Abstract
Osteosarcoma (OS), characterized by high morbidity and mortality, is the most common bone malignancy worldwide. MicroRNAs (miRNAs) play a crucial role in the initiation and development of OS. The purpose of this study was to investigate the roles of miR-1270 in OS. RT-qPCR and Western blot were applied to detect the mRNA and protein level, respectively. CCK-8, colony formation, and TUNEL assays were conducted to determine the cell viability, proliferation, and apoptosis of OS cells. Wound healing and transwell assay were performed to detect the migration and invasion ability of OS cells. Bioinformatics analysis and dual-luciferase reporter assay were used to predict the target genes of miR-1270. Tumor xenograft in vivo assay was carried out to determine miR-1270 effect on the tumor size, volume, and weight. In this study, miR-1270 was overexpressed in OS tissues and cells. However, miR-1270 knockdown inhibited the proliferation, migration and invasion, and promoted the OS cells’ apoptosis. Mechanistically, cingulin (CGN) was predicted and proved to be a target of miR-1270 and partially alleviated the effects of miR-1270 on the proliferation, migration and invasion ability of OS cells. Taken together, knockdown of miR-1270 may inhibit the development of OS via targeting CGN. This finding may provide a novel therapeutic strategy for OS.
Key words: Osteosarcoma, miR-1270, cingulin, proliferation, migration, invasion
Introduction
Osteosarcoma (OS) is the most prevalent bone malignancy frequently happening in children and adolescents worldwide.1 Despite significant advances in the treatment of OS over the past decades,2 the mortality and morbidity of OS remain high. Faraway metastasis and recurrence are the leading causes of OS-related death.3 The 5-year survival rate of OS patients with no metastasis is 75%, but no more than 30% with metastasis evidence.4 Therefore, investigating the underlying molecular mechanisms on the metastasis of OS has attracted increasing attention.
MicroRNAs (miRNAs) are a family of small non-coding RNAs at the length of 18-22 nucleotides.5 miRNAs degrade mRNA via binding to the 3’ untranslated region (3’ UTR) of its target. 6 MiRNAs have been shown to play important roles in many cancer types.7-12 Numerous studies have reported that miRNAs participate in various biological processes, such as cell proliferation, apoptosis, migration, autophagy, and invasion.13-16 Interestingly, miRNAs are collectively involved in the initiation and progression of cancer, including OS.17,18 For instance, miR-624-5p promotes the migration and invasion of OS via targeting protein tyrosine phosphatase receptor type B (PTPRB).19 MiR-509-3p suppresses the proliferation, migration, and invasion of OS and promotes the OS cells’ chemo-sensitivity to cisplatin.20 It has also been verified the potential of the LINC00588/miRNA-1972/TP53 axis as a therapeutic target in OS patients.21 The upregulation of miR-1270 in OS predicts poor prognosis, while downregulated miR-1270 inhibits the proliferation, migration and tumorigenesis of OS.22 However, the underlying molecular mechanism remains unclear.
Augmenting studies demonstrate that the dysfunction of tight junction (TJ) proteins contributes to the initiation and progression of human cancer through regulating TJ structure and associated signaling pathways.23,24 TJ formation facilitates the paracellular diffusion barrier and modulates cell polarity and permeability.25 In addition, TJ proteins suppress growth- and metastasis-related genes to modulate the progression of tumors, including OS.26-28 Cingulin (CGN), a transmembrane pore-forming barrier protein, plays a crucial role in TJ via peripheral scaffolding proteins, such as zona occludens-1 (ZO-1). Aberrantly expressed CGN induces the proliferation, migration and invasion of cancer, and acts as an anti-tumor gene in mesothelioma and ovarian cancer.29-31 However, the role of CGN in OS has not been elucidated.
The purpose of this study was to investigate the potential roles of miR-1270 in OS and the underlying molecular mechanisms, and our findings may provide a promising therapy for OS.
