Abstract
Campylobacter jejuni has been identified as the predominant cause of antecedent infection in Guillain-Barré syndrome (GBS) and Miller Fisher syndrome (MFS). The risk of developing GBS or MFS may be higher after infection with specific C. jejuni types. To investigate the putative clonality, 18 GBS- or MFS-related C. jejuni strains from The Netherlands and Belgium and 17 control strains were analyzed by serotyping (Penner and Lior), restriction fragment length polymorphism analysis of PCR products of the flaA gene, amplified fragment length polymorphism analysis, pulsed-field gel electrophoresis, and randomly amplified polymorphic DNA analysis. Serotyping revealed 10 different O serotypes and 7 different Lior serotypes, thereby indicating a lack of serotype clustering. Two new O serotypes, O:35 and O:13/65, not previously associated with GBS or MFS were found. Serotype O:19 was encountered in 2 of 18 strains, and none was of serotype O:41. The results of all genotypic methods also demonstrated substantial heterogeneity. No clustering of GBS- or MFS-related strains occurred and no molecular marker capable of separating pathogenic GBS or MFS from non-GBS- or non-MFS-related enteritis strains could be identified in this study. Sialic-acid-containing lipopolysaccharides (LPS) are thought to be involved in the triggering of GBS or MFS through molecular mimicry with gangliosides in human peripheral nerves. Therefore, further characterization of GBS- or MFS-related C. jejuni should target the genes involved in the synthesis of LPS and the incorporation of sialic acid.
The Guillain-Barré syndrome (GBS) is the most frequent form of acute inflammatory polyneuropathy. The Miller Fisher syndrome (MFS) is considered a rare variant of GBS. GBS and MFS patients demonstrate a heterogeneous clinical presentation and outcome (28). Campylobacter jejuni infections in GBS are associated with the presence of antibodies to GM1 and other peripheral nerve gangliosides (10, 11). These antibodies presumably are induced by the infectious agent since C. jejuni lipopolysaccharide (LPS) from GBS and MFS patients shows molecular mimicry with several gangliosides (17).
C. jejuni is the most frequent cause of bacterial diarrhea. Approximately 1 in every 1,000 C. jejuni infections will be followed by GBS (15). Several authors have hypothesized that GBS-related C. jejuni strains share specific features by which they induce antibodies cross-reactive with peripheral nerve tissue. GBS-related C. jejuni strains have been reported to be associated with the specific Penner serotypes O:19 and O:41, and these appeared to be clonally related (6, 12, 24). The risk of developing GBS may be higher after infection with serotype O:19 (15).
Unfortunately, the time between the preceding intestinal infection and the onset of GBS often exceeds the duration of excretion of viable C. jejuni cells in stools. Thus, the number of GBS-related C. jejuni isolates available for further study is limited.
The aim of the study was to investigate the genetic variation among these strains by using serotyping and various genotyping methods, including a flagellin typing method that determines polymorphisms in the flaA gene (3), amplified fragment length polymorphism (AFLP) analysis (which has recently been adapted for genotyping of Campylobacter spp. [4]), pulsed-field gel electrophoresis (PFGE), and randomly amplified polymorphic DNA (RAPD) analysis (3–5, 7).
MATERIALS AND METHODS
Bacterial strains.
In order to maximize the number of isolates, we prospectively cultured stool specimens of patients presenting with GBS or MFS, starting in 1994 and using a variety of sensitive and selective culture techniques, including broth enrichment and mechanical filtration. We collected 18 GBS- or MFS-related C. jejuni strains from patients in The Netherlands and Belgium. The 18 clinical C. jejuni strains analyzed in this study were isolated in the acute phase of the disease from the stools of 17 Dutch patients and 1 Belgian patient between 1991 and 1998. All patients had a history of diarrhea prior to the onset of GBS or MFS and/or anti-Campylobacter antibodies, which is suggestive of a recent infection (1). Four C. jejuni isolates were isolated from patients with MFS, and 12 were from patients with GBS. Two isolates came from the diarrheal stools of two family members of a GBS patient who remained culture negative throughout but showed a serological response highly suggestive of a recent Campylobacter infection (1). In addition, nine C. jejuni isolates from unrelated enteritis patients without neurological symptoms and eight reference C. jejuni O serotypes were included. All GBS patients fulfilled the diagnostic criteria (2). All MFS patients suffered from ophthalmoplegia, ataxia, and areflexia (14).
