Abstract
In a bid to contain the current COVID-19 (coronavirus disease 2019) pandemic, various countermeasures have been applied. To date, however, there is a lack of an effective drug for the treatment of COVID-19. Through molecular modelling studies, simeprevir, a protease inhibitor approved for the management of hepatitis C virus infection, has been predicted as a potential antiviral against SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), the causative agent of COVID-19. Here we assessed the efficacy of simeprevir against SARS-CoV-2 both in vitro in Vero E6 cells and in vivo in a human angiotensin-converting enzyme 2 (hACE2) transgenic mouse model. The results showed that simeprevir could inhibit SARS-CoV-2 replication in Vero E6 cells with a half-maximal effective concentration (EC50) of 1.41 ± 0.12 μM. In a transgenic hACE2 mouse model of SARS-CoV-2 infection, intraperitoneal administration of simeprevir at 10 mg/kg/day for 3 consecutive days failed to suppress viral replication. These findings collectively imply that simeprevir does not inhibit SARS-CoV-2 in vivo and therefore do not support its application as a treatment against COVID-19 at a dosage of 10 mg/kg/day.
Keywords: SARS-CoV-2, COVID-19, Simeprevir, Antiviral efficacy
1. Introduction
COVID-19 (coronavirus disease 2019) is a highly infectious respiratory disease caused by a novel coronavirus, SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2). The first COVID-19 outbreak was reported in China in December 2019 and it has since spread rapidly resulting in a global pandemic [1]. The fight against COVID-19 has been one filled with challenges, including the failure of experimental drugs. The most recent challenge is the emergence of variant strains that are capable of undermining the protective effect of vaccines that are being administered [2]. Hence, even with the availability of vaccines, there is still a pressing need for an effective anti-SARS-CoV-2 drug. Remdesivir was the first drug to be approved by the US Food and Drug Administration (FDA) for the treatment of COVID-19. Several clinical trials were conducted, but discordant findings were reported regarding its clinical efficacy [3]. Recently, analysis of real-world data from three retrospective studies on the treatment and outcome of remdesivir against COVID-19 have demonstrated that remdesivir-treated hospitalised patients have a significantly reduced risk of mortality compared with matched controls (https://www.gilead.com/news-and-press/press-room/press-releases/2021/6/gileads-veklury-remdesivir-associated-with-a-reduction-in-mortality-rate-in-hospitalized-patients-with-covid19-across-three-analyses-of-large-ret). Monoclonal antibody therapies, including casirivimab + imdevimab and bamlanivimab + etesevimab, have gained increasing attention, especially for use against variant strains [4]. The most recent advancement involves three investigational drugs (monulpiravir, paxlovid and favipiravir) that have performed well in clinical trials and shown promising results in reducing the risk of hospitalisation and death [5,6]. Despite the availability of numerous treatment approaches, there is still no definitive treatment for SAR-CoV-2 infection.
To rapidly develop an antiviral drug, researchers have resorted to repurposing pre-existing FDA-approved drugs. Simeprevir is a second-generation protease inhibitor approved for the management of hepatitis C virus (HCV) infection. The anti-HCV activity of this drug is mediated by inhibition of viral NS3/4A protease, thus preventing viral maturation by impeding protein synthesis [7]. In silico structural modelling studies predicted that simeprevir inhibits SARS-CoV-2 by targeting the viral main protease (Mpro) [8], RNA-dependent RNA polymerase (RdRp, also called Nsp12) [9] and Nsp13 (NTPase/helicase) [10]. Mpro cleaves viral polyproteins to generate non-structural proteins including Nsp12 and Nsp13, which are essential components of the viral replication-transcription complex. Molecular dynamics simulation analysis also established that simeprevir could inhibit interaction of the viral spike protein with the receptor angiotensin-converting enzyme 2 (ACE2) by binding to side chains of residues in the binding pocket of the receptor-binding domain (RBD), suggesting that the drug may be a multitarget inhibitor [11]. Although there is a great deal of evidence supporting the anti-SARS-CoV-2 efficacy of simeprevir in vitro [12], [13], [14], its in vivo potency has not yet been fundamentally evaluated. In the present study, the antiviral effects of simeprevir against SARS-CoV-2 were evaluated both in vitro and in a human angiotensin-converting enzyme 2 (hACE2) transgenic mouse model.