Materials and methods
Clinical samples
Sixty pairs of OS tissues and normal tissues were collected from OS patients at Renmin Hospital of Wuhan University from May 1, 2018 to December 31, 2019. Clinical samples were immediately stored at -80°C after surgery. The patients enrolled in this study had not received chemotherapy and radiotherapy before. All patients provided informed consent. This study was approved by the Ethics Committee of Renmin Hospital of Wuhan University.
Cell culture
OS cell lines MG-63, U2OS, Saos2, 143B, and normal human osteoblast cell line hFOB1.19 were purchased from ATCC (USA). Cells were cultured in DMEM medium containing 10% FBS and 1% penicillin/streptomycin (Gibco, USA), at 37 °C with 5% CO2.
Transfection
Saos2 and 143B were transfected with 50 nM of miRNA negative control (miR-NC) mimic, miR-1270 mimic, miR-NC inhibitor, miR-1270 inhibitor, NC shRNA (sh-NC), or sh-CGN (GenePharma, China) by using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) for 48 h.
RT-qPCR
Total RNA was collected from OS tissues or cells with TRIzol reagent (Invitrogen, Carslbad, CA, USA). Then the RNA was reversely transcribed into cDNA using PrimeScript 1st strand cDNA Synthesis kit (Takara Bio, San Jose, CA, USA). The PCR was performed using SYBR-Green qPCR assay on Biosystems 7500 Real-Time PCR System under the following thermocycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 60 s, 72°C for 1 min, and 72°C for 10 min. The expression levels were measured with the 2-ΔΔCt method. U6 and β-actin were used as the internal control for miRNAs or mRNA, respectively.
Western blot
Total protein was isolated from OS cells. The concentration of the protein was calculated with BCA Kit (Beyotime, Shanghai, China). An equal amount of the protein was separated with 12% SDS-PAGE. Afterward, the proteins were moved onto Polyvinylidene Fluoride membranes (Millipore, Burlington, MA; USA). Then the membranes were sealed with 5% skimmed milk and incubated with primary antibodies, anti-ICAM-1 (ab222736, 56 kDa, 1: 1000, Abcam, Cambridge, MA, USA), anti-VCAM-1 (ab134047, 81 kDa, 1: 2000, Abcam), anti-CGN (ab117796, 136 kDa, 1: 2000, Abcam), anti-Bcl-2 (ab32124, 26 kDa, 1: 1000, Abcam), anti-BAX (ab32503, 21 kDa, 1: 1000, Abcam), anticleaved Caspase3 (ab32042, 17 kDa, 1: 500, Abcam), anticleaved- Caspase9 (ab2324, 37 kDa, 1: 1000, Abcam), and anti-β- actin (ab8227, 42 kDa, 1: 1000, USA) overnight at 4°C in shade. Lately, the membranes were washed with PBS 3 times and incubated with goat-anti-rabbit secondary antibodies (ab6721, 1: 2000, Abcam) for 2 h at 37°C. The protein in each band was determined with ECL solution (Millipore) in the shade. The protein levels were analyzed with ImageJ software 1.6.
Dual luciferase reporter assay
The luciferase activity of OS cells was determined with a luciferase assay kit (Promega, Madison, WI, USA). CGN wild type (WT) or mutant (MUT) containing the miR-1270 binding site was introduced into pmirGLO dual-luciferase vector (Promega, USA). Then the cells were co-transfected with CGN 3’UTR WT or CGN 3’UTR mutant, and miR-NC mimic or miR-1270 mimic for 48 h. The luciferase activity was calculated and normalized to Renilla luciferase activity.
Clone formation assay
Briefly, we melt 1.4% (v/v) agarose (Sigma-Aldrich, St. Louis, MO, USA) in a microwave and cooled to room temperature. Then, we mixed equal volumes of the 1.4% melted agarose with the complete cell culture medium. After that, 2 ml of 0.7% (v/v) low melting point agar was added into each well of the 6-well plate and set aside to allow agarose to solidify. After 48 h transfection, cells (4×103 cells/well) were mixed with 1.4% agarose in complete culture medium, plated on top of the solidified layer to form colonies in 1-3 weeks. Cells were fed with complete culture medium every 3 d. The colony is denied to consist of at least 50 cells32. After the cells have formed sufficiently large clones, we removed the medium above the cells and rinsed carefully with PBS. Then, colonies were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) for 10 min and stained with crystal violet. Subsequently, the colonies were counted and images recorded by an inverted microscope linked up with camera.