Serotyping.
All strains were serotyped with the heat-stable (HS or O) and heat-labile (HL) serotyping schemes of Penner and Lior, respectively. The serotyping was performed at the National Laboratory for Enteric Pathogens, Canadian Science Centre for Human and Animal Health, Winnipeg, Canada, as described previously (13, 22).
Bacterial DNA isolation.
Chromosomal DNA was isolated with the Wizard Genomic DNA purification kit according to the manufacturer's instructions (Promega, Madison, Wis.).
PFGE.
PFGE was performed as previously described (26). In short, samples of genomic DNA extracted from overnight cultures of the strains were digested with SmaI (Boehringer GmbH, Mannheim, Germany). Electrophoresis was performed in 1% SeaKem agarose in 0.5× Tris-borate-EDTA buffer by using a Bio-Rad CHEF mapper programmed in the auto-algorithm mode (run time, 19 h; switch time, 6.75 to 25 s). Gels were stained with ethidium bromide for 15 min, destained in distilled water for 1 h, and photographed under UV radiation. The gels were inspected visually by two different investigators. The patterns were interpreted according to the criteria described by Tenover et al. (25). Isolates that differed by one to three bands, consistent with one single differentiating genetic event, were assigned the same capital letter, but with a numbered subtype. Four or more band differences between two strains were defined as distinct genotypes and were designated with different capital letters.
RAPD.
RAPD was done in volumes of 50 μl containing 200 μM concentrations of each deoxyribonucleoside triphosphate, 50 ng of template DNA, 2.5 U of Taq DNA polymerase (Promega, Southhampton, United Kingdom), 1.5 mM MgCl2, and 5 μl of reaction buffer (Promega). The primers used were the enterobacterial repetitive intergenic consensus sequences ERIC-1 (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC-2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) (18). Amplification was performed in a DNA thermal cycler (Perkin-Elmer 9600; Perkin-Elmer, Norwalk, Conn.). The PCR program consisted of 4 min at 94°C, followed by 40 cycles of 45 s at 94°C, 45 s at 25°C, and 1 min at 74°C. The PCR products were analyzed by electrophoresis in 1% agarose gels. After staining with ethidium bromide and destaining, photographs were made using UV transillumination. Banding patterns were analyzed visually by two independent examiners, and the profiles were designated by a different capital letter whenever a single band difference was observed.
AFLP.
AFLP has recently been adapted for genotyping of Campylobacter spp., and it generated fingerprints with 50 to 80 bands with sizes ranging from 50 to 500 bp.
The AFLP reactions were performed as described previously (4). The restriction enzymes used were HindIII and HhaI. For DNA amplification, the HhaI primer (5′-GATGAGTCCTGATCGCA-3′) and the fluorescently labelled HindIII primer (5′-GACTGCGTACCAGCTTA-3′) were used. For selective PCR amplification, both primers contained a single additional A nucleotide at their 3′ ends. AFLP fingerprints were analyzed on a 373A ABI DNA sequencer, followed by numerical analysis of patterns. Strains with similarity levels of >90% were defined as genetically related and assigned the same capital letter (4). Strains that belonged to the same cluster, with >80% similarity, were defined as genetic subtypes and assigned the same capital letter, but with a numbered subtype. Distinct AFLP fingerprints that showed <80% similarity were designated with different capital letters.
flaA PCR-RFLP.
