Abstract
In budding yeast, anaphase initiation is controlled by ubiquitin-dependent degradation of Pds1p. Analysis of pds1 mutants implicated Pds1p in the DNA damage, spindle assembly, and S-phase checkpoints. Though some components of these pathways are known, others remain to be identified. Moreover, the essential function of Pds1p, independent of its role in checkpoint control, has not been elucidated. To identify loci that genetically interact with PDS1, we screened for dosage suppressors of a temperature-sensitive pds1 allele, pds1-128, defective for checkpoint control at the permissive temperature and essential for viability at 37°C. Genetic and functional interactions of two suppressors are described. RAD23 and DDI1 suppress the temperature and hydroxyurea, but not radiation or nocodazole, sensitivity of pds1-128. rad23 and ddi1 mutants are partially defective in S-phase checkpoint control but are proficient in DNA damage and spindle assembly checkpoints. Therefore, Rad23p and Ddi1p participate in a subset of Pds1p-dependent cell cycle controls. Both Rad23p and Ddi1p contain ubiquitin-associated (UBA) domains which are required for dosage suppression of pds1-128. UBA domains are found in several proteins involved in ubiquitin-dependent proteolysis, though no function has been assigned to them. Deletion of the UBA domains of Rad23p and Ddi1p renders cells defective in S-phase checkpoint control, implicating UBA domains in checkpoint signaling. Since Pds1p destruction, and thus checkpoint regulation of mitosis, depends on ubiquitin-dependent proteolysis, we propose that the UBA domains functionally interact with the ubiquitin system to control Pds1p degradation in response to checkpoint activation.
When DNA is damaged or chromosomes are incompletely replicated, cells become checkpoint arrested. These checkpoints avoid replication of damaged template DNA and prevent aberrant segregation of damaged or partly replicated chromosomes. In budding yeast, proteolysis of anaphase inhibitors is regulated by these checkpoint systems. Progression from metaphase to anaphase is inhibited by Pds1p in Saccharomyces cerevisiae (6, 7, 29, 30). Before anaphase, Pds1p binds to Esp1p, inhibiting its anaphase-promoting activity (3). During an unperturbed cell cycle, Pds1p becomes polyubiquitinated at the metaphase-to-anaphase transition by multienzyme anaphase-promoting complex (APC)–cyclosome complexes. The modified forms are then recognized and degraded by 26S proteasomes (7). Once released from Pds1p, Esp1p activity induces the onset of anaphase.
pds1 mutants fail to execute checkpoint control in response to DNA damage, spindle poisons, or replication inhibition (4, 29, 30). Pds1p is required for replication checkpoint control only late in S phase, not in the context of an early S-phase replication block enforced by hydroxyurea (HU) (4, 29, 30). In the presence of 0.1 M HU, replication proceeds more slowly. Under these conditions, cells perform other aspects of cell cycle progression, budding, and spindle assembly as rapidly as in the absence of HU; cell cycle arrest in G2 is then necessary to delay anaphase while replication is completed. Growth in the presence of a non-replication-arresting concentration of HU therefore challenges the ability of cells to accurately couple S phase with mitosis. Although pds1 mutants can prevent mitotic progression when replication is blocked, defective S-phase checkpoint control can be observed when replication is partially inhibited. Such experiments revealed a novel checkpoint pathway that is essential late in S phase. Components of this pathway, other than Pds1p and Mec1p (4; D. J. Clarke, M. Segal, S. Jensen, and S. I. Reed, submitted for publication), have not been identified. Moreover, the essential function of Pds1p at high temperature is not known. To gain insight into this function, we screened for dosage suppressors of pds1-128 and have characterized two, encoded by RAD23 and DDI1. Ubiquitin-associated (UBA) domains located at the C termini of both Rad23p and Ddi1p are required for suppression of pds1-128. Interestingly, these domains are found in several proteins involved in ubiquitin-dependent proteolysis, though no function has been assigned to UBA domains. Further experiments implicate these UBA domains in checkpoint signaling: strains with mutant versions of the proteins lacking the UBA domains are S-phase checkpoint defective.
MATERIALS AND METHODS
Yeast strain construction and growth conditions.
All strains are derivatives of BF264-15DU MATa ade1 his2 leu2-3,112 trp1-1a ura3 Dns (20). Cultures were grown at 30°C, unless otherwise stated, in yeast extract-peptone (YEP) medium containing 2% dextrose, raffinose, or galactose. Strains were constructed according to standard procedures (22) except for gene disruptions (27). pds1Δ cells were temperature sensitive at 28°C on YEP-dextrose plates in the BF264-15DU genetic background. Rad23p and Ddi1p were epitope tagged at the C-terminal extremities of their genomic coding regions with either six Myc or six His epitopes. The C-terminal region of each coding sequence was amplified by PCR using oligonucleotides that included NotI or SalI sites and then cloned into tagging vectors in frame with the epitope. Such constructs were linearized by restriction enzyme digestion within the RAD23 or DDI1 open reading frame (ORF) and then integrated at the endogenous RAD23 and DDI1 loci. GAL1:RAD23 and GAL1:DDI1 strains were constructed by amplifying the RAD23 and DDI1 coding sequences, cloning into a yeast integration vector behind a GAL1 promoter and then integrating at the LEU2 locus. Epitope tags could then be integrated behind the GAL1:RAD23 and GAL1:DDI1 sequences. rad23Δuba2 and ddi1Δuba mutant alleles, expressed endogenously or under GAL1 promoter control, were constructed by integrating epitope tags directly 3′ of the UBA domain sequences, thus truncating the corresponding coding sequence. Temperature-sensitive esp1 strains were generated by PCR-based mutagenesis (13, 18).
Yeast strain genotypes.
Genotypes used are shown in Table 1. Oligonucleotides used are shown in Table 2.
TABLE 1.