2. Materials and methods
2.1. Cells, compounds and virus
Vero E6 cells (an African green monkey kidney cell line) were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 2% fetal bovine serum, 100 μg/mL streptomycin and 100 U/mL penicillin at 37°C and 5% CO2. SARS-CoV-2 strain (nCoV-2019BetaCoV/Wuhan/WIV04/2019) was propagated in Vero E6 cells and was titrated by standard plaque assay following the standard procedure. Simeprevir (MedChemExpress) was dissolved in fresh dimethyl sulfoxide (DMSO) to the desired concentrations. Experiments involving SARS-CoV-2 virus culture were performed at the Wuhan Institute of Virology Zhengdian Biosafety Level 3 (BSL-3) facility.
2.2. Cytotoxicity assay
We evaluated the cytotoxicity of simeprevir in Vero E6 cells using a Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China). Cells were seeded at a density of 5 × 103 cells/well in 96-well plates and were incubated overnight at 37°C and 5% CO2. Cells were exposed to a series of simeprevir concentrations (0–100 μM) for another 24 h. The contents of the wells were then replaced with fresh medium containing 10% CCK-8 solution and were incubated at 37°C for 1.5 h. The optical density at 450 nm was measured using a Synergy H1 microplate reader (BioTek, USA).
2.3. In vitro antiviral assay
An antiviral assay was performed to assess the antiviral effects of simeprevir on the replication of SARS-CoV-2. Vero E6 cells were seeded at a density of 1 × 105 cells/well in 24-well plates at 24 h prior to the experiment. Cells were inoculated with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01 in the presence of varying concentrations of simeprevir (0–10 μM). At 24 h post-infection (hpi), RNA was extracted from the cell supernatant using a QIAamp® Viral RNA Mini Kit (QIAGEN) following the manufacturer's instructions. Viral RNA was quantified by real-time quantitative reverse transcription PCR (RT-qPCR) using a Luna® Universal Probe One-Step RT-qPCR Kit (Invitrogen) with primers targeting the ORF1ab gene of SARS-CoV-2. The primer sequences were as follows: ORF1ab-F, 5′-CCCTGTGGGTTTTACACTTAA-3′ and ORF1ab-R, 5′-ACGATTGTGCATCAGCTGA-3′, combined with the probe 5′-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′.
2.4. Western blotting
To evaluate the effect of simeprevir on protein expression levels in SARS-CoV-2-infected Vero E6 cells, western blot analysis was performed. Vero E6 cells were infected with SARS-CoV-2 in the presence of 0, 3 and 10 μM of simeprevir. Cells were lysed with RIPA lysis buffer (Thermo Scientific) and were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) at 24 hpi. Protein bands were electroblotted onto nitrocellulose membranes (Bio-Rad, Shanghai, China). The membranes were blocked with phosphate-buffered Salome (PBS) containing 0.1% Tween 20 and 5% skimmed milk. Proteins were detected by probing the membranes with a mixture of rabbit anti-SARS-CoV-2 nucleocapsid protein (NP) polyclonal antibody/rabbit anti-SARS-CoV-2 spike protein (SP) polyclonal antibody and rabbit anti-GAPDH polyclonal antibody (Beyotime) for 1 h at room temperature. Membranes were then washed in PBS containing 0.05% Tween 20 for 20 min. Blots were detected with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Beyotime) followed by chemiluminescence reagents. Signals were detected by exposure to X-ray film for 1–3 min.
