations, fisheries and aquaculture are seriously affected by infectious disease outbreaks that have become more frequent and severe due to climate changes and human activities (Guo and Ford, 2016).
Dermo is a lethal disease of the eastern oyster Crassostrea virginica caused by the protozoan parasite Perkinsus marinus. The intracellular parasite is transmitted directly from oyster to oyster through the water. The infection is characterized by hemocyte infiltration followed by tissue fibrinolysis and blockages of hemolymph vessels, leading to inhibition of gonadal development and death of the oyster by emaciation (Faisal et al., 1999). Dermo causes extensive mortalities of the eastern oyster along the Atlantic coast of United States and threatens both aquacultural and wild populations (Levine, 1978; Reece et al., 2008). Mortality events associated with this disease cause tremendous losses in the oyster industry, and often wipe out up to 100% of harvestable naive stocks. Such mass mortalities negatively impact water quality and the overall health of the ecosystem (Newell, 1988). While P. marinus is a highly successful parasite of C. virginica, the Pacific oyster Crassostrea gigas, a sister species from Asia, is resistant to P. marinus infections (Meyers et al., 1991). The parasite can infect C. gigas at low intensity but does not cause serious pathologies or mortalities. How sister species without adaptive immunity differ in their susceptibility to the same pathogen is of fundamental interest. The difference in Dermo resistance between two Crassostrea oysters provides a good opportunity to study the evolution of host-parasite interactions in organisms without adaptive immunity.
The complex interaction between the host and parasite is fundamental to the infection process, the outcome of which often determines life or death of the host or parasite. The evolutionary race between a host and its parasites as best characterized by the Red Queen hypothesis is intense and everlasting (Van Valen, 1973). The ability of a particular pathogen to cause disease is determined by its virulence, and the total “cards of virulence” possessed by a pathogen determine its relative capacity to induce disease within a host (Winnicki, 2010; Kim et al., 2016). Host organisms need to evolve sophisticated defense mechanisms against parasites that constantly invent new ways to infect. Host-pathogen interactions are often dynamic, and the disease outcome is commonly dependent on multiple factors including the state of the host, the pathogenic potential of the pathogen and environmental conditions or stressors (Allam et al., 2013; Debasish et al., 2021).
While bivalve molluscs do not have the canonical adaptive immune system, they have evolved effective host-defense mechanisms to cope with biotic and abiotic stress (Guo et al., 2015; Wang et al., 2018). The bivalve host-defense system includes two major components, humoral and cellular immune responses. On the molecular level, the humoral immunity starts from recognizing conserved microbial-associated molecular patterns (MAMPS) and pathogen-associated molecular patterns (PAMP) (Aladaileh et al., 2007). Upon recognition, the pathogen-associated pattern recognition receptors (PRRs) activate immune signaling pathways and antimicrobial effectors (Akira et al., 2006). The cellular immune responses begin from phagocytosis and encapsulation by hemocytes, and then trigger the release of enzymes and oxygen metabolites (Wang et al., 2018). Oxidative bursts not only have a cytotoxic effect on invading pathogens but also serve as signals that activate further defense reactions (Anderson, 1999; Goedken et al., 2005). Besides immune reactions, apoptosis (Zhang et al., 2011; Qu et al., 2015; Li et al., 2017) and authophage (Moreau et al., 2015) also play an important role in host defense of bivalves.
Despite the devastating impact of Dermo, the molecular mechanisms of the host-parasite interplay between oysters and P. marinus are still largely unknown (Anderson, 1996). Tanguy et al. (2004) identified candidate genes from several pathways that were upregulated in response to P. marinus infection in C. virginica and C. gigas, including toll-like receptors, galectin, and genes involved in energy metabolism and metal binding. A galectin of C. virginica has been shown to function as a hemocyte receptor that recognizes P. marinus and facilitates phagocytosis (Tasumi and Vasta, 2007). Using a 12 K oligonucleotide microarray, Wang et al. (2010) revealed that genes involved in antimicrobial defense, pathogen recognition and uptake, oxidative stress and apoptosis are differentially expressed under P. marinus infection. Transcriptomic analyses in C. virginica identified a large set of differentially expressed genes and the upregulation of genes for proteolysis regulation and oxidation-reduction processes in resistant families (Proestou and Sullivan, 2020). Models have been developed to simulate and predict P. marinus transmission in C. virginica populations (Powell et al., 2011; Bidegain et al., 2017). It has been suggested that C. gigas may be more effective in degrading P. marinus than C. virginica (La Peyre et al., 1995), although the molecular mechanisms that underpin its effective defense are not understood.
In this study, we compared transcriptomic response to P. marinus infection in the susceptible eastern oyster with that of the resistant Pacific oyster, to better understand the evolutionary dynamics of host-parasite interactions. Comparative analyses revealed specific dynamic and orchestrated expression of a large set of innate immune genes in both species. Comparing infected gene expression profiles against P. marinus infection between two oysters identified previously undescribed differences in host defense, which may underpin differences in susceptibility and point to new mechanisms for the evolution of disease resistance. These results contribute to our understanding of host-defense and adaptation, as well as the development of disease-resistant stocks for aquaculture.
Materials and Methods
Experimental Oysters, P. marinus Challenge and Sample Collection
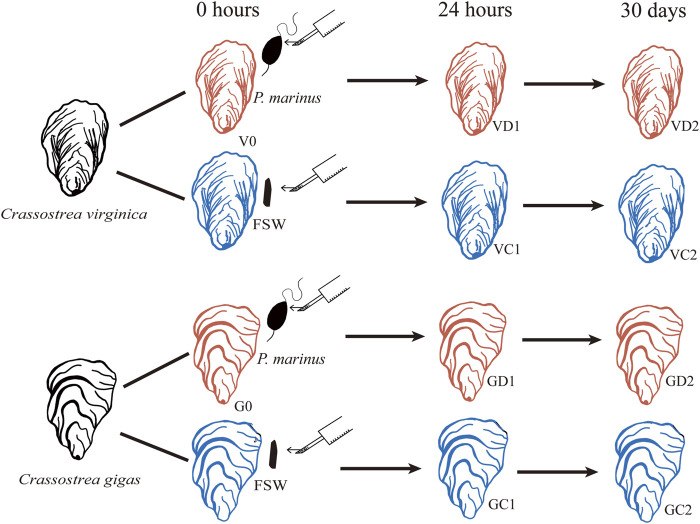
The eastern and Pacific oysters used in this study were obtained from oyster farms in Maine and Washington, respectively. Oysters were brought to the lab and acclimated for 24 h in 1 μm filtered seawater at 23°C before the challenge experiment. All oysters were notched at the ventral edge of the shell adjacent to the gills in preparation for shell cavity inoculation after previous studies using P. marinus harvested from infected oysters (Chintala et al., 2002; Ford et al., 2002). For the challenged group, the shell cavity of each oyster was inoculated with 1.2 × 106 P. marinus cells in 100 μL of seawater fortified with 100 U penicillin and 100 μg of streptomycin mL−1 (pen/strep). The control oysters were injected with 100 μL filtered seawater (FSW) similarly fortified with pen/strep (Figure 1). At the time of injection, ten oysters from each species were sampled, and a piece of gill was preserved in RNAlater as Time 0 samples. After injection, a wide rubber band was placed around each oyster to cover the notch and to minimize ejection of the inoculum. Eight hours after the injection, the oysters were returned to seawater and fed every day with algal paste. Seawater was changed twice per week. The infection by P. marinus was determined with Ray’s Fluid Thioglycollate Medium (RFTM, Bushek et al., 1994). At 24 h and 30 days post injection, ten oysters were sampled from challenged and control groups of each species by fixing a piece of gill in RNAlater. Samples were stored at −80°C before RNA extraction.
FIGURE 1.
Schematic diagram of experimental design. FSW, filtered seawater.
RNA Isolation and Library Construction
Total RNA of each sample was extracted using the Trizol-chloroform extraction method. The RNA quality and concentration were checked by Qubit™ RNA BR Assay Kit (Thermo Fisher Scientific) and Agilent Bioanalyzer 2100. All extracted samples had an A260/280 ratio greater than 1.8. RNA from the ten oysters was extracted separately and then pooled with equal amounts into one sample for library construction. RNA-sequencing libraries were constructed using the NEBNext Ultra RNA Library Prep Kit, following the manufacturer’s instructions. Briefly, 1 mg of total RNA from each sample was used for the double stranded cDNA synthesis. Then, size selection was performed using a 2% agarose gel to remove self-ligated adaptors, generating cDNA libraries with an average size of 200–250 bp. RNA sequencing of prepared libraries was carried out on high-throughput Illumina sequencing platforms. Libraries were quantified, multiplexed, and sequenced as 49 bp single-end reads. A total of 159 M reads were generated in this study (Supplemenatary Table S1).
Data Analysis, Identification and GO Enrichment Analysis of Differentially Expressed Genes
FastxToolkit pipeline (http://hannonlab.cshl.edu/fastx_toolkit/index.html) was used to process the raw reads to evaluate sequencing quality, remove low-quality reads (length threshold <50 bp and quality threshold <20), adaptor sequences, poly-N and known non-coding RNAs. The obtained clean reads were then mapped to the genomes of C. virginica (v3.0; NCBI Bioproject PRJNA379157) and C. gigas (v9; NCBI Bioproject PRJNA70283) by Tophat2 software (v2.1.1, Trapnell et al., 2009). Gene expression levels were measured by fragments per kilobase of exon per million fragments mapped (FPKM) using HT-seq (Anders et al., 2015). The differentially expressed genes (DEGs) were identified with the edgeR tool of R programming language with the threshold value |log2FC| ≥ 1 (multiple of Fold change, FC: difference) and FDR ≤ 0.05. The gene ontology (GO) enrichment analysis of DEGs was conducted with Blast2GO software. The annotated results with corrected FDR ≤ 0.05 were selected as the enrichment functions. Fisher’s LSD was used to test significant enrichment when the number of genes in a GO term was less than 5, and χ2 test was used when the number of genes ≥5.