Yeast strain genotypes
| Strain | Genotype |
|---|---|
| DCY1180 | MATabar1Δ |
| DCY1167 | MATα pds1-128 |
| DCY1283 | MATabar1Δ rad23::KANRddi1::KANR |
| DCY1270 | MATabar1Δ GAL1:RAD23(LEU2) |
| DCY1278 | MATabar1Δ GAL1:DDI1(LEU2) |
| DCY1228 | MATabar1Δ pds1::KANR |
| DCY1234 | MATabar1Δ pds1::KANR YEp-ESP1(URA3) |
| DCY1235 | MATabar1Δ pds1::KANR YEp-RAD23(URA3) |
| DCY1236 | MATabar1Δ pds1::KANR YEp-PDS1(URA3) |
| DCY1237 | MATabar1Δ pds1::KANR YEp-DDI1(URA3) |
| DCY1233 | MATabar1Δpds1::KANR YEp24(URA3) |
| DCY1195 | MATapds1-128 YEp-DDI1(URA3) |
| DCY1196 | MATapds1-128 YEp-ESP1(URA3) |
| DCY1199 | MATapds1-128 YEp-RAD23(URA3) |
| DCY1210 | MATapds1-128 YEp-PDS1(URA3) |
| DCY1790 | MATα pds1-128 YEp-pds1-128(URA3) |
| DCY1213 | MATapds1-128 YEp24(URA3) |
| DCY1384 | MATaesp1::KANR YCp-esp1-N5(TRP1) GAL1:RAD23(LEU2) |
| DCY1386 | MATaesp1::KANR YCp-esp1-B7(TRP1) GAL1:RAD23(LEU2) |
| DCY1786 | MATaesp1-N5(TRP) GAL1:RAD23(LEU2) |
| DCY1787 | MATaesp1-N5(TRP) GAL1:rad23Δuba2(LEU2) |
| DCY1788 | MATabar1Δ esp1-N5(TRP) GAL1:DDI1-His,)(TRP1)::LEU2 |
| DCY1789 | MATaesp1-N5(TRP) bar1Δ GAL1:ddi1Δuba-His,(URA3)::LEU2 |
| DCY1668 | MATaesp1::KANR YCp-esp1-B7(TRP1) GAL1:DDI1(LEU2) |
| DCY1264 | MATα bar1Δ rad23::KANR |
| DCY1258 | MATα bar1Δ ddi1::KANR |
| DCY1359 | MATα rad9::TRP1 |
| DCY1407 | MATapds1-128 GAL1:DDI1(LEU2) |
| DCY1272 | MATabar1Δ pds1-128 GAL1:RAD23(LEU2) |
| DCY1277 | MATabar1Δ pds1::KANR GAL1:DDI1(LEU2) |
| DCY1636 | MATapds1::KANR GAL1:RAD23(LEU2) |
| DCY1810 | MATapds1-128 chk1::KANR GAL1:RAD23(LEU2) |
| DCY1812 | MATapds1-128 chk1::KANRGAL1:DDI1(LEU2) |
| DCY1819 | MATα pds1-128 mec1-1 sml1Δ |
| DCY1826 | MATapds1-128 mec1-1 sml1Δ GAL1:DDI1(LEU2) |
| DCY1824 | MATapds1-128 mec1-1 sml1Δ GAL1:RAD23(LEU2) |
| DCY1838 | MATarad53-1 GAL1:RAD23(LEU2) |
| DCY1836 | MATapds1-128 rad53-1 GAL1:RAD23(LEU2) |
| GY1492 | MATα pds1-128 GAL1:rad23Δuba2-Myc, (URA3, LEU2) |
| GY1493 | MATα pds1-128 GAL1:DDI1-His6 (LEU2, TRP1) |
| GY1494 | MATα pds1-128 GAL1:ddi1Δuba-His6 (LEU2, URA3) |
| DCY1671 | MATabar1Δ GFP-TUB1::URA3 |
| DCY1672 | MATabar1Δ pds1-128 GFP-TUB1::URA3 |
| DCY1679 | MATabar1Δ rad23Δuba2-myc6::TRP1 ddi1Δuba-His6::LEU2 GFP-TUB1::URA3 |
| DCY1674 | MATabar1Δ rad23::KANRddi1::KANRGFP-TUB1::URA3 |
| DCY1859 | MATapds1-128 rad53-1 YEp-DDI1(URA3) |
| DCY1860 | MATapds1-128 rad53-1 YEp-RAD23(URA3) |
| DCY1858 | MATapds1-128 rad53-1 YEp-PDS1(URA3) |
| DCY1861 | MATapds1-128 rad53-1 YCp50 |
| DCY1865 | MATamec1-1 sml1 ΔYC50 |
| DCY1863 | MATamec1-1 sml1Δ YEp-DDI1(URA3) |
| DCY1864 | MATamec1-1 sml1Δ YEp-RAD23(URA3) |
| DCY1862 | MATamec1-1 sml1Δ YEp-PDS1(URA3) |
| DCY1869 | MATapds1-128 mec1-1 sml1Δ YEp-DDI1(URA3) |
| DCY1870 | MATapds1-128 mec1-1 sml1Δ YEp-RAD23(URA3) |
| DCY2078 | MATabar1Δ rad23Δuba2-Myc6::TRP1 ddi1Δuba-His6::LEU2 CUP1:tetR-GFP(KANR) trp1::tetO(TRP1) |
| DCY1662 | MATabar1Δ CUP1:tetR-GFP(KANR) trp1::tetO(TRP1) |
| DCY1866 | MATapds1-128 mec1-1 sml1Δ YEp-PDS1(URA3) |
| DCY1867 | MATapds1-128 mec1-1 sml1Δ YEp24 |
TABLE 2.
Oligonucleotide used in plasmid or strain construction
| PCR product | Oligonucleotide sequence (5′→3′) |
|---|---|
| RAD23 ORF for GAL1 expression (5′, BamHI site) | CGGGATCCAAAAATGGTTAGCTTAACCTTT |
| RAD23 ORF for GAL1 expression (3′, BamHI site) | CGGGATCCAATCTCAGTCGGCATGATCGCT |
| DDI1 ORF for GAL1 expression (5′, BamHI site) | CGGGATCCAAAGATGGATTTAACAATTTCA |
| DDI1 ORF for GAL1 expression (3′, BamHI site) | CGGGATCCTTGATCATTGGAAAAGGAGGGA |
| Rad23p C terminus for tagging (5′, SalI site) | GTCGACCATCCGGTGCGCTTGGCACAACTG |
| Rad23p C terminus for tagging (3′, NotI site) | GCGGCCGCCGTCGGCATGATCGCTGAATAG |
| Ddi1p C terminus for tagging (5′, SalI site) | GTCGACAATTATTGGGAGAATTCACCAAGC |
| Ddi1p C terminus for tagging (3′, NotI site) | GCGGCCGCCTTGGAAAAGGAGGGATGCAGC |
| Radp23p ΔUBA2 C terminus for tagging (3′, NotI site) | GCGGCCTCAATAGTCAACTTGGAAAGAACCTTC |
| Ddi1p ΔUBA C terminus for tagging (3′, NotI site) | GCGGCCTCACGTTGTAGCACCGGCTGTACTTCT |
Isolation of pds1-128 dosage suppressors.
pds1-128 suppressors were isolated by transformation with an S. cerevisiae genomic library contained in a vector which is maintained at high copy number in yeast cells (YEp24 library [2]). Transformants were replicated onto YEP-galactose plates, and suppressors were selected at 37°C. About 160,000 transformants were analyzed to identify 110 colonies rescued in a plasmid-dependent manner; 80 of these colonies appeared wild type (WT) at 37°C on YEP-galactose medium. Of the 13 clones examined further, all contained PDS1; 30 of the 110 colonies grew more poorly than WT at 37°C on YEP-galactose medium. Plasmids isolated from each of these clones contained either ESP1, RAD23, or DDI1.
