Abstract
In the human genome, retrotranspositionally competent long interspersed nuclear elements (L1Hs) are involved in the generation of processed pseudogenes and mobilization of unrelated sequences into existing genes. Transcription of each L1Hs is initiated from its internal promoter but may also be driven from the promoters of adjacent cellular genes. Here I show that a hitherto unknown L1Hs antisense promoter (ASP) drives the transcription of adjacent genes. The ASP is located in the L1Hs 5′ untranslated region (5′UTR) and works in the opposite direction. Fifteen cDNAs, isolated from a human NTera2D1 cDNA library by a differential screening method, contained L1Hs 5′UTRs spliced to the sequences of known genes or non-proteincoding sequences. Four of these chimeric transcripts, selected for detailed analysis, were detected in total RNA of different cell lines. Their abundance accounted for roughly 1 to 500% of the transcripts of four known genes, suggesting a large variation in the efficiency of L1Hs ASP-driven transcription. ASP-directed transcription was also revealed from expressed sequence tag sequences and confirmed by using an RNA dot blot analysis. Nine of the 15 randomly selected genomic L1Hs 5′UTRs had ASP activities about 7- to 50-fold higher than background in transient transfection assays. ASP was assigned to the L1Hs 5′UTR between nucleotides 400 to 600 by deletion and mutation analysis. These results indicate that many L1Hs contain active ASPs which are capable of interfering with normal gene expression, and this type of transcriptional control may be widespread.
Long interspersed nuclear elements (LINEs or L1s) are an abundant class of non-long terminal repeat or poly(A)-type retrotransposons found in all mammalian genomes (17, 30). The human genome contains about 100,000 to 500,000 copies of L1 retrotransposons, most of which have 5′-end truncations and are flanked by 7- to 20-bp target site duplications. Full-length human L1s (L1Hs) are about 6 kb long and possess a 910-bp 5′ untranslated region (5′UTR), two nonoverlapping open reading frames (ORFs), and a 205-bp 3′UTR. ORF1 encodes an RNA binding protein, a major component of the L1Hs ribonucleoprotein particles (15). ORF2 encodes at least two enzymatic activities, an endonuclease (11) and a reverse transcriptase (21), both of which are required for L1Hs autonomous retrotransposition (24). Of the roughly 4,000 full-length L1Hs (1), seven cloned retrotransposons have been shown to be capable of retrotransposition in cultured cells (24, 28). An estimated 30 to 60 copies of L1Hs may be active (28) and possibly involved in the mobilization of cellular mRNAs (10, 20) and Alu elements (5). Random insertion of the retrotranspositionally competent L1s into the human genome has resulted in genetic disease in 12 reported cases (16). Although most L1 retrotranspositions generated truncated and rearranged inactive copies of the progenitor elements, their insertion into genes has demonstrated that L1Hs can interfere with normal gene expression.
Full-length and polyadenylated L1Hs-specific mRNAs have been detected in the human teratocarcinoma cell line NTera2D1 but not in the differentiated cell line (31). The majority of these transcripts were derived from a specific subset of the genomic L1s, and their ORFs were frequently interrupted by stop codons (32). Low-level transcription of L1Hs in other cell lines (HeLa, HL60, and 293) has been indirectly revealed by the presence of ORF1-specific antiserum-positive products (19). Critical sequences necessary for the transcriptional initiation of L1Hs were located in the first 100 bp of the 5′UTR (22, 34). The region (+13 to +21) contains a binding site for the ubiquitous transcription factor YY1. Oligonucleotides containing this sequence formed a specific complex with YY1 protein produced in Escherichia coli or with the same protein present in NTera2D1 nuclear extracts (3). Primer extension studies demonstrated that L1Hs transcription starts from nucleotide (nt) +1 in both NTera2D1 and L1Hs-transfected HeLa cells (22, 32). Therefore, similar to jockey, an L1-like element of Drosophila melanogaster (23), L1Hs has an internal promoter, and its mRNA protein coding potential and polyadenylation predict RNA polymerase II-dependent transcription. Also, it has been demonstrated that L1Hs transcription in vitro may be dependent on RNA polymerase III and YY1 may be involved in both transcription systems (18). However, because of its ubiquitous nature, it is unlikely that YY1 is responsible for the elevated L1Hs transcription in NTera2D1 cells. Therefore, additional factors may be involved in the regulation of cell type specificity. Several such factors belonging to the testis-determining factor SRY or SOX family have also been shown to modulate L1Hs promoter activity in a transfection assay (36). Two binding sites for the SOX family members were located between nt +472 to +477 and +572 to +577. Although not proven, it is possible that SOX factors interacting with YY1 are involved in the regulation of L1Hs cell-specific transcription. Besides an internal promoter, the L1Hs 5′UTR contains an enhancer located around nt +500. As revealed by deletion, mutation, and DNase footprinting analyses, its activation involves Ets and possibly other transcription factors, including Sp1 (40). In the human genome, the presence of about 4,000 full-length copies of the L1Hs containing enhancer elements in the 5′UTR suggests an exceptional power of these retrotransposons to regulate the expression of nearby genes.
Historically, an abundant heterogeneous population of L1 nuclear transcripts has been detected in various human and monkey cell lines (31). These transcripts corresponded to both L1Hs strands, and some contained unrelated sequences apparently derived from readthrough transcription directed by adjacent cellular promoters (29). Recently, high-frequency retrotransposition of the engineered L1Hs into transcribed genes in HeLa cells yielded promiscuous mobilization of the L1Hs 3′ flanking sequences into new genomic regions (25). Such a sequence shuffling was due to the weak L1Hs transcription termination signals and was thought to be involved in the creation of new genes or altering the expression of existing ones.
In this paper I describe the potential of the L1Hs retrotransposon to control the expression of cellular genes by direct means, i.e., a hitherto unknown antisense promoter (ASP) driving transcription opposite to the L1Hs (sense) promoter and ORFs. Here I demonstrate that several known genes are transcribed from the L1Hs ASP.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa, JEG 3, and NTera2D1 cells (provided by H. Ilves, Systemix, Palo Alto, Calif.) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and antibiotics at 37°C and 5% carbon dioxide. Cells were passaged by standard procedures. HeLa S3 cells, grown in Iscove modified Dulbecco medium to 50 to 80% confluence, were transfected with SuperFect (Qiagen) according to the protocol supplied by the manufacturer. Briefly, 1 μg of the pGL3 construct containing luciferase reporter gene and 0.25 μg of the β-galactosidase pSV vector (Promega) were incubated with 3 μl of SuperFect reagent at room temperature in 50 μl of H2O to allow complex formation and thereafter used for cotransfection of the cells. All transfections were done in triplicate by using 24-well cell culture plates. After transfection, cells were grown for 24 to 30 h and then lysed in 60 μl of Tris-EDTA containing 0.2% Triton X-100. Luciferase and β-galactosidase activities were measured with a Dual Light kit (Tropix, Inc.), and the results were normalized with respect to transfection efficiency.
Library screening and DNA purification.
NTera2D1 cDNA library λgt10.1, obtained from J. Skowronski (32), was plated from 1 to 20,000 plaques per 83-mm-diameter plate and transferred to Hybond-N filters (Amersham). Filters were hybridized in the hybridization mix (27) with a 700-bp StyI-KpnI 32P-labeled ORF1 probe, derived from a representative L1Hs genomic clone, λms (my unpublished data). After washing at 50°C in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecylsulfate (SDS), filters were exposed to Hyperfilm MP film (Amersham). The ORF1 probe was removed by boiling in 0.1× SSC–0.1% SDS solution. Filters were then rehybridized with a 600-bp KpnI-BglII L1Hs promoter (λms) probe and autoradiographed. Only L1Hs promoter-positive and ORF1-negative plaques were selected, and their positions were aligned to the plates. Individual recombinant lambda phages were further purified to the homogeneity by a second screening. High-titer phage stocks, prepared from the plate lysates, were used for the preparation of λgt DNA in microgram amounts according to established methods (27). The presence of L1Hs promoter sequences in these DNAs was verified by EcoRI digestion and Southern blotting.
Genomic DNA PCR and cloning.
PCR of the L1Hs promoter-ORF1 region was carried out by using Pfu polymerase (Stratagene), total HeLa DNA isolated by a standard method (27), and primers 5′P-GGGAGGAGCCAAGATGGC and 3′P-GCTGGTGAGGAACTGCGT. Amplification for 20 cycles was done in a Thermal Cycler (PE Biosystems) with the following temperature profile: 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The resulting two DNA fractions of about 1.0 and 1.1 kb (the latter contained an insert of about 100 bp [see below and reference 14]) were further treated with Klenow polymerase (Boehringer Mannheim). The 1.0-kb fraction was gel purified from SeaPlaque low-melting-point agarose (FMC) and cloned into a SmaI-digested pBluescript (pBS)KS vector (Stratagene). Plasmid DNAs from hybridization-positive and randomly selected colonies were purified by alkaline lysis (4), and analyzed for the presence of KpnI, BglII, and PstI sites in their inserts. A DNA clone, named SFL1, containing all three sites was selected for further work. Its sequence, containing 903 bp of L1 promoter and 86 bp of ORF1, displayed 98% similarity to nt 1 to 996 of the published L1.2 sequence (8; GenBank accession no. M80343). Amplification and cloning of a 1.0-kb genomic fragment derived from the bacterial artificial chromosome (BAC) clone (accession no. AC006559; obtained from the BACPAC Resource Center, Oakland, Calif.) corresponding to the P5 cDNA was performed as described for HeLa DNA.
