Abstract
Chromatin assembly factor 1 (CAF-1) is a protein complex formed of three subunits, p150, p60, and p48, conserved from the yeast Saccharomyces cerevisiae to humans, which can promote nucleosome assembly onto newly replicated DNA. In S. cerevisiae, deletion of the genes encoding any of the three CAF-1 subunits (cacΔ mutants), although nonlethal, results in a silencing defect of genes packaged into heterochromatin. Here we report on a mammalian cell model that we devised to monitor gene silencing and its reversal in a quantitative manner. This model relies on the use of a cell line stably transfected with a reporter gene in a silenced state. Reversal of reporter gene silencing was achieved upon treatment of the cells with 5-azacytidine, which resulted in the demethylation of the reporter gene copies. We show that expression of a cDNA for the human p150 CAF-1 subunit harboring 5′ truncations, but not that of a cDNA encoding the full-length p150 CAF-1 subunit, increases by more than 500-fold the frequency at which transcriptional silencing of the reporter gene copies is reversed in these cells. Reversal of gene silencing is dependent upon expression of a truncated protein, possibly acting as a dominant negative mutant of the wild-type CAF-1, is associated with alterations in chromatin structure as measured by an endonuclease sensitivity assay and is not associated with detectable changes in the methylation status of the silenced genes. These results suggest that the role of CAF-1 in the epigenetic control of gene expression has been conserved between yeast and mammals, despite the lack of DNA methylation in yeast chromatin.
Chromatin assembly factor 1 (CAF-1) is a histone chaperone which promotes nucleosome assembly on DNA undergoing replication (11, 21, 23) and is involved in the repair of DNA damage (7, 8, 12, 14). It is a complex of three subunits, p150, p60, and p48 (11, 23, 25), which can form larger structures containing specific acetylated forms of histones H3 and H4 (11, 25). CAF-1 performs the first step of nucleosome assembly, bringing histones H3 and H4 to the replicating DNA; then histones H2A and H2B bind to this chromatin precursor to complete the histone octamer (11, 24, 25). CAF-1 differs from other assembly factors because it preferentially assembles nucleosomes onto DNA which has undergone replication. This coupling between DNA replication and CAF-1 activity could be explained by the fact that CAF-1 activity requires an interaction with the proliferating cell nuclear antigen, which specifically marks the newly replicated DNA (21). Although the N-terminal domains of histones H3 and H4 may not be absolutely required for nucleosome assembly (22), functional CAF-1 complexes isolated from cells in culture contain histones H3 and H4 with a specific acetylation pattern (25, 26). Consequently, chromatin structures newly assembled by CAF-1 could be marked by a distinctive acetylation pattern. In the budding yeast Saccharomyces cerevisiae, null mutations of any of the three genes encoding the CAF-1 subunits (cacΔ mutants) are not lethal, suggesting the existence of other replication-coupled chromatin assembly mechanisms (12, 13). However, all the cacΔ mutants display defects in the silencing of genes packaged into the heterochromatin (i.e., within regions of condensed chromatin fiber). Silencing of reporter genes packaged into the telomeric heterochromatin and that of the mating type genes present at the silent HM loci are impaired in these mutants (5, 6, 12, 13, 17). Such phenotypes suggest that, at least in yeast, there is a functional link between the ability to synthesize a functional CAF-1 complex and the inheritance of epigenetically determined chromosomal states conditioning gene silencing. The composition and chromatin assembly activity of CAF-1 are conserved among humans, Drosophila, and yeast (3, 11, 25), yet until now no evidence showing that CAF-1 may be similarly involved in the maintenance of gene silencing and/or the inheritance of epigenetic chromosomal states in mammalian cells has been provided. In fact, S. cerevisiae heterochromatin is biochemically very different from that of mammals. For instance, key components of yeast heterochromatin, such as Sir3 and Sir4 proteins, have no obvious ortholog in other eukaryotes, and the maintenance of gene silencing in mammals seems to rely, in part, on mechanisms that are lacking in yeast, such as DNA methylation. In fact, the characterization of the role of the CAF-1 complex in mammals has been hampered by the lack of available homozygously null CAF-1 mutants.
In the course of an extensive screening of a retroviral cDNA expression library, aimed at isolating factors which can activate transcription of a stably transfected human LINE-1 promoter, a cDNA of the CAF-1 p150 subunit mRNA with a 5′ truncation encompassing the first 1,178 nucleotides of this factor (CAFΔ1178) was isolated (Tchénio, unpublished data). Yet ectopic expression of this cDNA in transient-transfection assays failed to enhance to any significant extent the expression of a luciferase reporter gene placed under the control of the LINE-1 promoter, strongly suggesting that this cDNA was not acting as a transcription factor. As previous reports revealed a role for the CAF-1 complex in the maintenance of gene silencing in yeast (see references mentioned above), it was tempting to envision a similar role for the mammalian CAF-1 complex and to assay whether the expression of a defective human CAF-1 p150 subunit could reverse gene silencing in mammalian cells in the case of genes repressed by chromatin-dependent mechanisms. To address this question, we first devised a simple and well-controlled genetic test in mammalian cells in culture, which allowed us to assay in a quantitative manner the reversion of gene silencing. Accordingly, we were able to show that expression of a 5′-truncated p150 CAF-1 cDNA (CAFΔ1178), which encodes a truncated CAF-1 p150 subunit, increased the frequency at which transcriptional gene silencing can revert in these cells more than 500-fold.
MATERIALS AND METHODS
Cell culture, transfections, and infections.
Cells were grown in Dulbecco's modified Eagle's medium (4.5 g of glucose/liter) supplemented with 10% serum and antibiotics, in 6% CO2 at 37°C. The serum used (Life Technologies) was fetal calf serum for HeLa cells and newborn calf serum for PA12-derived cells. Selections were performed in growth medium supplemented with 500 μg of G418 (Life Technologies)/ml for neo expression and 150 U of hygromycin (Calbiochem)/ml and 1.5 μg of puromycin (Sigma)/ml for hygromycin and puromycin resistance, respectively. Transfections of dispersed cells were carried out with Lipofectamine Plus reagent (Life Technologies) according to the manufacturer's instructions. Cells were harvested 2 days posttransfection for transient-transfection assays. Luciferase activity was measured with a luciferase assay kit (Promega), and β-galactosidase activity was measured with chlorophenol red-β-d-galactopyranoside (Boehringer Mannheim) as a β-galactosidase substrate, using the same cell extract. Production of infectious recombinant retroviruses was performed by transient transfection of Bosc-23 packaging cells (19) with the plasmids carrying the retroviral expression vector and harvesting of cell supernatants 2 and 3 days posttransfection. Cells were infected with 1 ml of viral supernatant in the presence of Polybrene (8 μg/ml). The percentage of infected cells ranged from 10 to more than 50%. G418 selection was initiated 7 days after cell infection.