2.5. Immunofluorescence assay
To further verify the inhibitory impact of simeprevir on the expression of SARS-CoV-2 antigens, an indirect immunofluorescence assay was performed. Vero E6 cells were seeded on glass slides 24 h prior to the experiment. SARS-CoV-2 virus was then added to the cells in the presence or absence of 10 μM simeprevir. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature at 24 hpi. Cells were incubated with 1:1000 diluted primary antibody against the SP of SARS-CoV-2 for 1 h at room temperature. After a thorough wash for 15 min to remove unbound antibodies, cells were incubated with 1:500 diluted Alexa Fluor 594-conjugated goat anti-human IgG antibodies (Invitrogen, Thermo Fisher Scientific) for 1 h at room temperature. Slides were washed for another 15 min, followed by nuclei staining with 4,6-diamidino-2-phenylindole (DAPI) (Sigma), and were then visualised by confocal fluorescence microscopy (UltraVIEW® VoX; PerkinElmer, USA).
2.6. Mouse experiments
All experiments were performed in a BSL-3 facility with all experimental methods carried out in accordance with the regulations and guidelines set forth by the Animal Experiments Committee of Wuhan Institute of Virology, Chinese Academy of Sciences. All experimental protocols were approved by the Animal Experiments Committee of Wuhan Institute of Virology, Chinese Academy of Sciences.
For the in vivo antiviral assay, 12–14-week-old male hACE2 TgTn transgenic mice (CAG-human-ACE2-IRES-Luciferase; Shanghai Model Organisms, Shanghai, China) were intranasally inoculated with 105 TCID50 (median tissue culture infectious dose) of SARS-CoV-2 and were randomly divided into three experimental groups (n = 6). In one group, mice were injected intraperitoneally with 10 mg/kg simeprevir at 4, 24 and 48 hpi. Similarly, the second group received 15 mg/kg remdesivir at 4, 24 and 48 hpi. The intraperitoneal injection volume for the drugs was 100 μL. The third group was mock-treated with an equal volume of solvent buffer [saline containing 10% DMSO, 40% polyethylene glycol 300 (PEG300) and 5% Tween 80] as a placebo control. The body weight of each mouse was recorded daily. All mice were euthanised at 72 hpi and organs including, liver, lungs, kidney, brain and intestines were collected. Viral RNA was extracted from organ homogenates using an RNeasy® Mini Kit (QIAGEN) following the manufacturer's instructions. The viral load was quantified by RT-qPCR using a Luna® Universal Probe One-Step RT-qPCR Kit with primers and probe targeting the ORF1ab gene of SARS-CoV-2. Data analysis was performed using GraphPad Prism v.8 (GraphPad Software Inc., La Jolla, CA, USA).
3. Results and discussion
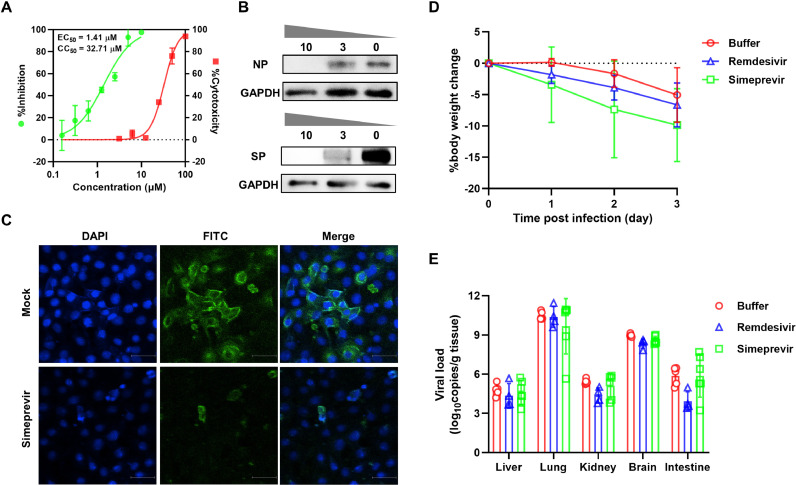
First, the cytotoxicity of simeprevir in Vero E6 was examined by CCK-8 assay. Consistent with previous studies, simeprevir exhibited moderate cytotoxicity with a 50% cytotoxic concentration (CC50) of 32 .71 ± 0.94 μM (Fig. 1 A). Next we assessed the inhibition potential of simeprevir on SARS-CoV-2 in Vero E6 cells pre-treated with different simeprevir doses ranging from 0–10 μM. The results showed that simeprevir inhibited SARS-CoV-2 replication in a dose-dependent manner, achieving a half-maximal effective concentration (EC50) value of 1.41 ± 0.12 μM and a selectivity index of >23 (Fig. 1A). These results are in agreement with previous studies that showed potent antiviral activity of simeprevir in human cell lines including human 293T cells [half-maximal inhibitory concentration (IC50) = 2.3 μM; CC50 ˃ 50 μM], A549-hACE2 cells (EC50 = 9 μM; CC50 = 56 μM) and human Huh7.5 cells (EC50 = 14 μM; CC50 = 33 μM) [12,14].