Plasmid constructions.
For cDNA subcloning and sequencing, recombinant λgt DNAs isolated from the cDNA library were digested with EcoRI, and the fragments obtained were subcloned into the pBS KS vector. Inserts of the lambda DNAs containing one or more internal EcoRI sites were first amplified by PCR using Pfu polymerase and λgt primers with engineered 5′-terminal SalI sites. After SalI digestion, the fragments were subcloned into pBS KS vector. Each subcloned DNA was compared to the parental λgt DNA for identity by restriction mapping and PCR amplification.
For riboprobe preparation, two in situ deletion constructs were prepared from SFL1 plasmid with the indicated regions retained and restriction enzymes: ΔP, 1 to 597) and PstI; and ΔBB, 659 to 989 and BamHI-BglII. Further deletions from the ΔP created constructs ΔAP (1 to 311 and PstI-AflII) and ΔAPB (308 to 597. and BamHI-AflII).
For transfection of cells and luciferase assays, DNA fractions of about 1.0 and 1.1 kb containing the L1Hs promoter and ORF1 region (86 bp), obtained by PCR as described above, were cloned into SmaI-digested pGL3-basic vector (Promega). Hybridization-positive colonies were selected, and the insert orientation for these DNAs was determined by colony PCR using a combination of vector- and insert-specific primers. Plasmids were further purified from a 1.5-ml overnight bacterial culture by a modified alkaline lysis protocol (4). To maximize the yield of supercoiled plasmid DNA, lysis solution containing 0.1 N (instead of 0.2 N) NaOH was used.
DNA sequencing.
Sequences of all plasmid clones and subclones were determined by cycle sequencing protocol using a BigDye kit (PE Biosystems). Sequences of up to 500 nt were obtained with the ABI 310 and 377 DNA sequencers (Applied Biosystems). To facilitate sequencing of the 1- to 3-kb-long cDNAs cloned in pBS KS, various in situ deletion constructs were prepared using different restriction enzymes. The remaining gaps were closed with the help of synthetic primers. L1Hs promoter regions containing putative transcription factor binding sites were sequenced twice from both strands. Sequences of the genomic L1Hs promoters cloned in pGL3 vector and used in transfection experiments are available upon request.
RNA isolation, hybridization, and detection.
Total RNA from different cell lines was isolated with RNAzol (Biotecx Laboratories, Inc.) according to the manufacturer's instructions. All RNA preparations were treated with proteinase K (Sigma), phenol extracted, and ethanol precipitated. Contaminating DNA was removed by digestion with RQ DNase (Promega). Total RNAs (0.12, 0.25, and 0.5 μg) were denatured as described elsewhere (27) and loaded manually to the six panels of a Hybond N+ filter (Amersham Pharmacia Biotech). Filters were hybridized in SDS-containing hybridization cocktail (6) at 68°C for 12 h, using strand-specific 32P-labeled riboprobes transcribed from appropriately linearized SFL1-ΔAP, -ΔAPB, and -ΔBB constructs. T3 and T7 RNA polymerases were used to generate probes complementary to the sense and antisense L1Hs transcripts encompassing the 5′UTR. Only full-length probes having same specific activity and a high yield (>90%), as judged by Northern blot analysis, were used. To minimize nonspecific hybridization of the vector-derived sequences included in the 5′ regions of probes, approximately 10 μg of the unlabeled small RNA identical to the 5′ portion of the probe was added to the hybridization cocktail. Two controls, unlabeled full-length RNAs (50, 100, and 200 pg) transcribed from the sense and antisense strands of SFL1 were used in each panel. Unlabeled RNAs identical to the probe sequences did not hybridize. Total DNAs (1, 3, and 10 ng) were from HeLa DNA and NTera2D1 cells. SFL1 (0.1, 0.3, and 1 ng) was used as a control for DNA panels. All data were normalized to the control RNA or DNA signals. DNA dot blotting was carried out as described for RNA except that before loading, DNAs were denatured in a solution containing 0.5 N NaOH and 1.5 N NaCl. Washing of the blots was done with 0.1× SSC–0.1% SDS at different stringencies (60 to 80°C) for 30 min each time. Blots were quantified with a PhosphorImager (Molecular Dynamics). RNase protection was carried out according to the published protocol (27), using 32P-labeled antisense riboprobes generated from cDNAs (see figure legends for details). Protected RNAs were analyzed on a 5% denaturing polyacrylamide gel, and their images were scanned by the PhosphoImager or detected by autoradiography.
Computer analysis.
All cDNA sequences were compared to entries in the GenBank divisions (nr, EST [expressed sequence tag], gss, and htgs) using an advanced BLASTN program run with default parameters (2). Repeated DNA sequences were identified and masked with Repeatmasker (A. F. A. Smit and P. Green, unpublished data; http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). ORF analysis was carried out with ORF Finder (T. Tatusov and R. Tatusov, unpublished data). Alignments of the two sequences (e.g., L1Hs 5′UTR and cDNA 5′ end) were done with BLAST2 sequences (35). BLAST and ORF Finder programs were run on the National Center for Biotechnology Information network service. Multiple sequence alignments were carried out with CLUSTAL W (version 1.7) (38; http://www.clustalw.genome.ad.jp/) and Multalin (version 5.4.1) (7; http://www.toulouse.inra.fr/multalin.html). Distribution of the L1Hs 5′UTR-ORF1 sense and antisense sequences in the human EST database (as of 6 Apr. 2000) was revealed by the WU-BLAST2 program (W. Gish, unpublished data; http://dove.embl-heidelberg.de/Blast2/).
Nucleotide sequence accession numbers.
Twelve cDNA sequences were submitted to GenBank with accession numbers AF279773 to AF279784.
RESULTS
Search for a cellular gene with an L1-like promoter yielded transcripts derived from the opposite strand of the L1Hs 5′UTR.
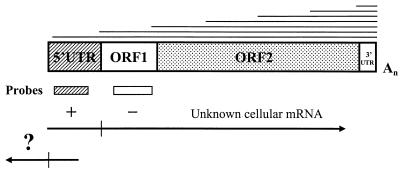
A common feature of all mammalian L1 retrotransposons is that they have highly similar coding regions (ORFs) but unrelated 5′UTRs or promoters. Therefore, it is possible that L1 promoters were originally derived from the host genes located close to L1 insertion sites by a mechanism of promoter capture. According to this scenario, the human genome may still contain an unknown gene which is driven by an L1-like promoter, and as the L1Hs promoter is active in NTera2D1 cells (31), it should be also expressed in these cells. Assuming that this gene has a different ORF, I have screened a human NTera2D1 cDNA library [made by oligo(dT) priming] with the L1 ORF1-specific probe to exclude known L1Hs transcripts (Fig. 1). To reveal transcripts originating from the promoter region, a second screening with L1Hs promoter-specific probe was carried out using the same set of filters. Unexpectedly, hybridization-positive signals for the promoter probe showed about five fold-greater frequency (5 of 1,000 plaques) than signals obtained for the ORF1 probe. The majority of the L1Hs ORF1 hybridization signals coincided with the promoter hybridization signals. Based on the results of differential screening, 15 L1Hs promoter-positive and ORF1-negative plaques were selected and purified to homogeneity, and their cDNA inserts were subcloned into pBS KS vector. Sequencing of these cDNAs revealed that they all contained a terminal region of about 100 to 400 bp 81 to 98% similar to the L1Hs promoter, as expected (Table 1). Surprisingly, however, 13 of these cDNAs were oriented as though they were derived from a promoter located on the antisense strand, i.e., from the ASP (Fig. 2A). Although the remaining two cDNAs (N9 and P2) showed the same polarity as the L1Hs (sense) promoter, neither of them was considered a candidate gene of interest (N9 displayed similarity to the L1Hs promoter region in its 3′ end; P2 had a small 5′-terminal fragment similar to the promoter followed by several Alu elements [see below]). Therefore, preliminary analysis of the cDNA sequences suggested that L1Hs has an ASP which is capable of driving transcription from the L1 5′UTR to the neighboring genomic sequences located upstream of the L1Hs retrotransposon.
FIG. 1.