Identification of Orthologous Genes Between C. virginica and C. gigas
OrthoFinder (Emms and Kelly, 2015) was used to identify orthologous genes between C. virginica and C. gigas with the following steps: 1) all-vs-all search through BLAST for potential orthologous genes with evalue ≤ 1e-3 as the threshold; 2) standardize the score of BLAST bit based on gene length and phylogenetic analysis; 3) use Reciprocal Best length-Normalised Hit (RBNHs) to determine the threshold of sequence similarity of homologous groups; and 4) Markov clustering (MCL) gene clustering and orthologous grouping.
Identification of Immune-Related Genes
We used domain prediction and sequence homology search to annotate genes that are potentially involved in immune response in C. virginica and C. gigas. First, InterProScan (Quevillon et al., 2005) and Pfam (Mistry et al., 2021) were used to annotate genes and gene models with characteristic immune domains or motifs. Second, BLASTP package (Mahram and Herbordt, 2015) was used to search against oyster gene datasets with canonical immune genes from model organism as inquiries. Third, TBLASTN package (Gertz et al., 2006) and Genewise (http://www.ebi.ac.uk/Tools/Wise2/index.html) were used to predict genes from genome sequences. Immune gene models predicted from these three methods were integrated to the primary immune geneset. Then, all the candidate gene models annotated with immune function by blast against NCBI nr database and predicted with immune domains by SMART (http://smart.embl-heidelberg.de/) were manually checked, resulting in the final immune geneset.
Phylogenetic Analysis
Multiple sequence alignments were performed using ClustalW with default parameters and the resulting alignments were refined with trimAl (Capella-Gutierrez et al., 2009). Phylogenetic trees were constructed with maximum likelihood (ML) analytical approaches. MEGA and PHYML were used to construct the NJ and ML trees, respectively. The robustness of the inferred trees was assessed using bootstrapping 1,000 in the phylogenetic tree.
PAML (v4) was used for comparing the rate per site of dN (non-synonymous) to the rate per site of dS (synonymous) mutations. The recommended subset of four M-series models of M1a (nearly neutral), M2a (positive selection), M7 (beta) and M8 (beta & ω) coupled with Bayesian Empirical Bayes (BEB) methods (Yang et al., 2005) were implemented. The Log-likelihood values (lnL) of M2a-M1a and M8-M7 were from explicit tests for the presence of positively selected sites. The p values were corrected by a multiple testing correction method (Benjamini and Hochberg, 1995). Furthermore, the probabilities of sites under positive selection were assessed by their posterior probabilities calculated with the BEB method. The amino acid site would be considered as a positively selected site if the value of dN/dS > 1 appears in the LRT and posterior probability exceeds 90% (Lan et al., 2018). Finally, SWISS-MODEL (http://swissmodel.expasy.org/) was used to locate and visualize the positively selected sites.
Results
P. marinus Challenge Experiment in the Oysters
No P. marinus was detected in C. virginica and C. gigas at Time 0, and in unchallenged control of either species on Day 30 (n = 10 for all). In the challenged groups, P. marinus was detected in 90% of C. virginica (n = 40) at an average infection intensity of 3.2 (∼160,938 P. marinus cells g−1 wet tissue, Choi et al., 1989), which is heavy and indicates that the challenge is successful. Infection was detected in all C. gigas (n = 15) but at a lower average infection intensity of 1.4 (∼11,202 P. marinus cells g−1 wet tissue, Choi et al., 1989), confirming a successful challenge and that C. gigas is resistant or less susceptible.
Transcriptome Analyses of C. virginica Response to P. marinus Challenge
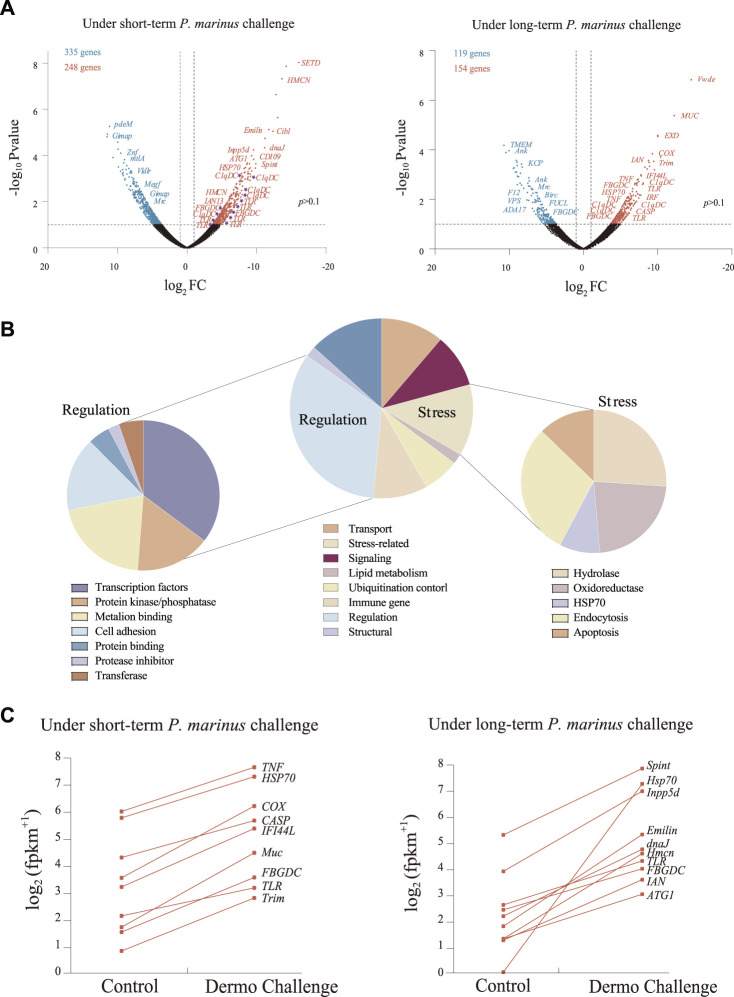
After filtering and cleaning, 65,537,432 high-quality reads were mapped to the reference genome for C. virginica, averaging 10 to 14 million per sample (Supplementary Table S1). The mapping ratio ranged from 80.8 to 84.2%. Of the 39,505 protein-coding genes in the C. virginica genome, 28,918 genes showed expression levels of FPKM ≥ 1. Overall, 759 genes were differentially expressed in oysters challenged with P. marinus (VD1 and VD2) compared with unchallenged controls (V0, VC1, and VC2) (Supplemenatary Table S2), including 583 DEGs from short-term challenge (24 h) and 273 DEGs from long-term challenge (30 days) (Figure 2A). Of these, 248 and 154 DEGs were up-regulated while 335 and 119 genes were down-regulated under short- and long-term P. marinus challenge, respectively (Figure 2A).
FIGURE 2.
Transcriptional changes induced by P. marinus in C. virginica. (A), Volcano plots of transcriptional change under short-term and long-term P. marinus challenge. Blue points represent downregulated genes, red points represent upregulated genes, and black points represent non-differentially expressed genes. (B), Functional annotation of differentially expressed genes under P. marinus challenge. (C), Immune-related genes significantly up-regulated after dermo challenge.
Functional analysis showed that the 759 DEGs induced by P. marinus challenge were mostly involved in transport, stress, signaling, lipid metabolism, ubiquitination control, immune response, regulation and structure (Figure 2B). The “regulation” term was further classified into categories of transcription factors, protein kinase/phosphatase, metal ion binding, cell adhesion, protein inhibitor and transferase. Similarly, the “stress” term included sub-categories of hydrolase, oxidoreductase, HSP70, endocytosis and apoptosis. The up-regulation of genes related to “regulation” and “stress” terms indicated that genes related to stress response system and metabolism were largely recruited under challenge.
A large number of genes encoding immune receptors (e.g., TLRs, C1qDCs, FBGDCs) and effectors were significantly upregulated during pathogen infection (Figures 2A,C), revealing their important role in the response to P. marinus challenge in C. virginica.
Comparative Transcriptomic Analyses of Resistant C. virginica and Susceptible C. gigas
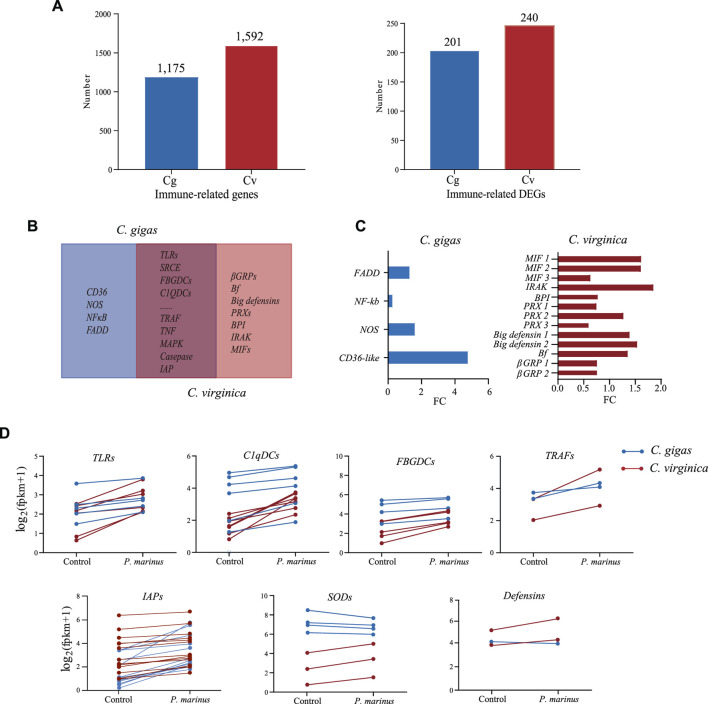
To understand the differences in host defense between the two species, we first annotated immune related genes in the two genomes: 1,175 in C. gigas and 1,592 in C. virginica (Figure 3A). Of these, 201 in C. gigas and 240 in C. virginica were differentially expressed during P. marinus infection (Figure 3A). Most of these genes were shared by the two oyster species (Figure 3B). These shared DEGs mainly consisted of genes for immune recognition receptors, effectors, lectins, and other proteins related to host-parasite interaction that play important roles in preventing infection or improving defense against pathogens. In addition, genes coding for CD36, NOS, NF-κB, and FADD were differentially expressed in C. gigas but not in C. virginica. Meanwhile genes coding for βGRP, Bf, Big defensin, PRX, BPI, IRAK, and MIF were differentially expressed only in C. virginica (Figure 3C). The gene Cvgal, a galectin consisted of four carbohydrate recognition domains (CRDs) and a hemocyte receptor of P. marinus (Tasumi and Vasta, 2007), was only significantly upregulated by P. marinus infection in C. virginica (Supplementary Figure S1).
