Abstract
Collaborative studies open doors to breakthroughs otherwise unattainable by any one laboratory alone. Here we describe the initial collaboration between the Griffith and de Lange laboratories that led to thinking about the telomere as a DNA template for homologous recombination, the proposal of telomere looping, and the first electron micrographs of t-loops. This was followed by collaborations that revealed t-loops across eukaryotic phyla. The Griffith and Tomáška/Nosek collaboration revealed circular telomeric DNA (t-circles) derived from the linear mitochondrial chromosomes of nonconventional yeast, which spurred discovery of t-circles in ALT-positive human cells. Collaborative work between the Griffith and McEachern labs demonstrated t-loops and t-circles in a series of yeast species. The de Lange and Zhuang laboratories then applied super-resolution light microscopy to demonstrate a genetic role for TRF2 in loop formation. Recent work from the Griffith laboratory linked telomere transcription with t-loop formation, providing a new model of the t-loop junction. A recent collaboration between the Cesare and Gaus laboratories utilized super-resolution light microscopy to provide details about t-loops as protective elements, followed by the Boulton and Cesare laboratories showing how cell cycle regulation of TRF2 and RTEL enables t-loop opening and reformation to promote telomere replication. Twenty years after the discovery of t-loops, we reflect on the collective history of their research as a case study in collaborative molecular biology.
Keywords: t-loop, R-loop, t-circle, super resolution microscopy, double strand breaks, DNA repair, telomeres
1. Introduction
Over 80 years ago, Herman Muller predicted that the terminal regions of linear chromosomes, which he termed telomeres, “…must have a special function, that of sealing the end of the chromosome,” and that “for some reason a chromosome cannot persist indefinitely without having its ends thus sealed” [1]. Contemporaneously, Barbara McClintock published her pioneering work on maize chromosomes demonstrating that unprotected chromosome ends undergo cycles of fusion and breakage resulting in gross karyotype rearrangements [2]. Today, it is well established that telomeres function as essential protective structures that prevent the erroneous identification of chromosome ends by the DNA repair machinery [3]. In addition, the telomeric machinery must counteract the end-replication problem caused by the inability of conventional DNA polymerases to complete replication of linear templates [4, 5].
Until the primary sequences of telomeres were elucidated, it was not possible to understand how chromosome ends are protected and replicated. The first telomeres to be sequenced and shown to consist of an array of short tandem repeats (TTGGGG) were from the ciliate Tetrahymena [6]. Subsequently, it was found that nuclear telomeres in a wide variety of eukaryotes have a very similar primary sequence organization. For example, in vertebrates, the chromosomes terminate with multi-kilobase arrays of the hexa-nucleotide repeat TTAGGG [7, 8], and plants frequently terminate with long blocks of TTTAGGG repeats [9]. The motif of one strand rich in Gs and the other thus rich in Cs appears to be nearly universal across species. Telomeres were shown to frequently contain an extension of the G-rich strand as a 3’ single stranded (ss) overhang, which in human cells is on the order of 50–200 nt [10]. Key discoveries included the observation that telomeres of primary mammalian cells undergo a progressive erosion during repeated passage, a process linked to the end replication problem and nucleolytic resection necessary to create the ss overhangs [11, 12]. Once telomeres shorten to a certain critically short length (around 3–5 kb in human cells), cells enter senescence and, with further shortening, apoptosis (reviewed in [13]). This observation led to a model in which the length of telomeres in vivo is monitored, possibly by a protein counting mechanism, whereby reaching a certain critical (short) length triggers entry into senescence. How this might occur was not clear, nor did it explain how chromosomes hide their ends from erroneous detection as double strand breaks (DSBs). It had been well established that in vivo, even a few DSBs, if not repaired, trigger potent DNA damage response pathways controlled by the ATM and ATR factors and can lead to apoptosis (reviewed in [14]). In humans, telomere shortening is a barrier to unlimited proliferation and serves as a potent tumor suppressor mechanism [15]. We now understand that telomere-dependent tumor suppression is regulated through loss of the protective telomere functions postulated by Muller and McClintock.
The early days of telomere molecular biology were largely focused on the budding yeast Saccharomyces cerevisiae. This led to breakthroughs including evidence for the telomere elongating enzyme telomerase [16], the discovery of telomere extension through homologous recombination (HR) [17, 18] and the identification of specific protein factors that regulate telomere function (reviewed in [19]). Unlike vertebrate telomeres, however, S. cerevisiae telomeres consist of irregular repeat arrays that are typically hundreds of base pairs as opposed to multiple kilobase pairs in length. Additionally, budding yeast lack telomeric proteins that are direct homologs of their vertebrate counterparts. While S. cerevisiae telomeres are not conducive to telomere looping as described here, nonetheless, it was suggested by Grunstein [20] that they may fold back through protein-protein interactions. Evidence supporting this hypothesis was subsequently described [21–23], although the telomere looping in S. cerevisiae is still a matter of debate [24].
Concurrent with the pioneering studies in budding yeast, research using the ciliated protozoan Oxytricha nova provided a protein-based model of chromosome end protection. In addition to a set of chromosomes in a micronucleus, Oxytricha contains a macronuclear pool of amplified linear single gene DNA fragments, each capped by a 36 bp tract of telomere repeats. The Zakian and Cech labs identified a protein complex that binds to Oxytricha macronuclear telomeres and hypothesized that this protected DNA ends from nucleases and covalent ligation [25, 26]. Based on this paradigm, it was suggested that higher eukaryotes would follow with specific proteins that bind and cap the chromosome ends.
The first mammalian telomere-specific protein was identified in the early 1990s by the de Lange laboratory. Titia de Lange developed an interest in telomeres during her doctoral studies on trypanosome surface antigens with Piet Borst in Holland. Following her post-doctoral work at UCSF with Harold Varmus, she established her own laboratory at the Rockefeller University. Searching for human proteins that bind specifically to double stranded (ds) telomeric DNA using (TTAGGG)n fragments in pull-down experiments, they identified a protein they termed TRF1 (telomere repeat binding factor 1) [27]. TRF1 was found to contain a single Myb domain conferring modest affinity for TTAGGG repeats in dsDNA. It also possesses a homodimerization domain that allows it to form stable dimers. With two Myb domains, the dimers exhibit a much higher affinity for telomeric dsDNA, and TRF1 was found to bind along the length of telomeric dsDNA rather than at the ends, arguing that it was not an end-capping protein.
Jack Griffith’s long term focus has been on developing and applying electron microscopic (EM) methods for the visualization of DNA-protein complexes and, in 1975, he applied them to provide the first EM images of nucleosomes where the amount of DNA in the nucleosome and the length between adjacent ones could be determined [28]. His laboratory at the University of North Carolina later focused efforts on in vitro homologous recombination (HR) reactions driven by the recA and UvsX proteins [29]. A collaboration with Robert Wells at the University of Texas led to studies of the structure of expanded triplet repeat DNAs. However, at that time, a limitation for the EM work was the lack of proteins with specificity for these DNAs. In a seminar at UNC, de Lange described her work on TRF1 which bound to a repeating di-triplet (TTA and GGG), and this suggested that an EM study of TRF1 and telomeric DNA might prove of interest. This initiated a highly productive collaboration that has spanned over 20 years.
2. Visualizing human telomere proteins in vitro
Visualization of TRF1 (purified from insect cells in the de Lange lab) bound to a plasmid DNA containing a long array of TTAGGG repeats [30] produced images of TRF1 dimers and tetramers spaced along the repeat blocks, with binding being relatively non-cooperative. At higher densities of TRF1, the tracts were fully coated (Fig. 1A) and, often, two TRF1-bound DNAs were seen synapsed via protein-protein interactions (Fig. 1B). Otherwise, there was no preference for unusual junctions or structures in the telomeric DNA.
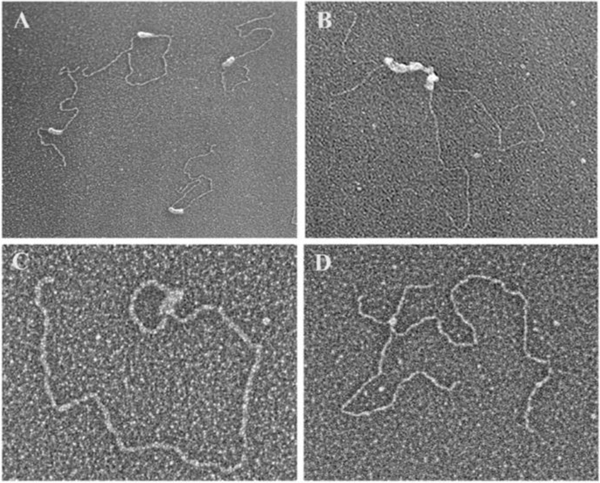
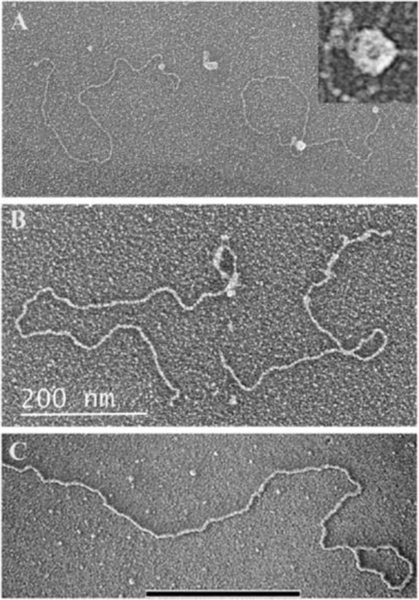
Fig. 1. TRF1 and TRF2 binding and sculpting DNA in vitro.