Strategy for differential screening of the L1Hs cDNAs. L1Hs transcripts with homogeneous 3′ ends and heterogeneous 5′ ends, shown at the top, are aligned to the general L1 retrotransposon structure. Below this structure, positions of DNA probes corresponding to the L1Hs promoter (5′UTR) and ORF1 are indicated. A search for an unknown gene or mRNA with an L1-like internal promoter by screening NTera2D1 cDNA library for promoter-positive and ORF1-negative cDNAs (marked by + and − signs, respectively) yielded novel cDNAs (marked by ?) containing the L1Hs 5′UTR linked to known and unknown sequences located upstream of the L1Hs (bottom structure). Vertical bars mark the 5′ and 3′ ends of the 5′UTR.
TABLE 1.
Sequencing and mapping data of 15 cDNAs isolated from the NTera2D1 library
| Clone | Lengtha (bp) | Similarity to L1.2 5′UTRb
|
Similarity/identity to known gene or regionc | BAC clone (accession no.) | Chrd | No. of exonse | Promoter distancef (kb) |
|---|---|---|---|---|---|---|---|
| Region (bp) % | |||||||
| N1 | 2,181 | 1–163 ≡ 163–2 95 | 171–284—Alu | AC020581 | 7 | 4 | >12 |
| 345–778—Tigger2a | AC009264 | ||||||
| 1752–2004—MIR | |||||||
| 1882–2161—AA084761 (EST) | |||||||
| N4 | 1,205 | 10–211 ≡ 461–261 90 | GAP-43 mRNA (NM_002045) | AC021376 | 3 | 4 | >50 |
| 210–324 ≡ 117–4 98 | 362–1205 = 122–965 | AC012565 | |||||
| N5 | 1,664 | 1–156 ≡ 414–260 94 | 398–508—Alu | AC010148 | NDg | 4 | ∼1 |
| 782–1151/1361–1664—MER4 | |||||||
| N7 | 1,670 | 1–190 ≡ 449–260 97 | 192–677—Hyp protein (47%) (Q12765) | AC018470 | 2 | 5 | >4 |
| N8 | 762 | 1–326 ≡ 329–5 95 | 339–762—L1 (M80343) | ND | ND | ND | ND |
| N9 | 1,956 | 1050–1632 ≡ 769–175 81 | mRNA, homologue of mouse | AL022400 | 1 | 9 | ND |
| 1636–1771 ≡ 146–12 90 | Hh1 mRNA (AL157958) | AL022171 | |||||
| 1–64 ≡ 913–976/63–989 ≡ 1011–1847 (99%) | AL021069 | ||||||
| N10 | 1,930 | 1–450 ≡ 456–7 98 | CHRM3 gene (U29589) | AC015901 | 1 | 5 | >62 |
| 760–1922 ≡ 183–1345 | AC013517 | ||||||
| N11 | 1,777 | 1–104 ≡ 449–346 97 | 103–244—FAM | AC015495 | 3 | ≥2 | >15 |
| 403–473—MIR | |||||||
| 1368–1777—AW513672 (EST) | |||||||
| N12 | 441 | 1–177 ≡ 434–259 93 | SPT3 mRNA (AF073930) | AL138880 | 6 | ≥4 | >40 |
| 176–441 ≡ 173–438 | AL161905 | ||||||
| L2 | ∼2,000 | 1–109 ≡ 453–346 94 | 105–280/651–893/3′ end—L1 | AC019310 | 4 | ND | ND |
| 469–527—MIR | |||||||
| P1 | 1,687 | 1–194 ≡ 455–261 93 | 189–292—MLT | AC023485 | 3 | ≥5 | >9 |
| 1026–1326/1373–1477—MER4 | |||||||
| P2 | ∼3,500 | 1–122 ≡ 23–142 93 | 5′ and 3′ ends/396–524—Alu | ND | ND | ND | ND |
| 275–395—Tigger1 | |||||||
| P3 | 1,818 | 1–221 ≡ 481–261 92 | 361–460/488–816/876–1320/1502–1616—MLT | AC021696 | >9 | >7 | |
| P5 | 2,881 | 1–429 ≡ 428–1 93 | OATP mRNA (U21943) | AC006559 | 12 | 16 | ∼60 |
| 669–2881 ≡ 1–2213 | AC022224 | ||||||
| P6 | ∼3,000 | 3–152 ≡ 153–5 96 | 223–319—MLT | ND | ND | ND | ND |
Complete and partial sequences (5′ and 3′ ends) of individual cDNAs are noted by exact and approximate sizes, respectively.
Similarity between regions of cDNA and L1.2 5′UTR (accession no. M80343) determined by BLAST2 sequences comparison and indicated by ≡ sign.
cDNA sequences similar to DNA repeats or EST sequences and identical to the corresponding regions of known genes (indicated after long dashes) are shown. Accession and ESTs numbers are indicated in parentheses.
Chr, chromosome number determined from BAC location.
Determined from cDNA and BAC sequence comparison using BLAST2.
Approximate distance of the ASP from the coding sequence is shown because of gaps and unordered fragments in unfinished BACs.
ND, not determined.
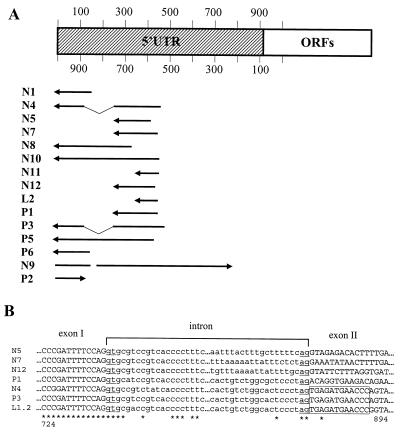
FIG. 2.
Mapping of the cDNA 5′-terminal sequences to the L1Hs 5′UTR. (A) cDNA sequences with the mRNA sequence polarity (5′ to 3′) indicated by arrows were mapped to the L1Hs 5′UTR. Numbering of the top L1Hs strand starts with +1 (34), and numbering of the bottom strand starts from nt 86 of ORF1. Splicing of the N4 and P3 cDNAs within the 5′UTR is indicated by thin lines. Two cDNAs (N9 and P2) showed opposite polarity. (B) Six cDNA sequences (N4, N5, N7, N12, P1, and P3) compared to the corresponding genomic sequences (Table 1) were aligned to show their exon and intron structures, indicated by upper- and lowercase letters, respectively. The conserved 5′ and 3′ intronic dinucleotide sequences (GT and AG) are underlined. Two cDNAs (N4 and P3) are spliced within the L1Hs 5′UTR, and their identical exon II sequences compared to the corresponding L1 sequence are boxed. The others are spliced to known or unknown sequences located further upstream of the L1Hs. Numbering is according to the L1Hs 5′UTR bottom strand. L1.2, accession no. M80343 (8). Identical nucleotides are marked below sequences by asterisks; sequences not shown are indicated by dots.
Many known genes are transcribed from the L1Hs 5′UTR, yielding chimeric transcripts.
Using an advanced BLASTN program (2), complete sequences of the 12 cDNAs and partial sequences of the remaining 3 cDNAs were searched for homologous sequences in the GenBank database. Table 1 summarizes the results of this search. Four cDNAs (N4, N10, N12, and P5) contained sequences identical to previously known mRNA or gene sequences. N4 encompassed a sequence identical to the mRNA of human neuronal growth protein 43 (GAP-43); N10 included a sequence identical to the 5′ two-thirds of the gene for the human m3 muscarinic acetylcholine receptor (CHRM3); N12 had a small 3′-terminal fragment identical to the 5′ region of the human histone acetylase complex subunit (SPT3) mRNA, and P5 sequence was identical to the mRNA of human organic anion transporting polypeptide (OATP). Graphical representations of these comparisons are shown in Fig. 3. In addition, several cDNAs (N1, N5, N8, N11, L2, P1 to P3, and P6) showed similarity to Alu, Tigger, MER, or other repeated DNAs; two of them (N1 and N11) displayed similarity to EST sequences (ESTs were listed only for those cDNAs for which no matches were found in GenBank's nr division). N7 encoded a hypothetical protein predicted from a coding sequence of a cDNA clone (26). N9 was identical to the novel human mRNA showing similarity to the mouse Hh1 mRNA. No statistically significant similarities, besides similarity to the L1Hs 5′UTR and various repeated DNAs, were detected for presumably unique sequences of seven cDNAs (N5, N8, L2, P1 to P3, and P6). In summary, the computer analysis showed that at least four previously known gene sequences were linked to the L1Hs 5′UTRs and thus were possibly transcribed from the L1Hs ASP. Because these transcripts were of major interest, each of them was studied in detail.