FIGURE 3.
Comparative transcriptome analysis between C. gigas and C. virginica under P. marinus challenge. (A), Number of immune-related genes identified (Left) and number of immune-related genes differentially expressed (Right) in C. gigas (Cg) and C. virginica (Cv). (B), Venn diagrams of differentially expressed immune genes under P. marinus challenge between the two oyster species. (C), Unique and differentially expressed immune genes induced by P. marinus in two oyster species. (D), Transcriptional change of differentially expressed immune genes shared by two oyster species, including pattern recognition receptors (e.g., TLRs) and immune effectors (e.g., TRAFs).
Although there was no significant difference in the numbers of differentially expressed immune-related genes between the two oyster species after P. marinus infection, the magnitude of up-/downregulation of the shared immune-related DEGs were different between the two oyster species. For instance, the upregulation of PRRs such as TLRs, FBGDCs and C1qDCs were much higher in C. virginica than that in C. gigas (Figure 3D). Similarly, immune adaptors (e.g., TRAF) and effectors (e.g., SOD and Big defensin) also showed stronger upregulation in C. virginica (Figure 3D). In contrast, stronger upregulation of inhibitors of apoptosis (IAPs) was observed in C. gigas than in C. virginica under P. marinus infection (Figure 3D). Thirteen of the expressed 23 IAPs showed 2-fold or more upregulation in C. gigas compared with only one of the 30 in C. virginica.
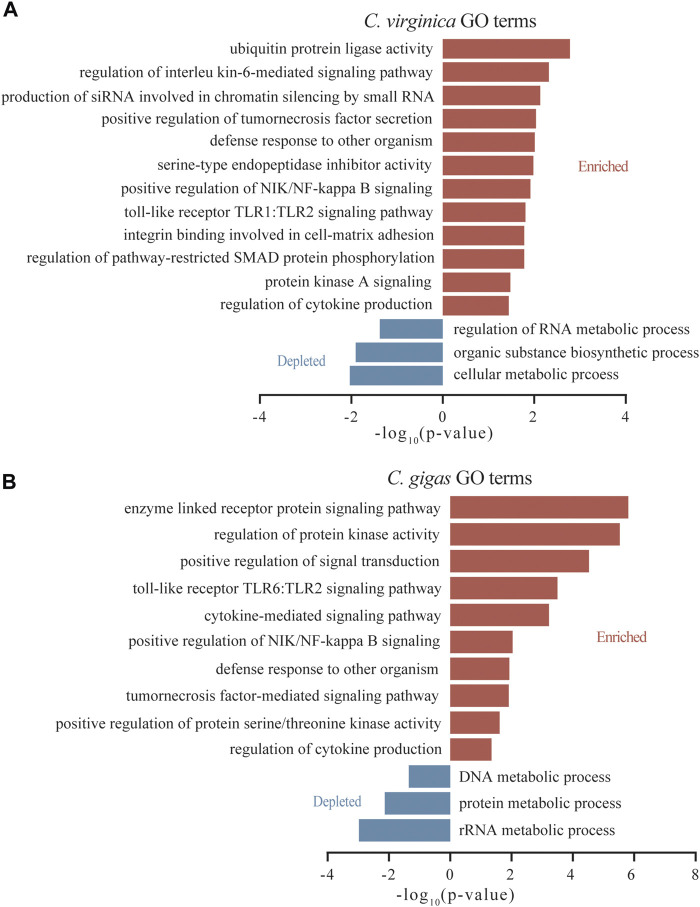
Among DEGs induced by P. marinus, GO terms for “interleukin-6-mediated signaling pathway,” “production of siRNA involved in chromatin silencing by small RNA,” and “serine-type endopeptidase inhibitor activity” were significantly enriched in C. virginica (Figure 4A), while GO terms for “positive regulation of protein serine/threonine kinase activity” were significantly enriched in C. gigas (Figure 4B). Interestingly, TLRs were significantly enriched in both species, but for different TLR pathways: “TLR1:TLR2” in C. virginica versus “TLR6:TLR2” in C. gigas (Figure 4). This finding indicates that different types of TLRs may play different roles in responding to P. marinus in the two oyster species.
FIGURE 4.
GO enrichment analysis of the differentially expressed genes after Dermo challenge between C. virginica (A) and C. gigas (B). Red and blue columns represent functional enrichments of up-regulated and down-regulated genes, respectively.
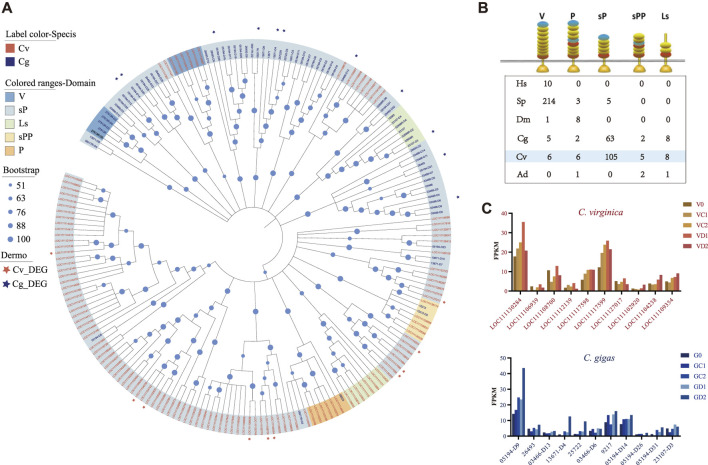
Positive Selection in P. marinus Responsive TLRs in C. virginica
Given that the pattern recognition receptor genes (TLRs, C1qDCs, and FBGDCs) showed significant differential expression in the two oyster species under P. marinus challenge, we examined molecular evolutionary history of these gene families in the two species. The TLRs can be separated into five types based on domain structures: V-, P-, sP-, sPP-, and Ls-type. The sP-type was greatly expanded in oysters, and to a greater extent in C. virginica (105) than in C. gigas (63) (Figures 5A,B). All P. marinus responsive TLRs of C. virginica were different from those of C. gigas (Figure 5A). These results indicated that these P. marinus responsive TLRs were from different lineage-specific expansions in the two oyster species and may have different functions in their response to P. marinus. All differentially expressed TLRs, 10 in C. virginica and 11 in C. gigas, belonged to sP-type (Figures 5A,C).
FIGURE 5.
Expansion of TLRs and divergent expression patterns under P. marinus challenge between C. gigas and C. virginica. (A), Phylogenetic tree constructed with the maximum likelihood method showing lineage-specific expansion of TLRs in C. gigas (red font) and C. virginica (blue font). Red and blue asterisks represent the differentially expressed TLRs in C. gigas and C. virginica, respectively. (B), Comparison of TLRs with different domain architectures in: human (Homo sapiens-Hs), sea urchin (Strongylocentrotus purpuratus-SP), fruit fly (Drosophila melanogaster-Dm), Pacific oyster (Crassostrea gigas-Cg), Eastern oyster (Crassostrea virginica-Cv), and staghorn coral (Acropora digitifera-Ad). LRRCT in red, LRRNT in blue and LRR in yellow. Toll/interleukin-1 receptor (TIR) domains are shown as gold triangles. (C), Diverse expression pattern of the differentially expressed TLRs, marked with asterisks in (A), during P. marinus challenge in C. gigas (Top) and C. virginica (Bottom).