(A) Purified TRF1 binding to a linear ~3 kb plasmid DNA containing a single tract of 162 TTAGGG repeats roughly 1/3rd the distance from one end [30]. TRF1 fully coats the repeat tract. (B) TRF1 coating a 780 bp tract of TTRGGG repeats in the ~3 kb plasmid frequently results in a parallel synapsis of two DNAs at the repeat tract. Image from [30]. (C) TRF2 forms a t-loop on a model telomere DNA. The DNA backbone is ~3 kb and contains a 576 bp TTAGGG tract at one end including a 3’ ss G-rich overhang. The t-loop junction is coated with TRF2. (D) In this experiment from [39] a 3.5 kb pRST5 plasmid containing a replication fork within a 585 bp block of TTAGGG repeats was allowed to generate a chicken foot structure via slippage of the replication fork within the telomeric repeat tract. Addition of TRF2 revealed a strong preference for its binding at the Holliday junction on the chicken foot structure. Electron micrographs shown in reverse contrast, samples were directly mounted onto thin carbon foils, rotary shadow-cast with tungsten in a high vacuum and examined at 40 kV in a transmission electron microscope. See papers for details.
In parallel, the de Lange lab and, concomitantly, Eric Gilson’s team, identified a second factor similar to TRF1 [31, 32]. This protein, telomere repeat binding factor 2 (TRF2), also contains a single Myb domain and a homodimerization domain which, together, generate dimers with an affinity for telomeric dsDNA similar to TRF1. The two proteins have 27% homology at an amino acid level; however, while TRF1 has an acidic N-terminal domain, the N-terminal domain of TRF2 is basic and, moreover, is similar to the basic C-terminus of p53.
Concurrent with the in vitro EM studies, the de Lange lab found that TRF2 conferred chromosome end-protection. They accomplished this through a dominant negative approach of over-expressing TRF2 alleles that contained a wild type homodimerization domain, but which lacked the Myb domain, thus eliminating their affinity for telomeric DNA [33]. When the mutant protein formed dimers with wild type protein in the cell, the heterodimers possessing only one Myb domain exhibited greatly reduced affinity for telomeric dsDNA. Inhibiting TRF2 induced covalent end-to-end chromosome fusions, cell death in transformed cell lines, and senescence in primary human cells [33–35]. Of note, the senescence phenotype induced in primary cells with dominant negative TRF2 expression was similar to replicative senescence in aged cells when telomeres had eroded to a critically short length [35]. This forced a reevaluation of existing models on how short telomeres trigger senescence. Because primary cells with long telomeres became senescent in the absence of functional TRF2, this suggested senescence was not regulated by telomere length and a protein counting mechanism per se. Instead, telomeres appeared to regulate proliferative arrest through alteration of a protective chromosome end-structure dependent on both elongated telomeres and adequate TRF2 binding.
EM images of plasmid DNA containing telomeric repeat tracts bound by TRF2 were confounding in that TRF2 did not show an even coverage along the tracts as had TRF1, but rather bound in patches and generated complex interactions between telomeric sequences within the same DNA molecule, and between two or more DNAs (Griffith and de Lange, unpublished results). These structures evoked images of the complex strand invasion/exchange structures Griffith had observed in studies with recA and UvsX proteins, when the interacting with DNAs contained shared homologies.
3. Rationale for proposing telomere looping and the first experiments
New ideas seldom appear fully fleshed-out, but rather evolve. Based on their results with TRF1, Griffith had suggested to de Lange that telomeres might fold back on themselves. However, the idea of a loop stabilized by a strand invasion event arose from pondering images of the structures generated by TRF2 and considering the nature of the telomeric DNA as seen through the lens of homologous recombination (HR). In specific, telomeres contain both elements required in recA/uvsX/Rad51-catalyzed reactions: a linear ss DNA with a 3’ terminus and a ds DNA with a sequence homologous to that of the ss DNA. For telomeres, both elements are present in the same molecule and HR would generate a lasso-like looped structure composed of a circle and a tail. Proposing this on a visit to the Rockefeller, it did not miss Griffith and de Lange’s attention that such looping would provide a simple and elegant way for the end of the telomere to be hidden from the DSB recognition and repair machinery. The overarching question, then, was how to combine their very different technologies to test this possibility. The Griffith laboratory contributed EM visualization and expertise in building large complex DNA substrates and the de Lange group provided knowledge of the isolation and characterization of telomeric DNA and had the purified telomeric factors.
A first effort focused on testing the idea with purified TRF2 and a model telomere DNA. Rachel Stansel, a graduate student in the Griffith laboratory, developed a clever slipped-structure replication scheme that added up to 2 kb of TTAGGG repeats to the end of a linear pGEM plasmid, which could then be resected to generate a long 3’ ss tail. When this artificial telomere was incubated with purified TRF2, a significant fraction (up to 20%) of the DNA, as seen by EM, was arranged into a lasso-like structure with the circular portion falling within the size range (1–2 kb) of the telomere repeat block. A distinct mass of TRF2 protein was present at the circle-tail junction (Fig. 1C). It could not be determined whether the loops were formed by a strand invasion event or just held back (fold-back structure) by the mass of TRF2. To distinguish between these possibilities, a psoralen/UV crosslinking strategy, commonly used in EM studies and originally developed by John Hearst, was employed. Indeed, it had been used by Tom Cech and Mary Lou Pardue in 1976 to probe for unusual structures in mammalian DNA [36]. Psoralen intercalates into duplex DNA and, in the presence of UV light, generates a covalent crosslink between the two strands of the duplex at A/T steps. If a D-loop is generated by insertion of the telomeric 3’ ss tail back into the preceding duplex sequence, psoralen should crosslink the tail to the duplex, freezing the telomere loop. Thus, the loops should remain fixed following removal of TRF2. Psoralen/UV crosslinking has remained a key step in all subsequent work. When the crosslinked DNA was deproteinized and spread on an air/water interface for EM, looped molecules were observed at the same frequency as the looped structures that formed in the presence of TRF2 [37]. As controls, incubations with TRF1 or tankyrase failed to generate loops. Stansel’s experiments provided the encouragement to tackle a much harder problem, which was to isolate and visualize telomere loops directly from mammalian cells.
The logistics of visualizing telomeres arranged into loops by EM are daunting. For every microgram of mammalian cell telomeric DNA there are 3000 to 6000 micrograms of genomic DNA. A fraction of a microgram is needed for EM analysis and this required starting with ~5×108 cells. The greatest challenge was separating this infinitesimal amount of telomeric DNA from the bulk genomic DNA. de Lange suggested the use of multiple 4-base cutters which would fragment the genomic DNA but not the telomeric DNA, together with the use of the HeLa 1.2.11 cell line, which has telomeres in the 15–20 kb range as contrasted to ~10 kb for most other HeLa lines. The Griffith lab had used gel filtration columns to separate DNAs of different sizes in the past, and this method was scaled up greatly by using 3-foot-long columns containing 200 mL of agarose beads. As verified by blotting with telomeric probes, this was effective in allowing the long telomeric DNAs to elute prior to the flood of short genomic fragments, and provided DNA in a clean buffer for EM examination [37].
The first experiment was carried out by Laurey Comeau in the Griffith laboratory. Examining DNA spreads from the early eluting fractions of the agarose bead column, Comeau observed looped molecules in the EM (Fig. 2A). The telomere loop structures were called t-loops in short, a term that has since become adopted by the telomere field. Had this first effort not been successful, the project may well have been left to just the in vitro studies. However, with exciting images of t-loops formed in vivo, the work continued, and results were replicated multiple times and extended to other cell lines and tissues (mouse liver, human white blood cells) [37]. This resulted in a large collection of t-loop examples. Key observations, to be confirmed later by super resolution light microscopy, were that only a fraction of the telomeric DNA is arranged into loops (at most 20% in the EM studies), and the location at which the end of the telomere is inserted back into the duplex DNA is highly variable, from close to the telomere end to proximal to the sub-telomeric sequences [37, 38]. T-loops from mouse tissue, as expected, were larger than ones from human sources, and HeLa 1.2.11 t-loops were larger than ones from HeLa S3 cells. Psoralen/UV crosslinking was important in retaining the loops, which were otherwise often lost, likely due to their exposure to prolonged incubation at 55 degrees during the purification. A mixing experiment involving non-crosslinked HeLa1.2.11 DNA (long telomeres, large t-loops) mixed with crosslinked HeLa S3 DNA (short telomeres, small t-loops), where only short t-loops were seen, argued against the loops being formed during the preparation [37]. This first study provided a tribute to the power of the collaboration between two groups with very different sets of expertise and suggested a structural solution for how telomeres protect the ends of chromosomes. The findings, however, raised many new questions, in particular the specific role of TRF2 in sculpting DNA.