DNA constructs. (i) Construction of tetOPneoIRESlacZ.
The plasmid tetOPneoIRESlacZ was constructed in two steps. First, a BglII-BamHI neo fragment excised from pMLV-SVtkneo (20) was inserted at the BamHI site of the linker sequence of pUHD10-3 (which contains the tetOP promoter, consisting of a minimal human cytomegalovirus [hCMV] promoter [sequence from position −53 to +75] linked to seven repeats of the tet operator sequence, upstream of a polylinker, and of the simian virus 40 [SV40] polyadenylation signal sequence) (10) to generate the tetOPneo plasmid. Then, the IRESlacZ sequence excised from plasmid 1520 (a gift from I. Ghattas [9]) by XbaI-NheI restriction and treated with Klenow fragment was inserted into tetOPneo cleaved by BamHI (just 3′ to neo) and Klenow treated.
(ii) Construction of tetOPLi-tTa.
The LINE-1 promoter sequence (Li) was excised from pL1.2A (which contains the L1.2A human LINE-1 element [4] subcloned into pGEM5Zf+) as a SacII (in pGEM5Zf+ polylinker, 5′ to L1.2A)-BstXII (position 902 in L1.2A) fragment. The tetOP promoter was excised from plasmid pUHD10-3 as a XhoI-SacII fragment. These two fragments were inserted by a single-step ligation in place of the excised hCMV promoter sequence in the tTA-encoding pUHD15-1 plasmid (10), cleaved by EcoRI (between hCMV and tTA sequences) and XhoI (just upstream of the hCMV promoter), after Klenow treatment of both vector and inserts.
(iii) Construction of the MoSV retroviral expression vectors.
pMoSV was derived from pMLV-SVtkneo (20) by replacing a HindIII (between the tandemly arranged SV40 and thymidine kinase promoters)-ClaI (just upstream of the 3′ Moloney murine leukemia virus long terminal repeat) fragment by a polylinker sequence containing the EcoRI and XhoI sites (this EcoRI site was unique, as the sequence between the EcoRI and ClaI sites in the pBR322-derived plasmid backbone had been previously deleted). MoSV-CAFΔ1178 was isolated from a retroviral cDNA expression library derived from human NTera2/D1 cells in the course of a genetic screen initially devised to select LINE-1 promoter transactivators (Tchénio, unpublished). MoSV-CAF-1 was constructed upon insertion of an EcoRI-EcoRI (both in linkers) CAF-1 cDNA fragment excised from pKK8 (see below) into the EcoRI site of MoSV.
(iv) Plasmids for in vitro translation.
pKS-CAFΔ1178 was constructed by inserting an EcoRI-XhoI CAFΔ1178 fragment excised from MoSV-CAFΔ1178 into pBluescript II KS(−) cleaved by EcoRI and XhoI. pKS-CAFΔ1437, pKS-CAFΔ1731, and pKS-CAFΔ1906 were derived from pKS-CAFΔ1178 upon excision of the sequences between the EcoRI (in the linker) and RsrII (nucleotide [nt] 1436), BssHII (nt 1731), and KpnI (nt 1902) sites in the CAFΔ1178 sequence, respectively, followed by Klenow treatment and religation of the plasmid. Plasmid pKK8 (a gift from P. D. Kaufman) is a complete and functional CAF-1 p150 cDNA inserted at the EcoRI site of pBluescript II SK(+).
Nucleic acids and run-on analyses.
Northern and Southern blot analyses were performed with Hybond N+ membranes (Amersham) according to the recommendations of the manufacturer. α-32P-labeled DNA probes were synthesized with a multiprime labeling kit (Amersham). Total RNA was extracted from cells (2 × 107) by the guanidinium thiocyanate-cesium chloride procedure. Isolation of polyadenylated RNA was performed with a Dynabeads mRNA purification kit (Dynal) according to the manufacturer's instructions. For the run-on analyses, cells (107) were washed three times with cold phosphate-buffered saline and lysed by a 5-min incubation on ice in 10 mM Tris HCl (pH 7.4)–10 mM NaCl–3 mM MgCl2–0.5% NP-40. Nuclei were isolated by centrifugation at 500 × g for 5 min and washed twice in 5 ml of cold lysis buffer. The transcription reaction was achieved by a 30-min incubation of the nuclei at 37°C in 60 μl of transcription buffer containing 20% glycerol, 30 U of RNasin (Promega), 33 mM Tris HCl (pH 8), 150 mM KCl, 0.5 mM dithiothreitol, 3 mM spermidine, 5 mM MgCl2, 3.3 mM concentrations of ATP, CTP, and GTP, and 100 μCi of [α-32P]UTP at 3,000 Ci/mmol. Transcription products were then purified by elution through a G50 Sephadex (Pharmacia) column after cellular DNA digestion by DNase RQ1 (Promega; 6 U with incubation at 37°C for 20 min) and protein removal (by the addition of 700 μg of proteinase K/ml and 3% sodium dodecyl sulfate [SDS] and incubation at 37°C for 30 min followed by phenol-chloroform extraction). The radioactive material was hybridized to Southern blots (Hybond N+; Amersham) of restricted phage or plasmid DNAs, in Church buffer (0.5 M phosphate buffer [pH 6.8], 7% SDS, 1 mM EDTA) at 65°C for 3 days. Washes were at 70°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate)–0.1% SDS.
Endonuclease protection assays.
For endonuclease protection assays, cells (5 × 106) were washed three times with cold phosphate-buffered saline and lysed by a 5-min incubation on ice in 10 mM Tris HCl (pH 7.8)–10 mM NaCl–3 mM MgCl2–0.5% NP-40–1 mM dithiothreitol–0.1 mM EDTA–0.1 mM EGTA–0.75 mM spermidine. Nuclei were isolated by centrifugation at 500 × g for 5 min and washed twice in 5 ml of cold lysis buffer. Nuclei were digested with 0, 33, or 100 U of restriction enzyme at 37°C for 1 h in 250 μl of 50 mM Tris HCl (pH 8)–4 mM MgCl2–50 mM NaCl, and then lysed in 50 mM EDTA–0.2% SDS–50 mM Tris HCl (pH 8)–300 μg of proteinase K/ml for 16 h at room temperature. DNAs were isolated by phenol-chloroform extraction, RNase A treatment, another phenol-chloroform extraction, and ethanol precipitation and then analyzed by Southern blotting as described above.