CCK-8 assay
Cell viability was determined with CCK-8 Kit. Saos2 and 143B were seeded into a 96-well plate (2×103 cells/well) and cultured for 0, 24, 48, and 72 h. Then the cells were added with CCK- 8 solutions. The absorbance at the wavelength of 450 nm was measured with a microplate reader.
TUNEL assay
The apoptosis of OS cells was determined with the TUNEL Apoptosis Kit (Thermo Fisher Scientific, Waltham, MA, USA). After transfection, cells were collected and stained with DAPI in darkness. The apoptotic cells were detected by fluorescence microscopy (Olympus, Tokyo, Japan). Each experiment was conducted in triplicate.
Wound healing assay
Cells were seeded in a 6-well plate (5×104 cells/well). A 20 μL micropipette tip was applied to make a scratch. Then the cell cultures were observed at 0 and 24 h, when the width of the scratch was measured and normalized to baseline values.
Cell migration and invasion assay
The cells were seeded into a 24-well plate (4×103 cells/well). The cells in the serum-free medium of the upper chamber were treated with or without Matrigel (BD Bioscience, Franklin Lakes, NJ, USA). Next, the lower chamber was supplemented with a culture medium containing 10% FBS, while the cells in the upper chamber were fixed with 4% ethanol and stained with 0.05% crystal violet. Finally, the cells were measured with a light microscope (200×, Olympus).
Xenograft assay
Saos2 cells transfected with miR-1270 mimic, and miR-NC mimic were collected and used in a xenograft assay. 9 C57BL/6 nude male mice (6 weeks old, 20-22 g) were purchased from Institute of Laboratory Animal Sciences (Beijing, China). For each mouse, 5×106 Saos2 cells were subcutaneously injected to the flanks. The subcutaneous OS xenografts were established within 3 weeks, with each tumor close to 100 mm3 in volume. Mice were randomly divided into 3 groups: blank group, miR-NC mimic (NC), and miR-1270 mimic group. Mice were kept at 20-26 °C with 55 ± 5% humidity, 12 h light/dark cycle, and ad libitum access to food and water. Mice body weight and bidimensional tumor measurements were recorded every 7 d. Then, the mice were killed by intraperitoneal injection of sodium pentobarbital, and the tumors’ volume and weight were measured. The experiment was authorized by the Ethics Committee of Renmin Hospital of Wuhan University.
Figure 1.
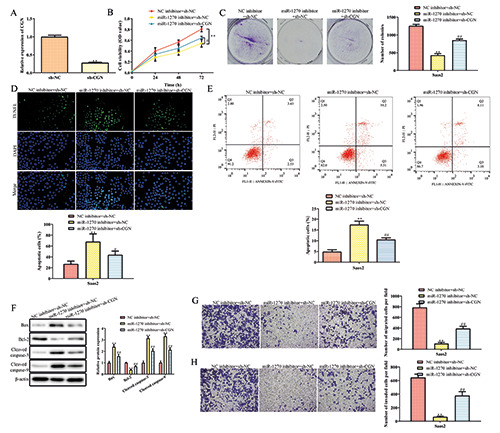
Downregulated miR-1270 inhibits the proliferation and promotes the apoptosis of OS cells. A) The expression of miR-1270 in OS tissues. B) The expression of miR-1270 in OS cells. C) Cell transfection efficiency detected by qRT-PCR. D) OS cell viability determined by CCK-8. E) OS cell proliferation detected by colony formation assay. F) The apoptosis rates of OS cells determined by TUNEL assay. G) The apoptosis rates determined by flow cytometry. H) The protein level of Bcl-2, Bax, cleaved-Caspase-3, and cleaved- Caspase-9 measured by Western blot. *p<0.05, **p<0.01 vs normal tissues, hFOB1.19, or NC inhibitor.