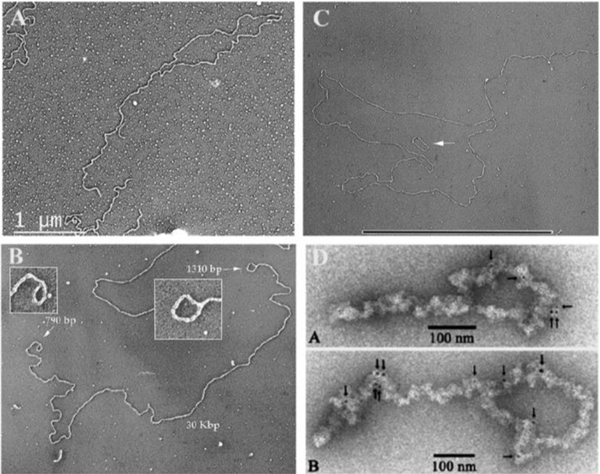
Fig. 2. Examples of t-loop from across the phyla.
(A). T-loop isolated from HeLa 1.2.11 cells using the methods described in [37]. (B) Mini-chromosome from Trypanosoma brucei showing t-loops at both ends. The mini-chromosomes contain long TTAGGG tracts at each end. Image from [44]. (C) t-loop isolated from shoots and roots of common garden peas. Image from [45]. Small circular DNA is a 3 kb plasmid. Bar is equivalent to 10 kb in length. (D) Figure from [47] showing t-loops from chicken erythrocyte cells isolated as chromatin and stained with gold tagged beads carrying antibodies to TRF1.
In vitro studies have shown that, in interactions with DNA, unlike TRF1 which binds along the length of duplex telomeric sequences, TRF2 is much more structure specific. This specificity may reside in part in the highly basic N-terminus which has a striking similarity to the basic C-terminus of p53. Both proteins show a high affinity for DNA junctions: ds/ss junctions, replication forks, and Holliday junctions [39, 40] (Fig. 1D). Indeed, as seen by EM, the p53 and TRF2 basic termini alone will bind such junctions, and, for TRF2, binding occurs even in the absence of TTAGGG repeats [39]. The combination of TRF2’s affinity for telomeric repeats conferred by the Myb domain, and its specificity for junctions conferred by the N-terminus, determines its strong affinity for the ss/ds junction at the telomere terminus. Additionally, Gilson and colleagues have examined TRF2 for other activities and found that TRF2 possesses a topoisomerase-like capability to wrap DNA, and that hindering TRF2 DNA wrapping impeded telomere protection [41, 42].
4. T-loops in diverse eukaryotic phyla
After the initial discovery of t-loops, several studies that confirmed the presence of t-loops in diverse eukaryotes followed. Immediately after the publication of the Cell study by Griffith and de Lange, David Prescott at the University of Colorado indicated in a personal communication that, several years earlier, his postdoctoral fellow K.G. Murti had obtained EM images showing looped DNA at the ends of Oxytricha micronuclear chromosomes. These micrographs were found and the study subsequently published [43]. The loops were relatively large and, of note, were observed without psoralen and UV crosslinking, arguing that t-loops were not an artifact of crosslinking. They noted that, while the linear single gene DNAs in Oxytricha macronuclei are capped by the heterodimeric telomeric binding protein previously found [25, 26], the long canonical micronuclei chromosomes may be protected by t-loops.
Trypanosoma brucei nuclei harbor multiple ~20–30 kb mini chromosomes. Each end is terminated with several kilobases of TTAGGG repeats and a 3’ ss overhang. In this collaboration between the Griffith lab and the Cross and de Lange laboratories at the Rockefeller, it was observed that, in the absence of crosslinking, T. brucei mini-chromosomes exhibit ss tails at both ends that stain with the E. coli single strand DNA binding protein. However, when psoralen/UV crosslinking was added to the isolation protocol, numerous examples of DNAs with loops at both ends were found (Fig. 2B) [44].
These observations provided an example of chromosomes with ss overhangs and loops at both ends, finding that had been surmised but not demonstrated directly in any system.
Plant telomeres can be extremely long, and in most species the repeat has an additional thymine base: TTTAGGG. Tony Cesare, then a graduate student in the Griffith lab, isolated nuclei from Pisum sativum (pea) cotyledons, purified and sequenced the telomeric DNA, identifying the TTTAGGG repeat sequence. Further analysis revealed telomeres of up to 100 kb in peas, with t-loops containing circular portions of up to 80 kb in circumference [45] (Fig. 2C). Additionally, the Griffith lab collaborated with the Karlseder lab at the Salk Institute and found t-loops in telomere enriched DNA from the nematode Caenorhabditis elegans [46]. Further, they observed that when the C. elegans telomere proteins CeOB1 and CeOB2 were added back to t-loop-enriched fractions, the proteins localized to t-loop junctions. Cumulatively, the findings from P. sativum (TTTAGGG repeats), C. elegans (TTAGGC), and Oxytricha (TTGGGG) demonstrated that t-loops are present in eukaryotic species with divergent telomere sequences and binding proteins, signifying the evolutionary importance of t-loop structures.
A further confirmation of t-loops appeared from Chris Woodcock’s laboratory at the University of Massachusetts in a study where they took advantage of the fact that chromatin in quiescent cells of avian erythrocytes and mouse splenocytes is not attached to the nuclear matrix as firmly as it is in most active cells. This allowed them to isolate telomeres in a chromatin state, following restriction cleavage and sucrose gradient sedimentation. Their images revealed looped chromatin molecules that stained with gold-tagged antibodies to TRF1, which were scattered along the length of the telomeric chromatin fragments [47] (Fig. 2D). This work provided further evidence that t-loops are not induced by crosslinking and added avian species to the growing list of eukaryotes with looped chromosome ends.
5. Discovery of t-circles and their link to t-loops
The vast biodiversity of yeasts began to be explored in the late 1960s by a handful of laboratories, including that of Ladislav Kovac at Comenius University in Bratislava, Slovakia. Kovac and his colleagues introduced several nonconventional yeast species as model organisms for addressing general biological processes [48, 49]. One area of Kovac’s research related to telomeres was his discovery that mitochondrial DNA (mtDNA) in the yeast Candida parapsilosis was linear [50]. The mitochondrial telomeres of C. parapsilosis were subsequently explored by two of Kovac’s students, Jozef Nosek and Ľubomir Tomáška. In collaboration with Hiroshi Fukuhara, Nosek found that (i) the terminal mtDNA sequences in C. parapsilosis consisted of a variable number (up to 12) of long (738 bp) tandem terminal repeats, (ii) the last repeat is incomplete (~400 bp), and (iii) the very end of the molecule contains a ~110 nucleotide long ss overhang with a 5’ terminus [51]. The Tomáška/Nosek lab later purified a protein that exhibited a preference for the C. parapsilosis telomeric oligonucleotide and termed it mitochondrial telomere-binding protein (mtTBP) [52]. mtTBP was found to belong to a large family of tetrameric SSB proteins, suggesting that mtTBP fulfills dual functions in C. parapsilosis mitochondria: participation in “standard” DNA transactions and protection of the telomeric overhang [53]. However, they had no proof that mtTBP binds to mitochondrial telomeres in vivo. In 1999, Tomáška visited the Griffith lab at UNC to visualize the interaction between mtTBP and mtDNA. Immunogold EM experiments showed mtTBP binding to the very end of C. parapsilosis mtDNA molecules demonstrating that, similar to their nuclear counterparts, mitochondrial telomeres are protected by a dedicated protein [54] (Fig. 3). This result initiated a collaboration that has spanned two decades and represents yet another example of a highly productive interaction between two laboratories with very different sets of expertise focusing on a single problem.
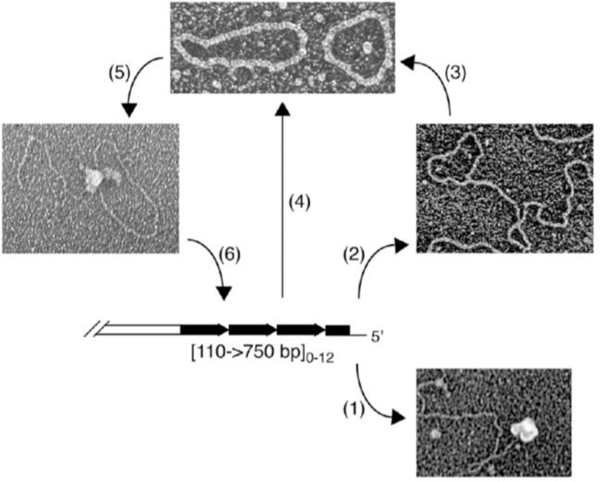
Fig. 3. T-loops and t-circles observed at yeast mitochondrial and nuclear telomeres.
(A) Several yeast species contain linear mitochondrial genomes with various types of organization of telomeres [116]. Here, mitochondrial telomeres are composed of an array of tandem repeats whose sequence and length is species-specific and a long (~100 nt) 5’ single-stranded overhang [51]. In C. parapsilosis this overhang can be protected by (1) mitochondrial telomere binding protein [52–54], or (2) form a protective t-loop [58]. (3) T-loops may be resolved into t-circles that can also arise by (4) recombination between tandem repeats [55]. Once formed, (5) t-circles can undergo rolling-circle replication [57] generating a long array of telomeric repeats that (6) can integrate back into the main genome [59]
A central question was how mitochondrial telomeres of C. parapsilosis are replicated. The results of two-dimensional electrophoresis experiments indicated that mitochondrial telomeric sequences in C. parapsilosis were present in both the linear mitochondrial genomes and as extrachromosomal DNA fragments. Indeed, EM visualization of the bulk mtDNA showed the presence of large linear molecules and a few small circles. Alkaline lysis of the purified mitochondria revealed very tightly twisted DNA rods, typical of highly supertwisted DNA and, upon nicking, open circular molecules (which the authors termed t-circles), whose sizes corresponded to multiples of the 738 bp telomeric repeat [55] (Fig. 3). T-circles were also detected in other yeast species whose linear mtDNAs also terminate in arrays of tandemly repeated sequences [55, 56]. Importantly, analysis of the C. orthopsilosis and C. metapsilosis strains lacking mitochondrial t-circles revealed that the original linear mitochondrial genome had circularized [56]. This suggested that mtDNA t-circles may participate in a mechanism of telomere maintenance.