Western blot and in vitro translation analyses.
For Western blot analyses, cells (107) were lysed by a 10-min incubation on ice in 0.5 ml of EBC buffer (0.5% NP-40, 10 mM Tris HCl [pH 7.5], 150 mM NaCl) supplemented with protease inhibitors (0.1 U of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml) and vigorous vortexing. The supernatant recovered after a 10-min centrifugation at 13,000 rpm (9,000 × g) was aliquoted for protein quantitation using a Bradford assay or mixed with 2× Laemmli buffer and boiled before Western blotting. Protein electrophoresis was carried out in 8 or 10% polyacrylamide gels with 0.1% SDS, using size markers (Sigma) for gel calibration. Proteins were blotted electrophoretically onto 0.2-μm-pore-size nitrocellulose membranes (Sigma) which were stained with Ponceau S solution (Sigma) to ensure that equal amounts of protein were transferred. Immunoblots were incubated for 16 h in 5% nonfat dried milk, and immunodetection was performed with a horseradish peroxidase detection kit (ECL+; Amersham), using a commercially available mouse monoclonal antibody (ab-3; Oncogene Research Products, Calbiochem) against the human CAF-1 p150 subunit. Preliminary studies had indicated that the epitope for ab-3 is located in the C-terminal half of the human CAF-1 p150 but not in the murine CAF-1 p150. In vitro translation analyses were performed with a TNT-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine according to the manufacturer's instructions, using the T7 RNA polymerase promoter. Translation products were analyzed by SDS-polyacrylamide gel electrophoresis, and radiolabeled peptides were detected upon autoradiography of the gels.
RESULTS
Mammalian cell model of transcriptional gene silencing.
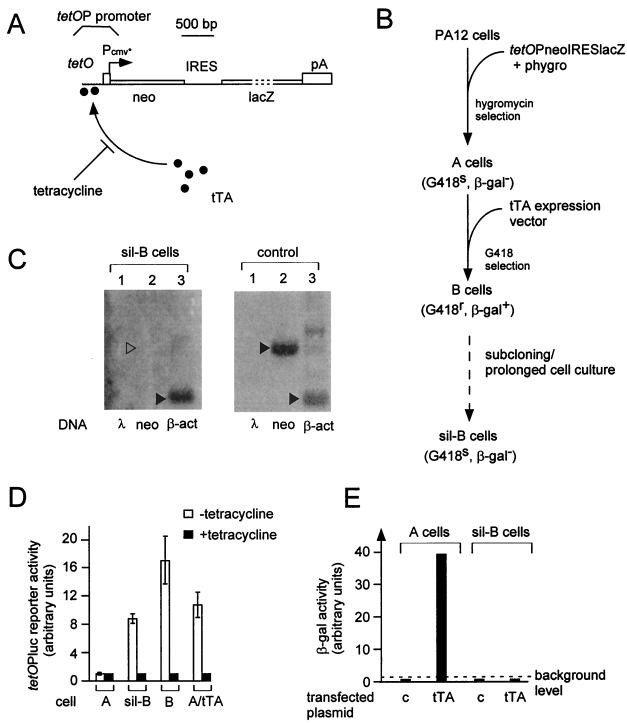
We first tried to develop a well-controlled genetic system in mammalian cells, displaying transcriptional gene silencing and allowing a quantitative assay for its reversion. To do so, we made use of the tetracycline regulatory system first developed by Gossen and Bujard (10). This system relies on the use of an engineered tetracycline-regulated promoter (tetOP) which combines a minimal promoter (derived from the hCMV promoter) and tandemly arranged tet operator sequences (tetO), each containing a binding site for a tetracycline-sensitive transactivator (tTA). Tetracycline can interact with tTA and prevent its binding to the tetO sequences, and thus, it strongly impairs transactivation of the tetOP promoter by tTA (Fig. 1A). We then took advantage of the observation that, in this system, a stably transfected tetracycline-regulated reporter gene may become stably silenced if the cells are maintained in culture for a prolonged period in the absence of selection for expression of the reporter gene or under repressive conditions in the presence of tetracycline (Tchénio, unpublished). Such a silenced cell line, sil-B, was obtained from a previously stably transfected cell population, following the procedure illustrated in Fig. 1 and described below. Murine PA12 fibroblast cells (15) were first cotransfected with a tetracycline-responsive reporter gene (tetOPneoIRESlacZ, which expresses both neomycin resistance and β-galactosidase activity in the presence of the tTA transactivator) (Fig. 1A) and a tk-hygro plasmid. A stably transfected clone (clone A) containing about 20 integrated copies of tetOPneoIRESlacZ was isolated upon hygromycin selection. Clone A was found to be β-galactosidase negative (β-Gal−) and G418 sensitive (G418s) but became strongly β-Gal+ when the tTA transactivator was introduced into the cells, for instance by transient transfection (Fig. 1E). Cells from clone A were then stably transfected with a tTA-expressing vector, and G418 selection of the transfected cells led to the isolation of a G418-resistant (G418r) and β-Gal+ cell clone (clone B, Fig. 1B). The sil-B cells (Fig. 1B) were finally obtained, by puromycin selection, as a subclone of B cells which had been transfected (for a purpose unrelated to the present study) with a luciferase reporter gene and a plasmid conferring resistance to puromycin. It subsequently turned out that the sil-B cells were G418s and β-Gal−: G418 selection of sil-B cells resulted in the isolation of G418r revertants at a very low apparent frequency (<3 × 10−7), and no β-galactosidase activity could be detected in sil-B cells by either an enzymatic assay on protein cell extracts or in situ histochemical staining (Fig. 1E and data not shown). Yet Southern blot analysis of the tetOPneoIRESlacZ reporter gene copies in the sil-B cells did not reveal any change compared to the A or B cells (data not shown). In fact, Northern blot analysis demonstrated that the sil-B cell phenotype is due to the lack of tetOPneoIRESlacZ transcript accumulation (Fig. 2C). Run-on analyses demonstrated that the tetOPneoIRESlacZ gene is silenced at the transcriptional level (Fig. 1C). This lack of transcription is remarkable, since the sil-B cells constitutively express a tTA transactivating activity at a level close to that measured in G418r B cells or in A cells transduced with a tTA retroviral expression vector (Fig. 1D). The lack of tetOPneoIRESlacZ gene expression upon transient transfection of the sil-B cells with a strong tTA expression vector, at variance with what is observed under identical conditions for transiently transfected A cells, confirmed that the tetOPneoIRESlacZ reporter gene had been switched to a silenced state which could not be reversed simply by the overexpression of a specific transactivator (Fig. 1E).