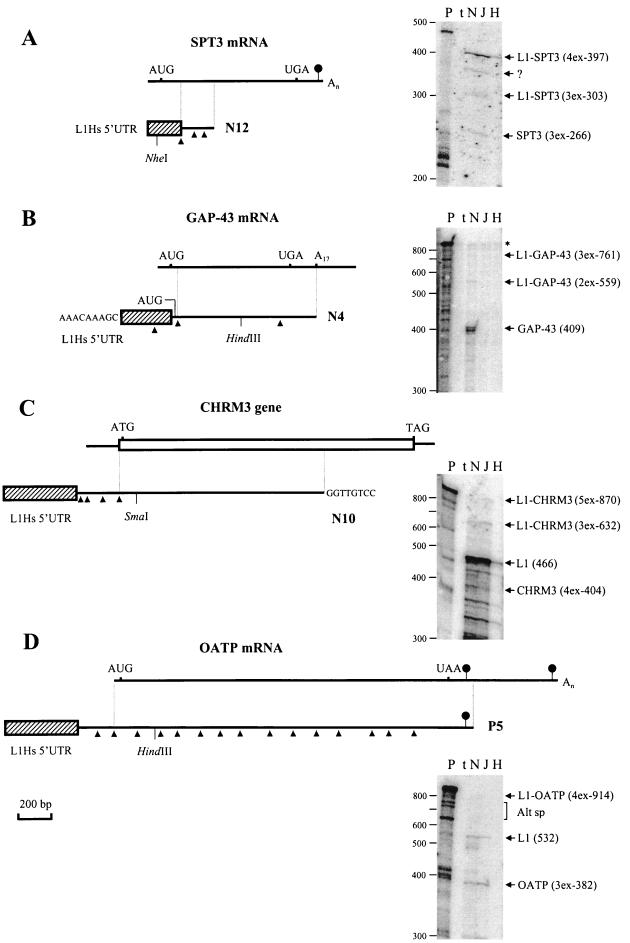
FIG. 3.
Graphical representation and detection in different cell lines of chimeric transcripts generated from the L1Hs ASP. Diagrams show alignments of N12 and SPT3 mRNA (A), N4 and GAP-43 mRNA (B), N10 and CHRM3 gene (C), and P5 and OATP mRNA (D). Chimeric cDNAs and mRNAs are indicated by lines, the CHRM3 gene (mRNA sequence unknown) is shown by an open box, and L1Hs 5′UTR sequences transcribed from the L1Hs ASP and linked to the known mRNA and gene sequences are indicated by hatched boxes. Identical regions between mRNA (gene) and chimeric cDNAs are shown by vertical lines. Introns, determined from the alignment of genomic and cDNA sequences (Table 1), are shown by arrowheads below cDNA structures only for aligned portions and 5′ regions of chimeric cDNAs. Initiator AUG/ATG and stop codons are shown above the lines; polyadenylation signals (not found for the structures of GAP-43 mRNA and CHRM3 gene depicted) are marked by oval arrows. Additional sequences were found at the 5′ end of N4 and 3′ end of N10 cDNAs. Their origin is unclear. Detection of the four chimeric transcripts in different cell lines by using RNase protection (27) is shown at the right. All protected RNAs were analyzed on a 5% denaturing polyacrylamide gel by using the following 32P-labeled probes. (A) A 465-nt riboprobe encompassing 131 nt of the L1Hs 5′UTR, three exons of SPT3 mRNA (85, 87, and 94 nt), and 68 nt of vector sequences was generated from N12, cut with NheI, and transcribed with T7 RNA polymerase. (B) A 829-nt riboprobe encompassing two exons of the L1 5′UTR (150 and 202 nt), 409 nt of 5′ region of the GAP-43 mRNA, and 59 nt of vector sequence was generated from a HindIII deletion subclone of N4, cut with EcoRI, and transcribed with T3 RNA polymerase. (C) A 934-nt riboprobe encompassing 450 nt of the L1 5′UTR plus 16 nt of non-L1 sequence, three exons (63, 103, and 127 nt) and 5′ coding region of the CHMR3 gene (111 nt), and 61 nt of vector sequences was generated from a SmaI deletion subclone of N10, cut with EcoRI, and transcribed with T7 RNA polymerase. (D) A 1,009-nt riboprobe encompassing 429 nt of L1 5′UTR plus 103 nt of non-L1 sequence, 5′ region of OATP mRNA (three exons, 382 nt), and 94 nt of vector sequences was generated from a HindIII deletion subclone of P5, cut with XhoI, and transcribed with T7 RNA polymerase. Restriction enzymes used to make riboprobes and deletion subclones are shown below cDNA structures. Chimeric and alternative mRNAs detected are shown on the right of each panel. For these mRNAs, the number of exons (xex) and their sizes in nucleotides are indicated in parentheses. Protected fragments of N10 and P5 predict three and one additional exons for the 5′ ends of CHMR3 and OATP mRNAs, respectively. Protections with the L1Hs 5′UTR of N10 (C) generate several fragments of <466 nt representing highly homologous transcripts derived other genomic L1Hs. Similar protections for N12 and N4 (fragments <200 and <300, respectively) are not shown (A and B). Traces of the undigested probe (B) are shown by the asterisk. A 32P-labeled 100-bp ladder (BRL) was used as a molecular weight marker. P, uncut probe; t, tRNA. In each experiment, 5 μg of total RNAs from NTera2D1 (N), JEG 3 (J), and HeLa cells (H) were used. Alt sp, possible alternatively spliced products; ?, unknown product.
A 441-bp N12 cDNA contained a 177-bp 5′-terminal region similar to the L1Hs 5′UTR (Table 1) followed by a 264-bp sequence identical to the 5′ sequence of the SPT3 mRNA coding region (Fig. 3A). A comparison with the two genomic sequences (BACs in Table 1) showed that N12 has at least four exons, and its 5′ end was mapped >40 kb upstream from its coding sequence (because of the gaps and unordered fragments of BACs, precise mapping was not possible). To reveal chimeric transcripts identical to N12 cDNA and to estimate the ratio of alternative transcripts derived from the L1Hs ASP and the SPT3 promoter, RNase protection with an antisense riboprobe was carried out using total RNA isolated from three different cell lines. As shown in Fig. 3A (right), chimeric L1-SPT3 transcripts containing (i) L1Hs 5′UTR antisense strand and two or three exons of SPT3 mRNA (303 and 397 nt, respectively) and (ii) SPT3 mRNA (three exons) were detected. In all cell lines analyzed, chimeric L1-SPT3 transcripts (four exons, 397 nt) were about fivefold more abundant than SPT3 transcripts (estimation based on visual inspection of band intensities and comparison with the known amounts of 32P-labeled marker), suggesting that transcription of SPT3 from the L1Hs ASP is more efficient than that from the SPT3 promoter.
A 1,205-bp N4 cDNA contained two 5′-terminal fragments similar to the L1Hs 5′UTR (Table 1; Fig. 2A) followed by a 38-bp unique sequence and a 843-bp 3′-terminal sequence identical to the GAP-43 mRNA coding region and a part of the 3′UTR (Fig. 3B). Comparison with the GAP-43 mRNA showed that the most 5′ region (30 nt) of GAP-43 ORF was missing in N4. The ORF of N4 started with four codons (ATG ACA TTA GCT) derived from the exon located upstream to the GAP-43 coding sequence. N4 cDNA was apparently derived from a chimeric GAP-43 mRNA by oligo (dT) priming from the A-rich region of 3′UTR. Further mapping to the BAC clones and other genomic sequences revealed that the missing 30 bp matched the first coding exon of GAP-43 mRNA. The most 5′ sequence AAACAAAGC of N4 was not found in the corresponding genomic region (including 13 kb upstream from the 5′UTR). Therefore, the question remained as to whether the 5′ nonanucleotide sequence of N4 was an artifact generated by cloning or, alternatively, whether it represented part of an upstream exon spliced to the remaining sequence. N4 has four exons with sizes from 150 to 598 bp. Its 5′-terminal sequence, similar to the L1Hs 5′UTR, was located >50 kb from its coding region. To reveal the presence of chimeric N4 transcripts in total RNA of different cell lines, RNase protection was carried out. As shown in Fig. 3B (right), faint bands (observed after careful inspection of the original autoradiogram) corresponding to alternative transcripts containing one or two exons of L1Hs 5′UTR linked to GAP-43 mRNA (559 or 761 nt) and a strong band corresponding to GAP-43 mRNA were detected only in NTera2D1 cells. The amount of chimeric transcripts accounted for <1% of that of the GAP-43 transcripts, suggesting that transcription from the ASP of N4 was rather inefficient.
A 1,930-bp N10 cDNA contained a 5′-terminal region highly similar to the L1Hs 5′UTR (Table 1) followed by a 309-bp unique sequence, encompassing three exons, and a 1,163-bp 3′-terminal sequence identical to the CHRM3 gene coding region, including the initiator ATG (Fig. 3C). Because the mRNA sequence of the CHRM3 gene was not known, comparison with the genomic structure was made (Table 1). N10 cDNA had five exons from 63 to 1,163 bp. Its last 8 bp were different from the corresponding gene sequence (no splice sites were found). The 5′ end of N10 was mapped >62 kb upstream from its coding sequence. Using RNase protection, two chimeric and alternatively spliced N10 transcripts, one containing five exons (870 nt), including 111 nt of the CHRM3 coding region, and the other containing three exons (632 nt), were detected in NTera2D1 and JEG 3 cell lines. Comparison with CHRM3 transcripts, containing four exons (404 nt), also produced in these cell lines, showed that the abundance of chimeric transcripts was comparable with that of CHRM3 transcripts in JEG 3 but significantly lower (about 10-fold) than that in NTera2D1.