Cell biology protocols.
Samples were prepared for FACScan analysis as described elsewhere (14). Spindles were visualized by fluorescence microscopy by using strains expressing a TUB1-GFP construct (23). Cells were gamma irradiated in liquid medium (3.5 Gy per min). For G1 arrest, strains were grown overnight to saturation and then diluted to an optical density of 0.15 to 0.2; then α-factor (200 ng/ml) was added; 90 to 100% arrest was complete within 2 to 2.5 h at 26 to 30°C. For experiments in which spindle morphology was analyzed, it was necessary to grow cells overnight in synthetic medium to allow good visualization of the spindles. When induction of genes from the GAL1 promoter was required, overnight cultures were grown in YEP-raffinose. Synchrony was carried out in rich medium as described in the text.
RESULTS
Dosage suppressors of pds1-128.
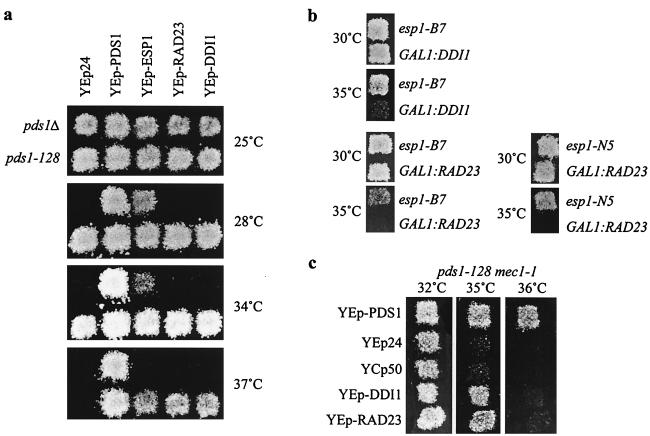
A genetic strategy was used to find proteins that functionally interact with Pds1p. We screened for genes whose increased dosage suppressed the temperature sensitivity of pds1-128. From a genomic S. cerevisiae library (maintained at high copy number in a yeast episomal vector), three dosage suppressors were identified: (i) ESP1 (13 independent isolates, representing six plasmid types), (ii) RAD23 (4 isolates, one plasmid type), and (iii) DDI1 (8 isolates, two plasmid types). Since a genetic interaction between ESP1 and pds1 was previously reported (7), two novel suppressors of pds1-128, RAD23 and DDI1, were identified (Fig. 1a).
FIG. 1.
Dosage suppressors of pds1-128. (a) Suppression of pds1-128 but not pds1Δ temperature sensitivity by episomal plasmids maintained at high copy number in yeast cells carrying YEp24(URA3), YEp-PDS1, YEp-RAD23, YEp-DDI1, and YEp-ESP1. Patches were grown at 25°C on synthetic medium containing dextrose but without uracil to prevent loss of the YEp24-based plasmids. These patches were replicated on to YEP-dextrose plates and grown at 25 to 37°C for 2 days. (b) Genetic interactions between GAL1:RAD23 or GAL1:DDI1 and temperature-sensitive esp1 strains. Strains carrying esp1 alleles (esp1-N5 and esp1-B7 [13]) with or without RAD23 and DDI1 alleles under control of the inducible GAL1 promoter were grown at 30°C on YEP-dextrose plates, then replicated to YEP-galactose plates, and grown for 2 days at 30 to 35°C. (c) Suppression of pds1-128 mec1-1 temperature sensitivity by episomal plasmids maintained at high copy number in yeast cells carrying YEp-PDS1, YEp-RAD23, YEp-DDI1, YEp24, and YCp50, grown as for panel a.
pds1-128 cells become aneuploid when grown at the restrictive temperature, as a result of unequal nuclear division. Dosage suppression by RAD23 and DDI1 could result from a specific effect at the time of nuclear division or could improve cell viability in a less specific manner, not directly relevant to the function of Pds1p. To address this, cycle progression of pds1-128 cells, with or without RAD23 overexpression, was monitored by FACScan analysis following release from G1 (data not shown). While most pds1-128 cells became aneuploid following nuclear division, the majority of cells overexpressing RAD23 maintained 1C or 2C DNA contents. Similar results were obtained when DDI1 was overexpressed. Therefore, dosage suppression of pds1-128 at least partially corrected the primary functional defect in nuclear division at 37°C.
Specificity of pds1-128 dosage suppression.
Additional criteria were used to assess the specificity of the genetic interactions with pds1-128: first, whether the suppressors could rescue a pds1 null mutant (pds1Δ); and second, whether genetic interactions with esp1 (encoding a protein that physically interacts with Pds1p) could be detected (Fig. 1a and b).
Neither DDI1 nor RAD23 could rescue the inviability of pds1Δ cells at 28°C, the restrictive temperature of pds1Δ (strains harboring 2 μm plasmids or integrated versions of RAD23 and DDI1 induced from the GAL1 promoter). Thus, high dosage of DDI1 or RAD23 cannot bypass the pds1Δ temperature sensitivity.
If Rad23p and Ddi1p are specific regulators of Pds1p, a genetic interaction with esp1 mutants should be detectable. To test this, for this purpose, we constructed WT or esp1 mutant strains which express RAD23 or DDI1 from the inducible GAL1 promoter. Expression of either gene in WT cells had little effect on cell growth (not shown), but the lethality of strains with temperature-sensitive esp1 alleles (esp1-N5 and esp1-B7 [13]) was enhanced by overexpression of RAD23 or DDI1 (Fig. 1b). This result can be rationalized in the context of the model described above; Pds1p inhibits the anaphase-promoting function of Esp1p, and Rad 23p and Ddi1p are putative positive regulators of Pds1p function. Therefore, high-dosage RAD23 and DDI1 would be expected to further compromise mutant esp1 function, resulting in increased temperature sensitivity.
The genetic interactions of RAD23 and DDI1 with pds1-128 were also examined in the context of Pds1p-dependent checkpoint pathways. The ability of RAD23 or DDI1 to rescue pds1-128 temperature sensitivity in mec1-1, rad53-1 or chk1Δ mutant backgrounds was examined (Fig. 1c). Experiments with the YEp24-based plasmids, maintained at high copy number in yeast cells, or with RAD23 and DDI1 expressed from the GAL1 promoter gave essentially the same results. pds1-128 rad53-1 and pds1-128 chk1Δ cells were temperature sensitive at 37°C, as are pds1-128 cells, and viability was restored by high dosage of the suppressors (not shown). Using the same analysis, pds1-128 mec1-1 cells were only partly rescued by RAD23 and DDI1 overexpression (up to 35°C). pds1-128 mec1-1 cells were temperature sensitive at 35°C, though mec1-1 single mutants were not temperature sensitive. The inviability of pds1-128 mec1-1 cells overexpressing RAD23 or DDI1 at 36°C suggests that rescue is partly dependent on Mec1p. Rad23p, Ddi1p, and Mec1p may function in a common pathway that targets Pds1p.