Fig. 1.
Effects of simeprevir on the replication of SARS-CoV-2. (A) Dose–response curve and cytotoxicity of simeprevir in Vero E6 cells. Cells were infected with SARS-CoV-2 at an MOI of 0.01 in the presence of various doses of simeprevir for 24 h. Viral yield in the supernatant was quantified by RT-qPCR. The cytotoxicity of simeprevir against Vero E6 cells was determined by Cell Counting Kit-8 (CCK-8) assay at simeprevir concentrations of 0–100 μM. (B) Western blot analysis of nucleocapsid protein (NP) and spike protein (SP) expression in SARS-CoV-2-infected cells pre-treated with 0, 3 and 10 μM simeprevir at 24 hpi. GAPDH was used as the reference gene. (C) Indirect immunofluorescence assay of SARS-CoV-2 in Vero E6 cells. Cells were infected with the virus at an MOI of 0.01 in the presence of 10 μM simeprevir for 24 h and were stained with a primary antibody specific for the SP. (D,E) Effects of simeprevir on the replication of SARS-CoV-2 in vivo. For the in vivo antiviral assay, 12–14-week-old male hACE2 transgenic mice were intranasally inoculated with 105 TCID50 virus and were distributed into three groups (n = 6): simeprevir-treated; remdesivir-treated; and mock-treated (buffer). In the drug-treated groups, simeprevir 10 mg/kg and remdesivir 15 mg/kg were administered intraperitoneally at 4, 24 and 48 h after SARS-CoV-2 infection. The intraperitoneal volume for both drugs was 100 μL. The control group was treated with an equal volume of drug solvent buffer. Mice were continuously monitored for change in body weight and were euthanised at 72 hpi for viral load determination. Rapid weight loss was observed in all treatment groups (D). No significant difference in viral load was detected between the simeprevir-treated, remdesivir-treated and control groups (E). SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MOI, multiplicity of infection; RT-qPCR, real-time quantitative reverse transcription PCR; EC50, half-maximal effective concentration; CC50, 50% cytotoxic concentration; hpi, hours post-infection; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; DAPI, 4,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; hACE2, human angiotensin-converting enzyme 2; TCID50, median tissue culture infectious dose.
To evaluate protein expression levels, whole-cell lysate of SARS-CoV-2-infected cells treated with 0, 3 and 10 μM of simeprevir were subjected to western blot analysis using a mixture of rabbit polyclonal antibodies specific for SARS-CoV-2 NP and SP. As shown in Fig. 1B, expression of viral NP and SP was significantly suppressed in cells treated with simeprevir in comparison with the drug-free control, with expression levels barely detectable in cells treated with 10 μM simeprevir. To further verify the antiviral efficacy of simeprevir, an indirect immunofluorescence assay was performed on Vero E6 cells infected with SARS-CoV-2 in the presence or absence of 10 μM simeprevir for 24 h. As shown in Fig. 1C, a significant reduction in fluorescence signal was observed in cells treated with simeprevir compared with mock-treated cells, indicating suppression of SARS-CoV-2 replication. Taken together, these data collectively show that simeprevir could efficiently inhibit the replication of SARS-CoV-2 in vitro.