A 2,881-bp P5 cDNA contained a 429-bp 5′-terminal region similar to the L1Hs 5′UTR (Table 1) followed by a 240-bp unique sequence and a 2,212-bp 3′-terminal sequence identical to the entire OATP coding region, including a 147-bp fragment of the 3′UTR (Fig. 3D). P5 cDNA has 16 exons from 65 to 532 bp, and its 5′ end was mapped ∼60 kb upstream of the OATP coding sequence. Using RNase protection, a series of low-abundance alternative P5 transcripts were detected in NTera2D1 (Fig. 3D, right) but not in other cell lines. The amount of these chimeric transcripts accounted for about 10% of that of the OATP transcripts, suggesting low-level transcription from the L1Hs ASP of P5.
In summary, cDNA mapping and RNase protection data suggest that the L1Hs 5′UTR may contain an ASP initiating transcription from within the 5′UTR and driving it into nearby genes, thus generating chimeric transcripts. Compared to the transcription of four known genes, variable levels of transcription from L1Hs ASP were observed. Chimeric transcripts accounted for roughly 1 to 500% of the known transcripts.
L1Hs chimeric transcripts are correctly spliced.
To understand whether the L1Hs ASP-driven transcripts were the unspliced products obtained by readthrough transcription or whether they represented correctly spliced and processed forms of pre-mRNAs, exon-intron junctions and tail regions of 10 cDNAs were analyzed. From the total of 96 donor and acceptor splice sites, 94 followed the GT-AG rule and had pyrimidine-rich 3′-terminal intronic sequences (data not shown). These data show that L1Hs cDNAs represented correctly spliced transcripts and thus were not the products of promiscuous readthrough transcription. A major donor splice site for six cDNAs was discovered at nt 736 within the L1Hs 5′UTR antisense strand (Fig. 2B). Interestingly, two cDNAs (N4 and P3) had both donor and acceptor splice sites within this region. Also, the other two cDNAs (N11 and L2) contained donor sites at nt 652 and acceptor sites located upstream to the L1Hs. Several splice sites were also found in the repeated DNA sequences of cDNAs N1, N5, and P3. It is likely that splicing in these cases has occurred at cryptic splice sites.
In contrast, polyadenylation of the cDNAs was less clear. Even though seven cDNAs had polyadenylation signals, only four of them contained poly(A) tails. It has been suggested that the lack of poly(A) tails in transcripts isolated from the NTera2D1 library may be related to cloning procedures (32). As noted above, at least for one cDNA (N4), an oligo(dT) priming of the initial transcript probably occurred from the oligo(A) stretch located in the 3′UTR. However, it is unclear why the 3′ end of cloned N4 had no poly(A) tail.
L1Hs ASP predicted from the distribution of EST sequences in the 5′UTR.
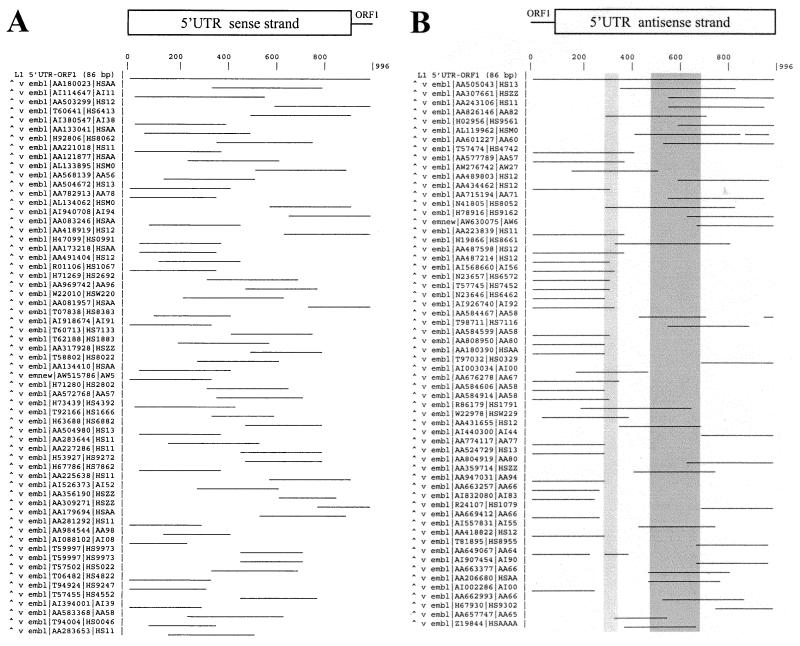
In an ideal case, ESTs corresponding to the contiguous sequences, such as mRNA coding sequence, should follow a uniform distribution because of the random priming used in their synthesis. Similarly, ESTs derived from mRNA 5′ and 3′ ends (e.g., initiation and termination regions) should tend to be overrepresented compared to the neighboring nontranscribed sequences (e.g., promoter). This is exactly what is shown by EST analysis of the L1Hs 5′UTR-ORF1 region (both strands). Figure 4A shows that the first 60 ESTs showing the highest similarity (85 to 95%), mapped to the L1Hs 5′UTR sense strand (including 86 nt of ORF1), are nearly randomly distributed over the entire region without any major preference. However, ESTs mapped to the antisense strand on the same region are overrepresented essentially in two regions (Fig. 4B). The first, rather broad region is located from nt 1 to 300, and its 3′ end predicts a transcriptional termination site for about 30% of ESTs (marked by a narrow zone), because no transcriptional start sites have been previously mapped to this region (transcription of the L1Hs sense strand starts from nt +1). This region is followed by a less represented region <200 nt in length (about 20% ESTs). The second, more diffuse region is located around nt 500 to 700 (Fig. 4B, marked by a wide zone), which probably marks the initiation region for multiple ESTs (about 30% from 60 ESTs analyzed). This result is consistent with the location of cDNA 5′ sequences in the L1Hs 5′UTR from nt 500 to 600 (Fig. 2A) and also suggests that if the promoter is located close to the initiation sites, it should lie between nt 300 and 500.
FIG. 4.
Graphical representation of EST sequences similar to the L1Hs 5′UTR-ORF1 region. Sense (A) and antisense (B) strands of a 996-bp L1.2 5′UTR-ORF1 sequence (accession. no. M80343) were searched for homologous sequences in EST databank using the WU-BLAST2 program. Only the 60 first sequences with the highest scores are shown. Numbering of the L1Hs sense strand starts from +1 (34), and numbering of the antisense strand starts from nt 86 of ORF1. A possible transcriptional initiation region from nt 500 to 700 and termination site around nt 300 are shaded by wide and narrow zones, respectively (see the text for explanation).
Based on the computer-generated map, analysis of the randomly chosen ESTs' flanking sequences confirmed the predicted termination and initiation pattern of the L1Hs ASP region. The ESTs terminating around position 300 were found in various normal and tumor cells. Similarly, the expression of ESTs whose transcription initiated from nt 500 to 700 was detected in a wide variety of cell lines and tissues.
Chimeric transcripts initiate from nt 300 to 500 of the L1Hs 5′UTR in different cell lines.
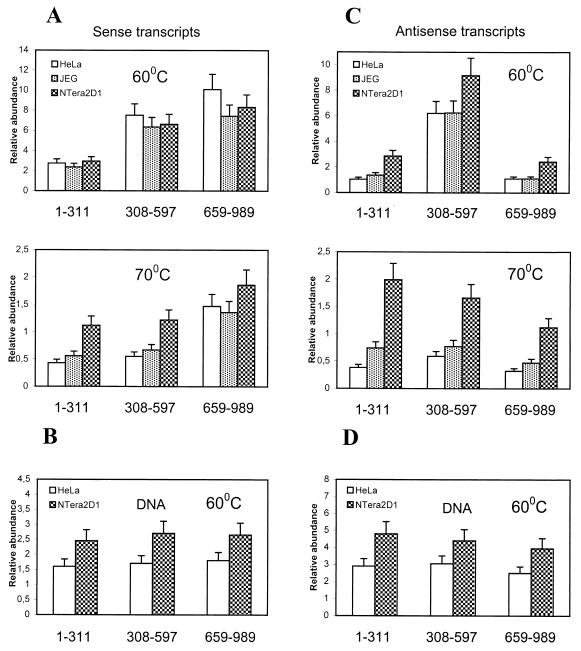
Distribution of the transcripts in the L1Hs 5′UTR and ORF1 region (nt 1 to 989) was studied by quantitative dot blot assay. Total RNAs from HeLa, JEG 3, and NTera2D1 cells and various controls were loaded to six identical panels (as described in Materials and Methods), which were then hybridized with sense and antisense riboprobes derived from different L1Hs 5′UTR-ORF1 regions. Distribution of the L1Hs sense transcripts shows that transcripts homologous to the 5′ end (nt 1 to 311) are less abundant than those derived from the central (nt 308 to 597) and 3′-end (nt 659 to 989) of L1Hs 5′UTR regions (Fig. 5A, top). Increasing the stringency by washing at 70°C (Fig. 5A, bottom) showed almost equal distribution of transcripts containing 5′-end and central regions of the L1Hs 5′UTR, whereas transcripts encompassing the 3′-end region were about twofold more abundant, suggesting transcriptional initiation from downstream sequences, nt 659 to 989. These data suggest that an additional promoter (besides the internal promoter) may be located in the central region of the L1Hs 5′UTR (P. Nigumann and M. Speek, unpublished data). Further washes at 80°C resulted about 10-fold reductions of signals for all blots (data not shown). Hybridization to the genomic DNAs followed a uniform distribution (Fig. 5B), also previously observed by others (13), confirming that differences in transcript distribution were not due to the distribution of different regions of genomic L1Hs 5′UTR.