Statistical analysis
The data were analyzed using SPSS 16.0. All data were expressed as mean ± standard deviation (SD). Paired student t test was applied to evaluate the difference between two groups, and one-way ANOVA was followed by Dunnett’s post hoc test to assess differences among multi-groups. Pearson analysis was used for evaluating the correlation between miR-1270 and CGN. A pvalue <0.05 was considered as statistical significance.
Results
MiR-1270 modulates the proliferation and apoptosis of OS cells
The qRT-PCR assay was performed to determine the expression level of miR-1270. As shown in Figure 1A, miR-1270 was overexpressed in OS tissues compared to healthy control. Moreover, the level of miR-1270 in OS cells was significantly higher than that in normal human osteoblast cell line hFOB1.19. Moreover, the expression in Saos2 and 143B was more remarkable (Figure 1B). Thence, they were used in the following experiment. In addition, the level of miR-1270 in cells transfected with miR- 1270 inhibitor was significantly decreased compared with the miR-NC inhibitor group, suggesting that cells had been successfully transfected (Figure 1C). CCK-8, colony formation, TUNEL and flow cytometry assays were conducted to detect the roles of miR- 1270 in OS cells proliferation and apoptosis. The results showed that downregulated miR-1270 inhibited the cell viability of Saos2 and 143B (Figure 1D). This result was paralleled with that from the colony formation assay (Figure 1E). Moreover, the apoptosis rates of OS cells treated with miR-1270 inhibitor were significantly increased in comparison with the NC inhibitor group (Figure 1 F,G). Additionally, the down-regulation of miR-1270 increased the protein level of proapoptotic genes, such as Bax, cleaved Caspase- 3, and cleaved Caspase-9, whereas decreased the protein level of Bcl-2 (Figure 1H).
Knockdown of miR-1270 inhibits the migration and invasion of OS cells
Wound healing and transwell chamber assays were utilized to determine the migratory and invasive capacity of OS cells. Knockdown of miR-1270 repressed the migration of Saos2 and 143B, compared with the NC inhibitor group (Figure 2A). These results were consistent with the transwell chamber assay (Figure 2B). Consistently, downregulation of miR-1270 inhibited the invasion ability of OS cells (Figure 2C). Moreover, the protein expression levels of ICAM-1 and VCAM-1 were significantly reduced in OS cells transfected with miR-1270 inhibitor (Figure 2D).
MiR-1270 promotes tumor growth of OS in-vivo
To study the potential role of the miR-1270 on OS cell growth in vivo, a xenograft mouse model was applied. The Saos2 cells were s.c. injected into the flanks of mice. Within 3 weeks, OS xenografts were established, with each tumor close to 100 mm3. As shown in Figure 2 E-G, the tumor size, volume, and weight were significantly increased in the miR-1270 mimic group, suggesting miR-1270 may be an onco-miRNA in OS.
Figure 2.
Downregulated miR-1270 suppresses the migration and invasion of OS cells and miR-1270 promotes tumor growth of OS in vivo. A) The migration ability of OS cells determined by wound healing assay. B) The migration ability determined by transwell chamber assay (magnification: 100x). C) The invasion ability of OS cells detected by transwell assay (magnification: 100x). D) The protein level of ICAM-1 and VCAM-1 determined by western blot. E) The size of OS tumor in vivo assays. F) The volume of OS tumor in vivo assays. G) The weight of OS tumor in vivo assays. *p<0.05 and **p<0.01 vs NC inhibitor or Blank group.