For DNA amplification, the flaA primer (5′-CGTATTAACACAAATGTTGCAGC-3′) and flaR primer (5′-GATTTGTTATAGCAGTTTCTGCTATATCC-3′) as described by Ayling et al. (3) were used. Reaction mixtures consisted of 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.01% (wt/vol) gelatin, 2 mM MgCl2, 0.2 μM deoxynucleoside triphosphates, 50 pmol of each primer, 50 pg of genomic DNA, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer, Gouda, The Netherlands), with a total reaction volume of 50 μl. Reaction conditions were 60 s at 94°C, followed by 45 cycles of 45 s at 94°C, 45 s at 55°C, and 2 min at 72°C, and a 5-min extension at 72°C. After verification of the PCR, 12.5 μl of the amplicon was digested for 2 h at 37°C using 10 U of DdeI (Boehringer) in a total volume of 15 μl. Restriction fragments were separated on a 2% (wt/vol) NuSieve (FMC, Rockland, Maine) and 0.5% (wt/vol) MP (Boehringer Mannheim, Germany) ethidium bromide-stained agarose gel in 1× Tris-acetate-EDTA. Gels were electrophoresed for 4 h at 80 V, and images of the gels were digitized and saved as TIFF files.
Distinct flaA fingerprints showing a single band difference were designated with different capital letters.
Data processing.
Levels of similarities between banding patterns were determined with the GelCompar v4.1 software (Applied Maths, Kortrijk, Belgium). For analysis of AFLP fingerprints, the Pearson product-moment correlation coefficient was used. The flaA, PFGE, and RAPD banding patterns were analyzed with the Dice band-based coefficient. Cluster analysis was performed with the unweighted pair group method with averages (29).
RESULTS
Serotyping.
Serotyping of 18 GBS- or MFS-related strains revealed 10 different O serotypes and 7 different HL serotypes (Table 1). C. jejuni O:19, Lior 77 was encountered in 2 of 18 (11%) patients. C. jejuni O:2 was found in one GBS patient, in two family members of another GBS patient, and in one MFS patient. C. jejuni O:4/64 was encountered in one GBS patient and one MFS patient; the related C. jejuni O:4/13/64 strain was found in one GBS patient. C. jejuni O:13/65 and O:35 were two new serotypes not earlier described in association with GBS or MFS (Table 1). Four patients with MFS had C. jejuni of different O serotypes (Table 1). All other strains had unique O serotypes.
TABLE 1.
Survey of serotyping (Penner and Lior) and genotyping data for C. jejuni strains associated with GBS and MFSa
| Strain code | O serotype | HL serotype | PFGE | RAPD
|
flaA | AFLP | |
|---|---|---|---|---|---|---|---|
| ERIC-1 | ERIC-2 | ||||||
| GBS or MFS | |||||||
| GB1 | 1 | UT | A | A | A | A | A |
| GB2 | UT | 36 | B | B | B | B | B |
| GB3 | 19/24 | 77 | C | A | C | B | B |
| GB4 | 37 | 28 | D | C | D | C | C |
| GB5 | 4/64 | 1 | E | D | E | D | D |
| MF6 | 4/64 | 1 | E1 | E | E1 | E | E |
| MF7 | 35 | UT | F | F | F | F | F |
| MF8 | 23/36 | 5 | G | G | G | G | G |
| GB11 | 2 | 4 | H | H | E1 | H | H1 |
| GB13 | 2 | 4 | H1 | H1 | H | E | H |
| GB14 | 2 | 4 | H1 | H1 | E1 | E | H |
| GB15 | 5/34 | UT | I | I | I | I | I |
| GB16* | 13/65 | UT | J | J | J | B | J |
| GB17 | 4/13/64 | UT | K | K | E | E | E |
| GB18 | 19 | 77 | L | L | G | B | B |
| GB19 | 4/50 | 7 | M | M | K | J | J |
| MF20 | 2 | 4 | N | N | L | K | K |
| GB21 | 13/65 | 7 | O | O | M | L | L |
| Serostrains | |||||||
| CCUG 10935 | 1 | P | P | N | D | A1 | |
| CCUG 10936 | 2 | Q | Q | O | E | H2 | |
| CCUG 10938 | 4 | R | R | O | M | M | |
| CCUG 10950 | 19 | L1 | S | C | N | B | |
| CCUG 10954 | 23 | S | T | P | D | N | |
| CCUG 10965 | 35 | S1 | U | Q | O | O | |
| CCUG 10966 | 36 | T | V | R | G | G | |
| CCUG 24868A | 64 | U | W | S | P | P | |
| Enteritis | |||||||
| 98-623 | V | X | T | G | Q | ||
| 98-624 | W | Y | U | Q | R | ||
| 98-652 | X | Z | V | R | K | ||
| 98-682 | Y | AA | W | S | A | ||
| 98-706 | Z | AA1 | X | I | I | ||
| 98-1033 | AA | BB | Y | T | S | ||
| 98-1039 | BB | CC | Z | U | T | ||
| 98-1040 | CC | DD | AA | V | U | ||
| 98-1087 | DD | EE | BB | T | V | ||
The following were serotyped and genotyped: strains from The Netherlands (n = 17) and Belgium (n = 1), enteritis control strains (n = 9), and Penner O serostrains (n = 8). *, Belgian strain; UT, untypeable.