FIG. 1.
A mammalian cell model of transcriptional gene silencing. (A) Structure of the tetOPneoIRESlacZ reporter gene. The tetO regulatory sequences for binding of the tetracycline-repressible tTA transactivator, the minimal PCMV* promoter, and the neo and lacZ ORFs are shown; an internal ribosomal entry site (IRES) allows the simultaneous translation of both neo and lacZ ORFs within the tetOP-initiated transcripts. (B) Construction of the silenced sil-B cells. The murine fibroblasts PA12 cells had originally been selected for their ability to package retroviral transcripts (15), a feature not used in the present assay and actually lost in the course of the hygromycin selection procedure. The successive cell clones obtained are illustrated, together with their phenotypes (in parentheses). The dotted line corresponds to a >2-month culture period following transfection of the cells with a luciferase reporter and a puromycin resistance-encoding vector (the activity of the former vector was not used in the present series of experiments); the tTA expression vector (tetOPLi-tTA) contains tTA under the control of the tandemly arranged tetOP and LINE-1 promoters (see Materials and Methods). (C) Nuclear run-on analysis of the tetOPneoIRESlacZ gene in sil-B cells and control (CAFΔ1178 G418r sil-B revertants [Fig. 2]) cells. Run-on transcripts were synthesized and hybridized to Southern blots of restricted lambda phage DNA as a control or plasmids containing neo or β-actin (β-act) DNA fragments (arrowheads). (D) tTA activity in the A, B, and sil-B cells and in control A cells that were made G418r upon infection with a tTA retroviral expression vector (A/tTA). The tTA activity was assayed upon transient transfection of the indicated cells with plasmid pUHC13-3 (which carries a luciferase reporter gene under the control of the tetOP promoter, tetOPluc), and measurement of luciferase activity was done in cells without and with tetracycline in the culture medium. The values are the means of more than five independent transfection experiments, with duplicate luciferase assays for each experiment and measurement of β-galactosidase activity of cotransfected CMVβ plasmids as a control of transfection efficiency (β-galactosidase activity of the resident tetOPneoIRESlacZ reporter genes was in all cases negligible compared to that of CMVβ). The bars indicate standard deviations. (E) Inducibility of the tetOPneoIRESlacZ reporter gene in sil-B and control A cells. Cells were transiently transfected with an expression vector for tTA (pUHD15-1, a tTA expression vector driven by the strong immediate early promoter of hCMV) or a control (c) vector with the same promoter, and β-galactosidase activity was measured 2 days posttransfection.
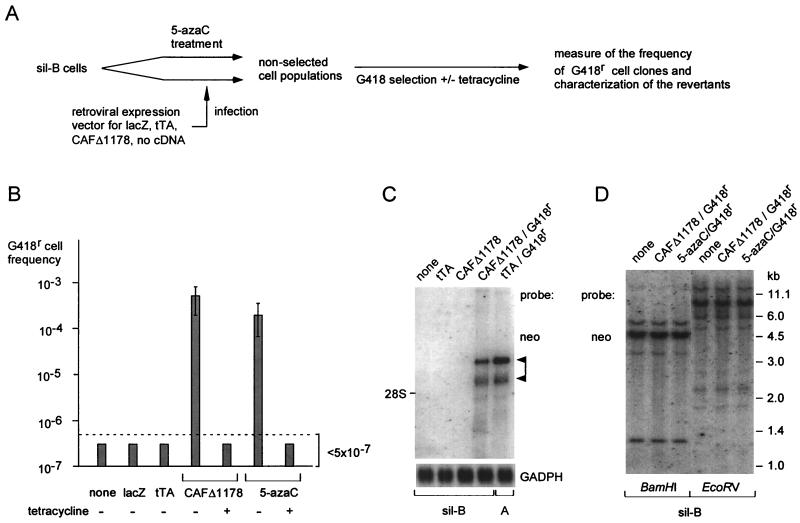
FIG. 2.
Reversion of tetOPneoIRESlacZ gene silencing in sil-B cells. (A) Rationale of the cell assay. Sil-B cells were either treated with 5-azacytidine (5-azaC; 10 μM for 1 day) or infected with retroviral expression vectors for the indicated genes. Seven days later, the resulting nonselected cell populations were then subjected to G418 selection to assay reversion of tetOPneoIRESlacZ silencing, as measured by the frequency (per cell) of emerging G418r cell clones. RNAs were extracted from both the nonselected cell populations and the G418r cells for Northern blot analysis. (B) Reversal of tetOPneoIRESlacZ gene silencing upon sil-B cell infection with the indicated expression vectors or 5-azacytidine treatment. The indicated frequencies are the means of four independent experiments, with standard deviation indicated by the bars. Control G418 selections were performed in the presence of 0.5 μg of tetracycline/ml added throughout the selection procedure. The functionality of the tTA expression vector was tested in a parallel infection experiment using A cells (not shown). (C) Northern blot analysis of tetOPneoIRESlacZ transcripts. Polyadenylated RNA (isolated from 25 μg of total RNA) was extracted from sil-B cells infected with the indicated expression vectors, from G418r cell revertants (CAFΔ1178 G418r), and from G418r A cells obtained after infection with the tTA expression vector (tTA/G418r). The blots were hybridized with a neo probe, or with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe as a control. The positions of two major tetOPneoIRESlacZ transcripts are indicated by arrowheads. (D) Southern blot analysis of the multiple tetOPneoIRESlacZ gene copies in the sil-B cells (none) and the derived G418r revertants (CAFΔ1178/G418r and 5-azaC/G418r). The extracted DNAs were cut with BamHI (two restriction sites in the transfected plasmid, both downstream of neo within tetOPneoIRESlacZ) or EcoRV (one restriction site, in lacZ). The blots were hybridized with a neo probe.