Dosage suppression of the pds1-128 S-phase checkpoint defect.
The genetic interactions of RAD23 and DDI1 with pds1-128 were examined in the context of documented pds1 checkpoint defects. We tested whether induction of GAL1:RAD23 or GAL1:DDI1 was able to suppress the sensitivity of pds1-128 to nocodazole, gamma irradiation, or HU to examine whether Rad23p and Ddi1p could function in the spindle assembly, DNA damage, or S-phase checkpoint pathways. There was no suppression of the gamma irradiation sensitivity of pds1-128 (data not shown). In contrast, a high-copy-number plasmid containing the pds1-128 allele (YEp-pds1-128) did rescue the gamma irradiation sensitivity. Thus, increasing the amount of the mutant Pds1-128p corrected the DNA damage checkpoint defect of these cells. Similarly, YEp-pds1-128 rescued the temperature sensitivity of pds1-128 cells. By Western blot analysis, pds1-128 cells have substantially less Pds1 than do WT cells (13). Therefore, in the context of the DNA damage checkpoint, RAD23 or DDI1 overexpression presumably cannot increase the amount of Pds1-128p. Overexpression of RAD23 or DDI1 was also unable to rescue the nocodazole sensitivity of pds1-128 cells (data not shown). In this case, however, YEp-pds1-128 was similarly unable to rescue lethality caused by nocodazole treatment. Therefore, it was not possible to infer whether or not RAD23 and DDI1 overexpression affects Pds1-128p levels in the context of the spindle assembly checkpoint.
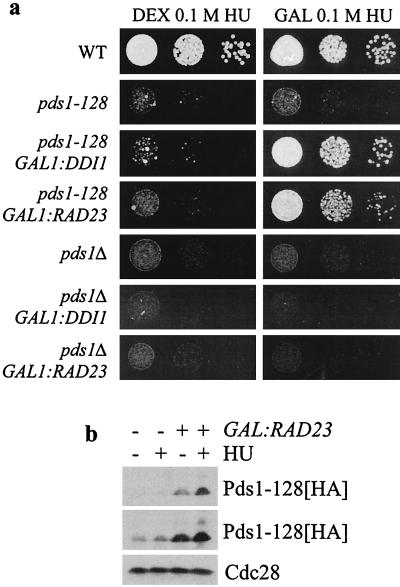
In contrast, induction of GAL1:RAD23 or GAL1:DDI1 did suppress the sensitivity of pds1-128 cells to HU (Fig. 2a). Growth on HU plates was also restored by YEp-pds1-128 (not shown). Crucially, GAL1:RAD23 and GAL1:DDI1 could not rescue the HU sensitivity of pds1Δ strains, revealing that rescue was dependent on the presence of Pds1-128p. High-copy-number RAD23 did increase Pds1-128p levels, and this effect was enhanced in the context of the S-phase checkpoint response (Fig. 2b). Moreover, overexpression of RAD23 was not able to rescue the HU sensitivity of a rad53-1 strain (another S-phase checkpoint-defective mutant), indicating that not all HU-sensitive checkpoint mutants are suppressed. The above experiments revealed that RAD23 and DDI1 are not dosage suppressors of the DNA damage checkpoint defect of pds1-128 mutant cells but can suppress the S-phase checkpoint defect. The data suggest that Rad23p and Ddi1p function specifically in a Pds1p-dependent S-phase checkpoint control.
FIG. 2.
Suppression of pds1-128 S-phase checkpoint defects. (a) Suppression of pds1-128 HU sensitivity by RAD23 and DDI1 overexpression. Strains were grown to midlog phase, and then serial dilutions were spotted on to YEP-dextrose or YEP-galactose plates (to repress or induce GAL1:RAD23 and GAL1:DDI1) containing HU and grown for 2 to 3 days. (b) Overexpression of RAD23 increases Pds1-128p levels in HU-treated cells. Cells containing a GAL1:RAD23 gene and a hemagglutinin (HA) epitope-tagged pds1-128 allele were grown to midlog phase (minus HU) or accumulated in S-phase with 100 mM HU for 3 h (plus HU) in dextrose (−) or galactose (+) medium to repress or induce GAL1:RAD23. Cellular protein extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Pds1-128-p-HA was detected after Western blotting. Cdc28p levels served as a loading control. A light exposure and a darker exposure of the same blot are shown.
Role of Rad23p and Ddi1p UBA domains in dosage suppression of pds1-128.
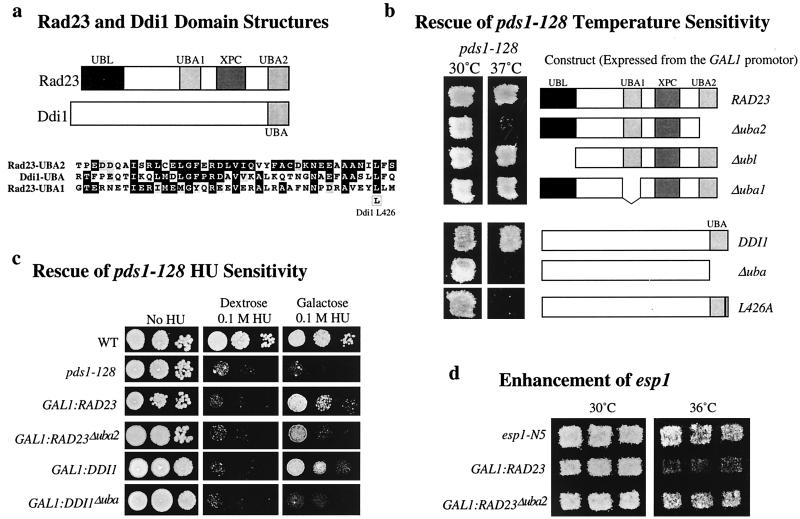
One striking similarity between the C termini of Rad23p and Ddi1p is that both contain a UBA domain (Rad23p has a second, internally located UBA domain [Fig. 3a]). These are small domains (each about 10% of the full-length protein) of unknown function. Six UBA domain-containing proteins have been identified in budding yeast.
FIG. 3.
Suppression of pds1-128 temperature and HU sensitivity depends on Rad23p and Ddi1p UBA domains. (a) Structural similarity between RAD23 and DDI1. Box diagrams show relative sizes of each ORF and the size and position of UBA, XPC, and UBL domains. C-terminal UBA domains (right) share 38% identity. The position of Ddi1p L426 is indicated in the alignment below. A hypothetical fission yeast protein (gb|Z69728 and gi|1204230) has an overall 32% identity to budding yeast Ddi1p. Probable Ddi1p homologues giving high identity scores are present in Homo sapiens (gb|AA406136 and gi|2064117; dbest database), Caenorhabditis elegans (gb|U50068 and gi|1208859) and Leishmania major (gb|AC002305 and gi|2429118). (b and c) Temperature sensitivity (b) and HU sensitivity (c) of pds1-128 strains overexpressing RAD23 and DDI1 or truncated versions of these genes expressing proteins lacking UBA domains or the RAD23 UBL. Similar results were obtained when the WT or truncated proteins were tested as (Rad23p-MYC6 and Ddi1p-His6) fusions. Strains were handled as described in the legends to Fig. 1b and 2a. (d) Lack of genetic interaction between GAL1:rad23Δuba2 and temperature-sensitive esp1. Strains were handled as described in the legend to Fig. 1b.