flaA PCR-RFLP.
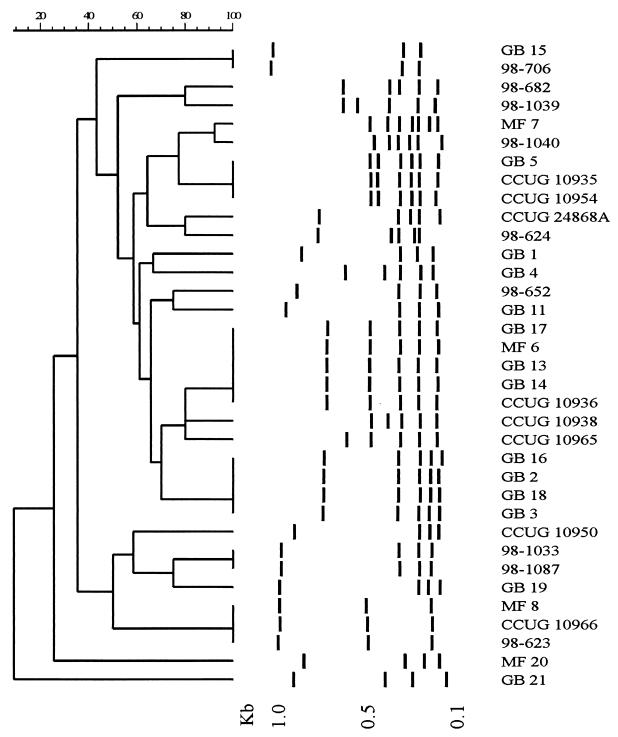
flaA PCR-RFLP analysis of 18 GBS- or MFS-associated C. jejuni strains identified 12 distinct patterns (Table 1; Fig. 1). Analysis of GB13 (O:2) and GB14 (O:2), both strains isolated from family members of a GBS patient, produced flaA patterns indistinguishable from those of GB17 (O:4/13/64) and MF 6 (O:4/64). The two O:19 strains (GB3 and GB18) showed identical flaA patterns that were also shared with strains GB2 (O:UT) and GB16 (O:13/65), but not with the O:19 reference strain. Other GBS strains had flaA patterns that were highly related to flaA patterns of enteritis-related strains and reference serotype strains (Table 1; Fig. 1). The heterogeneity of flaA patterns of the GBS and MFS strains was comparable to that of the enteritis and Penner reference strains. With cluster analysis, no specific flagellin type was present among the GBS- or MFS-related Campylobacter strains tested (Fig. 1).
FIG. 1.
FlaA PCR-RFLP patterns of 18 GBS- or MFS-related Campylobacter jejuni strains, 9 enteritis control strains, and 8 Penner O serostrains. The dendrogram was constructed with band-based analysis and unweighted pair group method with averages clustering. The sizes of standard DNA fragments (in kilobase pairs) are indicated below.
AFLP.
AFLP detected 12 distinct fingerprints within the 18 GBS or MFS C. jejuni strains. Two small clusters of strains were found (Table 1). The O:19 GBS strains (GB3 and GB18) and O:19 serostrain showed highly related AFLP fingerprints. A high level of homology was also observed when comparing AFLP fingerprints of GB13, GB14, GB16, GB19, and the O:2 serostrain. Although strain GB11 showed some band differences compared to the patterns of GB13 and GB14, a genetic relationship between these strains was evident. Two GBS strains, GB1 and GB15, and strain MF20 shared AFLP fingerprints with some of the enteritis strains (Table 1). Cluster analysis showed no separate clustering of GBS- and MFS-related strains. Within each cluster of AFLP fingerprints, strains from GBS and MFS patients as well as reference serotype strains and strains from enteritis patients were present.