CGN is a target of miR-1270
The online database TargetScan predicted CGN was a target of miR-1270. The binding sites of miR-1270 on CGN were shown in Figure 3A. Dual-luciferase reporter assay showed the luciferase activity of OS cells co-transfected with miR-1270 mimic, and CGN 3’UTR WT was significantly decreased. At the same time, there was no significant difference between CGN 3’UTR MUT groups (Figure 3B). MiR-1270 knockdown induced the up-regulation of CGN both in mRNA and protein level (Figure 3 C,D). Moreover, CGN was downregulated in OS tissues and cells (Figure 3E). The expression of miR-1270 was negatively correlated with CGN (Figure 3F).
CGN mediates the effects of miR-1270 inhibitor on the proliferation, apoptosis, migration, and invasion of OS cells
The OS cell line Saos2 was used in the rescue assay. As shown in Figure 4A, CGN shRNA significantly downregulated the mRNA level of CGN. In addition, CGN knockdown alleviated the increase in apoptosis and the decrease in proliferation of Saos2 induced by miR-1270 inhibitor (Figure 4 B-E). Moreover, the regulatory role of miR-1270 knockdown in the expressions of Bcl-2, Bax, and cleaved Caspase-3 and Caspase-9 were antagonized by silenced CGN (Figure 4F). Meanwhile, the silencing of miR-1270 restored the migration and invasion ability of Saos2 cells (Figure 4 G,H).
Discussion
It has been reported that miRNAs are involved in the pathogenesis of cancers.33 The aberrant expression of miRNA acts as a tumor suppressor or onco-miRNA.8-11 MiR-1270 serves as an onco-miRNA in bladder cancer, ovarian cancer, papillary thyroid cancer, as well as an anti-tumor miRNA in glioblastoma.22,34-37 Therefore, identifying the roles of miR -1270 in OS is of vital essence. In the present study, miR-1270 was overexpressed in OS tissues and cells, which indicates that miR-1270 might act as an onco-miRNA in OS.
MiR-1270 has been reported to be involved in the progression of cancer via regulating various biological processes, such as cell differentiation, proliferation, migration and invasion. For instance, overexpression of miR-1270 attenuates chemo-sensitivity and apoptosis in bladder cancer.34 However, down-regulation of miR- 1270 inhibits the proliferation and migration of papillary thyroid cancer.38 Interestingly, the downregulation of miR-1270 represses the proliferation and migration of OS.22 In the present study, downregulated miR-1270 suppressed the proliferation, migration and invasion of OS cells, while promoting their apoptosis. Moreover, downregulation of miR-1270 modulated the expression of apoptosis- related genes (Bcl-2, Bax, Caspase-3, and Caspase-9) and metastasis-related genes (ICAM-1 and VCAM-1).38-40 These results suggested that miR-1270 may be an effective target for the treatment of OS. However, the underlying mechanisms were still unclear. Metastasis and metastasis-induced recurrence are the main reasons for OS-related death.3,4 Generally, metastasis is accompanied by the degradation of TJ. CGN, as a junctional component, regulates scaffolding proteins and interacts with microtubules and actin.30 CGN, or coxsackievirus and adenovirus receptor (CXADR), is a transmembrane protein acting as a tumor suppressor in breast cancer via inhibiting its migration and invasion of breast cancer.41 MiR-16 and miR-125b have been reported to be involved in barrier function dysregulation through the modulation of claudin-2 and CGN expression in the jejunum in irritable bowel syndrome with diarrhea.42 However, the roles of CGN in OS have not been elucidated. In this study, CGN was downregulated in OS tissues and cells, suggesting that CGN may serve as an anti-tumor gene in OS. A previous study has demonstrated that post-translational degradation of junctional proteins disrupts the TJ structure and promotes cancer metastasis.43 miRNAs play a key post-translational role and regulate gene expression via binding to the 3’ untranslated region (3’UTR) of their target genes.44 In this study, CGN was predicted and proved to be a target of miR-1270, and its expression was negatively correlated with miR-1270. Intriguingly, the effects of miR-1270 on the proliferation, apoptosis, migration and invasion of OS cells were reversed by CGN. These results revealed that miR-1270 targeted junctional protein CGN to regulate the proliferation, apoptosis, migration and invasion of OS cells, thereby inhibiting the progression of OS.