FIG. 5.
Distribution of L1Hs transcripts in total RNAs of different cell lines. Hybridization to the transcripts and total DNA (HeLa and NTera2D1) encompassing different L1Hs 5′UTR-ORF1 regions (shown below the columns) was carried out with L1Hs antisense (A and B) and sense (C and D) riboprobes as described in Materials and Methods. Probes 1–311, 308–597, and 659–989, encompassing 5′, central, and 3′ portions of the L1 5′UTR (sense-strand numbering), were used. Columns show relative abundance of transcripts or genomic DNA regions normalized with respect to the hybridization signals obtained from synthetic sense and antisense control RNAs and control DNA (SFL1). Upper and lower parts of panels A and C represent the same blot washed at 60 and 70°C, respectively. Data for DNA blots washed at 60°C are shown in panels B and D.
Differently from sense transcripts, however, antisense transcripts showed about fivefold-higher abundances of transcripts homologous to the central region (nt 308 to 597) of the L1Hs 5′UTR (Fig. 5C, top), suggesting transcriptional initiation from this region. After increasing the stringency by washing at 70°C (or 80°C [data not shown]), the distribution of antisense transcripts changes. Transcripts homologous to the 5′ end are about twofold more abundant than transcripts homologous to the 3′ end (Fig. 5C, bottom), whereas DNA hybridization shows almost equal distribution of different L1Hs 5′UTRs (Fig. 5D). Therefore, transcription from the L1Hs 5′UTR antisense strand appears to be confined to a small region between nt 300 and 600, possibly <200 nt, which is consistent with the mapping data for cDNA 5′ ends (Fig. 2A). This also explains why the hybridization of transcripts, apparently initiating from the central region, was greatly reduced upon increasing the temperature, while the antisense transcripts encompassing the region from nt 1 to 311 showed much stronger hybridization. Since the probes used in this study had the same specific activity, and because of various controls used (see Materials and Methods), the results of hybridization with sense and antisense probes can be compared directly. This comparison suggests preferential transcription (about twofold) from the L1Hs 5′UTR antisense strand with the initiation sites located from nt 300 to 500 (Fig. 2A). Similar transcriptional patterns were observed for HeLa, JEG 3, and NTera2D1 cell lines.
Genomic L1Hs 5′UTR contain ASPs active in transient transfection assays.
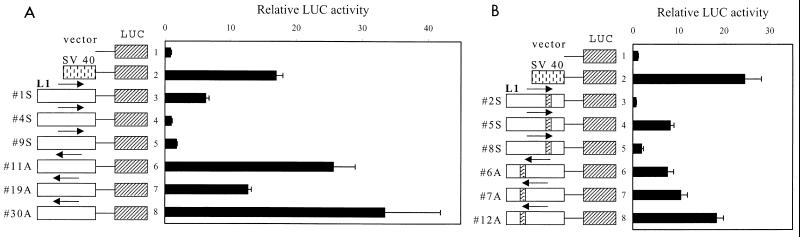
Because of the abundance of full-length L1 retrotransposons, and because no two identical cDNAs derived from the L1Hs 5′UTR were found, it seemed reasonable to assume that the human genome contains a large number of potentially active L1Hs ASPs. To test this hypothesis, 21 randomly amplified genomic L1Hs 5′UTRs were cloned in a pGL3 luciferase expression plasmid in the sense and antisense orientations and then used to transiently transfect HeLa cells. The results presented in Fig. 6A show that the promoter activity produced by the first three antisense constructs, 11A, 19A, and 30A, was considerably (about 2- to 30-fold, depending on the construct) higher than that for the three sense constructs, 1S, 4S, and 9S. The activity of each antisense construct was comparable to or exceeded the activity of a simian virus 40 (SV40) promoter control. Interestingly, the activity of sense construct 1S was about sixfold higher than that of the promotorless vector or sense construct 4S, suggesting that some of the genomic L1Hs (sense) promoters may have retained their activity. This result may explain the discrepancy between the results obtained by two different groups (22, 34). Further transfection and analysis of nine additional constructs containing the L1Hs 5′UTR in the antisense orientation and a similar construct containing the genomic region of P5 cDNA (derived from BAC) showed low activity (relative luciferase activity [RLA] = 2 to 5) for five constructs and high activity (RLA = 7 to 52) for the remaining constructs (see Fig. 8). In these assays, the activity of promotorless vector was set to 1.
FIG. 6.
L1Hs 5′UTR promoter activity test. Twelve randomly selected genomic L1Hs 5′UTR-ORF1 fragments cloned in the luciferase (LUC) expression vector were tested for sense and ASP activities in transfected HeLa cells. RLAs were measured for the sense (1S, 4S and 9S) and antisense (11A, 19A and 30A) L1Hs 5′UTR-ORF1 constructs (A) and the sense (2S, 5S, and 8S) and antisense (6A, 7A, and 12A) L1Hs 5′UTR-ORF1 constructs containing an insertion of 110 to 130 bp around nt 780 (B). Luciferase activity was normalized to β-galactosidase activity for each construct in three separate experiments. Vector, promotorless pGL3-basic plasmid; SV 40, pGL3-promoter plasmid.
FIG. 8.
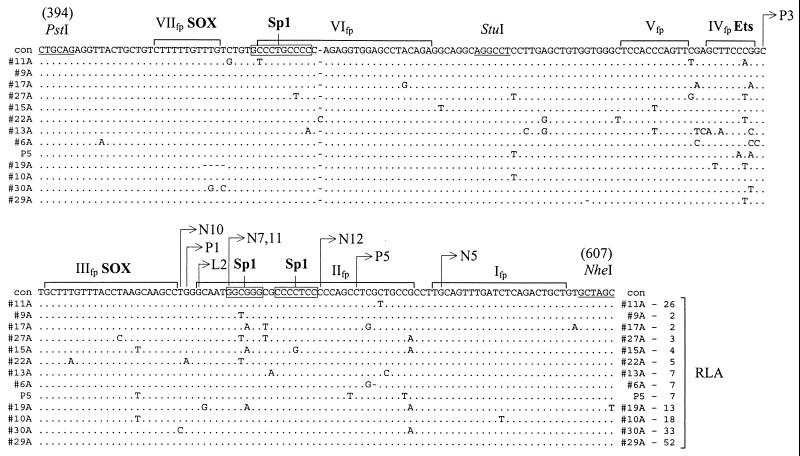
Multiple sequence alignment and L1Hs ASP activity of the L1Hs genomic clones. Sequences of the 12 randomly selected genomic clones and the P5 genomic clone encompassing a putative L1Hs ASP region (nt 394 to 607; numbering of the antisense strand starting from nt 86 of ORF1 [accession no. M80343]) were aligned and compared to the RLA data (shown at the end of the alignment) measured for each genomic clone as described in Materials and Methods. For the alignment, only sequence differences are shown. Identical nucleotides are marked by dots; dashes indicate deletions. Con, consensus sequence. DNase footprints, marked above the sequences by brackets and roman numerals I to VII, and binding sites for SOX and Ets associated with these footprints are from data of others (36, 40). 5′-terminal nucleotides of different cDNAs are mapped to the sequence positions by arrows. Possible binding sites for Sp1 in footprints II and VI are boxed. RLA for the promotorless vector pGL3 was set to 1.