RAPD and PFGE.
The PFGE analysis of 18 GBS- or MFS-related C. jejuni strains revealed the presence of 15 distinct types (Table 1). The two C. jejuni O:2, Lior 4 strains (GB13 and GB14), isolated from family members of a GBS patient, were indistinguishable from each other and were related to GB11. However, these strains were unrelated to MF20, a strain with the same O and HL serotypes. C. jejuni GB5, isolated from a GBS patient, appeared subclonally related to C. jejuni MF6, a strain obtained from a patient with MFS. With computer-aided analysis, no clustering of GBS- or MFS-related strains was found. With the RAPD analysis of the 18 GBS or MFS strains, 15 (ERIC-1) and 13 (ERIC-2) distinct types were obtained (Table 1). No clustering of GBS or MFS strains was found by computer analysis.
DISCUSSION
This study illustrates the substantial heterogeneity of C. jejuni strains associated with GBS or MFS in a restricted geographical area of the world. Of the GBS- or MFS-related strains in our study, 2 of 18 (11%) were of serotype O:19 and none were of serotype O:41. The most frequently observed serotype was O:2. Strains reacting with one or more of the antisera O:4, O:13, O:50, O:64, and O:65 are often related and classified as O:4-complex. C. jejuni O:4-complex was observed in four GBS patients and in one MFS patient. The O:2 serotype was found in two GBS- or MFS-related strains and in two strains from family members of a GBS patient. C. jejuni O:2 is the prevailing serotype in collections of strains from patients with enteritis and, according to Oosterom et al. (21), accounts for approximately 25% of the enteritis strains in The Netherlands. The Penner O serotyping scheme has been used in several previous studies to characterize C. jejuni strains isolated from patients with GBS or MFS, and those associated with these conditions include O:1, O:2, O:4, O:4/50, O:5, O:10, O:16, O:19, O:23, O:37, O:41, O:44, and O:64 (18). We report two new C. jejuni O serotypes, C. jejuni O:35 and O:13/65, that have not been described previously in association with GBS. The great variety of O serotypes that are found in the literature and in this study confirm our previous suggestion that GBS and MFS are not exclusively associated with a specific Penner O serotype (9). However, C. jejuni serotype O:19 appears to be overrepresented among strains isolated from patients with GBS or MFS from the United States and Japan (16, 24). In a Japanese study (24), serotype O:19 comprised 12 of 16 (75%) of the GBS-related C. jejuni isolates, while in a U.S.-based study (16), 2 of 7 (29%) were of serotype O:19. In both countries, this serotype is encountered in less than 3% of the C. jejuni strains from patients with enteritis in the absence of neurological involvement. In South Africa, 9 of 9 (100%) C. jejuni isolates from GBS patients were of serotype O:41, whereas this serotype was found in less than 2% of enteritis control strains (12). In addition, some authors suggested that Penner O:19 and O:41 strains, whether isolated from patients with GBS or from enteritis patients without neurological involvement, were clonally related, thereby indicating that these strains were particularly virulent (6, 12, 24). The data presented here demonstrate that the overrepresentation of specific O serotypes, as reported by others, is a phenomenon not seen in The Netherlands. Therefore, there would appear to be much variety in the distribution of O serotypes in different geographic locations. In addition to the more traditional phenotypic analysis, a variety of molecular typing techniques were used to unravel the genomic differences or similarities of the C. jejuni strains. The conclusions drawn from the results of serotyping were corroborated by the compiled data of the different molecular typing methods. In general, the analysis of the GBS- or MFS-related C. jejuni strains demonstrated substantial genetic heterogeneity. No clustering of GBS- or MFS-related strains was found, irrespective of the method used. Although small clusters of related strains were found, strains from GBS and MFS patients, as well as those from enteritis patients, were present in these clusters. In some cases, a remarkable correlation was found between the results of serotyping and the different molecular techniques. GB11, GB13, and GB14 were all C. jejuni O:2, Lior 4 and showed great homology by PFGE, RAPD, and RFLP. The two C. jejuni O:19, Lior 77 strains were highly related by flaA and AFLP but had different PFGE and RAPD patterns. Thus, although the discriminatory power of the different techniques varies significantly, the clustering of some strains was comparable.