Figure 3.
CGN is a target of miR-1270. A) The binding site of miR-1270 on CGN. B) The luciferase activity of OS cells determined by dual reporter luciferase assay. C) The mRNA level of CGN in OS tissues and cells calculated by qRT-PCR. D) The correlation between CGN and miR-1270. E: The mRNA level of CGN in OS cells. F) The protein level of CGN in OS cells. **p<0.01 vs normal tissues, hFOB1.19, or NC inhibitor group.
Figure 4.
CGN knockout mediates the proliferation, apoptosis, migration, and invasion of OS cells induced by the miR-1270 downregulation. A) The expression of CGN. B) OS cell viability of OS cells determined by CCK-8 assay. C) The proliferation of OS cells determined by colony formation assay. D) The apoptosis of OS determined by TUNEL assay. E) The apoptosis rates determined by flow cytometry. F) The expression of Bcl-2, BAX, cleaved-Caspase-3, and cleaved-Caspase-9 determined by Western blot. G,H) The migration and invasion ability determined by transwell assay (magnification: 100x). **p<0.01 vs NC inhibitor group; ##p<0.01 vs miR-1270 inhibitor group.
Nevertheless, there are several constraints to this study, such as the number of OS patients is limited, and more patients need to be recruited to make the results fully convincing. Additionally, further studies should be focused on the possible mechanism or signaling pathway downstream of miR-1270.
In conclusion, miR-1270 acted as an onco-miRNA in OS. Furthermore, knockdown of miR-1270 inhibited the proliferation, migration and invasion, and triggered apoptosis of OS cells via targeting CGN. Thus, our results indicated that the miR-1270/CGN axis may be a promising therapeutic target of OS.
Acknowledgments
We appreciate the language editing on our manuscript by Dr. Andrew Irving from Department of Life Science, Dell Medical School of the University of Texas at Austin, Austin, TX, USA.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res 2009;152:3-13. [DOI] [PubMed] [Google Scholar]
- 2.Fathizadeh H, Mirzaei H, Asemi Z. Melatonin: an anti-tumor agent for osteosarcoma. Cancer Cell Int 2019;19:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop Mw, Janeway Ka, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr 2016;28:26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Wang L, Zhang X. Knockdown of lncRNA HOXAAS2 Inhibits viability, migration and invasion of osteosarcoma cells by miR-124-3p/E2F3. Onco Targets Ther 2019;12:10851-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. Micrornas: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [DOI] [PubMed] [Google Scholar]
- 6.Dong S, Xiao Y, Ma X, He W, Kang J, Peng Z, et al. miR-193b increases the chemosensitivity of osteosarcoma cells by promoting FEN1-mediated autophagy. Onco Targets Ther 2019;12:10089-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salarinia R, Sahebkar A, Peyvandi M, Mirzaei HR, Jaafari MR, Riahi MM, et al. Epi-drugs and Epi-miRs: Moving beyond current cancer therapies. Curr Cancer Drug Targets 2016;16:773-88. [DOI] [PubMed] [Google Scholar]
- 8.Sadri Nahand J, Moghoofei M, Salmaninejad A, Bahmanpour Z, Karimzadeh M, Nasiri M, et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: A review. Int J Cancer 2020;146:305-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gholamin S, Pasdar A, Khorrami MS, Mirzaei H, Mirzaei HR, Salehi R, et al. The potential for circulating microRNAs in the diagnosis of myocardial infarction: a novel approach to disease diagnosis and treatment. Curr Pharm Des 2016;22:397-403. [DOI] [PubMed] [Google Scholar]