We tested whether the UBA domains of Rad23p and Ddi1p are required for suppression of pds1-128. Mutant versions of RAD23 and DDI1 that lack the C-terminal UBA domains (about 10% of each protein deleted) were created. To ensure that the mutant versions were stable in yeast cells, we added epitope tags at the C termini and confirmed endogenous expression to be at WT levels (or to be sufficiently elevated when under control of the GAL1 promoter) by Western blotting (not shown). When expressed from the GAL1 promoter, the mutant versions of Rad23, and Ddi1p (encoded by GAL1:rad23Δuba2 and GAL1:ddi1Δuba) were unable to rescue the temperature sensitivity of pds1-128, though rescue was achieved by full-length versions of either protein (Fig. 3b). Suppression was also achieved by a Rad23p construct that lacked the N-terminal ubiquitin-like (UBL) domain and also by a Rad23p mutant lacking the internal UBA domain. Thus, the C-terminal UBA domains of Rad23p and Ddi1p seem to be the critical domains for pds1-128 suppression. In the case of Ddi1p, we also constructed point mutants of the UBA domain. One such mutant, L426A, could be expected to disrupt UBA integrity, based on structure prediction. In yeast two-hybrid experiments, this mutant was able to form Ddi1p-Ddi1p homodimers (B. L. Bertolaet and S. L. Reed, unpublished data), demonstrating that other than the UBA domain, the protein must be structurally intact. L426A did not, however, have the ability to suppress pds1-128 temperature sensitivity (Fig. 3b). Thus, L426A specifically abolished the ability of Ddi1p to suppress pds1-128. The C-terminal UBA domains of Rad23p and Ddi1p were also required for enhancement of esp1 temperature sensitivity (Fig. 3d and data not shown).
The ability of the mutant forms to rescue the HU sensitivity of pds1-128 was also examined. Induction of GAL1:ddi1Δuba was unable to rescue pds1-128 HU sensitivity, whereas deletion of UBA2 from Rad23p produced a mutant that could only partially rescue the HU sensitivity (Fig. 3c). Therefore, the UBA domain of Ddi1p was essential for suppression of the pds1-128 HU sensitivity, and deletion of the C-terminal UBA domain of Rad23p reduced the ability of Rad23p to rescue the HU sensitivity of pds1-128 cells.
rad23Δ ddi1Δ null mutants are sensitive to HU and defective in cell cycle progression in the presence of HU.
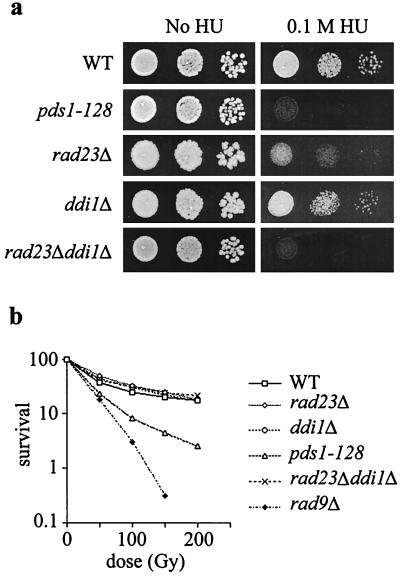
The genetic and functional interactions of RAD23 and DDI1 with pds1-128 suggested that Rad23p and Ddi1p may function in the S-phase checkpoint pathway and that the putative checkpoint function of Rad23p and Ddi1p depends on conserved UBA domains. It could therefore be expected that loss-of-function alleles of RAD23 or DDI1 would result in loss of S-phase checkpoint control. The common UBA domains of Rad23p and Ddi1p suggested that the two proteins may have some functional redundancy. We therefore constructed RAD23 and DDI1 deletion strains and tested their HU sensitivity. (Fig. 4a). rad23Δ ddi1Δ cells were as sensitive to HU as are pds1-128 cells, though rad23Δ and ddi1Δ single mutants were either much less sensitive (rad23Δ) or not sensitive at all (ddi1Δ). rad23Δ ddi1Δ cells were not sensitive to gamma irradiation (Fig. 4b) or nocodazole (not shown).
FIG. 4.
rad23Δ ddi1Δ cells are HU sensitive but not radiation sensitive. (a) Sensitivity of pds1 and rad23Δ ddi1Δ mutants to HU. Strains were grown to midlog phase, then serial dilutions were spotted on to YEP-dextrose or YEP-dextrose plates containing HU, and growth continued for 2 to 3 days at 30°C. (b) Sensitivity of pds1, rad23Δ, ddi1Δ, and rad9Δ mutants to gamma irradiation. Strains were grown to midlog phase at 30°C in YEP-dextrose medium, irradiated, and then plated on YEP dextrose. Colonies were counted after 2 to 4 days.
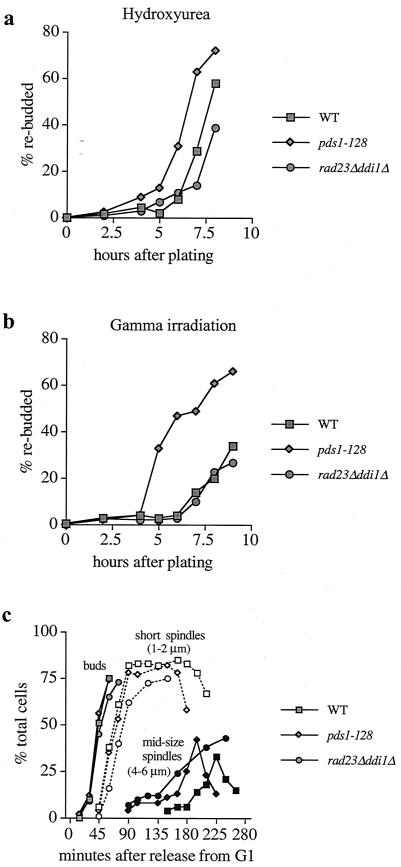
To test whether rad23Δ ddi1Δ cells are S-phase checkpoint defective, a rebudding assay was performed (Fig. 5a). Cells released from G1 following α-factor arrest were grown on solid medium containing 100 mM HU, and cell bodies per microcolony were counted. For WT pds1-128 and rad23Δ ddi1Δ cells, budding occurred with very similar timing (not shown). The premature appearance of colonies containing three or more cell bodies (rebudding) would indicate an uncoupling of the normal timing of cell cycle progression from S phase. In these experiments, pds1-128 cells rebudded before WT cells. However, if anything, rad23Δ ddi1Δ cells took longer to rebud than WT cells, suggesting a slight delay before or during mitosis (Fig. 5a). In a similar experiment, in which the strains were gamma irradiated before plating onto rich medium, WT and rad23Δ ddi1Δ cells rebudded with indistinguishable timing (Fig. 5b), whereas pds1-128 cells rebudded before WT and rad23Δ ddi1Δ cells, due to the DNA damage checkpoint defect of this strain (Fig. 5b and reference 29).