Fujimoto et al. (6) recently determined the extent of genetic variation among C. jejuni strains, including nine strains from GBS patients. Although the strains were isolated from patients residing in countries as diverse as the United States, Japan, and Germany, the authors found that by flaA-RFLP and RAPD analysis, at least five of nine GBS strains were closely related. The five strains, however, were all of serotype O:19. In addition, the data indicated that all O:19 strains, whether GBS related or not, were clonally related. RAPD and RFLP of other O serotypes, in contrast, were reported to be different (6). In a recent Japanese study, 12 of 16 (75%) of the GBS-related C. jejuni strains that had been serotyped were of serotype O:19 (24). flaA PCR-RFLP patterns of the 12 strains and of enteritis-related C. jejuni O:19 were identical and distinguishable from other O serotypes (20). In the present study, clonal relationships among the three O:19 strains were only observed by AFLP. The two GBS strains had identical flaA types, while the pattern was quite distinct from the O:19 serotype strain. Although this collection of Dutch GBS- or MFS-related C. jejuni strains is characterized by a high degree of heterogeneity, there were too few isolates from each type present in the study. Therefore, the existence of clonality among certain O serotypes cannot be excluded.
The heterogeneity among phenotypes and genotypes in the present study may reflect the heterogeneity of clinical symptoms and anti-ganglioside antibodies in GBS patients. C. jejuni infections are associated with a severe pure motor variant of GBS (31) but have been reported in MFS patients, GBS patients with ophthalmoparesis, and patients with isolated abducens paresis as well (23, 27). The clinical heterogeneity may be related to the variety of ganglioside-like structures in the LPS of GBS- or MFS-related C. jejuni, although this was not the subject of study. No common molecular markers separating pathogenic GBS or MFS from non-GBS- or non-MFS-related enteritis strains were identified. Based on the present results, it would be premature at this stage to dismiss the hypothesis of the “bad bug that causes GBS.” The methods used in the study may simply lack the power to detect the particular determinants of C. jejuni strains related to GBS-MFS. Several authors have provided evidence that the molecular basis of the mimicry between C. jejuni and sialidated gangliosides resides in the LPS fraction of Campylobacter (9, 17, 18). Analysis of genes involved in the synthesis of LPS and the incorporation of sialic acid may turn out to be of great importance to further clarify the particularity of the C. jejuni strains involved in the pathogenesis of GBS and MFS. Although ganglioside-like epitopes have recently been described to be present in a significant percentage of C. jejuni isolates from patients with uncomplicated enteritis (19), it seems likely that additional genotypic methods may detect markers of pathogenicity in the near future (32).
Finally, the results of serotyping and genotyping of the two strains from a family outbreak of C. jejuni enteritis followed by one case of GBS demonstrate a clonal relationship of the strains and, therefore, suggest the importance of host factors in the pathogenesis of GBS (1).
ACKNOWLEDGMENTS
We thank the microbiologists and neurologists participating in the Dutch Guillain-Barré trial for their efficient logistic support in providing fecal specimens and A. S. Lampe, P. M. Schneeberger, C. J. M. Persoons, and N. van Leeuwen for providing four C. jejuni strains.