Further EM examination of the mtDNA from C. parapsilosis revealed the frequent presence of tailed circles suggestive of rolling-circle intermediates [57] and, when the mitochondria were crosslinked with UV and psoralen paralleling the approach used for mammalian t-loop isolation, ~5% of molecules showed terminal loops, suggestive of telomeres undergoing recombination [58] (Fig. 3). These observations were complemented by electrophoretic analyses demonstrating a strong incorporation of nucleotides into the telomeric region of linear C. parapsilosis mtDNA. This led to a model in which t-circles are generated by intramolecular recombination between the repeats (or possibly via t-loops formed by the invasion of the 5’ overhang), followed by extrachromosomal amplification of telomeric repeats via rolling-circle replication of the t-circles, and finally the introduction of these sequences back into the main mtDNA molecules by recombination [59].
Tony Cesare in the Griffith lab became interested in the macromolecular structure of telomeres in human ALT cells [60]. Employing the psoralen/UV crosslinking and telomere enrichment protocol, he observed t-loops by EM at the expected frequency and size distribution in the telomere enriched fractions from GM847 and VA-13 ALT cell lines (Fig. 4A). He also observed a considerable concentration of variably sized duplex circles in the same DNA population. Southern blots of telomere restriction fragments separated by 2D pulsed-field gel electrophoresis confirmed the presence of circular telomeric DNA species in the VA-13 and GM847 ALT cells, with no evidence of t-circles in telomerase positive HeLa cultures or primary foreskin fibroblasts. Moreover, the size distribution of t-circles from ALT cells closely followed the distribution of the circular loop portion of the t-loops isolated in the same cell line (Fig. 4B). This was the first direct evidence that t-circles arise from t-loop cleavage. This also provided the first demonstration that t-circles exist in mammalian systems, and marked an important step in efforts to understand telomere length dynamics in ALT cells typified by rapid telomere extension and deletion events [61]. Thereafter, t-circles were observed in diverse eukaryotic species in the presence of ALT-like telomere extension [46, 62]. Additionally, multiple labs have since shown that altering any of several key factors in telomere maintenance, including TRF2 [63], WRN [64], BLM [65], and RTEL [66], increases t-circle abundance. T-circles were later identified as byproducts of telomere shortening events that normally regulate telomere length in germ cells, and which accompany the deleterious phenotypes of Hoyeraal-Hreidarsson syndrome [67, 68]
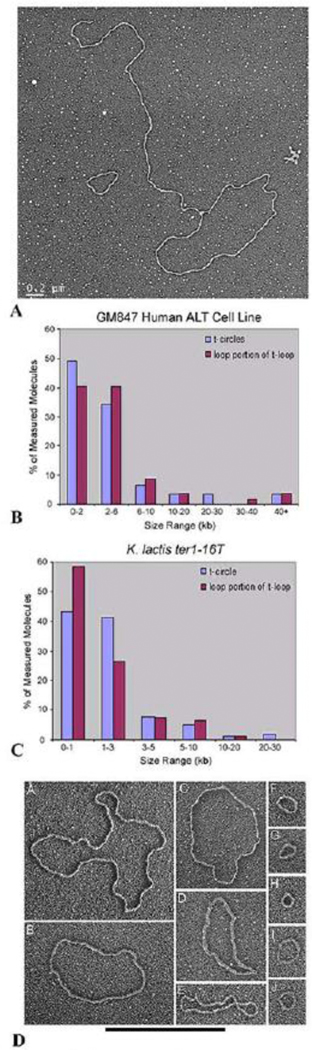
Fig. 4. T-circles from human ALT and K. lactis cells.
(A) Micrograph of a t-loop and t-circle isolated from GM847 ALT cells following psoralen/UV crosslinking as in [37]. (B) Distribution of circle sizes measured from the circular portions of t-loops isolated from GM847 cells (red) or t-circles (blue) from the same cells. Data from [60]. (C) Distribution of circle sizes measured from the circular portions of t-loops isolated from K lactis cells (red) or t-circles (blue) from the same cells. Data from Cesare et al (2008) [62]. (D). Panel of examples of t-circles isolated from K. lactis (ter1–16T) cells. Image from [62].
The original t-circle experiments in ALT cells revealed a mixture of closed, nicked, and gapped double strand DNA circles. Small and mostly ss circular DNAs of the G-rich or C-rich sequence were later reported in ALT cells [69]. These mostly ss species likely result from the exonucleolytic degradation of nicked duplex circles and, in the initial EM experiments, avoided detection due to their small size and fractionation with the bulk genomic DNA. The presence of circular telomeric DNAs is now commonly assayed through detection of Φ29 polymerase-mediated rolling circle amplification products [69, 70]. While it remains unclear exactly which circular species are extended by Φ29 polymerase, circular telomeric molecules have become the experimental standard for ALT positivity [69].
6. Yeast as a model organism for studying t-loop function
Schizosaccharomyces pombe (or fission yeast) provides a powerful model system for telomere studies as its chromosome ends are protected by a protein complex composed of essentially the same subunits as in the mammalian shelterin complex [3]. The principal component of the fission yeast shelterin is Taz1, a functional homologue of mammalian TRF1 and TRF2 [71]. The collaboration between the Griffith and Tomáška/Nosek groups continued with an examination of the DNA binding properties of Taz1. A model telomere template consisting of a plasmid backbone followed by 518 bp of the consensus Sch. pombe telomere repeat (5’-GGTTACA-3’) was generated. When this model telomere template, ending in a 3’ ss overhang, was incubated with purified Taz1, t-loops formed at a frequency approaching 13% of the DNA templates. Paralleling results with TRF2, templates with blunt ends or non-telomeric overhangs were deficient in t-loop formation. Taz1 also bound along the array of telomeric repeats, resembling the pattern of TRF1 binding. Some t-loops were larger in size than the telomeric tract. This observation, the high frequency of DNAs with Taz1 at the very end of the telomeric tract, together with a donut shape of the Taz1 oligomer, suggest that Taz1 binds the 3’ ss overhang and then extrudes a loop that grows in size as the donut slides along the duplex DNA [72] (Fig. 5A). These studies provided the first example of a yeast homolog of mammalian TRF1 and TRF2 able to promote formation of t-loops in vitro.
Fig. 5. T-loops in yeast: examples from in vitro and in vivo experiments.
(A) t-loops formed by purified Taz1 protein of S. pombe on a model telomere template containing a block of 518 bp of the S. pombe consensus sequence (5’-GGTTACA-3’) located at one end. Taz1 forms large donut-like oligomers which bind the telomeric repeats as described in [72]. Model DNAs measure 2.9 kb in length. Following end binding, the DNA appears to thread through the hole in the donut (inset panel A) followed by the donut sliding along the DNA to form a loop which can exceed the length of the telomeric tract. (B) Purified Tay1 protein from Y. lipolytica binds along a 810 bp tract of 5’-GGGTTAGTCA-3’ repeats at one end of a linear ~3 kb model telomere DNA. Tay1 coats the repeat tract in a mode similar to TRF1 binding, but also generates looped structures similar to the looping induced by TRF2. Image from [77]. (C) T-loop DNA isolated from ter1-16T mutant K. lactis cells containing long telomeres as described in [62]. Bar in C is equivalent to 3 kb.
Mining the wealth of yeast species with different telomeric repeats, the Tomáška /Nosek laboratory turned to Yarrowia lipolytica. This species is of particular interest as the telomeres consist of regular repeats of TTAGTCAGGG, where a 4 nt long spacer is embedded within a mammalian-type repeat. Y. lipolytica is located at the base of the phylogenetic tree, near the branch leading to basidiomyces such as Ustilago maydis, harboring a TTAGGG repeat, supposedly the ancestral telomeric repeat for fungi [73, 74]. Searching the genome of Y. lipolytica, the Tomáška/Nosek group found both the catalytic and RNA subunits of telomerase [75, 76], but failed to identify homologues for Rap1 or Taz1. Instead, they found a novel protein containing two Myb domains, each exhibiting a high degree of similarity to the Myb domains of human TRF1 and TRF2, and named it Tay1 (Yarrowia lipolytica 1). Tay1 was purified and tested on a model Y. lipolytica telomere template containing 810 bp of repeats. The EM studies showed that Tay1 binds along the telomeric tract as dimers and larger oligomers with a pattern resembling that of Taz1 and TRF1 (Fig. 5B). It also showed a preference for ss/ds junctions (as does TRF2), and an ability to synapse two or more DNAs together (as do TRF1 and Taz1). Finally, paralleling TRF2 and Taz1, Tay1 can remodel the templates, with up to 20% of the protein-bound DNAs arranged into t-loops [77]. Interestingly, the affinity of Tay1 for TTAGGG repeats is about 5-fold higher than for the Y. lipolytica repeats and 8-fold higher than that of either TRF1 or TRF2 for the mammalian repeats [78].