In fact, we could show that DNA methylation was involved in the maintenance of this silenced state. This is illustrated by the fact that 5-azacytidine treatment could reverse tetOPneoIRESlacZ gene silencing. Indeed, the frequency of G418r β-Gal+ revertants that could be isolated upon G418 selection of sil-B cells increased more than 500-fold after a 1-day treatment of the cells with 10 μM 5-azacytidine (average G418r revertant frequency, 2.1 × 10−4 ± 1.4 × 10−4 per cell; three independent experiments with duplicate or triplicate frequency measurements) (Fig. 2B). The revertants obtained upon 5-azacytidine treatment exhibited no visible rearrangement of the tetOPneoIRESlacZ reporter gene copies, at least as monitored by Southern blot analysis (Fig. 2D), thus suggesting that the transfected tetOPneoIRESlacZ gene in the sil-B cells had been inactivated solely by virtue of a DNA methylation-associated epigenetic change. It is worth mentioning that a similar inactivation of the tetOPneoIRESlacZ gene copies was also repeatedly observed in cell populations derived from the parental B cells upon prolonged cell culture (>3 weeks) in the presence of tetracycline (data not shown). Although the experiments reported in this work were essentially concerned with the reversion of the tetOPneoIRESlacZ gene silencing in sil-B cells, similar results were obtained with these silenced (G418s β-Gal−) B-cell-derived cell populations.
Expression of a 5′-truncated CAF-1 p150 cDNA reverses gene silencing.
We then tested whether the ectopic expression of the 5′-truncated cDNA of the CAF-1 p150 subunit mRNA (CAFΔ1178; see structure in Fig. 4D) enhanced, as observed for the 5-azacytidine treatment, the rate of reversion of the sil-B cells to a β-Gal+ G418r phenotype. To do so, the CAFΔ1178 cDNA was first introduced under the control of the early SV40 promoter into a recombinant retroviral vector (MoSV), and packaging cells were transfected with this construct to produce infectious viral particles which were finally used to infect the sil-B cells (Fig. 2A). Control vectors containing no cDNA or the lacZ or the tTA genes were used in parallel. As illustrated in Fig. 2B, infection of the sil-B cells with the CAFΔ1178 cDNA-containing retrovirus, but not with the control vectors, resulted in a dramatic increase in the frequency of G418r β-Gal+ cell revertants which could be isolated upon G418 selection started 7 days after infection of the cells: induction values ranged from 200- to 700-fold in four independent experiments (average G418r revertant frequency, 5.3 × 10−4 ± 3.2 × 10−4 per cell; a major cause of this fluctuation was the variable percentage of infected cells, from 10 to >50%). Analysis by Southern blot of the status of the transfected reporter gene copies in the revertants failed to detect any change in DNA structure, with identical patterns of bands upon restriction with a series of restriction enzymes (Fig. 2D). Rather, Northern blot analysis and run-on assays (Fig. 1C and 2C) provided evidence that the observed phenotypic reversion was the consequence of the transition of tetOneoIRESlacZ from a transcriptionally inactive to an active state. The tTA activity was required for the generation of G418r β-Gal+ sil-B cell revertants. Indeed, treatment of the sil-B cells with tetracycline after their infection with the CAFΔ1178-expressing retrovirus prevented the efficient recovery of revertants (Fig. 2B). In addition, all the G418r revertants derived from the CAFΔ1178-expressing sil-B cells turned out to be G418s when grown in the presence of tetracycline (data not shown). Yet infection of the sil-B cells with a tTA-expressing retrovirus had no effect on the frequency of occurrence of revertants, as expected, under conditions where infection of control A cells with the same tTA-expressing retrovirus resulted in a very high frequency of G418r β-Gal+ cells (>10−1; data not shown). Altogether, these results strongly suggest that CAFΔ1178 expression does not directly transactivate the tetOPneoIRESlacZ reporter gene but rather renders it competent for transactivation by the tTA protein. This interpretation is also in agreement with the observation that CAFΔ1178 expression vectors are unable per se to transactivate the tetOPneoIRESlacZ gene in transient-transfection assays using, for instance, A cells (data not shown). Similarly, CAFΔ1178 expression did not induce a general increase of cell transcription, as suggested by the similar amounts of total RNA extracted from sil-B cells and CAFΔ1178 G418r revertants (111 ± 10 μg and 103 ± 15 μg per 107 cells, respectively, for four independent assays), as well as by the similar levels of various mRNAs from expressed genes, including glyceraldehyde-3-phosphate dehydrogenase, β-actin, and endogenous intracisternal A particle (Fig. 2C and data not shown). In summary, we found two conditions resulting in tetOPneoIRESlacZ gene reactivation in the sil-B cells, one associated with CAFΔ1178 expression and the other with 5-azacytidine treatment, both most probably involving changes in the status of the tetOPneoIRESlacZ gene itself.
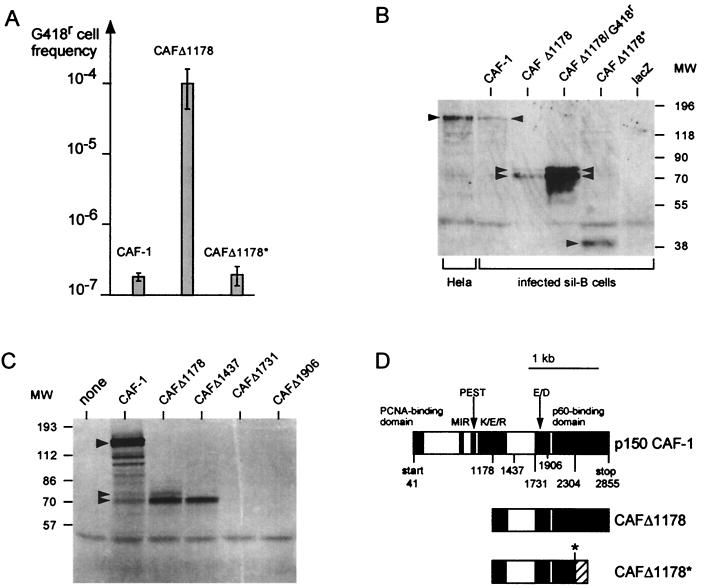
FIG. 4.