REFERENCES
- 1.Ang C W, van Doorn P A, Endtz H P, Martina I S J, Jacobs B C, van Koningsveld R, van der Meché F G A. A single case of Guillain-Barré syndrome in a family with Campylobacter jejuni enteritis. J Neurol. 1998;245:417. doi: 10.1016/s0165-5728(00)00369-6. [DOI] [PubMed] [Google Scholar]
- 2.Asbury A K, Cornblath D R. Assessment of current criteria for Guillain-Barré syndrome. Ann Neurol. 1990;27(Suppl.):21–24. doi: 10.1002/ana.410270707. [DOI] [PubMed] [Google Scholar]
- 3.Ayling R D, Woodward M J, Evans S, Newell D G. Restriction fragment length polymorphism of polymerase chain reaction products applied to the differentiation of poultry Campylobacters for epidemiological investigations. Res Vet Sci. 1996;60:168–172. doi: 10.1016/s0034-5288(96)90013-2. [DOI] [PubMed] [Google Scholar]
- 4.Duim B, Wassenaar T M, Rigter A, Wagenaar J. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 1999;65:2369–2375. doi: 10.1128/aem.65.6.2369-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endtz H P, Vliegenthart J S, Vandamme P, Weverink H W, van den Braak N P, Verbrugh H A, van Belkum A. Genotypic diversity of Campylobacter lari isolated from mussels and oysters in The Netherlands. Int J Food Microbiol. 1997;34:79–88. doi: 10.1016/s0168-1605(96)01174-9. [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto S, Mishu Allos B, Misawa N, Patton C, Blaser M J. Restriction fragment length polymorphism analysis and random amplified polymorphic DNA analysis of Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome. J Infect Dis. 1997;176(Suppl. 2):S1105–S1108. doi: 10.1086/516522. [DOI] [PubMed] [Google Scholar]
- 7.Hilton A C, Mortinboy D, Banks J G, Penn C W. RAPD analysis of environmental, food and clinical isolates of Campylobacter spp. FEMS Immunol Med Microbiol. 1997;18:119–124. doi: 10.1111/j.1574-695X.1997.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 8.Ho T W, Li C Y, Gao C Y, Cornblath D R, Griffin J W, Asburry A K, Blaser M J, McKhann G M. Guillain-Barré syndrome in northern China. Relationship to Campylobacter jejuni infection and anti-glycolipid antibodies. Brain. 1995;118:597–605. doi: 10.1093/brain/118.3.597. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs B C, Endtz H P, van der Meché F G A, Hazenberg M P, Achtereekte H A M, van Doorn P A. Serum anti-GQ1B IgG antibodies recognize surface epitopes on Campylobacter jejuni from patients with Miller Fisher syndrome. Ann Neurol. 1995;37:260–264. doi: 10.1002/ana.410370218. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs B C, van Doorn P A, Schmitz P I M, et al. Campylobacter jejuni infections and anti GM1 antibodies in Guillain-Barré syndrome. Ann Neurol. 1996;40:181–187. doi: 10.1002/ana.410400209. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs B C, Rothbarth P H, van der Meché F G A, Herbrink P, Schmitz P I M, Klerk M A, van Doorn P A. The spectrum of antecedent infections in Guillain-Barré syndrome, a case control study. Neurology. 1998;51:1110–1115. doi: 10.1212/wnl.51.4.1110. [DOI] [PubMed] [Google Scholar]
- 12.Lastovica A J, Goddard E A, Argent A C. Guillain-Barré syndrome in South Africa associated with Campylobacter jejuni O:41 strains. J Infect Dis. 1997;176(Suppl. 2):S139–S143. doi: 10.1086/513796. [DOI] [PubMed] [Google Scholar]
- 13.Lior H, Woodward D L, Edgar J A, Laroche L J, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller Fisher C. An unusual variant of acute idiopathic polyneuritis (syndrome of ophthalmoplegia, ataxia and areflexia) N Engl J Med. 1956;225:57–65. doi: 10.1056/NEJM195607122550201. [DOI] [PubMed] [Google Scholar]
- 15.Mishu Allos B. Association between Campylobacter infection and Guillain-Barré syndrome. J Infect Dis. 1997;176(Suppl. 2):S125–S128. doi: 10.1086/513783. [DOI] [PubMed] [Google Scholar]