- 10.Mirzaei H, Hamblin MR. Regulation of glycolysis by non-coding RNAs in cancer: Switching on the Warburg effect. Mol Ther Oncolytics 2020;19:218-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashemipour M, Boroumand H, Mollazadeh S, Tajiknia V, Nourollahzadeh Z, Rohani Borj M, et al. Exosomal microRNAs and exosomal long non-coding RNAs in gynecologic cancers. Gynecol Oncol 2021;161:314-27. [DOI] [PubMed] [Google Scholar]
- 12.Razavi ZS, Tajiknia V, Majidi S, Ghandali M, Mirzaei HR, Rahimian N, et al. Gynecologic cancers and non-coding RNAs: Epigenetic regulators with emerging roles. Crit Rev Oncol Hematol 2021;157:103192. [DOI] [PubMed] [Google Scholar]
- 13.Zhao ZY, Zhao YC, Liu W. Long non-coding RNA TUG1 regulates the progression and metastasis of osteosarcoma cells via miR-140-5p/PFN2 axis. Eur Rev Med Pharmacol Sci 2019;23:9781-92. [DOI] [PubMed] [Google Scholar]
- 14.Dou XQ, Chen XJ, Zhou Q, Wen MX, Zhang SZ, Zhang SQ. miR-335 modulates Numb alternative splicing via targeting RBM10 in endometrial cancer. Kaohsiung J Med Sci 2020;36:171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q, Xing S, Peng A, Yu Z. NORAD accelerates chemoresistance of non-small-cell lung cancer via targeting at miR- 129-1-3p/SOX4 axis. Biosci Rep 2020;40:BSR20193489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamali Z, Taheri-Anganeh M, Shabaninejad Z, Keshavarzi A, Taghizadeh H, Razavi ZS, et al. Autophagy regulation by microRNAs: Novel insights into osteosarcoma therapy. IUBMB Life 2020;72:1306-21. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Zhao M, Wang G. Hsa_circ_0051079 functions as an oncogene by regulating miR-26a-5p/TGF-β1 in osteosarcoma. Cell Biosci 2019;9:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Wei L, Sheng W, Kang B, Wang D, Zeng H. miR- 1225-5p functions as a tumor suppressor in osteosarcoma by targeting Sox9. DNA Cell Biol 2020;39:78-91. [DOI] [PubMed] [Google Scholar]
- 19.Luo Y, Liu W, Tang P, Jiang D, Gu C, Huang Y, et al. miR- 624-5p promoted tumorigenesis and metastasis by suppressing hippo signaling through targeting PTPRB in osteosarcoma cells. J Exp Clin Cancer Res 2019;38:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patil SL, Palat A, Pan Y, Rajapakshe K, Mirchandani R, Bondesson M, et al. MicroRNA-509-3p inhibits cellular migration, invasion, and proliferation, and sensitizes osteosarcoma to cisplatin. Sci Rep 2019;9:19089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu SM, Lee HP, Ham YW, Son DJ, Kim HY, Oh KW, et al. Piperlongumine improves lipopolysaccharide-induced amyloidogenesis by suppressing NF-KappaB pathway. Neuromolecular Med 2018;20:312-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong L, Zheng C, Fang H, Xu M, Chen B, Li C. MicroRNA- 1270 is associated with poor prognosis and its inhibition yielded anticancer mechanisms in human osteosarcoma. IUBMB Life 2018;70:625-32. [DOI] [PubMed] [Google Scholar]
- 23.Zhou W, Gong J, Chen Y, Chen J, Zhuang Q, Cao J, et al. Long noncoding RNA LINC00899 suppresses breast cancer progression by inhibiting miR-425. Aging (Albany NY) 2019;11:10144-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma D, Cao Y, Wang Z, He J, Chen H, Xiong H, et al. CCAT1 lncRNA promotes inflammatory bowel disease malignancy by destroying intestinal barrier via downregulating miR-185-3p. Inflamm Bowel Dis 2019;25:862-74. [DOI] [PubMed] [Google Scholar]
- 25.Turner JR, Buschmann MM, Romero-Calvo I, Sailer A, Shen LE. The role of molecular remodeling in differential regulation of tight junction permeability. Semin Cell Dev Biol 2014;36:204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo J, Wang H, Chen H, Gan G, Zheng Y. CLDN4 silencing promotes proliferation and reduces chemotherapy sensitivity of gastric cancer cells through activation of PI3K/Akt signaling pathway. Exp Physiol 2020;105:979-88. [DOI] [PubMed] [Google Scholar]