The in vitro data suggested that telomere looping might occur in fission and budding yeast. However, it remained difficult to isolate native telomeres from cultured yeast. Unlike mammalian telomere restriction fragments that are sufficiently long to be separated from bulk genomic restriction fragments using size exclusion, wild type yeast telomeres are only hundreds of base pairs and migrate with the genomic material. To circumvent this, the Griffith lab teamed up with Michael McEachern’s group at the University of Georgia to exploit the unusual telomere characteristics of the budding yeast Kluyveromyces lactis. As a post-doc with Elizabeth Blackburn, McEachern discovered that K. lactis telomeres are composed of 25 base pair repeats. Mutating the template sequence of the K. lactis telomerase RNA (Ter1) conferred inclusion of mutant telomere repeats that hindered telomere protein binding [79, 80]. One such mutation, ter1–16T, diminished telomere affinity for K. lactis Rap1 [81]. This resulted in highly recombinogenic and elongated telomeres approximating those in ALT-positive human cells. Similarly, mutation of the K. lactis stn1 gene also induced similar recombinogenic and elongated ALT-like telomeres [82].
Previous studies from the McEachern laboratory provided support for a telomerase-independent roll and spread model of telomere maintenance. Genetic evidence indicated that small, approximately 100 bp DNA circles acted as templates for rolling circle replication from chromosome ends in ter1–16T cells, followed by gene conversion events to extend adjacent telomeres [83]. The Griffith/McEachern collaboration also identified ds and small ss nuclear t-circles in low molecular weight DNA isolated from ter1–16T cells (Fig. 4D) [81]. These results were consistent with t-circles previously identified in ALT cells [60], and the ss telomere circles characterized in ALT cells years later [69]. Further, it was evident that K. lactis ter-16T cells had telomeres of 10 kb or more that were sufficiently long to enrich in gel filtration columns. This inspired using K. lactis for the first t-loop study in which cells were genetically altered, to probe for putative t-loop structures.
EM examination of K. lactis telomeres was a collaboration between Tony Cesare in the Griffith lab and several members of the McEachern lab [62]. By modifying the telomere enrichment protocol from mammalian cells, they observed a low percentage of t-loops in psoralen/UV crosslinked telomere enriched fractions from ter1–16T K. lactis cells (Fig. 5D). Consistent with ALT cells, ter1–16T cells contained t-circles whose diverse size correlated with the circular portion of the t-loops isolated from the same culture (Fig. 4C). Suppressing recombination in ter1–16T cells through RAD52 deletion prevented t-circle formation, consistent with t-circles arising due to HR-dependent t-loop resolution. Similar heterogeneously sized t-circles were observed in K. lactis stn1 mutant cells with recombinogenic and ALT-like telomeres. However, in contradiction to expectations, the percentage of t-loops observed in the telomere enriched fractions from ter1–16T cells was reduced when the ter1–16T phenotype was suppressed by RAP1 over-expression, or when HR was inhibited with RAD52 deletion. This suggested that t-loops in K. lactis were an outcome of telomere recombination, which, when left uncontrolled, resolves into t-circles. In other yeast species it was also found that formation of t-circles results from telomere recombination [84–86]. This finding presented a dichotomy between mammalian and yeast systems. In mammalian cells, t-loops were observed in the native telomeric DNA of primary and telomerase-positive human and murine cells that suppress telomere recombination. Ideally, a study examining the native yeast telomere structure would follow, to determine if t-loops are present in wild type budding yeast telomeres. However, it remained beyond our technology to isolate native yeast telomeric DNA for EM studies.
7. Visualization of t-loops by STORM microscopy
In the early to mid 2000s, members of the de Lange and Griffith labs tried several EM-based approaches for determining whether loss of TRF2 in vivo would lead to a reduction in t-loops. These attempts often utilized overexpression of the dominant negative TRF2 alleles, but, usually for technical reasons, results were not consistent. However, in the following years the de Lange group developed powerful murine knockout models to conditionally delete telomere proteins individually or in tandem in mouse embryonic fibroblasts. Further, they combined these conditional shelterin knockout models with deletions of genes functioning in DNA damage response and repair pathways. This identified critical protective functions for shelterin proteins at chromosome ends and novel information on DNA repair (expanded in detail below). Of note, this included showing that a TRF2 deletion resulted in chromosome end fusions only in the presence of a functional ATM pathway [87]. These powerful genetic models set the stage for a groundbreaking collaboration between the de Lange laboratory and Xiaowei Zhuang at Harvard.
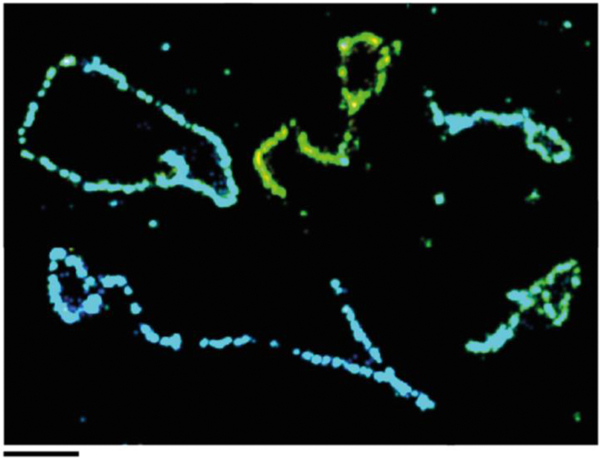
Zhuang had pioneered stochastic optical reconstruction microscopy (STORM): a method of super-resolution imaging of fluorescently labeled molecules capable of 20 nm resolution [88, 89]. Ylli Doksani, a postdoctoral fellow in the de Lange lab, devoted several years to adapting the t-loop isolation method used for EM to STORM imaging [38]. Key steps involved the lysis of cells in high salt with a trace of SDS and spreading the DNA over a glass slide by cyto-centrifugation. The telomeres were specifically labeled with a peptide nucleic acid probe using fluorescence in situ hybridization (Fig. 6). This identified the telomeric DNA within the vast excess of genomic DNA, thus eliminating one of the greatest barriers to the EM approach. Through STORM imaging, Doksani obtained results consistent with prior EM results: specifically, t-loops were present in approximately 20% of telomeres from psoralen/UV crosslinked samples, with similar variability in loop size and in the point of insertion of the chromosome end into the duplex telomeric DNA. The physical length of the fluorescently labeled telomeres also correlated with expectations based on telomere measurements using standard Southern blot methods.
Fig. 6.
Visualizing t-loops using light microscope STORM imaging. Montage of t-loop molecules imaged by STORM. From[38]. Courtesy of Ylli Doksani, IFROM, Milan, Italy.
With a visualization method in hand, Doksani combined STORM imaging with the murine cell lines to examine the effects of the elimination of telomere proteins, individually or in tandem. Consistent with prior studies, Doksani observed that TRF2 deletion resulted in covalent telomere-telomere fusions following activation of the ATM-dependent DNA damage response (DDR). Six days after a TRF2 deletion in ATM competent cells, STORM imaging revealed a reduction in looped telomeres but an increase in elongated linear molecules consistent with telomere-telomere fusions. At the same time, STORM imaging three days after TRF2 deletion in ATM null cells revealed linear telomere molecules the length of a single chromosome end. Overall, the loss of TRF2 resulted in a ~6-fold depletion of t-loop species. In other experiments, deletion of the remaining telomere proteins (TRF1, RAP1, Pot1a,b and Tin2) showed that only depletion of TRF2 led to a significant loss of t-loops. This definitively demonstrated, for the first time, that TRF2 functions to mediate t-loop formation in mammalian cells.
The importance of this collaboration cannot be underestimated. The unique capabilities of the de Lange and Zhuang laboratories combined with Doksani’s tenacity facilitated a new era in t-loop research. This directly enabled studies described in the final section of this review by the Boulton and/or Cesare labs that applied super resolution microscopy to directly probe questions of t-loop function in chromosome end protection and mechanisms of telomere replication.
8. Telomere transcription, TERRA, R-loops, and a new model for t-loop formation
While the STORM studies pointed to the essential role of TRF2 in observing t-loops in vivo, they did not distinguish between TRF2 actively forming the loops itself from a role in which loops may be formed via several routes, but in each case being stabilized by TRF2 within the full shelterin complex. The in vitro reactions driven by TRF2 alone are not efficient (at most 20% of the templates are arranged into t-loops). Furthermore, strand insertion and joint formation require that the helix be opened and TRF2 has not been shown to have ATPase or helicase activity. This raised the question of what other pathways in the cell might facilitate helix opening to allow strand insertion. The discovery of telomeric transcripts and telomeric R-loops suggested one explanation, specifically that the helix remains open at an R-loop.
Studies from the Lingner [90] and Blasco [91] groups showed that mammalian telomeres are transcribed, and reported transcripts as long as 9000 nt. The predominant transcript in mammalian cells is from the C-rich strand, producing a G-rich RNA termed TERRA. Subsequent studies from many laboratories (reviewed in [92, 93]) revealed this to be a commonality from yeast to human cells. TERRA plays a structural role in which it remains associated with the telomere, and interacts with a cadre of proteins, including TRF1, TRF2, HP1 and ORC [94], hnRNPA1 [95], and the chromatin remodeling complex NoRC [96]. Coincident with the discovery of TERRA, the importance of R-loops in the genome was being newly appreciated (reviewed in [97]. These structures consist of an RNA transcript remaining bound to the template strand following passage of the RNA polymerase, resulting in the non-transcribed strand being displaced as a ss loop. Transcription of TERRA leaves behind R-loops and their presence at telomeres has been detected using the S9.6 antibody specific for RNA/DNA hybrid structures [98]. Telomeric R-loops are particularly stable since the displaced G-rich strand contains runs of guanines with the potential of generating G quartets [99], the formation of which inhibits reannealing of the template and non-template strands.