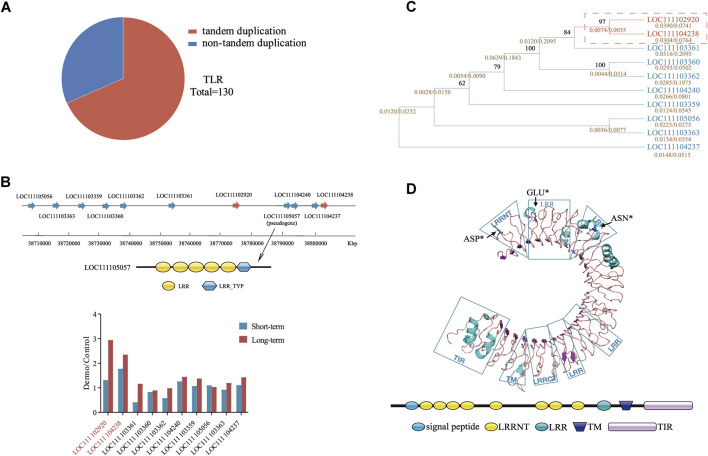
Mapping of TLRs to reference genomes revealed that most of the TLRs expansion in C. virginica and C. gigas was due to tandem duplication. For instance, of the 130 TLRs of C. virginica, 89 (68.46%) were found in tandemly duplicated clusters (Figure 6A). The largest cluster in C. virginica was located on chromosome 7 (NC_035786.1) and consisted of 11 TLRs (Figure 6B). In addition, a pseudogene LOC111105057 was detected within this cluster, suggesting that these expanded TLRs experienced rapid divergence including functional loss. Furthermore, two TLRs (LOC111102920 and LOC111104238) in this cluster that were significantly upregulated after the P. marinus challenge (Figure 6B) were clustered together on the phylogenetic tree (Figure 6C). Examination of synonymous (d S ) and nonsynonymous (d N ) nucleotide substitutions among TLRs of the cluster detected positive selection signals in these two upregulated TLRs (Figure 6C). The positive selection sites Asp 27 (p < 0.05), Glu 62 (p < 0.05) and Asn 199 (p < 0.05) were located in the LRR and LRRNT regions (Figure 6D) and likely involved in ligand/pathogen recognition. Similar analysis was performed in P. marinus responsive TLRs of C. gigas, but no positive selection sites were detected (Supplementary Figure S2). These results suggested that positive selection observed in P. marinus responsive TLRs may be important for the evolution of host-parasite interaction between C. virginica and P. marinus.
FIGURE 6.
Tandem duplication and positive selection of TLRs in C. virginica. (A), Tandem repeats as the major source of TLRs duplication in C. virginica. (B), The largest tandem repeat cluster (Top) and divergent expression patterns (Bottom) of tandemly linked TLRs in NC_035786.1 under P. marinus challenge. The wide arrows represent TLR genes, and genes with red background are differentially expressed under P. marinus challenge. A pseudogene LOC111105057 was detected in this repeat cluster. (C), Evolutionary relationships of duplicated TLRs. The calculated dN/dS (ω) values (gold) and bootstrap values are shown for each branch. The branches with ω values >1.0 are marked with red rectangle. The differential genes under P. marinus challenge that are positively selected are boxed with red dash lines. (D), Structural modeling of TLR depicting sites (blue) under positive selection. Positive selection sites (Asp27, Glu62, Asn199) are represented by black arrows.
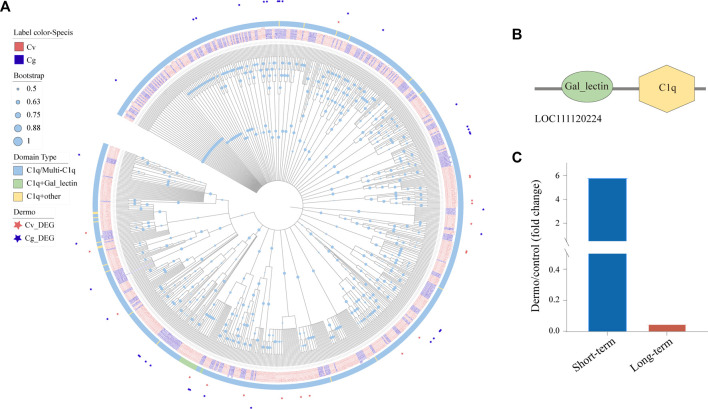
Upregulation of C1qDC With Galactose-Binding Lectin Domain by P. marinus
The globular C1q domain containing (C1qDC) proteins are a family of versatile PRRs that has been shown to play a significant role in a variety of processes including immune response, cell adhesion, inflammation, and apoptosis (Wang et al., 2012; Zong et al., 2019). We identified 477 and 333 C1qDCs in the genomes of C. virginica and C. gigas, respectively, and most of these C1qDCs belonged to the C1q/Multi-C1q type with two or more C1q domains (Figure 7A). Of note, phylogenetic analysis revealed a small cluster of 24 C1qDCs (3 C. gigas and 21 C. virginica) mostly with the “C1q + galactose-binding lectin” domain that has been shown to play an important role in immune response by binding to endogenous carbohydrates especially from P. marinus (Figures 7A,B). Interestingly, these “galactose-binding lectin” containing C1qDCs were only found in the genome of C. virginica (16 of 21 CvC1qDC), not in the C. gigas genome (Figure 7A). In addition, a C. virginica C1qDC with “galactose-binding lectin” domain (LOC111120224) was significantly upregulated in C. virginica after short-term induction of P. marinus challenge (Figure 7C). We speculate that this gene plays a significant role in P. marinus recognition during early infection stage in C. virginica.
FIGURE 7.
Expansion of C1qDCs and divergent expression patterns under P. marinus challenge between C. virginica and C. gigas. (A), Phylogenetic tree constructed with the maximum likelihood method showing lineage-specific expansion of C1qDCs in C. virginica (red font) and C. gigas (gray font). Different color backgrounds represent different types of C1qDC domain composition, and the red and gray asterisks represent differentially expressed C1qDCs under P. marinus challenge in C. virginica and C. gigas, respectively. (B), Domain composition of the “galactose-binding lectin” containing C1qDC gene. (C), A C1qDC gene (LOC111120224) with “galactose-binding lectin” domain was significantly upregulated in C. virginica after P. marinus challenge.
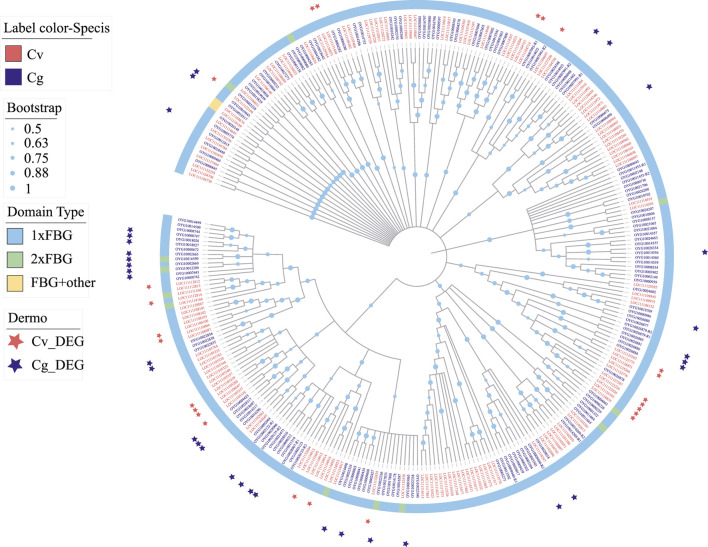
P. marinus Responsive FBGDCs Are Species Specific
FBGDCs (FBG-domain containing proteins) with one or more fibrinogen-like (FBG) domains are important pattern recognition receptors for pathogens recognition in vertebrates (Matsushita et al., 1996), urochordates (Kenjo et al., 2001) and invertebrates (Zhang et al., 2004; Zhang et al., 2009). We detected significant expansion of FBGDCs in both C. virginica (158 genes) and C. gigas (169 genes) (Figure 8). Following P. marinus challenge, differential expression was observed in 24 and 30 FBGDCs in C. virginica and C. gigas, respectively (Figure 8). Phylogenetic analysis of the FBG domain showed that most of the P. marinus responsive FBGDCs were derived from species-specific expansions in the two oyster species (Figure 8). Such results suggested that species-specific expansion of FBGDCs may enhance diversity and specificity in PAMP recognitions, and therefore play an important role in the evolution of host-parasite interaction.
FIGURE 8.
Expansion and phylogeny of FBGDC gene family in C. virginica (red font) and C. gigas (gray font). The phylogenetic tree is constructed with the maximum likelihood method showing lineage-specific expansion of FBGDC in C. virginica (red font) and C. gigas (gray font). Different color backgrounds represent different domain composition types, and the red and gray asterisks represent differentially expressed FBGDCs under P. marinus challenge in C. virginica and C. gigas, respectively.
Discussion
The protist P. marinus is a lethal pathogen of the eastern oyster C. virginica, while its sister species C. gigas is strongly resistant. The mechanisms responsible for this remarkable difference in resistance are not understood. In this study, we investigated transcriptomic response to P. marinus in two sister species that has diverged for 83 million years (Ren et al., 2010). Our work shows how comparative transcriptomics and genomics analyses contribute to our understanding of evolutionary dynamics of host-parasite interaction lophotrochozoans without adaptative immunity.
Although our discussion below focuses on differences in host defense between the two oyster species, two general observations are worth noting. First, results of this study are highly consistent with a previous study in C. virginica (Wang et al., 2010) where DEGs induced by P. marinus were determined with a 12 k microarray (challenges were conducted by the same team). For 37.8% of DEGs (389/1,029) from the previous study that had a one-to-one correspondence (BLAST, E-value < 1e-5 and identity >95%) to DEGs of this study (389/759), 83% of theme (323/389) showed the same trend of change in response to P. marinus in both studies (Supplemenatary Table S3), indicating that these results are robust. On the other hand, this study and a recent transcriptomic study (Proestou and Sullivan, 2020) shared only 39 DEGs (Supplemenatary Table S4), which could be caused by differences in challenge condition, genetic background or physiological state of oysters used. Second, this study identified 759 and 568 P. marinus induced DEGs in C. virginica and C. gigas (Supplementary Figure S3), respectively, with significant overlapping. The finding of many shared DEGs support a growing body of literature suggesting that there is a conversed set of gene families involved in host defense in molluscs (Gerdol and Venier, 2015; Figueras et al., 2019). In the present study, many immune-related genes (e.g., immune recognition receptors, effector genes, lectins, host-pathogen interaction genes) responded to P. marinus exposure in both species. For instance, TLRs are known to play a key role in PAMP recognition, and its activation is the first step in activation of mitogen activated protein kinase (MAPK) and tumor necrosis factor (TNF) during infection of vertebrates, which is pivotal in many cellular processes such as cell growth, differentiation, and apoptosis (Royle et al., 2003). The differential expression of these genes observed in this study is consistent with previous results (Tanguy et al., 2004; Wang et al., 2010), and may reflect a general response or cellular disorder caused by infections.