Given the promising antiviral potency of simeprevir in vitro, we further assessed its performance in vivo in a hACE2 transgenic mouse model. As shown in Fig. 1D, rapid weight loss of mice was observed in all three treatment groups, a typical sign of SARS-CoV-2 infection in transgenic mice [15]. High copies of viral RNA were observed in multiple organs of virus-infected mock-treated mice, including the lung, brain, liver, kidney and intestine (Fig. 1E), indicating active viral replication and distribution in these organs. The highest viral burden was observed in the lungs of infected mice (Fig. 1E). In contrast to findings reported in in vitro antiviral activity studies, no significant difference in viral load was observed between the simeprevir-treated and buffer-treated groups (Fig. 1E), indicating that simeprevir, at a dose much higher than that recommended in the treatment of HCV (150 mg/day, ∼2.5 mg/kg) [16], has little or no effects on the replication of SARS-CoV-2 in vivo. Similarly, the remdesivir-treatment group showed no significant reduction in virus titres compared with the control group (Fig. 1E).
Simeprevir is a strong HCV NS3/4A protease inhibitor and has been suggested as a potent inhibitor of SARS-CoV-2 Mpro protease. While HCV and SARS-CoV-2 proteases share some structural similarities, they are fundamentally different thus limiting the potency of simeprevir against SARS-CoV-2 [14]. Additionally, cytochrome P450 3A4 (CYP3A4) enzyme, which is involved in the biotransformation of simeprevir, is preferentially found in the liver, which may result in an uneven distribution of the active metabolite into relevant tissues such as the lungs [17]. Moreover, hepatic absorption of simeprevir is mediated by organic anion-transporting polypeptide 1B3 (OATP1B3), which is not expressed in our current mouse model. Factors such as uneven drug distribution from preferential metabolism in various organs and the complexity of pharmacokinetics, which do not affect standard cell culture protocols, may explain the discrepancies between the in vitro and in vivo performance of simeprevir. Poor pharmacokinetics of remdesivir in mouse models have been reported, which may explain its low efficacy in this study. The presence of high levels of serum esterase vastly affects the plasma stability of remdesivir in mice [18]. In vitro studies have demonstrated that concomitant use of remdesivir and simeprevir results in a synergistic antiviral response against SARS-CoV-2 [12,13]. We cannot rule out with certainty synergy between the two drugs in vivo solely based on our study findings. Further in vivo antiviral evaluation of this combination in a model that can adequately metabolise remdesivir should be considered.
Similar cases of prospective drugs inhibiting SARS-CoV-2 replication in vitro but failing to reflect the same in vivo have been reported for other compounds, including chloroquine and hydroxychloroquine [19]. However, we acknowledge that factors such as dosage and route of administration may influence the bioavailability of a drug and thereby significantly impact its efficacy. In the present study, we tested the in vivo antiviral potency of simeprevir at 10 mg/kg, a dose we considered to be within the approved therapeutic margins for the treatment of HCV. However, a major challenge in drug repurposing is the requirement of higher doses than those that effectively treat the original indication. Therefore, a limitation of this study is that simeprevir at the current dosage under investigation may have been too low to provide sufficient exposure in relevant tissues. Additionally, pharmacokinetics and drug exposure profiles of intraperitoneally administered simeprevir were not extensively studied to rule out poor metabolism as a reason for failed efficacy. Further investigation of the reasons for the lack of in vitro to in vivo translation of antiviral efficacy of simeprevir against SARS-CoV-2 should be considered. Future studies may explore the in vivo antiviral efficacy of simeprevir at higher doses or as a combination therapy with other drugs such as remdesivir that have been reported to augment its antiviral effects.
In summary, we report here that simeprevir shows inhibitory effects against SARS-CoV-2 in cell culture but shows little inhibitory activity in transgenic mice. Although the mechanisms of failed protection by simeprevir in vivo require further study, the current data suggest that simeprevir may not be an effective antiviral candidate for SARS-CoV-2 at a dosage of 10 mg/kg.
Declaration of Competing Interest
None declared.
Acknowledgments
Acknowledgments
The authors thank Tao Du, Lun Wang and Jin Xiong (Zhengdian Biosafety Level 3 Laboratory of Wuhan Institute of Virology) for their critical support. The authors also thank the Core Facility and Technical Support, Wuhan Institute of Virology.