A model based on transcription-mediated t-loop formation can be imagined (Fig. 7A), in which a single shelterin complex tethered to the ss terminus by POT1, engages an internal R-loop via binding of the TRF2 component of shelterin to TERRA. Alternatively, two shelterin complexes, one at the terminus and one internal, could associate to generate a t-loop. Details of how strand insertion would occur at the R-loop remain unclear. In either case, since the R-loops are assumed to be located at random along the telomere, a broad spectrum of loop sizes would be generated, which is what has been observed.
Fig. 7. Transcription drives t-loop formation.
(A) As described in [100] the ~3 kb plasmid pRST5 containing a 576 bp block of TTAGGG repeats at one end and immediately preceded by a T7 RNA polymerase promoter was transcribed in vitro followed by deproteinization and preparation for EM. Fields of DNAs showed up to 60% of the DNAs with a small t-loop at the end containing the telomeric repeats. (B) In [100] a rolling circle replication scheme was used to generate double stranded telomeric DNA exceeding 10 kb in length and containing a T7 promoter at one end. Its transcription in vitro frequently generated t-loop structures with dimensions very similar to ones isolated from human cells. (C) Model of how t-loops may form due to transcription and R-loop formation [100].
To test this hypothesis, members of the Griffith lab utilized a model telomere template consisting of a ~3 kb linear plasmid DNA terminating in 576 bp of TTAGGG repeats and various ss overhangs. The DNA contains a T7 RNA polymerase promoter at the beginning of the telomere block (Fig. 7A), allowing transcription of the C-rich strand to produce G-rich TERRA [100]. Following transcription, up to 60% of the templates exhibited t-loops at the telomeric end (Fig. 7B), a far higher percentage than had been achieved with the in vitro TRF2-driven reactions. The loops were unusually stable, and blunt-ended templates or ones with a 4-base 5’ overhang at the telomere were also efficient substrates for loop formation. This suggested that t-loops generated by transcription involve the insertion of both terminal strands in the preceding duplex. Furthermore, RNA frequently remained tightly bound at the circle-tail junction of the t-loop. A rolling circle replication scheme was then developed that generates duplex telomeric DNAs the size of human telomeres (5–15 kb), with each DNA containing a T7 promoter at the beginning. Transcription generated t-loops the size of those isolated from cells (Fig. 7C) and it was most efficient with DNAs consisting of TTAGGG repeats, as contrasted to mutated versions which disrupt the runs of 3 Gs. Together, these results point to a more stable, complex junction than had been envisioned before, with features of both replication forks and Holliday junctions (Fig. 7A). Indeed, data was recently presented [101] showing that the 3’ terminus inserted at the junction can prime replication, in a reaction that extends telomere length in the absence of telomerase, a result that provides an exciting palate for further studies.
9. Understanding t-loop functions in telomere protection and replication
From their conception, t-loops provided an elegant solution to chromosome end protection. T-loops also presented an obstacle to replication forks originating from sub-telomeric regions that would collide and stall when encountering the t-loop junction. If t-loops contribute to end protection, how the loops open and close during replication and transcription remained unclear for almost twenty years.
Determining how telomere proteins regulate chromosome end protection was driven primarily by the de Lange lab. Through a series of studies, they showed that deleting TRF1, POT1 or TPP1 activated an ATR-dependent DDR at chromosome ends with minimal covalent telomere ligation [87, 102, 103]. Conversely, deleting TRF2 induced a robust ATM-dependent telomere DDR and a striking phenotype where all chromosome ends eventually fuse through the canonical non-homologous end joining (NHEJ) DSB repair pathway [104–106]. Consistent with ATM-dependent DDR activation, TRF2 deletion elicits phosphorylation of the histone variant H2AX at chromosome ends and telomere localization of the DDR factor 53BP1 [87, 107]. Because TRF2 suppresses both ATM activation and NHEJ, it was suggested that TRF2-dependent t-loop formation simultaneously suppressed DDR activation and end-joining [38]. However, results from human and murine cells provided functional evidence that TRF2 suppressed ATM and NHEJ through distinct mechanisms.
Following completion of this Ph.D. in the Griffith lab, Tony Cesare undertook post-doctoral fellowships with Roger Reddel and Jan Karlseder. During this time, Cesare demonstrated that telomeres in aged and cancerous human cells could activate the ATM-dependent DDR without inducing end-to-end chromosome fusions [108, 109]. Similarly, teams led by Eros Lazzerini Denchi and Eric Gilson identified TRF2 separation of function alleles that promoted telomere DDR activation without accompanying NHEJ [110, 111]. Cesare also demonstrated similar DDR-positive and fusion-resistant telomeres following partial TRF2 depletion using shRNAs [112]. The role of t-loops within the continuum of protected telomeres, DDR-positive telomeres, and fused telomeres remained unclear.
After establishing his independent laboratory at CMRI (Sydney, Australia) in 2013, Cesare started a collaboration with super-resolution microscopy expert Katharina Gaus from the neighboring UNSW to implement Doksani’s super-resolution protocols. The Cesare and Gaus labs were able to replicate Doksani’s imaging; however, because few STORM imaging apparatuses were available locally, Cesare lab members David Van Ly and Sonja Frölich worked with the Gaus lab to optimize the Doksani protocol for use with alternative super-resolution modalities. While super-resolution stimulated emission depletion microcopy (STED), structured illumination microscopy (SIM), and AiryScan microscopy captured images with a lower resolution than STORM, they provided exceptional gains in imaging speed: 60 seconds or less for SIM, STED and AiryScan, as opposed to 30 minutes for STORM. Direct comparison of the imaging modalities revealed that the percentage of looped telomeres was not compromised with the rapid imaging methods. Further, reduced photobleaching with AiryScan enabled the Cesare lab to perform the first super-resolution light microscopic imaging of t-loops from human cells. They applied this method to a series of molecular biology models of telomere protection [113]. Through a combination of experiments in human and murine cells, using TRF2 separation of function alleles, differential TRF2 depletion with RNAi, and induction of non-canonical mitotic arrest-dependent telomere deprotection, the Cesare and Gaus collaboration showed that ATM activation at chromosome ends occurs specifically with t-loop unwinding and exposure of the linear chromosome end. Furthermore, their results revealed that ATM activation occurred in a mechanistically distinct step from NHEJ-dependent telomere fusions. This provided the first mechanistic evidence of a protective t-loop activity by indicating that t-loops specifically suppress activation of ATM by the naturally occurring chromosome end, independent from TRF2-mediated NHEJ inhibition.
While the de Lange and Cesare laboratories were working on techniques to visualize t-loops, Simon Boulton’s laboratory at the Francis Crick Institute in London was elucidating functions of the RTEL1 helicase at chromosome ends. The Boulton lab found that RTEL1 directly interacts with TRF2 and unwinds D-loop substrates equivalent to t-loop junctions [114]. Cellular and molecular experiments from the Boulton lab showed that preventing RTEL1 activity at telomeres induced rapid telomere shortening and free t-circles, outcomes consistent with replication fork collapse following collision with the invading t-loop strand [66, 114]. Moreover, they found that reciprocal CDK phosphorylation and PP6R3 mediated dephosphorylation of TRF2-S365 regulated RTEL1 association with dephosphorylated TRF2 specifically in S-phase. A collaboration between the Cesare and Boulton labs was established to apply Cesare’s rapid imaging of t-loops in the context of altered RTEL1 function.
Using AiryScan imaging of TRF2−/− murine cells complemented with WT or TRF2-S365A alleles, the Boulton/Cesare collaboration discovered that sequestering RTEL1 at a phospho-null TRF2 throughout the cell cycle promoted spurious t-loop unwinding, linear chromosome ends, and activation of the telomere DDR without accompanying telomere fusions [115]. This revealed RTEL1 as the bona fide t-loop helicase. Further, the study demonstrated how PP6R3 and CDK activity regulated S-phase specific RTEL1 interaction with TRF2 to unwind t-loops and facilitate effective telomere replication. The data suggest t-loops reform after telomere replication, when TRF2 is phosphorylated, and remain in this protective state until the following S-phase. Additionally, these data indicate persistent t-loop resolution by RTEL1 is deleterious, resulting in telomere DDR-activation, thereby confirming the protective nature of the t-loop structure. The Boulton/Cesare collaboration, where the genetics and molecular biology capability of the Boulton lab was complemented by the imaging capacity of the Cesare lab, is another example of collaborative work enabling a breakthrough not possible by either lab alone.
10. Conclusion
It has been 8 decades since Muller and McClintock recognized that chromosome ends must be sealed in some unique manner, but it required another 6 decades before a unifying insight into how this may occur was proposed, along with the first electron micrographs illustrating telomere looping. This structural solution has now been observed from yeast to human cells. This could not have been elucidated by any one laboratory alone but rather was the result of a series of collaborative efforts between research groups contributing unique expertise, beginning with the seminal collaboration between the Griffith and de Lange groups. Our current understanding is also a matter of experimental skill, good fortune, and tenacity: skill in the early experiments of Rachel Stansel and Laurey Comeau in the Griffith lab, good fortune that the first experiment to isolate and visualize t-loops from HeLa cells worked perfectly providing exciting images, and, over a decade later, the tenacity of Ylli Doksani in the de Lange laboratory in adapting the t-loop isolation methods for super resolution light microscopy. His work enabled a new generation of experiments that combine this new visualization tool with powerful genetics, as applied in his work and in the recent studies from the Cesare and Boulton groups.