Our comparative transcriptomics and genomics analyses identified four previously undescribed molecular mechanisms that may account for differences in P. marinus resistance in two oyster species and illustrate possible evolutionary dynamics in host-parasite interactions. First, the magnitude of upregulation of the immune-related DEGs (e.g., TLRs, FBGDCs, C1qDCs SODs and Defensins) were much higher in C. virginica than in C. gigas. The stronger response of C. virginica is also reflected in the upregulation of serine-type protease inhibitors, which have been shown to inhibit proteases from invading P. marinus, limit proliferation of P. marinus in vitro and confer resistance (He et al., 2012; Gutiérrez-Rivera et al., 2015). In this study, the GO term for “serine-type endopeptidase inhibitor activity” was enriched in upregulated DEGs of C. virginica, not C. gigas. All these findings suggest that the host response to P. marinus in C. virginica is more dramatic. Clearly, C. virginica recognizes P. marinus as a pathogen and is highly sensitive to its infection. Strong immune response does not equate resistance. Over reactive immune response, whether orchestrated by the host or parasite, can lead to pathology and mortality of the host, as it has been shown in Ostreid herpesvirus infections of the Pacific oyster (He et al., 2012). The susceptibility of C. virginica does not mean P. marinus is a new pathogen. The fact that P. marinus responsive TLRs of C. virginica has been under positive selection and C1qDCs with the P. marinus binding “galactose-binding lectin” domain is found in C. virginica, not in C. gigas (see below), suggest that the C. virginica and P. marinus have a long history of coevolution, and P. marinus is not a new pathogen of C. virginica. If true, the susceptibility and heavy mortality observed in C. virginica may be caused by recently increased virulence of P. marinus during the coevolutionary race between the host and parasite (Van Valen, 1973). It has been suggested that increased virulence possibly associated with warm winters due to global warming played major roles in outbreaks and northward range extension of P. marinus (Ford, 1996; Carnegie et al., 2021).
In contrast to the stronger response of many immune response genes in C. virginica, P. marinus induced stronger upregulation of IAPs in C. gigas than in C. virginica. Inhibitors of apoptosis play critical roles in immune and stress responses, and the expansion of IAPs underpins the remarkable environmental resilience of bivalves (Guo et al., 2015; Song et al., 2021). It has been suggested that, as an intracellular parasite, P. marinus induces inhibition of apoptosis in the host as a survival strategy (Hughes et al., 2010). The observation of heightened upregulation of IAPs in the resistant C. gigas in this study challenges the idea that inhibition of apoptosis of host cells is orchestrated by the parasite. Rather, it suggests that inhibition of apoptosis is initiated by the host as an active defense mechanism and may play a role in limiting parasite proliferation or release, making C. gigas less susceptible.
A second mechanism is underscored by dramatic differences in “galactose-binding lectin” domain containing genes between two oyster species. In a previous study (Tasumi and Vasta, 2007), a novel galectin with a unique carbohydrate recognition domain structure had been shown to be associated with the recognition of P. marinus in C. virginica hemocytes. Galectins is an evolutionarily conserved family of β-galactoside-binding lectins that play important roles in early development and immune response of organisms by binding to endogenous carbohydrates (Rabinovich et al., 2002; Ahmed et al., 2009). In addition, galectins have also been shown to act as PRRs in innate immunity by binding to exogenous glycans on the surface of potentially pathogenic microbes, parasites, and fungi (Kim et al., 2008; Wang et al., 2011; Bao et al., 2013). It is not surprising to find significant upregulation of galectins in C. virginica but not in C. gigas. Further, we annotated 16 “galactose-binding lectin” containing C1qDCs in the genome of C. virginica, but none in the genome of C. gigas. More importantly, a CvC1qDC that composed of “galactose-binding lectin” domain was significantly upregulated in C. virginica after P. marinus challenge. Together, these findings suggest that the presence and upregulation of genes with “galactose-binding lectin” domain in C. virginica may be an important factor contributing in its susceptibility to P. marinus. Again, these results support the theory that C. virginica and P. marinus have a long history of coevolution. The lack of galectin upregulation and absence of ‘galactose-binding lectin’ containing C1qDCs in C. gigas may limit the ability of P. marinus to bind and enter host hemocytes. A more complete understanding of the evolution and functional integrations of multigene families in a broad range of host responses and defenses may emerge from further investigations of the innate immune recognition systems found in invertebrate species.
A third notable mechanism rests on the duplication and functional divergence of immune receptors that contributes to differential recognition and response to P. marinus in the two oyster species. As more genomes and transcriptomes across diverse marine invertebrates are sequenced, it is becoming clear that gene duplication and subsequent diversification of expanded members of gene families driven by selection are a recognized mechanism of genome innovation and adaptation (Conant and Wolfe, 2008; Zhang et al., 2012; Zhang et al., 2014; Guo et al., 2015; Zhang et al., 2015). In our study, multigene families encoding innate immune receptors, such as TLRs, C1qDCs, FBGDCs, have experienced large-scale expansion and functional diversification in both C. virginica and C. gigas, which may play an important role in the immune response to different pathogens (Zhang et al., 2015). While the gene families are conserved, distinct gene members of the expanded families are involved in parasite defense in the two oyster species. For example, TLRs showed greater expansion in C. virginica, and the majority of TLRs upregulated by P. marinus are derived from distinct species-specific expansions in the two species. Similarly, species-specific expansion was also detected in gene families of C1qDC and FBGDC, which is consistent with previous findings for major histocompatibility complex (MHC) and immune globlin (Ig) genes in vertebrates, TLRs in sea urchin, TNFs in amphioxus and FREPs in freshwater snail (Nei et al., 1997; Rast et al., 2006; Huang et al., 2008; Hanington and Zhang, 2010). Moreover, the lineage-specific expansion of immune genes is associated not only with differential responses to pathogens but also with differential expression under environmental stress conditions that emulate those of its natural habitat (Zhang et al., 2015). These findings suggest that lineage-specific expansions of multigene families encoding innate immune receptors in present study may have led to divergence and species-specific functions between C. gigas and C. virginica.
The fourth mechanism worth noting is that positive selection might be an important driving force for immune recognition of P. marinus. Classical evolutionary studies of protein-coding genes have established that genes in the canonical immune system tend to be the most rapidly evolving genes within and among species (Sackton, 2018). Many immune-related molecules directly interact with rapidly evolving pathogenic structures causing strong parasite-mediated natural selection at particular binding positions consistent with the Red Queen hypothesis (Woolhouse et al., 2002). PRRs are evolutionary conserved proteins and believed to be under strongly functional constraint. However, recent studies have found evidence of positive selection in some vertebrate groups, such as positive selection sites (PSSs) in TLRs of birds, wild rodents, domestic pig and cetacean (Alcaide and Edwards, 2011; Fornůsková et al., 2013; Darfour-Oduro et al., 2015; Xu et al., 2018). Similarly, positive selection effects on PRRs have also been found in molluscs, including PSSs detected at peptidoglycan recognition proteins (PGRPs) in C. gigas (Zhang and Yu, 2013), and fibrinogen-related proteins (FREPs) in the fresh-water snail, Biomphalaria glabrata (Hanington and Zhang, 2010). All these studies suggested that positive selection has played an important role in the evolution of metazoan PRRs, which is consistent with co-evolution of host and pathogens (Dodds and Thrall, 2009; Phillips et al., 2018). The TLRs of C. virginica may have experienced greater selection pressure than that of C. gigas based on the positive selection analysis, which suggests that the TLRs of C. virginica may be actively evolving for recognition of P. marinus, while C. gigas may possess TLRs with more effective defense capability against P. marinus infection. This is consistent with the finding that large expansions of PRRs are due to lineage-specific selective pressure from pathogens, and pathogen-mediated selection may play a role in the evolution of these genes across a variety of taxa (Hanington and Zhang, 2010; Grueber et al., 2014). As expected, all positive selection sites identified in P. marinus responsive TLRs of C. virginica were located in the extracellular LRR domain that has been shown to be important for pathogen recognition in the host (Khan et al., 2019), consistent with the phenomenon found in Areal’s study (Areal et al., 2011). The evolved TLRs may play an important role in the specific recognition of P. marinus by C. virginica, adding support for coevolution, but further experimental verification is needed.
In summary, comparative analyses in this study revealed previously undescribed molecular mechanisms underlying remarkably differences in P. marinus resistance between two sister oyster species. Immune responses to P. marinus are more dramatic in C. virginica, and the over-reacting immune responses may contribute to increased stress and mortality of the host. In addition, the findings of C1qDCs with “galactose-binding lectin” domain that are implicated in P. marinus recognition and positive selection in P. marinus responsive TLRs in C. virginica indicate that C. virginica and P. marinus have a history of coevolution. These observations indicate that P. marinus is not a new pathogen of C. virginica, and the recent outbreaks may be due to increased virulence of P. marinus. Inhibitors of apoptosis showed stronger upregulation in resistant C. gigas suggesting it is an active defense of the host, rather than a passive response orchestrated by the parasite as previously suggested. Our results also suggest that gene duplication, functional divergence, and positive selection are important in shaping immune response and host-parasite interaction in the two oyster species. Understanding the molecular basis of host-parasite interaction and coevolution is one of the key goals of immunology, and this study provides an example for using non-model species in comparative analyses.