It should be noted that about 40% of the L1Hs 5′UTRs contain a 112- to 144-bp fragment of unknown function inserted at position 780 (my unpublished data; see also reference 14). Inspection of the genomic sequences available for the eight cDNAs (Table 1) revealed that only N4 had the 128-bp insertion located upstream of the cDNA 5′ end. However, computer analysis of the 10 randomly chosen genomic sequences matching the ESTs, similar to the cDNAs analyzed in this study, showed that seven of them had an insertion in the L1Hs 5′UTR upstream of the EST sequence. This result raised the question of whether the inserted fragment can function as an ASP. To test this possibility, six randomly selected constructs in two different orientations, each having an insertion around nt 780, were subjected to the transient expression test as described above. Again, antisense constructs 6A, 7A, and 12A showed maximum of 2- to 20-fold-higher activity than sense constructs 2S, 5S, and 8S (Fig. 6B). Therefore, the presence of an insert in the L1Hs 5′UTR inhibited rather than activated the expression of both sense and ASPs (ASP activity was reduced about twofold compared to the SV40 promoter activity [compare Fig. 6A and B). In summary, although the 15 randomly cloned genomic L1Hs ASPs demonstrated a large variation in activity nine of them displayed activities 7- to 50-fold greater than the background, and about one-third of them were comparable with that of the SV40 promoter.
L1Hs ASP region defined by deletion and mutation analysis.
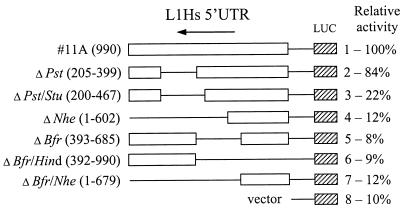
To define the L1Hs ASP region in the 5′UTR, a series of deletions were made in the genomic construct 11A, which showed high promoter activity in the previous experiments. These deletion constructs were transfected into HeLa cells and tested for luciferase activity. Figure 7 shows that deletion of a 194-bp region (205 to 399; construct 2) had only a small effect, while extension of the deletion in the 3′ direction by 68 bp caused a major (about four- to fivefold) decrease of activity (construct 3). Further deletions in both directions resulted in about a twofold reduction of activity (constructs 4 to 7), with some preference for activity loss in the 3′ direction (construct 4). These data show that the elements critical for ASP activity are located in a small region (nt 399 to 467) and also suggest that some additional elements in its 3′ flanking region which may play a stimulatory role. This result is consistent with the mapping of the cDNA 5′ ends immediately downstream to the region from nt 500 to 600 where possibly multiple transcriptional initiation sites are located (Fig. 2A and 8).
FIG. 7.
Definition of the L1Hs ASP region by deletion analysis. Various deletion constructs were prepared from 990-bp genomic clone 11A containing L1Hs 5′UTR-ORF1 ligated to the luciferase (LUC) expression vector in antisense orientation. Deleted regions are indicated in parentheses (except for 11A) and marked with thin lines. Numbering of the antisense strand starts at nt 86 of the ORF1 (accession no. M80343). The restriction enzymes used to make these deletions are shown at the left. Each construct was cotransfected into HeLa cells with β-galactosidase vector, and both luciferase and galactosidase activities were measured. RLA, normalized to β-galactosidase activity, is shown at the right. Activity of construct 11A was set to 100%.
To define the L1Hs ASP more precisely, I took advantage of the previous promoter activity data for 12 randomly selected genomic L1Hs 5′UTRs (described above) and the DNase footprinting data of others (40). Figure 8 shows multiple sequence alignment of the L1Hs ASP region from nt 394 to 607 derived from the genomic clones. The most critical region of 68 bp (from PstI-StuI site) required for the ASP activity determined here encompasses two previously known footprints, VI and VII, sequences of which contain binding sites for Sp1 (40) and SOX transcription factors (36). Four of the 12 genomic ASP constructs had substitution mutations in the footprint VI region (with the exception of construct 11A) and showed reduced promoter activity (constructs 17A, 27A, 15A, and 22A [Fig. 8]), suggesting that Sp1 and possibly some other transcription factors bound to this region may be responsible for the ASP activity. The marginal location of a mutation in construct 11A apparently had no major impact on promoter activity. It should be noted, however, that the Pst-Nhe region contains several DNA footprints (40), which makes the precise assignment of transcription regulatory regions rather difficult. In addition, binding sites for SOX transcription factors involved in the sense L1Hs promoter activity (36) and a binding site of the Ets enhancer (40) have been previously determined. Therefore, it is unclear which regions, besides nt 399 to 467, may be involved in the binding of transcription factors. Nevertheless, the L1Hs genomic ASP activity data suggest that a 15-bp region located from nt 544 to 558, containing two Sp1 binding sites (footprint II), may also contribute to the overall L1Hs ASP activity. For example, C-T substitution in the first Sp1 binding site reduced the activity >5-fold for constructs 9A, 27A, and 22A (Fig. 8). Alternatively, G-A substitution in the same region reduced the activity >6-fold for constructs 15A and 17A. The location of L1Hs ASP immediately upstream of and/or overlapping with the 5′ ends of cDNAs (Fig. 8) is consistent with the computer-predicted (Fig. 4) and hybridization (Fig. 5) data obtained in this study. Taken together, the results of deletion and mutation analysis show that two regions, one of them located about 60 nt upstream and the other partially overlapping with the transcriptional initiation region, are important for L1Hs ASP activity.
DISCUSSION
In search of a human cellular gene with an L1-like promoter, I have discovered antisense transcripts initiated from the L1Hs 5′UTR around nt 500 and spliced to known and unknown gene sequences, thus producing chimeric transcripts. Four of these transcripts containing sequences of known genes, selected for detailed analysis, were detected mostly in total RNA of NTera2D1 cells. Some of them were also present in RNAs isolated from JEG 3 and HeLa cells. As determined by RNase protection, the abundance of chimeric transcripts accounted for roughly 1 to 500% of that of the transcripts of four known genes (Fig. 3). Analysis of the EST sequences mapped to the L1Hs 5′UTR-ORF1 region (Fig. 4) and hybridization of the strand-specific probes to the transcripts derived from this region (Fig. 5) not only confirmed the existence of antisense transcripts in different cells but also predicted the L1Hs ASP driving transcription opposite L1Hs (sense) transcription. Indeed, about one-third of randomly selected genomic L1Hs 5′UTRs showed the high ASP activities in transiently transfected cells, suggesting that the human genome may contain a large number of potentially active L1 ASPs. Therefore, through its ASP, L1Hs has a powerful and a direct means to alter the normal gene transcription. Transcription of the adjacent genes by L1Hs ASP is orientation dependent, which means that only genes having the same transcriptional orientation as the ASP can be transcribed and processed (Fig. 9). Interestingly, the ASP can be located as close as 1 kb or can be as distant as >60 kb from the protein coding sequences (Table 1), suggesting that the transcriptional power of the ASP is not limited by distance between the ASP and the transcribed gene. Similarly, active ASPs are scattered all over the chromosomes, as exemplified by the mapping of the 10 cDNAs to seven different chromosomes (Table 1). Due to its unique property (transcription directed away from the L1Hs retrotransposon), almost any sequence located upstream of the L1Hs 5′UTR and containing no termination signals can be transcribed by the ASP. Therefore, it is not surprising that several cDNAs (N1, N5, and P3) containing repeated DNAs are produced and spliced at cryptic splice sites. However, the biological significance of the transcription driven by the L1Hs promoter is enigmatic. It could be that the L1Hs ASP interferes with transcription of many known genes by occluding their promoters with the readthrough transcription. It is known that transcriptional activation at an upstream promoter reduces transcription from a downstream promoter (12). Introduction of the poly(A) signal and downstream transcriptional pause sites is required for efficient transcriptional termination for RNA polymerase II (9). Therefore, to prevent readthrough transcription, termination signals must be placed upstream of the promoter sequences. Analysis of the distribution of EST sequences matching the L1Hs 5′UTR predicts the termination region to be around nt 300 (antisense strand), suggesting that the ASP itself may be protected from upstream readthrough transcription (Fig. 4B).
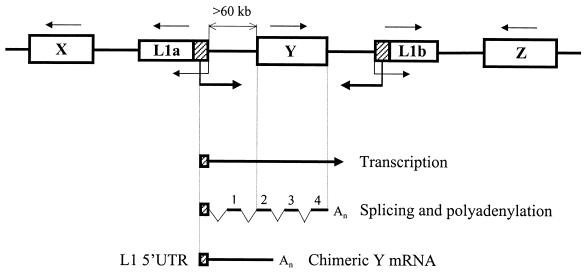
FIG. 9.
Transcriptional regulation by the L1Hs ASP. A fragment of the human genome containing two randomly positioned full-length L1 retrotransposons (L1a and L1b) and genes X, Y, and Z is shown. Direction of transcription for each gene is indicated by a thin arrow above the gene. Transcriptions driven by the L1a and L1b internal promoters (34) and ASPs, driving in opposite direction, are indicated by thin and thick arrows, respectively. Note that gene Y has the same direction of transcription as the L1a ASP but is located >60 kb further downstream. Genes X and Z do not match the L1a ASP orientation. Transcription of gene Y from the L1a ASP can generate a long precursor mRNA which upon splicing and polyadenylation according to the gene Y structure yields a chimeric Y mRNA. This mRNA contains the 5′-terminal L1a 5′UTR in antisense orientation spliced to the exon 1 derived from the intergenic region and exons 2 to 4 of gene Y. The latter structure is depicted at the bottom.