Highlights:
Review of the discovery of telomere looping (t-loops)
Review of the discovery of telomeric DNA circles (t-circles)
Relation of t-loop open and closed states to DNA repair response
Importance of diverse yeast species in t-loop and t-circle discover
Acknowledgements
We wish to thank Ms. Smaranda Willcox for help in the development and writing of this review. The work described here was supported by grants from the NIH to JDG (ES013373, 1R01ES031635–01). Research in the LT laboratory is supported by the grants from VEGA (1/0061/20) and APVV (19–0068). AJC is supported by grants from the Australian National Health and Medical Research Council (1162886 and 1185870) and The Neil and Norma Hill Foundation.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Dedication: The authors wish to dedicate this review to Dr. Samuel Wilson for his seminal contributions to our understanding of the basic mechanisms of DNA repair and his longstanding dedication to promoting this field including editorship of this journal.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Muller HJ, The remaking of chromosomes. Collecting net, 1938. 13: p. 181–198. [Google Scholar]
- [2].McClintock B, The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics, 1941. 26(2): p. 234–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].de Lange T, Shelterin-Mediated Telomere Protection. Annu Rev Genet, 2018. 52: p. 223–247. [DOI] [PubMed] [Google Scholar]
- [4].Olovnikov AM, A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol, 1973. 41(1): p. 181–90. [DOI] [PubMed] [Google Scholar]
- [5].Watson JD, Origin of concatemeric T7 DNA. Nat New Biol, 1972. 239(94): p. 197–201. [DOI] [PubMed] [Google Scholar]
- [6].Blackburn EH and Gall JG, A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol, 1978. 120(1): p. 33–53. [DOI] [PubMed] [Google Scholar]
- [7].Meyne J, Ratliff RL, and Moyzis RK, Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A, 1989. 86(18): p. 7049–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Moyzis RK, et al. , A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A, 1988. 85(18): p. 6622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Richards EJ and Ausubel FM, Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell, 1988. 53(1): p. 127–36. [DOI] [PubMed] [Google Scholar]
- [10].Wright WE, et al. , Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev, 1997. 11(21): p. 2801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Harley CB, Futcher AB, and Greider CW, Telomeres shorten during ageing of human fibroblasts. Nature, 1990. 345(6274): p. 458–60. [DOI] [PubMed] [Google Scholar]
- [12].Wu P, Takai H, and de Lange T, Telomeric 3’ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell, 2012. 150(1): p. 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Palm W. and de Lange T, How shelterin protects mammalian telomeres. Annu Rev Genet, 2008. 42: p. 301–34. [DOI] [PubMed] [Google Scholar]
- [14].Maréchal A. and Zou L, DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor perspectives in biology, 2013. 5(9): p. a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cesare AJ and Karlseder J, A three-state model of telomere control over human proliferative boundaries. Curr Opin Cell Biol, 2012. 24(6): p. 731–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Szostak JW and Blackburn EH, Cloning yeast telomeres on linear plasmid vectors. Cell, 1982. 29(1): p. 245–55. [DOI] [PubMed] [Google Scholar]
- [17].Lundblad V. and Blackburn EH, An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell, 1993. 73(2): p. 347–60. [DOI] [PubMed] [Google Scholar]
- [18].Pluta AF and Zakian VA, Recombination occurs during telomere formation in yeast. Nature, 1989. 337(6206): p. 429–33. [DOI] [PubMed] [Google Scholar]
- [19].Wellinger RJ and Zakian VA, Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics, 2012. 191(4): p. 1073–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Grunstein M, Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol, 1997. 9(3): p. 383–7. [DOI] [PubMed] [Google Scholar]
- [21].de Bruin D, et al. , Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol, 2000. 20(21): p. 7991–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].de Bruin D, et al. , Telomere looping permits gene activation by a downstream UAS in yeast. Nature, 2001. 409(6816): p. 109–13. [DOI] [PubMed] [Google Scholar]
- [23].Poschke H, et al. , Rif2 promotes a telomere fold-back structure through Rpd3L recruitment in budding yeast. PLoS Genet, 2012. 8(9): p. e1002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pasquier E. and Wellinger RJ, In vivo chromatin organization on native yeast telomeric regions is independent of a cis-telomere loopback conformation. Epigenetics & Chromatin, 2020. 13(1): p. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gottschling DE and Zakian VA, Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell, 1986. 47(2): p. 195–205. [DOI] [PubMed] [Google Scholar]
- [26].Price CM and Cech TR, Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes Dev, 1987. 1(8): p. 783–93. [DOI] [PubMed] [Google Scholar]
- [27].Zhong Z, et al. , A mammalian factor that binds telomeric TTAGGG repeats in vitro. Molecular and cellular biology, 1992. 12(11): p. 4834–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Griffith JD, Visualization of prokaryotic DNA in a regularly condensed chromatin-like fiber. Proc Natl Acad Sci U S A, 1976. 73(2): p. 563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Griffith JD and Harris LD, DNA strand exchanges. CRC Crit Rev Biochem, 1988. 23 Suppl 1: p. S43–86. [PubMed] [Google Scholar]
- [30].Griffith J, Bianchi A, and de Lange T, TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol, 1998. 278(1): p. 79–88. [DOI] [PubMed] [Google Scholar]
- [31].Bilaud T, et al. , Telomeric localization of TRF2, a novel human telobox protein. Nat Genet, 1997. 17(2): p. 236–9. [DOI] [PubMed] [Google Scholar]
- [32].Broccoli D, et al. , Comparison of the human and mouse genes encoding the telomeric protein, TRF1: chromosomal localization, expression and conserved protein domains. Hum Mol Genet, 1997. 6(1): p. 69–76. [DOI] [PubMed] [Google Scholar]
- [33].van Steensel B, Smogorzewska A, and de Lange T, TRF2 protects human telomeres from end-to-end fusions. Cell, 1998. 92(3): p. 401–13. [DOI] [PubMed] [Google Scholar]
- [34].Karlseder J, et al. , p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science, 1999. 283(5406): p. 1321–5. [DOI] [PubMed] [Google Scholar]
- [35].Karlseder J, Smogorzewska A, and de Lange T, Senescence induced by altered telomere state, not telomere loss. Science, 2002. 295(5564): p. 2446–9. [DOI] [PubMed] [Google Scholar]
- [36].Cech TR and Pardue ML, Electron microscopy of DNA crosslinked with trimethylpsoralen: test of the secondary structure of eukaryotic inverted repeat sequences. Proc Natl Acad Sci U S A, 1976. 73(8): p. 2644–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Griffith JD, et al. , Mammalian telomeres end in a large duplex loop. Cell, 1999. 97(4): p. 503–14. [DOI] [PubMed] [Google Scholar]
- [38].Doksani Y, et al. , Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell, 2013. 155(2): p. 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fouche N, et al. , The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem, 2006. 281(49): p. 37486–95. [DOI] [PubMed] [Google Scholar]
- [40].Stansel RM, Subramanian D, and Griffith JD, p53 binds telomeric single strand overhangs and t-loop junctions in vitro. J Biol Chem, 2002. 277(14): p. 11625–8. [DOI] [PubMed] [Google Scholar]
- [41].Amiard S, et al. , A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol, 2007. 14(2): p. 147–54. [DOI] [PubMed] [Google Scholar]
- [42].Benarroch-Popivker D, et al. , TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Molecular cell, 2016. 61(2): p. 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Murti KG and Prescott DM, Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc Natl Acad Sci U S A, 1999. 96(25): p. 14436–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Muñoz-Jordán JL, et al. , t-loops at trypanosome telomeres. The EMBO journal, 2001. 20(3): p. 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cesare AJ, et al. , Telomere looping in P. sativum (common garden pea). Plant J, 2003. 36(2): p. 271–9. [DOI] [PubMed] [Google Scholar]
- [46].Raices MC, et al. , elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell, 2008. 132(5): p. 745–57. [DOI] [PubMed] [Google Scholar]
- [47].Nikitina T. and Woodcock CL, Closed chromatin loops at the ends of chromosomes. J Cell Biol, 2004. 166(2): p. 161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Subík J, Kolarov J, and Kovác L, Bongkrekic acid sensitivity of respiration-deficient mutants and of petite-negative species of yeasts. Biochim Biophys Acta, 1974. 357(3): p. 453–6. [DOI] [PubMed] [Google Scholar]
- [49].Subík J, Kolarov J, and Kovác L, Anaerobic growth and formation of respiration-deficient mutants of various species of yeasts. FEBS Lett, 1974. 45(1): p. 263–6. [DOI] [PubMed] [Google Scholar]
- [50].Kovác L, Lazowska J, and Slonimski PP, A yeast with linear molecules of mitochondrial DNA. Mol Gen Genet, 1984. 197(3): p. 420–4. [DOI] [PubMed] [Google Scholar]
- [51].Nosek J, et al. , Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol Gen Genet, 1995. 247(1): p. 61–72. [DOI] [PubMed] [Google Scholar]
- [52].Tomáska L, Nosek J, and Fukuhara H, Identification of a putative mitochondrial telomere-binding protein of the yeast Candida parapsilosis. J Biol Chem, 1997. 272(5): p. 3049–56. [DOI] [PubMed] [Google Scholar]
- [53].Nosek J, et al. , Mitochondrial telomere-binding protein from Candida parapsilosis suggests an evolutionary adaptation of a nonspecific single-stranded DNA-binding protein. J Biol Chem, 1999. 274(13): p. 8850–7. [DOI] [PubMed] [Google Scholar]
- [54].Tomaska L, et al. , Electron microscopic analysis supports a dual role for the mitochondrial telomere-binding protein of Candida parapsilosis. J Mol Biol, 2001. 305(1): p. 61–9. [DOI] [PubMed] [Google Scholar]