Acknowledgments
The late Susan E. Ford contributed to the design and execution of the challenge experiments. Shan Wang, Iris Burt and Emily McGurk assisted with P. marinus challenge and quantification. We thank the high-performance computing center of Institute of Oceanology, CAS.
Data Availability Statement
All sequencing data associated with this project were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database, BioProject Accession Numbers: PRJNA778545.
Authors’ Contributions
Conceived and designed experiments: XG, GZ, LZ, and LL. Performed P. marinus challenge experiments: DB and XG. Conducted RNA extraction: LZ. Analyzed data: LW, JC, LZ, KM, YX, and XG. Contributed reagents/materials/computer resources: GZ, LZ, XG, and LL. Wrote the paper: JC, LW, LZ, and XG.
Funding
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences XDB42000000 to LZ, National Natural Science Foundation of China 41976088 to LZ, Key Development Project of Centre for Ocean Mega-Research of Science, Chinese academy of science COMS2019R01 to LZ and DB was supported in part by the USDA NIFA Hatch project 1009201-NJ32114. XG was supported in part by the USDA NIFA Hatch/Animal Health project 1004475-NJ32920.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.795706/full#supplementary-material
Domain structure of galectin (LOC111130470, Top) and its upregulation (Bottom) under P. marinus challenge in C. virginica.
Phylogenetic tree and positive selection analysis of the TLRs from the largest tandem repeat cluster in C. gigas. No positively selected branch or site was detected among these TLRs.
Volcano plots of transcriptional changes under short-term and long-term P. marinus challenge in C. gigas. Blue points represent downregulated genes, red points represent upregulated genes, and black points represent non-differentially expressed genes.
References
- Ahmed H., Du S.-J., Vasta G. R. (2009). Knockdown of a Galectin-1-like Protein in Zebrafish (Danio rerio) Causes Defects in Skeletal Muscle Development. Glycoconj J. 26 (3), 277–283. 10.1007/s10719-008-9178-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. (2006). Pathogen Recognition and Innate Immunity. Cell 124, 783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Aladaileh S., Nair S. V., Raftos D. A. (2007). Induction of Phenoloxidase and Other Immunological Activities in Sydney Rock Oysters Challenged with Microbial Pathogen-Associate Molecular Patterns. Fish Shellfish Immunol. 23 (6), 1196–1208. 10.1016/j.fsi.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Alcaide M., Edwards S. V. (2011). Molecular Evolution of the Toll-like Receptor Multigene Family in Birds. Mol. Biol. Evol. 28 (5), 1703–1715. 10.1093/molbev/msq351 [DOI] [PubMed] [Google Scholar]
- Allam B., Carden W. E., Ward J. E., Ralph G., Winnicki S., Pales Espinosa E. (2013). Early Host-Pathogen Interactions in marine Bivalves: Evidence that the Alveolate Parasite Perkinsus Marinus Infects through the Oyster Mantle during Rejection of Pseudofeces. J. Invertebr. Pathol. 113 (1), 26–34. 10.1016/j.jip.2012.12.011 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). Htseq--a python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 31 (2), 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. S. (1996). Interactions of Perkinsus Marinus with Humoral Factors and Hemocytes of Crassostrea Virginica . J. Shellfish Res. 15 (1), 127–134. [Google Scholar]
- Anderson R. S. (1999). Perkinsus Marinussecretory Products Modulate Superoxide Anion Production by Oyster (Crassostrea virginica) Haemocytes. Fish Shellfish Immunol. 9 (1), 51–60. 10.1006/fsim.1998.0174 [DOI] [Google Scholar]
- Areal H., Abrantes J., Esteves P. J. (2011). Signatures of Positive Selection in Toll-like Receptor (TLR) Genes in Mammals. BMC Evol. Biol. 11 (1), 368. 10.1186/1471-2148-11-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Shen H., Zhou H., Dong Y., Lin Z. (2013). A Tandem-Repeat Galectin from Blood Clam Tegillarca Granosa and its Induced mRNA Expression Response against Bacterial challenge. Genes Genom 35, 733–740. 10.1007/s13258-013-0123-3 [DOI] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodological) 57 (1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bibby R., Widdicombe S., Parry H., Spicer J., Pipe R. (2008). Effects of Ocean Acidification on the Immune Response of the Blue Mussel Mytilus edulis . Aquat. Biol. 2 (1), 67–74. 10.3354/ab00037 [DOI] [Google Scholar]
- Bidegain G., Powell E. N., Klinck J. M., Hofmann E. E., Ben-Horin T., Bushek D., et al. (2017). Modeling the Transmission of Perkinsus Marinus in the Eastern Oyster Crassostrea Virginica . Fish. Res. 186, 82–93. 10.1016/j.fishres.2016.08.006 [DOI] [Google Scholar]
- Bossart G. D. (2007). Emerging Diseases in Marine Mammals: from Dolphins to Manatees. Microbe 2, 544–549. 10.1128/microbe.2.544.1 [DOI] [Google Scholar]
- Burge C. A., Mark Eakin C., Friedman C. S., Froelich B., Hershberger P. K., Hofmann E. E., et al. (2014). Climate Change Influences on marine Infectious Diseases: Implications for Management and Society. Annu. Rev. Mar. Sci. 6 (1), 249–277. 10.1146/annurev-marine-010213-135029 [DOI] [PubMed] [Google Scholar]
- Bushek D., Ford S. E., Allen S. K. (1994). Evaluation of Methods Using Ray's Fluid Thioglycollate Medium for Diagnosis of Perkinsus Marinus Infection in the Eastern Oyster, Crassostrea Virginica . Annu. Rev. Fish Dis. 4, 201–217. 10.1016/0959-8030(94)90029-9 [DOI] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M., Gabaldon T. (2009). trimAl: a Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 25 (15), 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnegie R. B., Ford S. E., Crockett R. K., Kingsley-Smith P. R., Bienlien L. M., Safi L. S. L., et al. (2021). A Rapid Phenotype Change in the Pathogen Perkinsus Marinus Was Associated with a Historically Significant marine Disease Emergence in the Eastern Oyster. Sci. Rep. 11 (1), 12872–12879. 10.1038/s41598-021-92379-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintala M., Bushek D., Ford S. (2002). Comparison of In Vitro-cultured and Wild-type Perkinsus Marinus. II. Dosing Methods and Host Response. Dis. Aquat. Org. 51, 203–216. 10.3354/dao051203 [DOI] [PubMed] [Google Scholar]
- Choi K. S., Wilson E. A., Lewis D. H., Powell E. N., Ray S. M. (1989). The Energetic Cost of Perkinsus Marinus Parasitism in Oysters: Quantification of the Thioglycollate Method. J. Shellfish Res. 8, 125–131. Available From: http://hdl.handle.net/1969.3/19281 . [Google Scholar]
- Conant G. C., Wolfe K. H. (2008). Probabilistic Cross-Species Inference of Orthologous Genomic Regions Created by Whole-Genome Duplication in Yeast. Genetics 179 (3), 1681–1692. 10.1534/genetics.107.074450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfour-Oduro K. A., Megens H.-J., Roca A. L., Groenen M. A. M., Schook L. B. (2015). Adaptive Evolution of Toll-like Receptors (TLRs) in the Family Suidae. PLoS One 10 (4), e0124069. 10.1371/journal.pone.0124069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P., Thrall P. (2009). Recognition Events and Host-Pathogen Co-evolution in Gene-For-Gene Resistance to Flax Rust. Funct. Plant Biol. 36 (5), 395–408. 10.1071/fp08320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., Kelly S. (2015). Orthofinder: Solving Fundamental Biases in Whole Genome Comparisons Dramatically Improves Orthogroup Inference Accuracy. Genome Biol. 16 (157), 157. 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisal M., La Peyre J. F., Elsayed E., Wright D. C. (1999). Bacitracin Inhibits the Oyster PathogenPerkinsus Marinusin Vitro and In Vivo . J. Aquat. Anim. Health 11 (2), 130–138. [DOI] [Google Scholar]
- Figueras A., Moreira R., Sendra M., Novoa B. (2019). Genomics and Immunity of the Mediterranean Mussel Mytilus galloprovincialis in a Changing Environment. Fish Shellfish Immunol. 90, 440–445. 10.1016/j.fsi.2019.04.064 [DOI] [PubMed] [Google Scholar]
- Ford S., Chintala M., Bushek D. (2002). Comparison of In Vitro-cultured and Wild-type Perkinsus Marinus. I. Pathogen Virulence. Dis. Aquat. Org. 51, 187–201. 10.3354/dao051187 [DOI] [PubMed] [Google Scholar]
- Ford S. E. (1996). Range extension by the oyster parasite Perkinsus marinus into the northeastern United States: response to climate change?. J Shellfish Res. 15, 45–56. Available at: https://www.biodiversitylibrary.org/page/3034357 . [Google Scholar]
- Fornůsková A., Vinkler M., Pagès M., Galan M., Jousselin E., Cerqueira F., et al. (2013). Contrasted Evolutionary Histories of Two Toll-like Receptors (TLR4 and TLR7) in Wild Rodents (MURINAE). BMC Evol. Biol. 13 (1), 194. 10.1186/1471-2148-13-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdol M., Venier P. (2015). An Updated Molecular Basis for Mussel Immunity. Fish Shellfish Immunol. 46 (1), 17–38. 10.1016/j.fsi.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Gerrodette T. (2007). Marine Conservation Biology: the Science of Maintaining the Sea's Biodiversity. Bioscience 57, 536–537. 10.1641/B570611 [DOI] [Google Scholar]