Characterization of the translational products involved in the CAFΔ1178-mediated reversal of gene silencing. (A) Requirement for a truncated translational product. sil-B cells were infected with a retroviral expression vectors (MoSV) for the complete CAF-1 p150, for CAFΔ1178, and for a frameshift mutant (CAFΔ1178*) derived from CAFΔ1178 by deletion of four nucleotides at positions 2304 to 2307 (asterisk in panel D). The indicated frequencies of G418r cells isolated for each population of infected cells (percentage of infected cells close to 10%) are the means of three independent experiments, with standard deviations indicated by bars. (B) Western blot analysis of cell extracts from the murine sil-B cells infected with the indicated retroviral expression vectors and from control human HeLa cells for the endogenous CAF-1, using a monoclonal antibody specific for the human CAF-1 p150 protein. Amounts of protein extracts were 60 μg for sil-B cells infected with MoSV-CAFΔ1178, both before (CAFΔ1178) and after (CAFΔ1178/G418r) G418 selection, and 120 μg for all other cell extracts. (C) In vitro translation assay of CAF-1 and 5′-truncated derivatives (see panel D). Arrowheads indicate the 150-kDa and the 70- to 74-kDa products. (D) Structure of the complete p150 CAF-1 subunit and derivatives. The previously identified CAF-1 functional domains are schematized, according to references 11, 16, and 18. Numbers refer to nucleotide positions in the cDNA sequence. The asterisk (at position 2304) corresponds to a unique SacII site used to introduce a frameshift within CAFΔ1178 (the frameshifted codons are indicated by a hatched domain). MW, molecular weight (weights are in thousands); PCNA, proliferating cell nuclear antigen; MIR, MOD1 interaction region.
Comparative analysis of the methylation and chromatin accessibility status of the tetOPneoIRESlacZ gene.
We first analyzed the level of methylation of tetOPneoIRESlacZ when in a silenced and in a reactivated state. Methylation was assayed by Southern blot analysis, after digestion of cellular DNAs with the isoschizomer pair of restriction enzymes MspI/HpaII (which display differential sensitivities to DNA methylation of their common restriction sites). As illustrated in Fig. 3A, this analysis revealed that the tetOPneoIRESlacZ gene is methylated in the sil-B cells whereas it undergoes a demethylation in the G418r revertants isolated following 5-azacytidine treatment. We also found that tetOPneoIRESlacZ is hypomethylated in the A cells, which correlates with its strong inducibility by tTA (Fig. 1 and 3A). Conversely, and rather unexpectedly, analysis of four independent G418r sil-B revertant cell populations expressing CAFΔ1178 did not reveal any significant DNA demethylation of the bulk of the tetOPneoIRESlacZ gene copies (Fig. 3A). As an internal control for the DNA restriction enzyme treatment, rehybridization of the blots for the probing of the hypomethylated tTA expression vector disclosed bands of similar intensities for all cell types (data not shown).
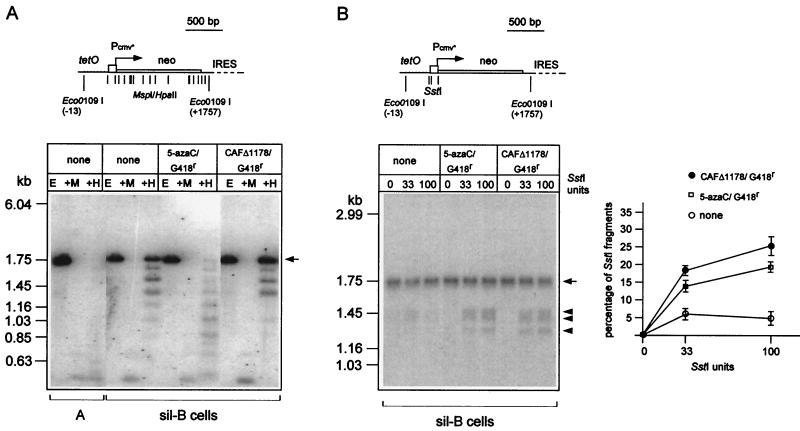
FIG. 3.
Methylation and chromatin accessibility of the tetOPneoIRESlacZ gene. (A) Methylation status of the tetOPneoIRESlacZ gene. Cellular DNAs from the indicated cells were extracted and digested with Eco0109I alone (E) or with Eco0109I plus the methylation-insensitive MspI (+M) or the methylation-sensitive HpaII (+H) enzymes (same recognition sequence). The resulting fragments were analyzed by Southern blotting and hybridization with a neo probe. The structure of the corresponding region of the tetOPneoIRESlacZ gene is schematized in the upper part of the figure, with the positions of the restriction sites for MspI and HpaII and for Eco0109I indicated by vertical bars. The cells tested include A cells, sil-B cells, and revertants obtained after treatment of sil-B cells with 5-azacytidine (5-azaC; 10 μM for 1 day) or infection with MoSV-CAFΔ1178 and G418 selection. G418r cell populations were continuously maintained under G418 selection pressure until cell lysis for cellular DNA extraction. (B) Endonuclease protection assay of the tetOPneoIRESlacZ gene. About 5 × 106 nuclei from the indicated cells were isolated as described in Materials and Methods and digested with 0, 33, and 100 U of SstI restriction enzyme. The DNA was then purified, digested with Eco0109I to release a 1.75-kb parent fragment, and analyzed by Southern blotting with a neo probe (B, left). The position of the 1.75-kb fragment is indicated by an arrow and those of the three neo-hybridizing fragments after SstI restriction by arrowheads (see the scheme in the upper part of the panel, with the positions of the SstI and Eco0109I sites indicated by vertical bars). The percentage of the tetOPneoIRESlacZ gene copies cut by SstI was quantitated by phosphorimager analysis (B, right). The ordinates are the percentages of total signal contained in the SstI-cut fragments at each enzyme concentration. IRES, internal ribosomal entry site.
To characterize the chromatin accessibility of tetOPneoIRES lacZ, we performed endonuclease protection assays on nuclei isolated from the sil-B cells and the derived G418r revertants obtained following CAFΔ1178 expression or 5-azacytidine treatment of the cells. Nuclei were isolated and treated with increasing amounts of SstI, which possesses three restriction sites in the tetOP promoter (Fig. 3B). DNA was then purified, digested with Eco0109I to release a 1.75-kb parent fragment, and analyzed by Southern blot with a neo probe. As illustrated in Fig. 3B, three bands of the expected size were observed with nuclei derived from G418r revertants obtained following CAFΔ1178 expression or 5-azacytidine treatment. In contrast, these bands are barely visible with nuclei of the parent sil-B cells in which all of the tetOPneoIRESlacZ gene copies are in a silenced state. These assays show that at least some of the tetOPneoIRESlacZ gene copies have gained an increased sensitivity to endonuclease digestion in the revertant cells. Similar results were obtained upon measurement of the chromatin accessibility of the two Eco0109I restriction sites bordering the neo sequence (data not shown).
CAFΔ1178-mediated reversal of gene silencing requires translation of a truncated CAF-1 product.