FIG. 5.
S-phase checkpoint defect of rad23Δ ddi1Δ mutants (a) Rebudding assay on HU plates. WT, pds1-128, and rad23Δ ddi1Δ cells were arrested in G1 at 30°C (α-factor), washed, and plated on YEP-dextrose plates containing 100 mM HU. Each strain budded with similar timing (not shown). Rebudding (the appearance of microcolonies containing three or more cell bodies) was scored at various time intervals (see also Fig. 6). (b) Rebudding assay following gamma irradiation. The procedure was as described for Fig. 5a. The ability of cells to rebud on YEP-dextrose plates following gamma irradiation in G1 was scored. (c) Timing of budding, G2 spindle formation, and spindle elongation in WT and mutant cells grown in liquid YEP-dextrose medium containing 100 mM HU following release from G1 (α-factor).
To further examine the phenotype of rad23Δ ddi1Δ cells in the presence of HU, we directly compared S-phase progression with the onset of anaphase following release from G1 (scoring budding, mitotic spindle formation using a TUB1-GFP construct, and DNA replication by FACscan analysis [Fig. 5c]). Budding, spindle formation, and DNA replication (FACScan data not shown) occurred with similar timing in each strain. Short G2 spindles were defined as those between 1 and 2 μm long in these experiments. Strikingly, spindle elongation began in the rad23Δ ddi1Δ cells at the same time as in pds1-128 cells, when about two-thirds of the DNA had been replicated. At this time, while spindles of WT cells remained 1 to 2 μm in length, spindles of rad23Δ ddi1Δ cells elongated to 4 to 6 μm. However, while spindles fully elongated (to 8 to 12 μm) and then disassembled in a timely manner in pds1-128 cells, rad23Δ ddi1Δ spindles remained about 4 to 6 μm in length (midsized spindles), consistent with a midanaphase delay. rad23Δ ddi1Δ cells eventually elongated their spindles fully and exited mitosis (not shown), but later than WT cells did, in agreement with the rebudding experiments. These data suggest that rad23Δ ddi1Δ cells may be defective for restraining anaphase onset in HU but are unable to complete spindle elongation and exit mitosis rapidly.
Role of Rad23p and Ddi1p UBA domains in S-phase checkpoint control.
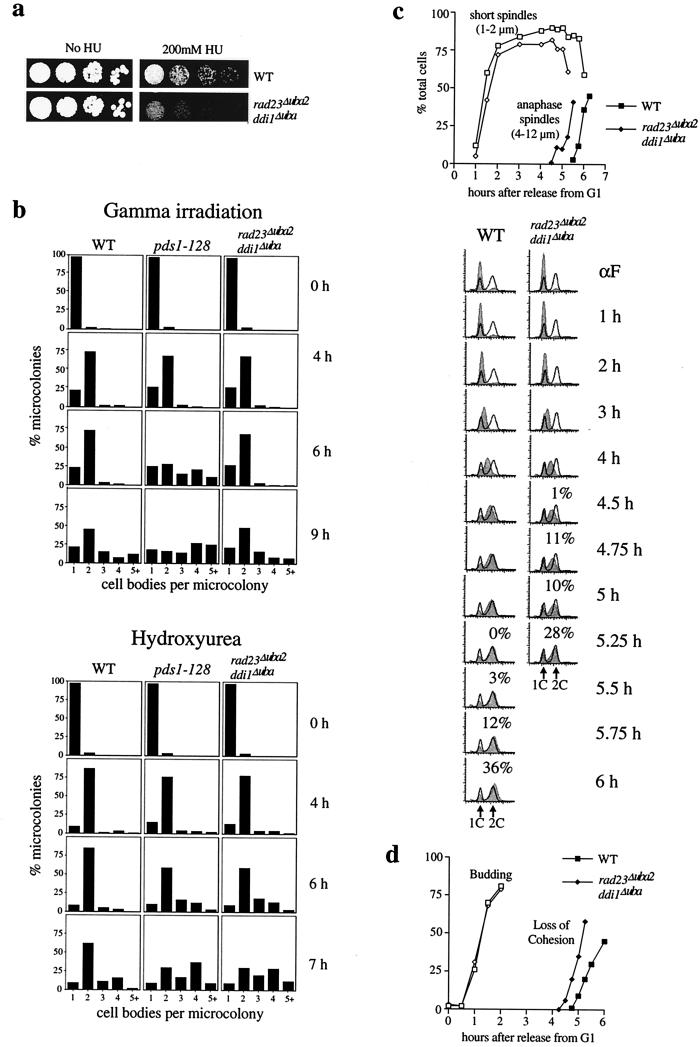
The potential dual roles of Rad23p in nucleotide excision repair (NER) and checkpoint control might result in a somewhat ambiguous checkpoint-defective phenotype in the rad23Δ ddi1Δ mutant. Although these cells appeared to initiate anaphase before S phase was complete, they also appeared to be delayed for progression through the latter parts of mitosis. To resolve this issue, we attempted to create strains with mutant versions of Rad23p and Ddi1p that would be defective for checkpoint control but not compromised in NER or in the ability to progress through mitosis. We constructed strains in which endogenous RAD23 and DDI1 were replaced with mutant versions lacking the C-terminal UBA domains (rad23Δuba2 and ddi1Δuba), based on the genetic evidence that these mutant versions cannot suppress pds1-128. The UBA domain deletion alleles were epitope tagged to confirm that expression levels of the resulting products were similar to those of the WT proteins (not shown). We reasoned that since these domains are at least partly required for suppression of pds1-128 HU sensitivity, it may be possible to detect a checkpoint defect in strains with UBA domain deletions. Either a rad23Δuba2 or a ddi1Δuba mutation showed only a marginal sensitivity to HU (not shown). In contrast, strains with both mutated alleles were HU sensitive (Fig. 6a), though not as sensitive as pds1-128 strains. This suggested that rad23Δuba2 ddi1Δuba cells might be partially defective in S-phase checkpoint regulation. rad23Δuba2 ddi1Δuba cells were not sensitive to nocodazole, gamma irradiation, or UV irradiation (not shown), correlating with the failure of high dosage RAD23 and DDI1 to rescue the sensitivity of pds1-128 to these agents and confirming that the rad23Δuba2 ddi1Δuba strain is NER competent.