Funding
This work was financially supported by the Youth Innovation Promotion Association CAS, the National Natural Science Foundation of China [Nos. 32070187 and 31770192], and Sino-Africa Joint Research Center (151542KYSB20200010).
Ethical approval
All experimental protocols were approved by the Animal Experiments Committee of Wuhan Institute of Virology, Chinese Academy of Sciences [WIVA17202005].
Editor: Dr Jim Gray
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 3.Gyselinck I, Janssens W. Remdesivir, on the road to DisCoVeRy. Lancet Infect Dis. 2021 Sep 14 doi: 10.1016/S1473-3099(21)00559-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falcone M, Tiseo G, Valoriani B, Barbieri C, Occhineri S, Mazzetti P, et al. Efficacy of bamlanivimab/etesevimab and casirivimab/imdevimab in preventing progression to severe COVID-19 and role of variants of concern. Infect Dis Ther. 2021;10:2479–2488. doi: 10.1007/s40121-021-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer W, Eron JJ, Holman W, Cohen MS, Fang L, Szewczyk LJ, et al. Molnupiravir, an oral antiviral treatment for COVID-19. medRxiv 2021 Jun 17 [preprint]. doi: 10.1101/2021.06.17.21258639.
- 6.Agrawal U, Raju R, Udwadia ZF. Favipiravir: a new and emerging antiviral option in COVID-19. Med J Armed Forces India. 2020;76:370–376. doi: 10.1016/j.mjafi.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenquist Å, Samuelsson B, Johansson PO, Cummings MD, Lenz O, Raboisson P, et al. Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor. J Med Chem. 2014;57:1673–1693. doi: 10.1021/jm401507s. [DOI] [PubMed] [Google Scholar]
- 8.Hosseini FS, Amanlou M. Anti-HCV and anti-malaria agent, potential candidates to repurpose for coronavirus infection: virtual screening, molecular docking, and molecular dynamics simulation study. Life Sci. 2020;258 doi: 10.1016/j.lfs.2020.118205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM. The potential of paritaprevir and emetine as inhibitors of SARS-CoV-2 RdRp. Saudi J Biol Sci. 2021;28:1426–1432. doi: 10.1016/j.sjbs.2020.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurung AB. In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Rep. 2020;21 doi: 10.1016/j.genrep.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trezza A, Iovinelli D, Santucci A, Prischi F, Spiga O. An integrated drug repurposing strategy for the rapid identification of potential SARS-CoV-2 viral inhibitors. Sci Rep. 2020;10:13866. doi: 10.1038/s41598-020-70863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gammeltoft KA, Zhou Y, Duarte Hernandez CR, Galli A, Offersgaard A, Costa R, et al. Hepatitis C virus protease inhibitors show differential efficacy and interactions with remdesivir for treatment of SARS-CoV-2 in vitro. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.02680-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo HS, Hui KPY, Lai H-M, He X, Khan KS, Kaur S, et al. Simeprevir potently suppresses SARS-CoV-2 replication and synergizes with remdesivir. ACS Central Sci. 2021;7:792–802. doi: 10.1021/acscentsci.0c01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bafna K, White K, Harish B, Rosales R, Ramelot TA, Acton TB, et al. Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verbinnen T, Fevery B, Vijgen L, Jacobs T, De Meyer S, Lenz O. In vitro activity of simeprevir against hepatitis C virus genotype 1 clinical isolates and its correlation with NS3 sequence and site-directed mutants. Antimicrob Agents Chemother. 2015;59:7548–7557. doi: 10.1128/AAC.01444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouwerkerk-Mahadevan S, Snoeys J, Peeters M, Beumont-Mauviel M, Simion A. Drug–drug interactions with the NS3/4A protease inhibitor simeprevir. Clin Pharmacokinet. 2016;55:197–208. doi: 10.1007/s40262-015-0314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheahan TP, Sims AC, Leist SR, Schäfer A, Won J, Brown AJ, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon β against MERS-CoV. Nat Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maisonnasse P, Guedj J, Contreras V, Behillil S, Solas C, Marlin R, et al. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020;585:584–587. doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]