- 16.Mishu Allos B, Lippy F T, Carlsen A, Washburn R G, Blaser M J. Campylobacter jejuni strains from patients with Guillain-Barré syndrome. Emerg Infect Dis. 1998;4:263–268. doi: 10.3201/eid0402.980213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran A P. Structure and conserved characteristics of Campylobacter jejuni lipopolysaccharides. J Infect Dis. 1997;176(Suppl. 2):S115–S121. doi: 10.1086/513781. [DOI] [PubMed] [Google Scholar]
- 18.Nachamkin I, Mishu Alos B, Ho T. Campylobacter jejuni and Guillain-Barré syndrome. Clin Microbiol Rev. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nachamkin I, Ung H, Moran A P, Yoo D, Prendergast M M, Nicholson M A, Sheikh K, Ho T, Asbury A K, McKhann G M, Griffin J W. Gangloside GM1 mimicry in Campylobacter strains from sporadic infections in the United States. J Infect Dis. 1999;179:1183–1189. doi: 10.1086/314725. [DOI] [PubMed] [Google Scholar]
- 20.Nishimura M, Nukina M, Kuroki S, Obayashi H, Ohta M, Jun Ma J, Saida T, Uchiyama T. Characterization of Campylobacter jejuni isolates from patients with Guillain-Barré syndrome. J Neurol Sci. 1997;153:91–99. doi: 10.1016/s0022-510x(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 21.Oosterom J, Bänffer J R J, Lauwers S, Busschbach A E. Determination of serotype and hippurate hydrolysis for Campylobacter jejuni isolates from human patients, poultry and pigs in The Netherlands. Antonie Leeuwenhoek J Microbiol. 1995;51:65–70. doi: 10.1007/BF00444229. [DOI] [PubMed] [Google Scholar]
- 22.Penner J L, Hennessy J N, Congi R V. Serotyping of Campylobacter jejuni and Campylobacter coli on the basis of thermostable antigens. Eur J Clin Microbiol Infect Dis. 1983;2:378–383. doi: 10.1007/BF02019474. [DOI] [PubMed] [Google Scholar]
- 23.Roberts T, Shah A, Graham J G, McQueen I N. The Miller Fisher syndrome following Campylobacter enteritis: a report of two cases. J Neurol Neurosurg Psychiatry. 1987;50:1557–1558. doi: 10.1136/jnnp.50.11.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saida T, Kuroki S, Hao Q, Nishimura M, Nukina M, Obayashi H. Campylobacter jejuni isolates from Japanese patients with Guillain-Barré syndrome. J Infect Dis. 1997;176(Suppl. 2):S129–S134. doi: 10.1086/513798. [DOI] [PubMed] [Google Scholar]
- 25.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosome DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Belkum A, van Leeuwen W, Verkooyen R, Can Saçilik C, Cokmus C, Verbrugh H. Dissemination of a single clone of methicillin-resistant Staphylococcus aureus among Turkish hospitals. J Clin Microbiol. 1997;35:978–981. doi: 10.1128/jcm.35.4.978-981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Kruijk R A C, Lampe A S, Endtz H P. Bilateral abducens paresis following Campylobacter jejuni enteritis. J Infect. 1992;24:215–216. doi: 10.1016/0163-4453(92)93136-e. [DOI] [PubMed] [Google Scholar]
- 28.van der Meché F G A, van Doorn P A. Guillain-Barré syndrome and chronic inflammatory demyelinating polyneuropathy: immune mechanisms and update on current therapies. Ann Neurol. 1995;37(Suppl. 1):S14–S31. doi: 10.1002/ana.410370704. [DOI] [PubMed] [Google Scholar]
- 29.Vauterin L A, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur J Clin Microbiol Infect Dis. 1992;1:37–40. [Google Scholar]
- 30.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:406–409. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visser L H, van der Meché F G A, van Doorn P A, Meulstee J, Jacobs B C, Oomes P G, Kleyweg R P the Dutch Guillain-Barré Study Group. Guillain-Barré syndrome without sensory loss. A subgroup with specific clinical, electrodiagnostic and laboratory features. Brain. 1995;118:841–847. doi: 10.1093/brain/118.4.841. [DOI] [PubMed] [Google Scholar]
- 32.Wassenaar T M, Newell D G. Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000;66:1–9. doi: 10.1128/aem.66.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]