FIG. 6.
S-phase checkpoint defect of rad23Δuba2 ddi1Δuba mutant cells. (a) Sensitivity of rad23Δuba2 ddi1Δuba mutant cells to HU. Strains were grown to midlog phase, then serial dilutions were spotted onto YEP-dextrose or YEP-dextrose plates containing HU, and growth continued for 2 to 3 days at 30°C. (b) Rebudding assay on HU plates or following gamma irradiation (see legend to Fig. 5 for details). Microcolonies were scored based on the number of cell bodies per microcolony. (c) Timing of short spindle formation, S-phase progression, and spindle elongation in WT and mutant cells grown in liquid YEP-dextrose medium containing 100 mM HU following release from G1 (α-factor). S-phase progression was monitored by FACScan analysis; histogram plots show DNA content throughout the experiment (in grey), and overlaid solid lines indicate positions of 1C and 2C DNA peaks taken from the asynchronous cultures prior to G1 arrest. The percentage of the total cells that had elongated spindles above each histogram. (d) Timing of loss of sister chromatid cohesion in WT and rad23Δuba2 ddi1Δuba mutant cells grown in liquid YEP-dextrose medium containing 100 mM HU following release from G1 (α-factor). Both strains budded and replicated DNA with similar timing (FACScan analysis not shown), but loss of cohesion occurred prematurely in the mutant cells. Cohesion was monitored at the TRP1 locus using the tetR-GFP/tetO system as previously described (4).
To determine whether rad23Δuba2 ddi1Δuba cells are S-phase checkpoint defective, the rebudding assay was performed on medium containing 100 mM HU (Fig. 6b). WT, pds1-128, and rad23Δuba2 ddi1Δuba cells budded with similar timing, but pds1-128 and rad23Δuba2 ddi1Δuba cells clearly rebudded prematurely, well before WT cells did. As a control, the rebudding assay was performed following gamma irradiation of G1 cells (as described for Fig. 5). In this case, the behavior of rad23Δuba2 ddi1Δuba cells was indistinguishable from that of WT cells, demonstrating that the DNA damage checkpoint is intact in this mutant strain.
Next we compared S-phase progression with the onset of anaphase (measuring spindle elongation or loss of sister chromatid cohesion) following release from G1 (Fig. 6c and d). Spindle formation and progression through S-phase occurred with similar timing in WT and rad23Δuba2 ddi1Δuba cells. WT cells reached G2 about 5 h following release from G1 and then initiated anaphase ∼30 to 40 min later. In contrast, spindle elongation in the rad23Δuba2 ddi1Δuba mutant began earlier than in WT cells and before the cells reached a 2C DNA content. The rad23Δuba2 ddi1Δuba mutant cells did not become delayed partway through spindle elongation, with 4- to 6-μm spindles, as was the case for the rad23Δ ddi1Δ mutant. Compared directly to pds1-128 cells (not shown), anaphase began at an intermediate time in rad23Δuba2 ddi1Δuba cells, later than in pds1-128 cells but consistently earlier than in WT cells. When loss of sister chromatid cohesion was used as an indicator of anaphase onset, similar results were consistently obtained (Fig. 6d). These kinetics correlate well with the relative sensitivities of rad23Δuba2 ddi1Δuba and pds1-128 cells to HU and suggest that rad23Δuba2 ddi1Δuba cells are partially defective in S-phase checkpoint control. The advancement of anaphase onset and of rebudding in the rad23Δuba2 ddi1Δuba mutant clearly implicates the UBA domains of Rad23p and Ddi1p in cell cycle control.
DISCUSSION
Genetic interactions of pds1-128 with RAD23 and DDI1.
The key role of Pds1p in M-phase cell cycle control has been well documented, but less is known about factors regulating Pds1p in response to checkpoint signals. We have used a genetic screen to identify proteins that interact with Pds1p. High dosage of RAD23 or DDI1 suppressed the temperature sensitivity of pds1-128 and rescued the HU sensitivity of pds1-128 at the permissive temperature. Neither the temperature sensitivity nor the HU sensitivity was rescued by RAD23 or DDI1 overexpression in pds1Δ strains; there was no bypass of the cellular requirement for Pds1p. In agreement, high-dosage RAD23 and DDI1 enhanced the temperature sensitivity of esp1 mutants. This would be expected of proteins that positively regulate Pds1p, since Esp1p separin activity, required for progression through mitosis, is negatively regulated by Pds1p. These interactions implicate Rad23p and Ddi1p in promoting Pds1p-dependent functions. Moreover, RAD23 and DDI1 specifically corrected the S-phase checkpoint defect of pds1-128 cells. Neither the gamma irradiation sensitivity nor the nocodazole sensitivity of pds1-128 was rescued by overexpressing RAD23 and DDI1. Thus, these data implicate Rad23p and Ddi1p in subset of the Pds1p-dependent checkpoint pathways.
UBA domains of Rad23p and Ddi1p are required for S-phase checkpoint control.
Rad23p is a highly conserved protein with a NER function. The role of Rad23 in NER depends on binding to the NER complex via its XPC-binding domain and on an interaction with the proteasome cap via its UBL domain (9–11, 16, 17, 24, 28). Almost nothing is known about the function of Ddi1p. It has an upstream activation domain identical to that of RAD23 and is induced by DNA damage and HU, but unlike rad23Δ mutants, ddi1Δ strains are not sensitive to UV light (data not shown) and are therefore not required for NER.
Since both Rad23p and Ddi1p have UBA domains at their C termini, we tested whether these domains are required for suppression of pds1-128 defects. rad23Δuba2 ddi1Δuba cells were not sensitive to gamma irradiation or nocodazole but were HU sensitive and underwent an advanced loss of sister chromatid cohesion and spindle elongation compared to WT cells when grown in the presence of 100 mM HU. rad23Δuba2 ddi1Δuba mutant cells also exited mitosis and rebudded prematurely in the presence of 100 mM HU. It is not clear whether this rapid exit from mitosis could contribute to the HU sensitivity of these cells. Still, rapid exit from mitosis could be accounted for by deregulation of Pds1p proteolysis during anaphase in the rad23Δuba2 ddi1Δuba mutant. As well as regulating anaphase onset, Pds1p plays a role in controlling exit from mitosis (1, 8, 15, 25). Although some Pds1p is degraded at the onset of anaphase (7), a subfraction of Pds1p remains and is localized on anaphase spindles (13). After anaphase onset, Pds1p inhibits exit from mitosis (5, 26). Thus, rad23Δuba2 ddi1Δuba cells may be unable to efficiently regulate Pds1p proteolysis to control the onset of anaphase and to ensure a timely disassembly of anaphase spindles and subsequent exit from mitosis.
Regulation of Pds1p by Rad23p and Ddi1p?