To determine whether the CAFΔ1178 cDNA reversed gene silencing by translating a 5′-truncated p150 CAF-1 product and to unambiguously identify the corresponding gene product, we measured the ability of several CAF-1-derived expression vectors to reverse gene silencing in sil-B cells. We used a full-length CAF-1 p150 cDNA (11), CAFΔ1178, and CAFΔ1178*. CAFΔ1178* includes a frameshift introduced by deletion of four nucleotides at positions 2304 to 2307, which results in a C-terminal truncation (Fig. 4D). In four independent experiments in which we introduced (by retroviral infection) the indicated expression vectors into the sil-B cells, we failed to detect any effect of either the complete p150 CAF-1 cDNA or the CAFΔ1178* cDNA on tetOPneoIRESlacZ gene reactivation. As illustrated in Fig. 4A, expression of these cDNAs was at least 200-fold less efficient than CAFΔ1178 expression in gene silencing reversion. Western blot analyses of the corresponding cell extracts (Fig. 4B), using a mouse monoclonal antibody specific for the human CAF-1 p150 subunit (theoretical molecular mass, 105 kDa; see control p150 detection in HeLa cell extracts), revealed a 150-kDa product in cells transduced with the full-length p150 CAF-1 cDNA expression vector and a 40-kDa product for CAFΔ1178*. For the sil-B cells transduced with the CAFΔ1178 expression vector, products of approximately 70 and 74 kDa were detected. The level of these 70- to 74-kDa products was higher in the sil-B cell revertants isolated upon G418 selection than in the primary infected cells (Fig. 4B), consistent with the efficiency of viral infection (approximately 10%; see the legend to Fig. 4) and the correlation between CAFΔ1178 expression and gene silencing reversion. The 70- and 74-kDa gene products most probably correspond to distinct initiation sites and should lack most, if not all, of the K/E/R domain (which extends from nucleotides 974 to 1378) (11) but still contain complete E/D and CAF-1 p60-binding domains (11) (Fig. 4D). In vitro translation assays using reticulocyte lysate (Fig. 4C) confirmed this interpretation, with the full-length p150 CAF-1 cDNA encoding a 150-kDa product and CAFΔ1178 cDNA encoding two major products of 70 and 74 kDa; additional 5′ truncations (down to nt 1437, 1731, and 1906) (Fig. 4D) resulted in the sequential disappearance of the 74- and 70-kDa products (Fig. 4D). Interestingly, the frameshift mutant CAFΔ1178*-encoded protein differs from that encoded by CAFΔ1178 by the lack of the terminal domain, which is known to bind the p60 subunit of CAF-1, thus suggesting that the ability to bind p60 CAF-1 is important for reversal of gene silencing.
DISCUSSION
To analyze the role of human CAF-1 p150 derivatives in the maintenance of gene silencing in mammalian cells, we have devised a genetic assay which relies on the use of a stably transfected tetracycline-responsive reporter gene (tetOPneoIRESlacZ) which can adopt three distinct states: an activated state in the presence of the specific tetracycline-sensitive tTA transactivator, a fully reversible inactive state in the absence of a functional tTA transactivator, and an almost irreversible inactive state in which the reporter gene is unresponsive to tTA transactivation. The latter state correlates with an extensive DNA methylation of the tetOPneoIRESlacZ copies and can be reversed upon treatment of the cells with the DNA-demethylating agent 5-azacytidine. We show that transcription of a truncated cDNA of the human p150 CAF-1, namely CAFΔ1178 cDNA lacking the first 1,178 nucleotides of the p150 CAF-1 mRNA, is at least as efficient as 5-azacytidine in reversing the transcriptional silencing of the methylated tetOPneoIRESlacZ gene.
CAFΔ1178 restores response to specific transcription factors.
Silencing of the tetOPneoIRESlacZ reporter gene in the sil-B cells is not due to the absence of appropriate transcription factors. Indeed, an advantage of the reporter gene devised here is that its transactivating factor is known, as it is specifically and de novo introduced into the cells. Furthermore, its presence in a functional state has been demonstrated, as even in sil-B cells, where the tetOPneoIRESlacZ reporter is silent, we have shown that a tTA activity is present at a level similar to that observed in fully activated B cells. Accordingly, the lack of transcription of the tetOPneoIRESlacZ reporter in the sil-B cells cannot be ascribed to an absence of the appropriate transcription factor. Rather, it is due to a lack of susceptibility of the tetOPneoIRESlacZ reporter gene to present specific transcription factors, a susceptibility which is restored by CAFΔ1178 expression. Consistent with this view on the CAFΔ1178 mode of action, we found that the CAFΔ1178-encoded protein is not acting as a transcription factor, as its expression had no effect on the transcription of various reporter genes when these genes are in a state allowing their direct transactivation by appropriate transcription factors. For instance, CAFΔ1178 expression had no effect on transiently transfected luciferase reporters under the control of various promoters (including the tetOP and SV40 immediate early promoters) (Tchénio, unpublished), nor can it transactivate in A cells the stably transfected tetOPneoIRESlacZ reporter, which is in an inducible state, as demonstrated by its ability to be transactivated upon transient transfection of a tTA expression vector.
CAFΔ1178-induced gene reactivation may not require DNA demethylation but involves changes in chromatin accessibility.
An intriguing issue of the present investigation is the apparent lack of DNA demethylation of the bulk of the tetOPneoIRESlacZ copies, observed in the G418r cell revertants isolated following CAFΔ1178 transduction. This is in contrast with the hypomethylated state of the tetOPneoIRESlacZ copies in the 5-azacytidine-induced sil-B cell revertants. In addition, the tTA-inducible but silent state of the tetOPneoIRESlacZ gene in the A cells also correlates with a hypomethylated state of the bulk of the reporter gene copies. Two nonexclusive interpretations could account for this paradoxical observation. It remains possible that one, or a few, of the ∼20 copies of the tetOPneoIRESlacZ gene have undergone a discrete DNA demethylation at some critical cytosine residue in the CAFΔ1178-induced sil-B cell revertants that would have escaped detection. Alternatively, CAFΔ1178 gene expression may overcome a requirement of DNA demethylation for gene reactivation. Indeed, several studies suggest that the repressive effect of DNA methylation involves changes in the nucleosomal structure, in particular through histone deacetylation which could interfere with the binding of transcription factors either directly or through the compaction of chromatin (reviewed in references 1 and 2). Then, a CAFΔ1178-induced alteration of the nucleosomal structure, through for instance a disruption of the inheritance of deacetylated histones, could bypass the need of DNA demethylation for gene reactivation. This hypothesis is in agreement with our finding that CAFΔ1178-induced tetOPneoIRESlacZ gene reactivation correlates with an increased chromatin accessibility of the tetOPneoIRESlacZ gene copies. Taking into account the fact that the increase in chromatin accessibility of the tetOPneoIRESlacZ gene copies is similar in the G418r revertants obtained following 5-azacytidine treatment and CAFΔ1178 expression, it is therefore likely that the CAFΔ1178-induced chromatin effects do not require DNA demethylation. Finally, it is noteworthy that CAFΔ1178-induced chromatin effects may be restricted to silenced genes; indeed, in preliminary experiments, we did not observe any change in chromatin accessibility of the SstI restriction site located in the second intron of the active c-myc gene (nearly maximal SstI cutting efficiencies close to 30% at a 33-U enzyme dose for the sil-B cell and its derived CAFΔ1178 G418r and 5-azacytidine G418r revertants in an assay similar to that of Fig. 3B; data not shown). Altogether, these observations suggest that CAFΔ1178 expression promotes a transition towards an open chromatin structure, which is already present in actively transcribed genes.