Another possibility for biological significance of the ASP-driven chimeric transcripts is that as they contain necessary translational signals and coding regions, they may be translated similarly to the bona fide mRNAs. For example, P5 cDNA containing the entire coding region of OATP gene, including the initiator AUG and stop codons (Fig. 3D), may be translationally competent. Similarly, N4 cDNA has recruited another initiator AUG from the upstream exon spliced to its coding sequence (N4 lacks the same initiator AUG as GAP-43 mRNA [Fig. 3B]) and thus may be translated. However, analysis of the N1 and N12 cDNA sequences suggests that the majority of ASP-driven chimeric transcripts encompass only fragments of coding regions (depending on the exon-intron structure of the gene) and cannot be translated because of the lack of initiation codons. Nevertheless, if a chimeric mRNA is transcribed from two or more nearby genes, there is a chance that the resulting mRNA, when processed properly, could encode a hybrid protein depending on which exons are spliced together. It has been suggested (33) that fortuitous splicing of cellular genes into the coding regions of DNA transposons could yield the hybrid proteins. I propose that transcription from the L1Hs ASP could provide a direct means for this type of exon combining or shuffling. In summary, I believe that the L1Hs chimeric transcripts, such as N10 and P5 determined in this study, have structures identical to the known cellular mRNAs and survive in the cell to be translated.
Activation of the L1Hs ASP may have a negative influence on gene transcription or expression. Although the ASP seems to be functional in different cells and tissues, it is not known in what cell types its activity is maximal. For example, chimeric N10 and N12 transcripts were more abundant in JEG 3 rather than NTera2D1 cells, suggesting more efficient transcription from the L1Hs ASP in the former cell line. It should be emphasized that activation and repression of L1Hs transcription and presumably its antisense transcription depend on the methylation status of the genomic L1Hs (37, 39). An interesting aspect of L1Hs transcription is that at least in NTera2D1 cells where sense transcription is activated, ASP-directed transcripts complementary to the sense transcripts of about 500 nt are also produced. If a single L1Hs 5′UTR contains both active promoters, competition between them could result in transcriptional interference. Here, another aspect of interest is a compact and overlapping location of transcription factor binding sites in the 5′UTR. It has been determined that the 5′UTR from nt 400 to 600 contains two binding sites for SOX factors involved in the L1Hs sense transcription (36) and an enhancer element, containing binding sites for Ets and Sp1 factors, capable of functioning in either orientation (40). For antisense transcription, binding sites located in this region, determined from deletion and mutation analysis, can be found for transcription factor Sp1 (Fig. 8). The deletion and mutation analysis revealed that the region between PstI and StuI sites (footprint VI) is apparently most critical for ASP activity. Nevertheless, not all data are in agreement with this result (genomic clones 9A and 15A show low activity despite having no mutations in this region). This discrepancy might be explained by the interference between the sense and antisense L1Hs promoters. For example, a mutation in one promoter could enhance transcription from another promoter, or vice versa. Therefore, additional experiments are needed to study the promoter interference as well as to identify the factors involved in the regulation of ASP-directed transcription.
If the L1Hs ASP-directed transcription is not confined to undifferentiated cells but also occurs in various other cells, as shown in this study, why has it remained unnoticed by others? Part of the reason may be due to the promiscuous readthrough transcription encompassing both strands of L1Hs and obscuring L1Hs-derived transcription (31). In fact, these authors did observe an increase of transcription from the L1Hs 5′UTR but did not discuss it. It cannot be denied that readthrough transcription represented by a heterogeneous population of nuclear RNAs is a common phenomenon observed in all cells, especially when total RNAs are analyzed. However, L1Hs ASP-directed transcription is clearly distinguishable from the background of the readthrough transcription when hybridization with strand-specific probes is used (for example, Fig. 5).
Successful amplification of L1Hs retrotransposons occurs largely due to their unusual property, transcription from an internal promoter. This promoter could be regenerated after retrotransposition, thus making newly transposed copies competent for further transcription, reverse transcription, and transposition. It is difficult to imagine that for its retrotransposition, L1Hs could require another promoter (ASP). It could be that L1Hs has borrowed its bidirectional promoter (5′UTR) from an unknown cellular gene or, perhaps, from two genes, expressed coordinatively from this promoter. Isolation of these genes and their regulatory sequences may help to reveal the expression properties of the L1Hs bidirectional promoter and also to shed the light on the formation and amplification of L1Hs retrotransposons.
ACKNOWLEDGMENTS
I thank J. Skowronski for the cDNA library and H. Ilves for NTera 2D1 cells and RNA, Külli Samuel and Pilvi Nigumann for excellent technical assistance, Riho Meier for the help in DNA sequencing, and members of the Laboratory of Molecular Genetics for valuable suggestions on the manuscript.
This work was supported by grant 3070 from the Estonian Science Foundation.
REFERENCES
- 1.Adams J W, Kaufman R E, Kretschmer P J, Harrison M, Nienhuis A W. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res. 1980;8:6113–6128. doi: 10.1093/nar/8.24.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker K G, Swergold G D, Ozato K, Thayer R E. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2:1697–702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke J D. LINEs and Alus—the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- 6.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dombroski B A, Mathias S L, Nanthakumar E, Scott A F, Kazazian H H., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 9.Eggermont J, Proudfoot N J. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 1993;12:2539–2548. doi: 10.1002/j.1460-2075.1993.tb05909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 11.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 12.Greger I H, Demarchi F, Giacca M, Proudfoot N J. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 1998;26:1294–1301. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimaldi G, Skowronski J, Singer M F. Defining the beginning and end of KpnI family segments. EMBO J. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattori M, Hidaka S, Sakaki Y. Sequence analysis of a KpnI family member near the 3′ end of human beta-globin gene. Nucleic Acids Res. 1985;13:7813–7827. doi: 10.1093/nar/13.21.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohjoh H, Singer M F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 16.Kazazian H H., Jr Mobile elements and disease. Curr Opin Genet Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 17.Kazazian H H, Jr, Moran J V. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 18.Kurose K, Hata K, Hattori M, Sakaki Y. RNA polymerase III dependence of the human L1 promoter and possible participation of the RNA polymerase II factor YY1 in the RNA polymerase III transcription system. Nucleic Acids Res. 1995;23:3704–3709. doi: 10.1093/nar/23.18.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leibold D M, Swergold G D, Singer M F, Thayer R E, Dombroski B A, Fanning T G. Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci USA. 1990;87:6990–6994. doi: 10.1073/pnas.87.18.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maestre J, Tchénio T, Dhellin O, Heidmann T. mRNA retroposition in human cells: processed pseudogene formation. EMBO J. 1995;14:6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathias S L, Scott A F, Kazazian H H, Jr, Boeke J D, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 22.Minakami R, Kurose K, Etoh K, Furuhata Y, Hattori M, Sakaki Y. Identification of an internal cis-element essential for the human L1 transcription and a nuclear factor(s) binding to the element. Nucleic Acids Res. 1992;20:3139–3145. doi: 10.1093/nar/20.12.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizrokhi L J, Georgieva S G, Ilyin Y V. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988;54:685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- 24.Moran J V, Holmes S E, Naas T P, DeBerardinis R J, Boeke J D, Kazazian H H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 25.Moran J V, DeBerardinis R J, Kazazian H H., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 26.Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sassaman D M, Dombroski B A, Moran J V, Kimberland M L, Naas T P, DeBerardinis R J, Gabriel A, Swergold G D, Kazazian H H., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 29.Shafit-Zagardo B, Brown F L, Zavodny P J, Maio J J. Transcription of the KpnI families of long interspersed DNAs in human cells. Nature. 1983;304:277–280. doi: 10.1038/304277a0. [DOI] [PubMed] [Google Scholar]
- 30.Singer M F, Krek V, McMillan J P, Swergold G D, Thayer R E. LINE-1: a human transposable element. Gene. 1993;135:183–188. doi: 10.1016/0378-1119(93)90064-a. [DOI] [PubMed] [Google Scholar]
- 31.Skowronski J, Singer M F. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc Natl Acad Sci USA. 1985;82:6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skowronski J, Fanning T G, Singer M F. Unit-length LINE-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smit A F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 34.Swergold G D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusova T A, Madden T L. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 36.Tchénio T, Casella J F, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thayer R E, Singer M F, Fanning T G. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene. 1993;133:273–277. doi: 10.1016/0378-1119(93)90651-i. [DOI] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodcock D M, Lawler C B, Linsenmeyer M E, Doherty J P, Warren W D. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem. 1997;272:7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem. 1998;273:891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]