- Gertz E. M., Yu Y.-K., Agarwala R., Schäffer A. A., Altschul S. F. (2006). Composition-based Statistics and Translated Nucleotide Searches: Improving the Tblastn Module of Blast. BMC Biol. 4 (1), 41. 10.1186/1741-7007-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedken M., Morsey B., Sunila I., Guise S. D. (2005). Immunomodulation of Crassostrea gigas and Crassostrea Virginica Cellular Defense Mechanisms by Perkinsus Marinus . J. Shellfish Res. 24, 487–496. 10.2983/0730-8000(2005)24[487:IOCGAC]2.0.CO;2 [DOI] [Google Scholar]
- Groner M. L., Maynard J., Breyta R., Carnegie R. B., Dobson A., Friedman C. S., et al. (2016). Managing marine Disease Emergencies in an Era of Rapid Change. Phil. Trans. R. Soc. B 371 (1689), 20150364. 10.1098/rstb.2015.0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber C. E., Wallis G. P., Jamieson I. G. (2014). Episodic Positive Selection in the Evolution of Avian Toll-like Receptor Innate Immunity Genes. PLoS ONE 9 (3), e89632. 10.1371/journal.pone.0089632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Ford S. E., Zhang F. (1999). Molluscan aquaculture in China. J. Shellfish Res. 18 (1), 19–31. Available at: https://www.biodiversitylibrary.org/item/23130#page/21/mode/1up [Google Scholar]
- Guo X., Ford S. E. (2016). Infectious Diseases of marine Molluscs and Host Responses as Revealed by Genomic Tools. Phil. Trans. R. Soc. B 371 (1689), 20150206. 10.1098/rstb.2015.0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., He Y., Zhang L., Lelong C., Jouaux A. (2015). Immune and Stress Responses in Oysters with Insights on Adaptation. Fish Shellfish Immunol. 46 (1), 107–119. 10.1016/j.fsi.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Rivera J., Arcos-Ortega G., Luna-González A., Rodríguez-Jaramillo M., Arechiga-Carvajal E., Vázquez-Juárez R. (2015). Differential Expression of Serine Protease Inhibitors 1 and 2 in Crassostrea Corteziensis and C. virginica Infected with Perkinsus Marinus . Dis. Aquat. Org. 112 (3), 185–197. 10.3354/dao02808 [DOI] [PubMed] [Google Scholar]
- Hanington P. C., Zhang S.-M. (2011). The Primary Role of Fibrinogen-Related Proteins in Invertebrates Is Defense, Not Coagulation. J. Innate Immun. 3 (1), 17–27. 10.1159/000321882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell C. D., Kim K., Burkholder J. M., Colwell R. R., Epstein P. R., Grimes D. J., et al. (1999). Emerging marine Diseases-Cclimate Links and Anthropogenic Factors. Science 285 (5433), 1505–1510. 10.1126/science.285.5433.1505 [DOI] [PubMed] [Google Scholar]
- Harvell D., Aronson R., Baron N., Connell J., Dobson A., Ellner S., et al. (2004). The Rising Tide of Ocean Diseases: Unsolved Problems and Research Priorities. Front. Ecol. Environ. 2 (7), 375–382. 10.1890/1540-9295(2004)002[0375:trtood]2.0.co;2 [DOI] [Google Scholar]
- He Y., Yu H., Bao Z., Zhang Q., Guo X. (2012). Mutation in Promoter Region of a Serine Protease Inhibitor Confers Perkinsus Marinus Resistance in the Eastern Oyster (Crassostrea Virginica). Fish Shellfish Immunol. 33 (2), 411–417. 10.1016/j.fsi.2012.05.028 [DOI] [PubMed] [Google Scholar]
- Huang S., Yuan S., Guo L., Yu Y., Li J., Wu T., et al. (2008). Genomic Analysis of the Immune Gene Repertoire of Amphioxus Reveals Extraordinary Innate Complexity and Diversity. Genome Res. 18 (7), 1112–1126. 10.1101/gr.069674.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes F. M., Foster B., Grewal S., Sokolova I. M. (2010). Apoptosis as a Host Defense Mechanism in Crassostrea Virginica and its Modulation by Perkinsus Marinus. Fish Shellfish Immunol. 29 (2), 247–257. 10.1016/j.fsi.2010.03.003 [DOI] [PubMed] [Google Scholar]
- Kenjo A., Takahashi M., Matsushita M., Endo Y., Nakata M., Mizuochi T., et al. (2001). Cloning and Characterization of Novel Ficolins from the Solitary Ascidian, Halocynthia Roretzi . J. Biol. Chem. 276 (23), 19959–19965. 10.1074/jbc.M011723200 [DOI] [PubMed] [Google Scholar]
- Khan I., Maldonado E., Silva L., Almeida D., Johnson W. E., O’Brien S. J., et al. (2019). The Vertebrate TLR Supergene Family Evolved Dynamically by Gene Gain/Loss and Positive Selection Revealing a Host-Pathogen Arms Race in Birds. Diversity 11 (8), 131. 10.3390/d11080131 [DOI] [Google Scholar]
- Kim J. Y., Kim Y. M., Cho S. K., Choi K. S., Cho M. (2008). Noble Tandem-Repeat Galectin of Manila Clam Ruditapes Philippinarum Is Induced upon Infection with the Protozoan Parasite Perkinsus Olseni . Dev. Comp. Immunol. 32, 1131–1141. 10.1016/j.dci.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Kim S., An S., Park B., Oh E.-G., Song K. C., Kim J.-W., et al. (2016). Virulence Factors and Antimicrobial Susceptibility of vibrio Parahaemolyticus Isolated from the Oyster Crassostrea gigas . Korean J. Fish. Aquat. Sci. 49 (2), 116–123. 10.5657/KFAS.2016.0116 [DOI] [Google Scholar]
- La Peyre J. F., Chu F.-L. E., Meyers J. M. (1995). Haemocytic and Humoral Activities of Eastern and pacific Oysters Following challenge by the Protozoan Perkinsus Marinus . Fish Shellfish Immunol. 5 (3), 179–190. 10.1016/S1050-4648(05)80012-9 [DOI] [Google Scholar]
- Lan Y., Sun J., Xu T., Chen C., Tian R., Qiu J.-W., et al. (2018). De Novo transcriptome Assembly and Positive Selection Analysis of an Individual Deep-Sea Fish. BMC Genomics 19 (1), 394. 10.1186/s12864-018-4720-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine N. D. (1978). Perkinsus Gen. N. And Other New Taxa in the Protozoan Phylum Apicomplexa. J. Parasitol. 64 (3), 549. 10.2307/3279807 96245 [DOI] [Google Scholar]
- Li Y., Zhang L., Qu T., Tang X., Li L., Zhang G. (2017). Conservation and Divergence of Mitochondrial Apoptosis Pathway in the pacific Oyster, Crassostrea gigas . Cell Death Dis 8 (7), e2915. 10.1038/cddis.2017.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahram A., Herbordt M. C. (2015). Ncbi Blastp on High-Performance Reconfigurable Computing Systems. ACM Trans. Reconfigurable Technol. Syst. 7 (4), 1–20. 10.1145/2629691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M., Endo Y., Taira S., Sato Y., Fujita T., Ichikawa N., et al. (1996). A Novel Human Serum Lectin with Collagen- and Fibrinogen-like Domains that Functions as an Opsonin. J. Biol. Chem. 271 (5), 2448–2454. 10.1074/jbc.271.5.2448 [DOI] [PubMed] [Google Scholar]
- Meyers J. A., Burreson E. M., Barber B. J., Mann R. (1991). Susceptibility of Diploid and Triploid pacific Oysters, Crassostrea gigas (Thunberg, 1793) and Eastern Oysters, Crassostrea Virginica (Gmelin, 1791), to Perkinsus Marinus . J. Shellfish Res. 10, 433–437. [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G. A., Sonnhammer E. L. L., et al. (2021). Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 49, D412–D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P., Moreau K., Segarra A., Tourbiez D., Travers M.-A., Rubinsztein D. C., et al. (2015). Autophagy Plays an Important Role in Protecting Pacific Oysters from OsHV-1 andVibrio Aestuarianusinfections. Autophagy 11 (3), 516–526. 10.1080/15548627.2015.1017188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gu X., Sitnikova T. (1997). Evolution by the Birth-And-Death Process in Multigene Families of the Vertebrate Immune System. Proc. Natl. Acad. Sci. 94 (15), 7799–7806. 10.1073/pnas.94.15.7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell R. I. (1998). Ecological changes in Chesapeake Bay: are they the result of overharvesting the American oyster, Crassostrea virginica. Understanding the estuary: advances in Chesapeake Bay research 129, 536–546. 10.1073/pnas.94.15.7799 [DOI] [Google Scholar]
- Phillips K. P., Cable J., Mohammed R. S., Herdegen-Radwan M., Raubic J., Przesmycka K. J., et al. (2018). Immunogenetic novelty Confers a Selective Advantage in Host-Pathogen Coevolution. Proc. Natl. Acad. Sci. USA 115 (7), 1552–1557. 10.1073/pnas.1708597115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proestou D. A., Sullivan M. E. (2020). Variation in global transcriptomic response to Perkinsus marinus infection among eastern oyster families highlights potential mechanisms of disease resistance. Fish and shellfish immunology 96, 141–151. 10.1016/j.fsi.2019.12.001 [DOI] [PubMed] [Google Scholar]
- Powell E. N., Klinck J. M, Guo X., Ford S. E, Bushek D. (2011). The Potential for Oysters, Crassostrea virginica, to Develop Resistance to Dermo Disease in the Field: Evaluation using a Gene-Based Population Dynamics Model. Journal of Shellfish Research 30 (3), 685–712. 10.2983/035.030.0310 [DOI] [Google Scholar]