CAFΔ1178 may encode a dominant negative mutant of p150 CAF-1.
We have shown that the CAFΔ1178-mediated effect requires expression of a specific CAFΔ1178 translational product that might be defective for correct interaction with some components of the heterochromatin and/or for chromatin assembly activity. The 5′-truncation in CAFΔ1178 encompasses the first 380 codons of the p150 CAF-1 subunit and thus includes a previously identified binding domain for proliferating cell nuclear antigen (16), a putative PEST domain, possibly regulating protein stability, and part of the K/E/R domain, which extends from nucleotides 974 to 1376 and is required for the chromatin assembly activity of the p150 CAF-1 subunit (11). Furthermore, recent studies have shown that the N-terminal domain of the murine and human CAF-1 p150 subunit is involved in the association of CAF-1 with members of the heterochromatin-binding protein-1 family (18), including structural components of the pericentromeric heterochromatin. In fact, Western blot analyses of CAFΔ1178-transduced sil-B cells, using a monoclonal antibody specific for the human CAF-1 p150, as well as in vitro translation analyses provided evidence for major translation products with an apparent molecular mass close to 70 kDa, which retain only the CAF-1 E/D and p60-binding domains.
Although further studies will be required to determine the precise mechanism by which expression of the 5′-truncated CAF-1 p150 product affects gene silencing in mammals, an attractive hypothesis is that the CAFΔ1178 cDNA is a dominant negative mutant encoding products that impair the normal function of the wild-type CAF-1 complex. This hypothesis would indeed be in agreement with our result showing that expression of a cDNA encoding a complete CAF-1 p150 subunit has no effect on the frequency of gene silencing reversion. Thus, CAFΔ1178 products could impair the formation of the wild-type CAF-1 complex by binding to and titrating out components of the CAF-1 complex, such as the CAF-1 p60 subunit, which binds to p150 (11). Alternatively, CAFΔ1178 could form a defective CAF-1 complex that competes with the wild-type one for its interaction with other specific components of the cell machinery. Accordingly, it would be interesting to test whether overexpression of the CAF-1 p60 subunit can counteract the effect of CAFΔ1178 on gene silencing. In any case, CAFΔ1178 would mimic the reported effects of null mutations of CAF-1 in yeast, which result in the impairment of the inheritance of repressive nucleosomal structures and in gene silencing disruption.
In conclusion, the results presented suggest that the CAF-1-dependent mechanisms for gene silencing maintenance and/or inheritance may have been conserved from yeast to mammals. They also provide new molecular and cellular tools to elucidate the functional links between CAF-1 and other factors that most probably cooperate for chromatin-dependent gene silencing and epigenetic control of gene expression.
ACKNOWLEDGMENTS
We acknowledge H. Bujard and M. Gossen for the gift of plasmids pUHD10-3, pUHC13-3, and pUHD15-1, I. Ghattas for plasmid 1520, and P. D. Kaufman for plasmid pKK8. We thank C. Lavialle for comments and critical reading of the manuscript.
This work was supported by a grant from the Association pour la Recherche sur le Cancer.
Footnotes
Corresponding author. Present address for Thierry Tchénio: CNRS UPR 1983, Institut de Recherche sur le Cancer André Lwoff, 94801 Villejuif, France. E-mail: tchenio@infobiogen.fr. Mailing address for Thierry Heidmann: Unité des Rétrovirus Endogènes et Elements Rétroïdes des Eucaryotes Supérieurs, CNRS UMR 1573, Institut Gustave Roussy, 94805 Villejuif, France. Phone: 33/1 42 11 49 70. Fax: 33/1 42 11 53 42. E-mail: heidmann@igr.fr.
REFERENCES
- 1.Bestor T H. Methylation meets acetylation. Nature. 1998;393:311–312. doi: 10.1038/30613. [DOI] [PubMed] [Google Scholar]
- 2.Bird A P, Wolffe A P. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 3.Bulger M, Ito T, Kamakaka T T, Kadonaga J T. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone binding protein. Proc Natl Acad Sci USA. 1995;92:11726–11730. doi: 10.1073/pnas.92.25.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dombroski B A, Mathias S L, Nanthakumar E, Scott A F, Kazazian H H. Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 5.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 7.Gaillard P H, Martini E M, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 8.Gaillard P H, Moggs J G, Roche D M, Quivy J P, Becker P B, Wood R D, Almouzni G. Initiation and bidirectional propagation of chromatin assembly from a target site for nucleotide excision repair. EMBO J. 1997;16:6281–6289. doi: 10.1093/emboj/16.20.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghattas I, Sanes J, Majors J. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufman P D, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 12.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman P D, Cohen J L, Osley M A. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martini E, Roche D M, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller A D, Law M F, Verma I M. Generation of helper-free amphotropic retroviruses that transduce a dominant-acting, methotrexate-resistant dihydrofolate reductase gene. Mol Cell Biol. 1985;5:431–437. doi: 10.1128/mcb.5.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moggs J, Grandi P, Ouivy J, Jonsson Z, Hubscher U, Becker P, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monson E, de Bruin D, Zakian V. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 19.Pear W, Nolan G, Scott M, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubenstein J L, Nicolas J F, Jacob F. Construction of a retrovirus capable of transducing and expressing genes in multipotential embryonic cells. Proc Natl Acad Sci USA. 1984;81:7137–7140. doi: 10.1073/pnas.81.22.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 22.Shibahara K, Verreault A, Stillman B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1-mediated nucleosome assembly onto replicated DNA in vitro. Proc Natl Acad Sci USA. 2000;97:7766–7771. doi: 10.1073/pnas.97.14.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 24.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 26.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]