- 27.Zhang K, Yang L, Wang J, Sun T, Guo Y, Nelson R, et al. Ubiquitin-specific protease 22 is critical to in vivo angiogenesis, growth and metastasis of non-small cell lung cancer. Cell Commun Signal 2019;17:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Wang H, Li Q, Li T. CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int 2018;18:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Itallie CM, Anderson JM. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol 2014;36:157-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveto S, Alfieri R, Miluzio A, Scagliola A, Secli RS, Gasparini P, et al. A polysome-based microRNA screen identifies miR-24-3p as a novel promigratory miRNA in mesothelioma. Cancer Res 2018;78:5741-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Feng T, Spicer LJ. The role of tight junction proteins in ovarian follicular development and ovarian cancer. Reproduction 2018;155:R183-98. [DOI] [PubMed] [Google Scholar]
- 32.Kim N, Do J, Bae JS, Jin HK, Kim JH, Inn KS, et al. Piperlongumine inhibits neuroinflammation via regulating NFkappaB signaling pathways in lipopolysaccharide-stimulated BV2 microglia cells. J Pharmacol Sci 2018;137:195-201. [DOI] [PubMed] [Google Scholar]
- 33.Silva-Vaz P, Abrantes AM, Castelo-Branco M, Gouveia A, Botelho MF, Tralhao JG. Multifactorial scores and biomarkers of prognosis of acute pancreatitis: applications to research and practice. Int J Mol Sci 2020;21:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan W, Zhou R, Wang J, Han J, Yang X, Yu H, et al. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol Oncol 2019;13:1559-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids 2019;18:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Yi T, Zhou X, Sang K, Zhou J, Ge L. MicroRNA-1270 modulates papillary thyroid cancer cell development by regulating SCAI. Biomed Pharmacother 2019;109:2357-64. [DOI] [PubMed] [Google Scholar]
- 37.Wei L, Li P, Zhao C, Wang N, Wei N. Upregulation of microRNA-1270 suppressed human glioblastoma cancer cell proliferation migration and tumorigenesis by acting through WT1. Onco Targets Ther 2019;12:4839-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan M, Watari H, Abualmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int 2014;2014:150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Giampazolias E, Tait SWG. Caspase-independent cell death: An anti-cancer double whammy. Cell Cycle 2018;17:269-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura A, Akeda K, Matsubara T, Kusuzaki K, Matsumine A, Masuda K, et al. Transfection of NF-κB decoy oligodeoxynucleotide suppresses pulmonary metastasis by murine osteosarcoma. Cancer Gene Ther 2011;18:250-9. [DOI] [PubMed] [Google Scholar]
- 41.Nilchian A, Johansson J, Ghalali A, Asanin St, Santiago A, Rosencrantz O, et al. CXADR-mediated formation of an AKT inhibitory signalosome at tight junctions controls epithelialmesenchymal plasticity in breast cancer. Cancer Res 2019;79:47-60. [DOI] [PubMed] [Google Scholar]
- 42.Ginzburg S, Golovine KV, Makhov PB, Uzzo RG, Kutikov A, Kolenko VM. Piperlongumine inhibits NF-kappaB activity and attenuates aggressive growth characteristics of prostate cancer cells. Prostate 2014;74:177-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cen J, Feng L, Ke H, Bao L, Li Lz, Tanaka Y, et al. Exosomal thrombospondin-1 disrupts the integrity of endothelial intercellular junctions to facilitate breast cancer cell metastasis. Cancers (Basel) 2019;11:1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu ZJ, Chen SG, Yang YZ, Lu SJ, Zhao XM, Hu B, et al. miR- 29a inhibits adhesion, migration, and invasion of osteosarcoma cells by suppressing CDC42. Int J Clin Exp Pathol 2019;12:4171-80. [PMC free article] [PubMed] [Google Scholar]