- Qu T., Zhang L., Wang W., Huang B., Li Y., Zhu Q., et al. (2015). Characterization of an Inhibitor of Apoptosis Protein in Crassostrea gigas Clarifies its Role in Apoptosis and Immune Defense. Dev. Comp. Immunol. 51, 74–78. 10.1016/j.dci.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., et al. (2005). Interproscan: Protein Domains Identifier. Nucleic Acids Res. 33, W116–W120. 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich G. A., Rubinstein N., Toscano M. A. (2002). Role of Galectins in Inflammatory and Immunomodulatory Processes. Biochim. Biophys. Acta 1572 (2-3), 274–284. 10.1016/S0304-4165(02)00314-8 [DOI] [PubMed] [Google Scholar]
- Rast J. P., Smith L. C., Loza-Coll M., Hibino T., Litman G. W. (2006). Genomic Insights into the Immune System of the Sea Urchin. Science 314 (5801), 952–956. 10.1126/science.1134301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece K., Dungan C., Burreson E. (2008). Molecular Epizootiology of Perkinsus Marinus and P. Chesapeaki Infections Among Wild Oysters and Clams in chesapeake bay, USA. Dis. Aquat. Org. 82 (3), 237–248. 10.3354/dao01997 [DOI] [PubMed] [Google Scholar]
- Ren J., Liu X., Jiang F., Guo X., Liu B. (2010). Unusual conservation of mitochondrial gene order in Crassostrea oysters: evidence for recent speciation in Asia. BMC evolutionary biology 10 (1), 1–14. 10.1186/1471-2148-10-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle M. C. J., Tötemeyer S., Alldridge L. C., Maskell D. J., Bryant C. E. (2003). Stimulation of Toll-like Receptor 4 by Lipopolysaccharide during Cellular Invasion by LiveSalmonella typhimuriumIs a Critical but Not Exclusive Event Leading to Macrophage Responses. J. Immunol. 170 (11), 5445–5454. 10.4049/jimmunol.170.11.5445 [DOI] [PubMed] [Google Scholar]
- Sackton T. B. (2018). Comparative Genomics and Transcriptomics of Host-Pathogen Interactions in Insects: Evolutionary Insights and Future Directions. Curr. Opin. Insect Sci. 31, 106–113. 10.1016/j.cois.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Saha D., Ota M. O. C., Pereira P., Buchy P., Badur S. (2021). Rotavirus Vaccines Performance: Dynamic Interdependence of Host, Pathogen and Environment. Expert Rev. Vaccin. 20, 945–957. 10.1080/14760584.2021.1951247 [DOI] [PubMed] [Google Scholar]
- Shumway S. (2021). (Edition)Molluscan Shellfish Aquaculture: A Practical Guide. 5m Books Ltd. [Google Scholar]
- Song H., Guo X., Sun L., Wang Q., Han F., Wang H., et al. (2021). The Hard Clam Genome Reveals Massive Expansion and Diversification of Inhibitors of Apoptosis in Bivalvia. BMC Biol. 19 (1), 15. 10.1186/s12915-020-00943-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A., Thieltges D., Poulin R. (2010). Parasites and Global Warming: Net Effects of Temperature on an Intertidal Host-Parasite System. Mar. Ecol. Prog. Ser. 415 (15), 11–22. 10.3354/meps08742 [DOI] [Google Scholar]
- Sutherland K. P., Berry B., Park A., Kemp D. W., Kemp K. M., Lipp E. K., et al. (2016). Shifting white Pox Aetiologies Affecting Acropora Palmata in the florida Keys, 1994-2014. Phil. Trans. R. Soc. B 371 (1689), 20150205. 10.1098/rstb.2015.0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy A., Guo X., Ford S. E. (2004). Discovery of Genes Expressed in Response to Perkinsus Marinus challenge in Eastern (Crassostrea Virginica) and pacific (C. gigas) Oysters. Gene 338 (1), 121–131. 10.1016/j.gene.2004.05.019 [DOI] [PubMed] [Google Scholar]
- Tasumi S., Vasta G. R. (2007). A Galectin of Unique Domain Organization from Hemocytes of the Eastern Oyster (Crassostrea Virginica) Is a Receptor for the Protistan Parasite Perkinsus Marinus . J. Immunol. 179 (5), 3086–3098. 10.4049/jimmunol.179.5.3086 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). Tophat: Discovering Splice Junctions with RNA-Seq. Bioinformatics 25 (9), 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. (1973). A New Evolutionary Law. Evol. Theory 1 (1), 1–30. [Google Scholar]
- Wang L., Song X., Song L. (2018). The Oyster Immunity. Dev. Comp. Immunol. 80, 99–118. 10.1016/j.dci.2017.05.025 [DOI] [PubMed] [Google Scholar]
- Wang L., Wang L., Zhang H., Zhou Z., Siva V. S., Song L., et al. (2012). A C1q Domain Containing Protein from Scallop Chlamys Farreri Serving as Pattern Recognition Receptor with Heat-Aggregated IgG Binding Activity. Plos One 7 (8), e43289. 10.1371/journal.pone.0043289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Peatman E., Liu H., Bushek D., Ford S. E., Kucuktas H., et al. (2010). Microarray Analysis of Gene Expression in Eastern Oyster (Crassostrea Virginica) Reveals a Novel Combination of Antimicrobial and Oxidative Stress Host Responses after Dermo (Perkinsus Marinus) challenge. Fish Shellfish Immunol. 29 (6), 921–929. 10.1016/j.fsi.2010.07.035 [DOI] [PubMed] [Google Scholar]
- Wang Z., Jian J., Lu Y., Wang B., Wu Z. (2011). A Tandem-Repeat Galectin Involved in Innate Immune Response of the Pearl Oyster Pinctada Fucata . Mar. Genomics 4, 229–236. 10.1016/j.margen.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Wilson B. A., Ho M. (2015). The Biosecurity Threat Posed by Biological Toxins. Biol. Toxins Bioterror. 4, 141–183. 10.1007/978-94-007-5869-8_18 [DOI] [Google Scholar]
- Winnicki S. M. (2010). The Influence of Pallial Mucus from the Oyster, Crassostrea Virginica, on the Virulence of its Pathogenic Alveolate, Perkinsus Marinus . Doctoral dissertation. New York: State University of New York at Stony Brook. [Google Scholar]
- Woolhouse M. E. J., Webster J. P., Domingo E., Charlesworth B., Levin B. R. (2002). Biological and Biomedical Implications of the Co-evolution of Pathogens and Their Hosts. Nat. Genet. 32 (4), 569–577. 10.1038/ng1202-569 [DOI] [PubMed] [Google Scholar]
- Xu S., Tian R., Lin Y., Yu Z., Zhang Z., Niu X., et al. (2019). Widespread Positive Selection on Cetacean Tlr Extracellular Domain. Mol. Immunol. 106, 135–142. 10.1016/j.molimm.2018.12.022 [DOI] [PubMed] [Google Scholar]
- Yang Z., Wong W. S. W., Rasmus N. (2005). Bayes Empirical Bayes Inference of Amino Acid Sites under Positive Selection. Mol. Biol. Evol. 22 (4), 1107–1118. 10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., et al. (2012). The Oyster Genome Reveals Stress Adaptation and Complexity of Shell Formation. Nature 490 (7418), 49–54. 10.1038/nature11413 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang L., Song L., Song X., Wang B., Mu C., et al. (2009). A Fibrinogen-Related Protein from bay Scallop Argopecten irradians Involved in Innate Immunity as Pattern Recognition Receptor. Fish Shellfish Immunol. 26 (1), 56–64. 10.1016/j.fsi.2008.07.019 [DOI] [PubMed] [Google Scholar]
- Zhang L., Li L., Guo X., Litman G. W., Dishaw L. J., Zhang G. (2015). Massive Expansion and Functional Divergence of Innate Immune Genes in a Protostome. Sci. Rep. 5, 8693. 10.1038/srep08693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li L., Zhang G. (2011). Gene Discovery, Comparative Analysis and Expression Profile Reveal the Complexity of the Crassostrea gigas Apoptosis System. Dev. Comp. Immunol. 35 (5), 603–610. 10.1016/j.dci.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Zhang L., Li L., Zhu Y., Zhang G., Guo X. (2014). Transcriptome Analysis Reveals a Rich Gene Set Related to Innate Immunity in the Eastern Oyster (Crassostrea Virginica). Mar. Biotechnol. 16 (1), 17–33. 10.1007/s10126-013-9526-z [DOI] [PubMed] [Google Scholar]
- Zhang S.-M., Adema C. M., Kepler T. B., Loker E. S. (2004). Diversification of Ig Superfamily Genes in an Invertebrate. Science 305 (5681), 251–254. 10.1126/science.1088069 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu Z. (2013). The First Evidence of Positive Selection in Peptidoglycan Recognition Protein (PGRP) Genes of Crassostrea gigas . Fish Shellfish Immunol. 34 (5), 1352–1355. 10.1016/j.fsi.2013.01.018 [DOI] [PubMed] [Google Scholar]
- Zong Y., Liu Z., Wu Z., Han Z., Wang L., Song L. (2019). A Novel Globular C1q Domain Containing Protein (C1qDC-7) from Crassostrea gigas Acts as Pattern Recognition Receptor with Broad Recognition Spectrum. Fish Shellfish Immunol. 84, 920–926. 10.1016/j.fsi.2018.10.079 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Domain structure of galectin (LOC111130470, Top) and its upregulation (Bottom) under P. marinus challenge in C. virginica.
Phylogenetic tree and positive selection analysis of the TLRs from the largest tandem repeat cluster in C. gigas. No positively selected branch or site was detected among these TLRs.
Volcano plots of transcriptional changes under short-term and long-term P. marinus challenge in C. gigas. Blue points represent downregulated genes, red points represent upregulated genes, and black points represent non-differentially expressed genes.
Data Availability Statement
All sequencing data associated with this project were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database, BioProject Accession Numbers: PRJNA778545.