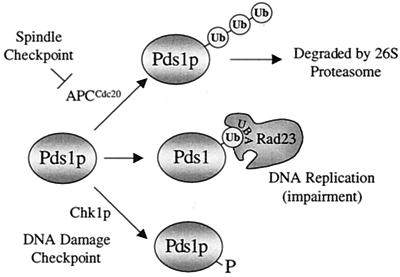
DNA damage, spindle perturbations, and replication inhibition increase Pds1p stability, which in turn inhibits the onset of anaphase. We have shown that RAD23 overexpression increases the level of Pds1-128p in a manner that is enhanced by S-phase checkpoint activation. The proposal that Rad23p and Ddi1p regulate Pds1p stability is an attractive one given the dependence of rescue on UBA domains. The function of UBA domains is not known, though they are present in different classes of enzyme involved in ubiquitin-dependent proteolysis (12), an intriguing coincidence given the dependence of Pds1p proteolysis on the ubiquitin system. Rad23p also has a UBL domain at its N terminus; this domain was recently shown to be required for a physical interaction with 26S proteasomes, the particles that degrade ubiquitinated Pds1p to initiate the onset of anaphase. However, deletion of this domain did not abrogate the ability of Rad23p to rescue pds1-128 when overproduced, and other data implicate the UBL domain in the NER function of Rad23p (21, 28). An alternative model for suppression of pds1-128 by RAD23 and DDI1 was recently suggested by the finding that both Rad23p and Ddi1p could bind to ubiquitin and that this interaction was dependent on the UBA domains (Bertolaet and Reed, unpublished). These data make it tempting to propose an intriguing mechanism for Rad23p- and Ddi1p-dependent checkpoint regulation. Rad23p and Ddi1p could regulate Pds1p stability by binding to ubiquitinated Pds1p or to the ubiquitin ligase complex that ubiquitinates Pds1p via the UBA domains (Fig. 7). Indeed, a recent report described a novel activity of Rad23p as an inhibitor of ubiquitin chain elongation (19). Rad23p was shown to bind to a mono- or a diubiquitinated substrate and prevent extension of the ubiquitin chain. A similar interaction with ubiquitinated Pds1p, or the ubiquitin ligase complex, stimulated by ongoing DNA replication, may provide one mechanism through which the S-phase checkpoint is enforced. How UBA domains participate in such control systems is currently being addressed.
FIG. 7.
Model for control of Pds1p stability. Pds1p can become polyubiquitinated by APCCdc20, a multienzyme E3 ubiquitin ligase. The ubiquitin chain enables a 26S proteasome to recognize and degrade Pds1p. These events must occur prior to the onset of anaphase, since Pds1p inhibits the activity of separin Esp1p, needed for anaphase spindle elongation and loss of sister chromatid cohesion. To block anaphase onset, checkpoint controls prevent Pds1p degradation. The spindle assembly checkpoint directly inhibits APCCdc20, and the DNA damage checkpoint is thought to promote Pds1p stability by a Chk1p-dependent phosphorylation of Pds1p. We propose a novel mechanism for anaphase control, in which Rad23p and Ddi1p are able to recognize ubiquitinated Pds1p, or the APC (not shown), and thereby inhibit the ubiquitin chain elongation that is required for targeting Pds1p to the 26S proteasome.
In summary, we propose that Rad23p and Ddi1p function in Pds1p-dependent S-phase checkpoint control in budding yeast. Rad23p and Ddi1p are likely to be universal checkpoint proteins. Their homologues in mammals and other eukaryotes are highly conserved, particularly within the UBA domains. Our data provide a first clue as to the cellular function of UBA domains in eukaryotes.
ACKNOWLEDGMENTS
We thank T. Weinert for strains, A. Straight for the TUB1-GFP construct, and L. Prakash for anti-Rad23p antibodies.
M.S. was supported by EMBO and HFSP fellowships, D.J.C. was supported by an EMBO fellowship and a U.S. Army Medical Research Materiel Command Breast Cancer Research Fellowship, B.L.B. was supported by fellowships from the NIH and University of California Office of the President, and S.J. was supported by a fellowship from the Danish Medical Research Council. This work was supported by NIH grant GM38328 (S.I.R.).
REFERENCES
- 1.Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 3.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 4.Clarke D J, Segal M, Mondesert G, Reed S I. The Pds1 anaphase inhibitor and Mec1 kinase define distinct checkpoints coupling S phase with mitosis in budding yeast. Curr Biol. 1999;9:365–368. doi: 10.1016/s0960-9822(99)80163-8. [DOI] [PubMed] [Google Scholar]
- 5.Cohen F O, Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen-Fix O, Koshland D. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci USA. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 8.Fraschini R, Formenti E, Lucchini G, Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 10.Guzder S N, Sung P, Prakash L, Prakash S. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J Biol Chem. 1998;273:31541–31546. doi: 10.1074/jbc.273.47.31541. [DOI] [PubMed] [Google Scholar]
- 11.He Z, Wong J, Maniar H S, Brill S J, Ingles C J. Assessing the requirements for nucleotide excision repair proteins of Saccharomyces cerevisiae in an in vitro system. J Biol Chem. 1996;271:28243–28249. doi: 10.1074/jbc.271.45.28243. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 13.Jensen S, Segal M, Clarke D J, Reed S I. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol. 2001;152:27–40. doi: 10.1083/jcb.152.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lew D J, Marini N J, Reed S I. Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells in the budding yeast Saccharomyces cerevisiae. Cell. 1992;69:317–327. doi: 10.1016/0092-8674(92)90412-6. [DOI] [PubMed] [Google Scholar]
- 15.Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc Natl Acad Sci USA. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masutani C, Araki M, Sugasawa K, van der Spek P J, Yamada A, Uchida A, Maekawa T, Bootsma D, Hoeijmakers J H, Hanaoka F. Identification and characterization of XPC-binding domain of hHR23B. Mol Cell Biol. 1997;17:6915–6923. doi: 10.1128/mcb.17.12.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller J P, Smerdon M J. Rad23 is required for transcription-coupled repair and efficient overrall repair in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2361–2368. doi: 10.1128/mcb.16.5.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 19.Ortolan T G, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 20.Richardson H E, Wittenberg C, Cross F R, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 21.Russell S J, Reed S H, Huang W, Friedberg E C, Johnston S A. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- 22.Sherman F, Fink G, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 23.Straight A F, Marshall W F, Sedat J W, Murray A W. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 24.Sugasawa K, Masutani C, Uchida A, Maekawa T, van der Spek P J, Bootsma D, Hoeijmakers J H, Hanaoka F. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol Cell Biol. 1996;16:4852–4861. doi: 10.1128/mcb.16.9.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavormina P A, Burke D J. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics. 1998;148:1701–1713. doi: 10.1093/genetics/148.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tinker K R, Morgan D O. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 28.Watkins J F, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathways(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]