- [55].Tomaska L, et al. , Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res, 2000. 28(22): p. 4479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kosa P, et al. , Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res, 2006. 34(8): p. 2472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nosek J, et al. , Amplification of telomeric arrays via rolling-circle mechanism. J Biol Chem, 2005. 280(11): p. 10840–5. [DOI] [PubMed] [Google Scholar]
- [58].Tomaska L, et al. , t-Loops in yeast mitochondria. Mitochondrion, 2002. 1(5): p. 455–9. [DOI] [PubMed] [Google Scholar]
- [59].Tomaska L, et al. , Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol, 2009. 16(10): p. 1010–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cesare AJ and Griffith JD, Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol, 2004. 24(22): p. 9948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Perrem K, et al. , Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol Cell Biol, 2001. 21(12): p. 3862–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cesare AJ, et al. , Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol Cell Biol, 2008. 28(1): p. 20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang RC, Smogorzewska A, and de Lange T, Homologous recombination generates T-loop-sized deletions at human telomeres. Cell, 2004. 119(3): p. 355–68. [DOI] [PubMed] [Google Scholar]
- [64].Laud PR, et al. , Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev, 2005. 19(21): p. 2560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sarkar J, et al. , SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates. Nucleic acids research, 2015. 43(12): p. 5912–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Vannier JB, et al. , RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell, 2012. 149(4): p. 795–806. [DOI] [PubMed] [Google Scholar]
- [67].Deng Z, et al. , Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proceedings of the National Academy of Sciences of the United States of America, 2013. 110(36): p. E3408-E3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Touzot F, et al. , Extended clinical and genetic spectrum associated with biallelic RTEL1 mutations. Blood advances, 2016. 1(1): p. 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Henson JD, et al. , DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol, 2009. 27(12): p. 1181–5. [DOI] [PubMed] [Google Scholar]
- [70].Zellinger B, et al. , Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell, 2007. 27(1): p. 163–9. [DOI] [PubMed] [Google Scholar]
- [71].Cooper JP, et al. , Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature, 1997. 385(6618): p. 744–7. [DOI] [PubMed] [Google Scholar]
- [72].Tomaska L, et al. , Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J Biol Chem, 2004. 279(49): p. 50764–72. [DOI] [PubMed] [Google Scholar]
- [73].Sanchez-Alonso P. and Guzman P, Predicted elements of telomere organization and function in Ustilago maydis. Fungal Genet Biol, 2008. 45 Suppl 1: p. S54–62. [DOI] [PubMed] [Google Scholar]
- [74].Yu EY, et al. , Brh2 and Rad51 promote telomere maintenance in Ustilago maydis, a new model system of DNA repair proteins at telomeres. DNA Repair (Amst), 2013. 12(7): p. 472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Červenák F, et al. , Identification of telomerase RNAs in species of the Yarrowia clade provides insights into the co-evolution of telomerase, telomeric repeats and telomere-binding proteins. Sci Rep, 2019. 9(1): p. 13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kinsky S, et al. , Lack of the catalytic subunit of telomerase leads to growth defects accompanied by structural changes at the chromosomal ends in Yarrowia lipolytica. Curr Genet, 2010. 56(5): p. 413–25. [DOI] [PubMed] [Google Scholar]
- [77].Kramara J, et al. , Tay1 protein, a novel telomere binding factor from Yarrowia lipolytica. J Biol Chem, 2010. 285(49): p. 38078–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Visacka K, et al. , Synergism of the two Myb domains of Tay1 protein results in high affinity binding to telomeres. J Biol Chem, 2012. 287(38): p. 32206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].McEachern MJ and Blackburn EH, Runaway telomere elongation caused by telomerase RNA gene mutations. Nature, 1995. 376(6539): p. 403–9. [DOI] [PubMed] [Google Scholar]
- [80].McEachern MJ and Blackburn EH, Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev, 1996. 10(14): p. 1822–34. [DOI] [PubMed] [Google Scholar]
- [81].Groff-Vindman C, et al. , Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Molecular and cellular biology, 2005. 25(11): p. 4406–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Iyer S, Chadha AD, and McEachern MJ, A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol, 2005. 25(18): p. 8064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Natarajan S. and McEachern MJ, Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol, 2002. 22(13): p. 4512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Larrivée M. and Wellinger RJ, Telomerase- and capping-independent yeast survivors with alternate telomere states. Nature Cell Biology, 2006. 8(7): p. 741–747. [DOI] [PubMed] [Google Scholar]
- [85].Lin CY, et al. , Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot Cell, 2005. 4(2): p. 327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yu EY, et al. , Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Molecular and cellular biology, 2010. 30(5): p. 1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Denchi EL and de Lange T, Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature, 2007. 448(7157): p. 1068–71. [DOI] [PubMed] [Google Scholar]
- [88].Huang B, Babcock H, and Zhuang X, Breaking the diffraction barrier: super-resolution imaging of cells. Cell, 2010. 143(7): p. 1047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Rust MJ, Bates M, and Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods, 2006. 3(10): p. 793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Azzalin CM, et al. , Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science, 2007. 318(5851): p. 798–801. [DOI] [PubMed] [Google Scholar]
- [91].Schoeftner S. and Blasco MA, Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol, 2008. 10(2): p. 228–36. [DOI] [PubMed] [Google Scholar]
- [92].Bettin N, Oss Pegorar C, and Cusanelli E, The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells, 2019. 8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Maicher A, Lockhart A, and Luke B, Breaking new ground: digging into TERRA function. Biochim Biophys Acta, 2014. 1839(5): p. 387–94. [DOI] [PubMed] [Google Scholar]
- [94].Deng Z, et al. , TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell, 2009. 35(4): p. 403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].López de Silanes I, Stagno d’Alcontres M, and Blasco MA, TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat Commun, 2010. 1: p. 33. [DOI] [PubMed] [Google Scholar]
- [96].Postepska-Igielska A, et al. , The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO reports, 2013. 14(8): p. 704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Toubiana S. and Selig S, DNA:RNA hybrids at telomeres - when it is better to be out of the (R) loop. Febs j, 2018. 285(14): p. 2552–2566. [DOI] [PubMed] [Google Scholar]
- [98].Arora R, et al. , RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun, 2014. 5: p. 5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Randall A. and Griffith JD, Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J Biol Chem, 2009. 284(21): p. 13980–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kar A, Willcox S, and Griffith JD, Transcription of telomeric DNA leads to high levels of homologous recombination and t-loops. Nucleic acids research, 2016. 44(19): p. 9369–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tomaska L, et al. , A New View of the T-Loop Junction: Implications for Self-Primed Telomere Extension, Expansion of Disease-Related Nucleotide Repeat Blocks, and Telomere Evolution. Front Genet, 2019. 10: p. 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hockemeyer D, et al. , Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell, 2006. 126(1): p. 63–77. [DOI] [PubMed] [Google Scholar]
- [103].Sfeir A, et al. , Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell, 2009. 138(1): p. 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Celli GB and de Lange T, DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol, 2005. 7(7): p. 712–8. [DOI] [PubMed] [Google Scholar]
- [105].Celli GB, Denchi EL, and de Lange T, Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol, 2006. 8(8): p. 885–90. [DOI] [PubMed] [Google Scholar]
- [106].Heacock ML, et al. , Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic acids research, 2007. 35(19): p. 6490–6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Takai H, Smogorzewska A, and de Lange T, DNA damage foci at dysfunctional telomeres. Curr Biol, 2003. 13(17): p. 1549–56. [DOI] [PubMed] [Google Scholar]
- [108].Cesare AJ, et al. , Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol, 2009. 16(12): p. 1244–51. [DOI] [PubMed] [Google Scholar]
- [109].Kaul Z, et al. , Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep, 2011. 13(1): p. 52–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Benarroch-Popivker D, et al. , TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Mol Cell, 2016. 61(2): p. 274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Okamoto K, et al. , A two-step mechanism for TRF2-mediated chromosome-end protection. Nature, 2013. 494(7438): p. 502–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Cesare AJ, et al. , The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol Cell, 2013. 51(2): p. 141–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Van Ly D, et al. , Telomere Loop Dynamics in Chromosome End Protection. Mol Cell, 2018. 71(4): p. 510–525 e6. [DOI] [PubMed] [Google Scholar]
- [114].Sarek G, et al. , TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol Cell, 2015. 57(4): p. 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sarek G, et al. , CDK phosphorylation of TRF2 controls t-loop dynamics during the cell cycle. Nature, 2019. 575(7783): p. 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Nosek J. and Tomaska L, Mitochondrial genome diversity: evolution of the molecular architecture and replication strategy. Curr Genet, 2003. 44(2): p. 73–84. [DOI] [PubMed] [Google Scholar]